Abstract
Background
A great diversity of multi-pass membrane receptors, typically with 7 transmembrane (TM) helices, is observed in the eukaryote crown group. So far, they are relatively rare in the prokaryotes, and are restricted to the well-characterized sensory rhodopsins of various phototropic prokaryotes.
Results
Utilizing the currently available wealth of prokaryotic genomic sequences, we set up a computational screen to identify putative 7 (TM) and other multi-pass membrane receptors in prokaryotes. As a result of this procedure we were able to recover two widespread families of 7 TM receptors in bacteria that are distantly related to the eukaryotic 7 TM receptors and prokaryotic rhodopsins. Using sequence profile analysis, we were able to establish that the first members of these receptor families contain one of two distinct N-terminal extracellular globular domains, which are predicted to bind ligands such as carbohydrates. In their intracellular portions they contain fusions to a variety of signaling domains, which suggest that they are likely to transduce signals via cyclic AMP, cyclic diguanylate, histidine phosphorylation, dephosphorylation, and through direct interactions with DNA. The second family of bacterial 7 TM receptors possesses an α-helical extracellular domain, and is predicted to transduce a signal via an intracellular HD hydrolase domain. Based on comparative analysis of gene neighborhoods, this receptor is predicted to function as a regulator of the diacylglycerol-kinase-dependent glycerolipid pathway. Additionally, our procedure also recovered other types of putative prokaryotic multi-pass membrane associated receptor domains. Of these, we characterized two widespread, evolutionarily mobile multi-TM domains that are fused to a variety of C-terminal intracellular signaling domains. One of these typified by the Gram-positive LytS protein is predicted to be a potential sensor of murein derivatives, whereas the other one typified by the Escherichia coli UhpB protein is predicted to function as sensor of conformational changes occurring in associated membrane proteins
Conclusions
We present evidence for considerable variety in the types of uncharacterized surface receptors in bacteria, and reconstruct the evolutionary processes that model their diversity. The identification of novel receptor families in prokaryotes is likely to aid in the experimental analysis of signal transduction and environmental responses of several bacteria, including pathogens such as Leptospira, Treponema, Corynebacterium, Coxiella, Bacillus anthracis and Cytophaga.
Background
Cells have evolved several strategies to recognize and respond to diverse stimuli that constantly bombard their cell surfaces. The most common strategy involves receptors that are embedded in the cell membranes [1,2]. Typically, these receptors comprise of an external sensory surface, a membrane-spanning module, and an intracellular surface that transmits signals to the internal cellular machinery. Numerous receptors, which are constructed on this basic architectural principle, are known from all the three domains of life. Particularly common, in both eukaryotes and prokaryotes, are the receptors that combine an extracellular ligand-binding domain with a single transmembrane segment followed by an intracellular signaling module [1,2]. In bacteria, the most frequently occurring intracellular signaling domain is the histidine kinase domain that ultimately catalyzes phosphotransfer to a receiver domain, as part of a two-component relay system [3-5]. In the more complex crown group eukaryotes, receptors with an intracellular kinase domain that catalyzes the phosphorylation of serine, threonine or tyrosine, are the most common receptors [6,7]. In both eukaryotes and prokaryotes, receptors with intracellular catalytic domains that signal via diverse cyclic nucleotides are also fairly widespread. In contrast, certain classes of receptors are relatively limited in their distribution. For example, the classic bacterial-type chemotaxis and temperature receptors are thus far restricted to prokaryotes [8,9].
Amongst the crown group eukaryotes, such as slime molds, fungi and animals, serpentine or seven-transmembrane receptors (7TMR) are a very widely used class of receptors. Members of this class are characterized by seven membrane-spanning segments, which are arranged approximately in two-layers [10,11]. In some cases such as rhodopsin, a light receptor, they may covalently bind a prosthetic group like retinal in the cavity formed by the helices. Alternatively, they bind to a variety of soluble or surface-anchored ligands such as odorants, neurotransmitters and peptides [11]. In certain cases, such as the animal metabotrobic glutamate receptors, frizzled and latrophilin-like receptors, the 7TMRs possess additional extracellular globular domains that specifically interact with their ligands. The structural scaffold of the 7TMRs apparently possesses a great degree of flexibility that allows them to sense a remarkable diversity of ligands, such as odorants, in animals [12]. As a result, the 7TMRs form some of the largest multigene families in the genomes of vertebrates and nematodes [13]. In animals the 7TMRs predominantly function via heterotrimeric GTPases (G-proteins), which in turn relay a signal to a variety of effectors, such as adenylyl cyclases, phospholipases and ion channels. In the fungi, the 7TMRs additionally activate signaling via Ras-like small GTPases, while in Dictyostelium they may also directly activate MAP kinase cascades and calcium channels though alternative pathways [11]. There is also some evidence for G-protein-independent pathways downstream of 7TMRs in animals and plants [11,14].
Though 7TMRs are currently unknown in eukaryotes other than animals, slime molds, fungi and plants, distantly related proteins, namely the prokaryotic rhodopsins, are encountered in bacteria and archaea [15,16]. The animal and the prokaryotic rhodopsins widely differ from each other in the residues that bind retinal and the actual location of the ligand in the internal pocket. However, structural comparisons between the animal rhodopsins and the prokaryotic proteins reveal that they adopt essentially the same topology and three-dimensional fold [10,17,18]. This suggests that they have most probably descended from a common ancestor despite extensive divergence of their sequence. The prokaryotic rhodopsins perform several different functions: 1) Classical bacteriorhodopsin and halorhodopsin from halophilic archaea and the proteorhodopsins from uncultured marine γ-proteobacteria act as photon-dependent proton or chloride transporters [19]. 2) The sensory rhodopsins from halophilic archaea function as light sensors that transmit a signal in the form of a light-induced conformational change to the transmembrane helices of receptors of the chemotaxis receptor family [16]. 3) The signaling rhodopsins from cyanobacteria, like Anabaena, function as light receptors that transduce a signal via a small intracellular conserved protein that is only found in bacteria [20].
Additionally, relatives of these prokaryotic rhodopsins are also found in several eukaryotes such as chlorophytes, dinoflagellates and fungi. While they appear to be light sensors in these organisms, their exact mode of action is poorly understood [21].
The prevalence of prokaryotic rhodopsins raises the question as to whether other, as-yet-uncharacterized 7TMRs might be deployed in prokaryotic signaling. The availability of prokaryotic genome sequences from across a wide phyletic spread allows one to address this question by using comparative genomics. Comparative genomics has extensively aided the detection of novel domains involved in signal transduction [22-27]. Furthermore, the use of contextual information that emerges from gene neighborhoods or predicted operons in prokaryotes and domain or gene fusions has provided several functional leads regarding the novel signaling domains [28]. Conserved gene neighborhoods or operons are often indicative of the products of those genes interacting physically to form complexes, or their involvement in successive steps of biochemical pathways [29,30]. Likewise, gene fusions also suggest the close physical interactions between the products of the fused genes. Recurrent fusions of uncharacterized domains with other functionally characterized domains also help in elucidating the functions of the former through the principle of "guilt by association" [31,32]. New genomic information coming from diverse organisms often improve these analyses, because they provide newer contextual connections and allow testing of previously observed connections. The increasing flow of genomic information also helps in the identification of new domains that are absent or infrequent in the proteomes of well-studied organisms.
In this work, we apply the tools of sequence profile analysis and comparative genomics to the wealth of new information from prokaryotic genomes to identify novel membrane-associated receptors. We identify new types of bacterial 7TMRs, and show that they are far more prevalent than previously suspected. They transduce downstream signals via various intracellular pathways and are likely to play an important regulatory role in several pathogenic and free-living bacteria. These bacterial 7TMRs are also associated with novel, extracellular, ligand-binding domains, some of which appear to have undergone lineage specific radiation to recognize diverse ligands. These bacterial receptors may also provide a model for the generalized principles of 7TMR function, and even help in understanding non-G protein linked signaling mechanisms via analogous receptors in eukaryotes. We also identified two other groups of widespread membrane-associated receptors, with five and eight membrane-spanning segments respectively, in diverse bacterial lineages.
Results and Discussion
Identification of novel putative receptors in bacterial proteomes
In order to characterize potential novel domains that may play a role in bacterial signal transduction we collated all available predicted proteomes of prokaryotes from across the entire phyletic spectrum (For details see Methods section below). We laid particular emphasis on including all the recently sequenced proteomes that had not been subjected to sensitive comparative sequence analysis by others or us. Using sensitive PSI-BLAST derived profiles, we collected all the proteins in these proteomes that contained one or more of the commonly occurring domains involved in signal transduction, such as the histidine kinase, chemotaxis receptor, GGDEF, EAL, HD hydrolase, PAS and GAF domains [3,22,33]. In order to identify different kinds of novel signaling receptors, we isolated all proteins in this set which satisfied at least one of the following criteria: 1) They possessed multiple (three or more) membrane-spanning seqments that could be predicted in them using the TOPRED [34], TMPRED [35], TMHMM2.0 [36] and PHDhtm [37] programs. This allowed us to enrich potential multi-TM signaling receptors that are distinct from the common single-pass (1TM) or double-pass (2TM) receptors. 2) They showed large globular extracellular regions that could not be mapped to any other previously characterized domains. This allowed us to identify potential uncharacterized extracellular domains that may function as extracellular sensors.
The regions from signaling proteins fulfilling the above-specified criteria were then clustered based on gapped-BLAST bit-score densities in the range of 0.8 to 0.4 per position, using the BLASTCLUST program. We specifically concentrated on those regions that formed distinct clusters with multiple representatives from the same or different organisms because they were likely to represent evolutionarily conserved domains with functional relevance in a wide range of organisms. We then used representative versions of each these regions of similarity as seeds in PSI-BLAST searches of the non-redundant protein database (NR database, National Center for Biotechnology Information). Through these searches, we were able to identify all currently available occurrences and characterize the diverse domain contexts in which they occurred. In searches involving membrane-spanning regions, we took care to avoid the inclusion of false positives arising due to their bias towards hydrophobicity. To achieve this, all searches were conducted using the correction for PSI-BLAST-statistics based on sequence composition [38] and the e-value threshold for inclusion in the profile was set at .001. We also ensured that all the detected TM domains were approximately the same size and adopted the same topology in predictions with the above-mentioned algorithms for TM prediction. Finally, we used reciprocal searches to determine whether a consistent set of proteins were recovered from different starting points with significant e-values (e < .001), and examined the sequence alignments for characteristic patterns that could distinguish them from other membrane proteins.
We describe below the novel classes of bacterial membrane receptors that were identified as a result of this analysis and the potential gleanings regarding their functions.
Characterization of a bacterial family of seven transmembrane receptors with diverse intracellular signaling modules
The proteins PA4856 from Pseudomonas aeruginosa and TP0040 from Treponema pallidum emerged as representatives of a large cluster of proteins identified in our receptor-search procedure. These proteins shared a homologous transmembrane domain with 7 predicted membrane-spanning helices (Figure 1) fused to histidine kinase catalytic domains and receiver domains in the case of PA4856, and a chemotaxis receptor domain in the case of TP0040 (Figure 2). An examination of their predicted membrane-spanning topology showed that it was identical to that observed in the eukaryotic 7TM receptors and the prokaryotic rhodopsins, with the N-terminus projecting into the extracellular (or periplasmic) space and the C-terminus into the intracellular space (Figure 2). Furthermore, they were approximately the same size as the prokaryotic rhodopsins and eukaryotic 7TMRs (250–300 residues) and did not deviate in terms of the size distribution of the inter-helix loops from the latter class of proteins. In order to further investigate their affinities and phyletic spread, we initiated PSI-BLAST searches with these proteins. These searches recovered numerous homologous 7TM domains from several proteobacterial lineages, such as Azotobacter, Rhizobia, Pseudomonas, Vibrio, Coxiella and Xylella, spirochetes like Treponema and Leptospira, Gram positive bacteria, like Clostridia and Bacillus halodurans, and other bacterial lineages, such as Cytophaga and Chlorobium (Figure 1, Table 1). In particular, several proteins with these 7TM domains were seen in Cytophaga hutchinsonii (13 copies) and Leptospira interrogans (14 copies) (Table 1). All complete versions of these domains were predicted to possess a characteristic topology with an outward facing N-terminal region and cytoplasmic C-terminus. The seven predicted TM helices corresponded precisely with the seven hydrophobic segments that were strongly conserved in all these proteins. When a profile including all the above-detected bacterial 7TMR domains was used to search the NR database, the eukaryotic 7TMR receptor domains of latrophilin (gi: 4185804), ETL (gi: 4423362) and a 7 TMR domain encoded by the prawn nidovirus (gi: 9082017) were recovered as the best hits (e-values = 10-2-3 × 10-3) outside of the bacterial family.
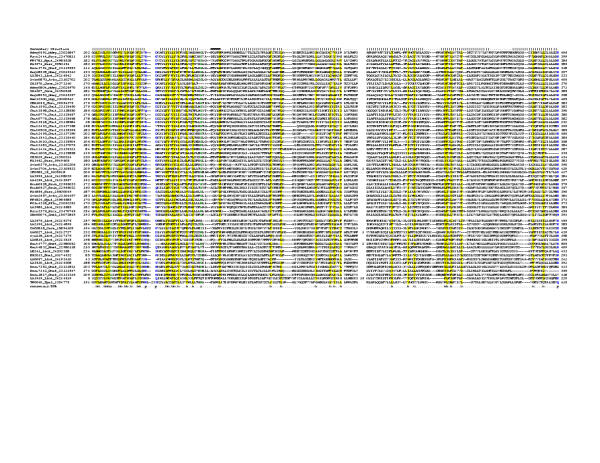
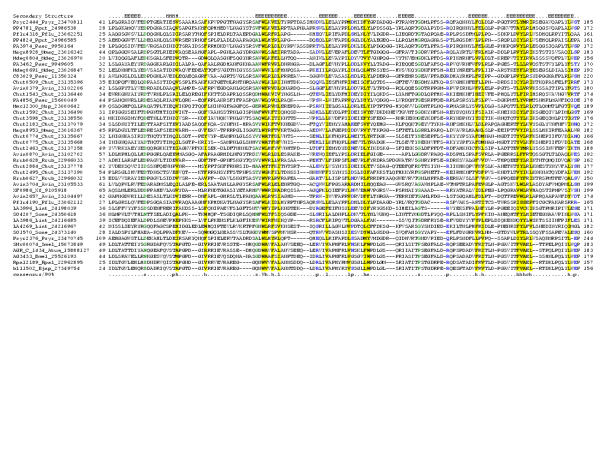
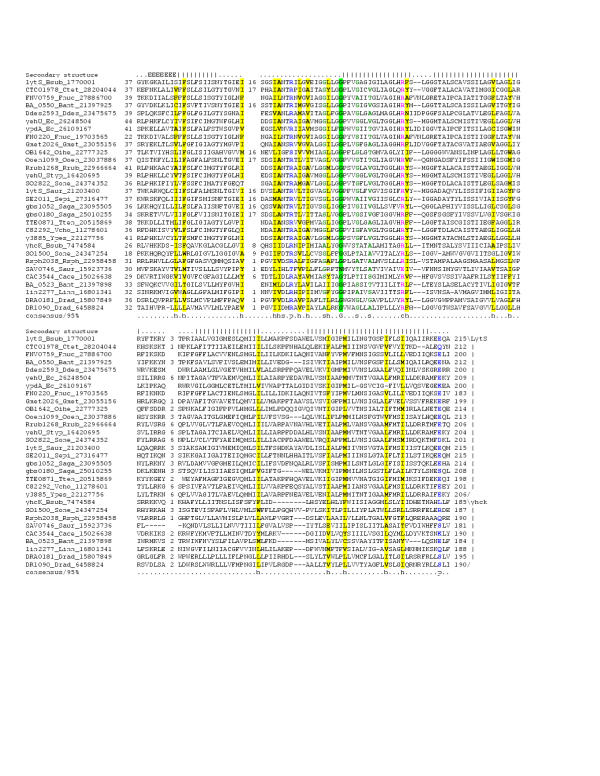
Figure 1.
Multiple sequence alignment of the 7TM domains of the 7TMR-DISM family. Multiple sequence alignment the 7TMR-DISM family was constructed using T-Coffee [73] after parsing high-scoring pairs from PSI-BLAST search results. The PHD-secondary structure [78] is shown above the alignment with | representing an α-helix. The 85% consensus shown below the alignment was derived using the following amino acid classes: hydrophobic (h: ALICVMYFW, yellow shading); small (s: ACDGNPSTV, green) and polar (p: CDEHKNQRST, blue). The limits of the domains are indicated by the residue positions, on each end of the sequence. The two major groups of these receptors, typically associated with either 7TMR-DISMED2s (upper group) or 7TMR-DISMED1s (lower group), are separated by a spacer. The numbers within the alignment are non-conserved inserts that have not been shown. The sequences are denoted by their gene name followed by the species abbreviation and GenBank Identifier (gi). The species abbreviations are as provided in Table 1. This alignment is provided as an additional file in the MS-WORD format (additional file 1).
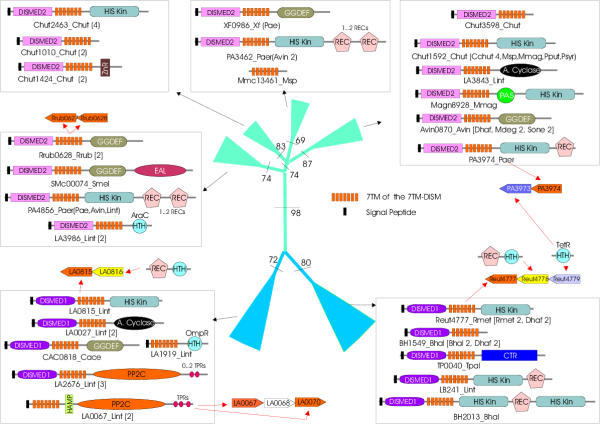
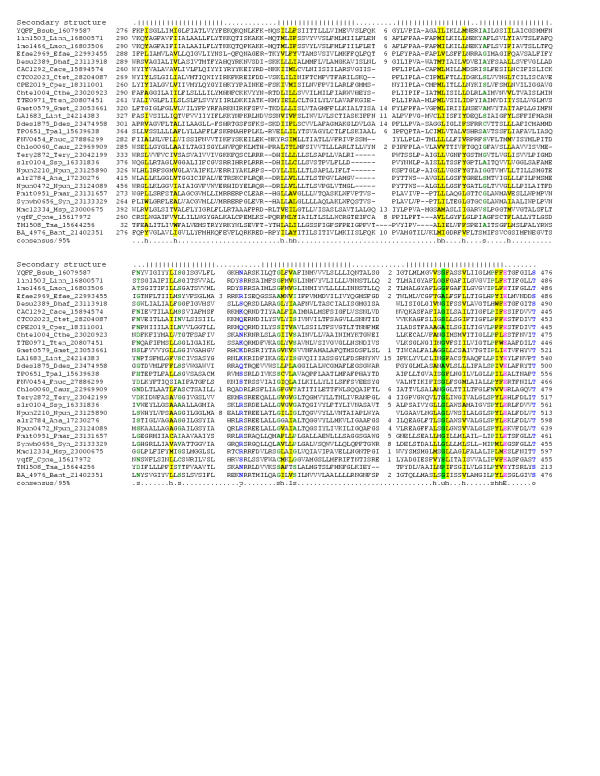
Figure 2.
Phylogenetic tree, domain architectures and gene neighborhoods of the 7TMR-DISM family. Phylogenetic relationships of the 7TMR-DISM domain containing proteins along with the domain architectures are shown. The seed alignment used for constructing the tree was one similar to that shown in Fig. 1. The RELL bootstrap values for the major branches are shown at their base. The thickness of a given branch is approximately proportional to the number of proteins contained within it. Domain architectures of the proteins in each branch of the tree are shown in boxes pointed to by the black arrows. The phyletic pattern of each family is shown, along with the number of proteins (if there are more than one). The gene neighborhood data for some of the genes encoding 7TMR-DISM encoding genes is depicted using block arrows. A red arrow indicates the domain architectures of proteins encoded by each gene. The species abbreviations are as shown in Table 1. Domain abbreviations are: DISMED1 – 7TMR-DISMED1; DISMED2 – 7TMR-DISMED2; A. cyclase-Adenylyl cyclases; GGDEF-GGDEF-motif-containing nucleotide cyclase domains; His Kin – Histidine Kinase; EAL-EAL motif containing cyclic nucleotide phosphodiesterases; REC – Receiver domain; PAS-Ligand binding domain found in Drosophila Period clock proteins, vertebrate Aryl hydrocarbon receptor nuclear translocator and Drosophila Single minded proteins; ZR, Zinc Ribbon HTH; Helix-Turn-Helix domain (of AraC, OmpR and TetR variety); PP2C – Sigma factor PP2C-like phosphatases ; TPR – etratricopeptide repeats; CTR – Chemotaxis receptor domain; HAMP – domain present in Histidine kinases, Adenylyl cyclases, Methyl-accepting proteins and Phosphatases.
Table 1.
Phyletic patterns and number of proteins *
| Domain | Firmicutes | Proteobacteria | Actinomycetes | Cyanobacteria | Spirochetes | other |
| 7TMR | Bhal (2); Cace; Ddeh; Dhaf (5) | Alpha– Atum; Bjap; Bmel; Ccre; Mmag (2); Rpal; Rrub (2); Smel. Beta– Rmet (2); Rsol. Gamma– Avin (4); Cbru; Mdeg (2); Paer (4); Pflu (2); Pput (2); Psyr (2); Sone (2); Xcam (2); Xf; Vvul Delta– Ddes. Unclassified– Msp (2) | - | - | Lint (14); Tpal. | Chut (13); Ctep. |
| 7TMR-HD | Bsub; Bant; Cace (2); Cper; Ctet; Cthe; Dhaf; Efae (2); Linn; Lmon; Oihe; Tten. | Delta– Gmet. Unclassified– Msp. | - | Ana; Npun (2); Pmar (2); Syn; Ssp; Tery | Lint; Tpal. | Cpne; Caur; Fnuc (2); Tmar. |
| 5TMR-LYT | Bsub (2); Bant (2); Cace; Ctet; Linn; Lmon; Oihe; Ooen; Saga (2); Sau (2); Sepi; Smut; Tten. | Alpha– Rsph; Rrub. Gamma– Ec (2); Styp; Sone (2); Vcho; Ypes. Delta– Ddes; Gmet. | - | - | - | Drad (2); Fnuc (2) |
| 8TMR-UT | Llac | Alpha– Bjap (2); Ccre; Mlot; Naro; Rsph; Smel (4). Beta– Bfun; Rmet (2); Rsol. Gamma– Ec (8); Mdeg (3); Pmul; Paer; Pput (2); Styp (4); Sone(2); Sfle (2) Vcho; Vpar; Xcam (2); Ypes Delta– Ddes; Mxan Unclassified– Msp. | Cglu; Tfus; Scoe. | Ana; Syn; Ssp (3). | Lint | Caur ESV |
| 7TMR-DISMED1 | Bhal (3); Cace; Ddeh; Dhaf (4) | Alpha– Ccre Beta– Rmet (2); Rsol. Gamma– Mdeg; Xcam (2). | - | - | Lint (9) | - |
| 7TMR-DISMED2 | Dhaf | Alpha– Atum; Bjap; Bmel; Mmag (2); Rpal; Rrub (2); Smel. Gamma– Avin (4); Cbru; Mdeg (2); Paer (4); Pput (2); Psyr (2); Sone (2); Xf; Vvul. Delta– Ddes. Unclassified– Msp. | - | - | Lint (4) | Chut (13) |
*Firmicutes: Bant – Bacillus anthracis; Bsub – Bacillus subtilis; Bhal – Bacillus halodurans; Cace – Clostridium acetobutylicum; Cper – Clostridium perfringens; Ctet – Clostridium tetani; Cthe – Clostridium thermocellum; Ddeh – Desulfitobacterium dehalogenans; Dhaf – Desulfitobacterium hafniense; Efae – Enterococcus faecium; Llac – Lactococcus lactis; Linn – Listeria innocua; Lmon – Listeria monocytogenes; Oihe – Oceanobacillus iheyensis; Ooen – Oenococcus oeni; Saga – Streptococcus agalactiae; Saur – Staphylococcus aureus; Sepi – Staphylococcus epidermidis; Smut – Streptococcus_mutans; Tten – Thermoanaerobacter tengcongensis. Alphaproteobacteria: Atum – Agrobacterium tumefaciens; Bjap – Bradyrhizobium japonicum; Bmel – Brucella melitensis; Ccre – Caulobacter crescentus; Mmag – Magnetospirillum magnetotacticum; Mlot – Mesorhizobium loti; Naro – Novosphingobium aromaticivorans; Rsph – Rhodobacter sphaeroides; Rpal – Rhodopseudomonas palustris; Rrub – Rhodospirillum rubrum; Smel – Sinorhizobium meliloti. Betaproteobacteria: Bfun – Burkholderia fungorum;Rmet – Ralstonia metallidurans; Rsol – Ralstonia solanacearum. Gammaproteobacteria: Avin – Azotobacter vinelandii; Cbru – Coxiella brunettii; Ec – Escherichia coli; Mdeg – Microbulbifer degradans; Pmul – Pasteurella multocida; Paer – Pseudomonas aeruginosa; Pflu – Pseudomonas fluorescens; Pput – Pseudomonas putida; Psyr – Pseudomonas syringae; Styp – Salmonella typhimurium; Sone – Shewanella oneidensis; Sfle – Shigella flexneri; Vcho – Vibrio cholerae; Vpar – Vibrio parahaemolyticus; Vvul – Vibrio vulnificus; Xcam – Xanthomonas campestris; Xf – Xylella fastidiosa; Ypes – Yersinia pestis. DeltaProteobacteria: Ddes – Desulfovibrio desulfuricans; Gmet – Geobacter metallireducens; Mxan – Myxococcus xanthus. Unclassified Proteobacteria: Msp – Magnetococcus sp. Cyanobacteria: Ana – Anabaena sp.; Npun – Nostoc punctiforme; Pmar – Prochlorococcus marinus; Syn – Synechococcus sp.; Ssp – Synechocystis sp.; Tery – Trichodesmium erythraeum. Spirochetes: Lint – Leptospira interrogans; Tpal – Treponema pallidum. Actinobacteria: Cglu – Corynebacterium glutamicum; Tfus – Thermobifida fusca; Scoe – Streptomyces coelicolor. Other:Cpne – Chlamydophila pneumoniae; Ctep – Chlorobium tepidum; Caur – Chloroflexus aurantiacus; Chut – Cytophaga hutchinsonii; Drad – Deinococcus radiodurans; ESV – Ectocarpus siliculosus virus; Fnuc – Fusobacterium nucleatum; Tmar – Thermotoga maritima. The number of proteins (if more than one) is given in parenthesis. Incomplete genomes are underlined.
A sequence alignment of the 7TM domains that were recovered in these searches showed that they shared a characteristic pattern of sequence conservation (Figure 1) including two well-conserved polar residues at the C-termini of the first and the last helix (typically basic residues). A comparison of these 7TM domains against a library of PSI-BLAST profiles and hidden Markov models (See Methods for details) for previously characterized membrane proteins gave the nematode 7TM receptor family and the prokaryotic rhodopsins as the top-scoring hits (e-values ~.01–.05), suggesting a closer relationship with the classic 7TM receptor families to the exclusion of various other membrane-associated proteins (e-values ~.9–3.5). An examination of the domain architectures of these 7TM proteins reveals considerable diversity around a shared basic architectural blue print. At their N-terminus, these 7TM domains were either directly preceded by a predicted signal peptide, or by different extracellular globular modules, analogous to the domain organization of the animal metabotropic glutamate receptors. At their C-termini they were typically fused a range of catalytic and non-catalytic signaling domains (Figure 2). The former category includes the quintessential bacterial two-component-system-modules, namely the histidine kinase and receiver domains, cyclic diguanylate signaling enzymes such as the GGDEF-type cyclase and EAL-type phosphodiesterase domains, cNMP generating cyclases and PP2C phosphatases [3,22,39,40]. The non-catalytic domains include the PAS, chemotaxis receptor, TPR, and HAMP domains [8,26,27,41-43]. Interestingly, three DNA binding domains, the AraC-type HTH [44], the OmpR-type HTH [45,46] and the bacterial IS1-like Zn finger domains (VA & LA unpublished), are also fused C-termini of certain 7TM receptors from Leptospira and Cytophaga (Figure 2). While a small subset of the receptors lack any intracellular domains, they could non-covalently associate with soluble catalytic domains on their intracellular surface. Hereinafter, we refer to these bacterial receptors as 7TMR-DISM (for 7TMreceptors with diverse intracellular signaling modules).
The great diversity of domain architectures of the 7TMR-DISM family, particularly in terms of their intracellular modules, suggests that they activate number of different intracellular signals in response to different external ligands. With respect to transmitting different cytoplasmic signals via their intracellular regions they resemble the eukaryotic 7TMRs, rather than the prokaryotic bacteriorhodpsins, which are mainly photon-dependent ion pumps. The presence of intracellular DNA-binding domains in certain 7TMR-DISMs suggests that they may take advantage of the non-compartmentalized state of the chromosome in bacteria to directly bind DNA and regulate transcription in response to ligand-induced conformational changes. Additionally, analysis of the gene neighborhoods reveals that in Ralstonia, Pseudomonas and Leptospira the genes encoding 7TMR-DISMs form predicted operons with HTH-transcription factors with receiver domains. These are likely to represent two-component systems in which the transcription factors are modulated by the signal-activated 7TMR-DISM proteins (Figure 2). The presence of TPR repeats in the intracellular regions of certain 7TMR-DISMs is reminiscent of components of eukaryotic signaling systems [42]. These repeats may act as structural scaffolds that link the 7TMR-DISMs to intracellular protein complexes.
The identification and functional analysis of the novel extracellular ligand-binding domains of 7TMR-DISM proteins
The N-termini of most 7TMR-DISMs are linked to large extracellular regions that are predicted to assume a globular structure. As these regions were also recovered in our procedure for identifying novel extracellular ligand-binding domains of receptors, we investigated them in greater detail. Clustering using BLASTCLUST showed that most of these extracellular domains associated with the 7TMR-DISMs fell in either of two distinct clusters. While some of the extracellular regions did not initially fall into any of the clusters, iterative PSI-BLAST searches with representative seed sequences unified all these extracellular regions with one or the other cluster. This suggested there are two distinct varieties of extracellular domains associated with the bacterial 7TMR-DISMs, which we accordingly refer to as 7TMR-DISMED1 and 7TMR-DISMED2 (for 7TMR-DISM extracellular domains 1 and 2).
Iterative PSI-BLAST searches of the NR database with 7TMR-DISMED1 additionally recovered a globular domain inserted in the middle of the sialate acetylesterase domain from various proteobacteria (e-value = 10-3-10-5 iteration 3), and in subsequent iterations carbohydrate metabolism enzymes (e-value = 10-2-10-4 iteration 4–5) such as β-galactosidases, β-mannosidases and β-glucuronidases [47,48]. For example, a search with the 7TMR-DISMED1 from the protein LA2676 from Leptospira recovered the insert domain of the sialate acetylesterases in iteration 3 (eg. Mdeg0217, Microbulbifer degradans, e = 10-4), β-galactosidase in iteration 4 (LacZ, E. coli, e = 10-5) and glucuronidases in iteration 6 (eg. GUS, Homo sapiens, e = 10-2). The 7TMR-DISMED1 corresponds precisely to a distinct domain in the galactosidases and the glucuronidases, which is seen to adopt a β-jelly roll topology in their crystal structures [49] (Figure 3A). These domains function as accessory carbohydrate binding domains, rather than catalytic domains of the enzymes in which they occur [49]. An examination of the sequence alignment (Figure 4) shows that the 7TMR-DISMED1s and the β-jelly roll domain of the carbohydrate-metabolism enzymes share several conserved residues, including certain characteristic aromatic positions. The sequence alignment also suggests that the 7TMR-DISMED1s are likely to preserve the spacious cavity of these jellyrolls with a characteristic triangular outline (Figure 3A). The projection of some of the highly conserved residues into this cavity (Figures 3A and 4) suggests that the core structure of the ligand-binding pocket is also likely to be conserved across all these proteins. These observations imply that the 7TMR-DISMs with the 7TMR-DISMED1 are most likely to function as receptors for carbohydrates or related derivatives.
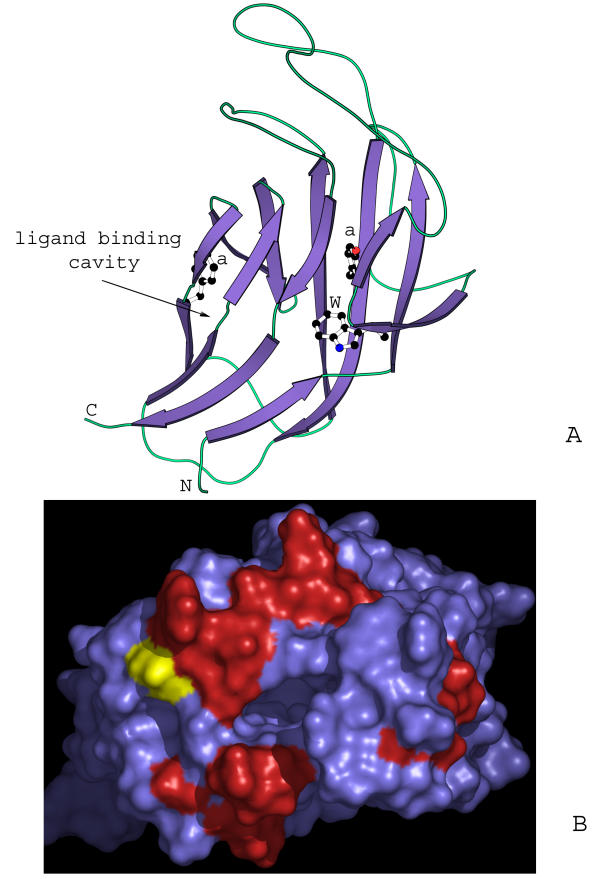
Figure 3.
Models of 7TMR-DISMED1 and the TM domain of the 7TMR-DISMs. (A) Prototype of the β-jellyroll seen in 7TMR-DISMED1, sialate 9-O-acetylesterases, β-glucoronidases and β-glucosidases. The β-jellyroll domain shown here is a cartoon representation of the domain from the crystal structure of β-galactosidase (PDB:1GHO). Conserved residues typical of the 7TMR-DISMED1 are shown in ball stick representation. "a" stands for a conserved aromatic position. (B) A homology model of the TM domain of the 7TMR-DISMs showing the distribution of conserved residues in the 7TMR-DISMs with 7TMR-DISMED1 domains. The model was constructed using bacteriorhodopsin (PDB: 1C3W) and bovine retinal rhodopsin (PDB:1F88) as templates. The N terminus of the 7TMR-DISM domain, where the extracellular domain is attached, is shown in yellow. The red color shows the distribution of residues on the external surface, which are uniquely conserved in 7TMR-DISMs with 7TMR-DISMED1s. This set of proteins essentially corresponds to the lower group of sequences in Fig. 1.
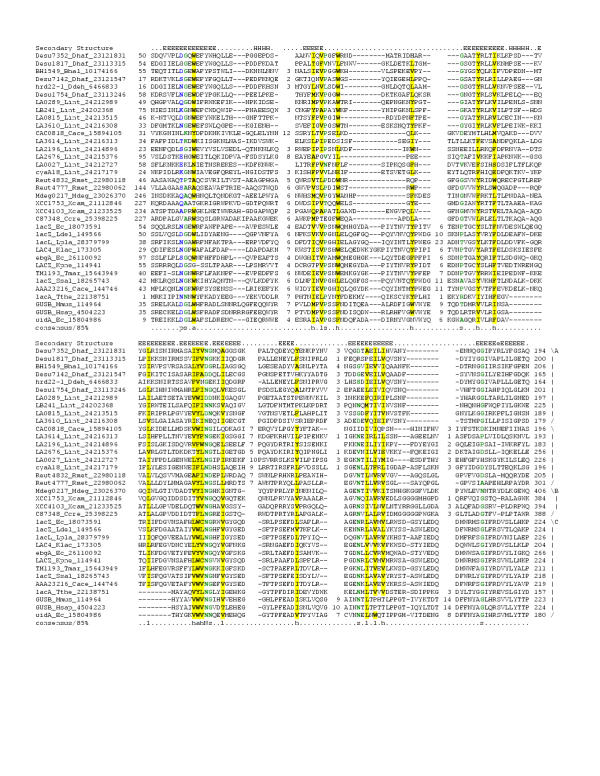
Figure 4.
Multiple sequence alignment of the 7TMR-DISMED1 and accessory domains of sialate 9-O-acetylesterases, β-glucoronidases and β-glucosidases.Multiple sequence alignment the 7TMR-DISMED1 was constructed as detailed in the legend to Figure 1. The PHD-secondary structure [78] is shown above the alignment with E representing a β strand, and H an α-helix. In addition to the convention described in Fig. 1 the consensus also shows the aliphatic subset of the hydrophobic class (l; ALIVMC, yellow shading) and the aromatic subset of the hydrophobic class (a; FHWY, yellow shading). The families shown to the right are A – 7TMR-DISMED1, B – accessory domains of sialate 9-O-acetylesterases and C – accessory domains of β-glucoronidases and β-glucosidases. The species abbreviations are as shown in Table 1 and Kpne – Klebsiella pneumoniae; Klac – Kluyveromyces lactis; Ldel – Lactobacillus delbrueckii; Lpla – Lactobacillus plantarum; Ssal – Streptococcus salivarius; Tthe – Thermoanaerobacterium thermosulfurigenes; Hsap – Homo sapiens; Mmus – Mus musculus.
In contrast to the 7TMR-DISMED1s, the 7TMR-DISMED2 domains did not recover any statistically significant hits to sequences with known structures. However, secondary structure prediction based on the multiple sequence alignment predicted that the 7TMR-DISMED2s are likely to adopt an all β-fold with at least 8 extended regions (Figure 5). The average size of the domain and the distribution of the lengths of the extended regions matched that of several carbohydrate-binding domains, such as the discoidin domain, the cellulose binding domains of cellulases and the fucose-binding domain, which share a common jelly roll topology with the 7TMR-DISMED1s [50,51]. Hence, it is plausible that the 7TMR-DISMED2s represent yet another distinct superfamily of the carbohydrate binding jelly roll fold. This would imply that the 7TMR-DISMED2s could also potentially function as sensors for carbohydrate or related ligands.
Figure 5.
Multiple sequence alignment of the 7TMR-DISMED2s.Multiple sequence alignment the 7TMR-DISMED2 domain was constructed as detailed in the legend to Figure 1. The 90% consensus follows the same convention as in Figure 1 and 2. The species abbreviations are as shown in Table 1.
Previous studies have shown that mapping of residues conserved in specific subgroups of a protein family on the surface view of a representative structure of that domain may throw light on regions involved in specific interactions of those subgroups [52,53]. As such analysis could throw more light on the mechanisms of action of 7TMR-DISMs, we constructed a homology model for the 7TM domain of a representative bacterial receptor using the vertebrate visual rhodopsin and the bacteriorhodopsin as templates. We then plotted the residues conserved in the two major subgroups (see below) of 7TMR-DISMs on to the surface view of this model. Several residues that were specifically conserved in the 7TMR-DISMs, which possessed 7TMR-DISMED1s, formed distinctive patches on the rim of the tubular 7TM structure (Figure 3B). These regions could represent regions of contact between the extracellular (or periplasmic) 7TMR-DISMED1 and the outer surface of the 7TM domain. This would imply that the alterations of the contacts between the extracellular domain and the 7TM domain upon ligand-binding, are likely generate the necessary conformational change for propagating an internal signal. The domain-architectural organization of most of the 7TMR-DISMs closely resembles that of the animal glutamate receptors and vertebrate taste receptors [11,54]. Hence, it is possible that these receptors could act through a similar mechanism in which the signal is relayed via an interplay between the extracellular ligand-binding domain and the 7TM domain.
Evolutionary diversification of the 7TMR-DISMs in bacteria
We analyzed the evolutionary history of the 7TMR-DISMs by constructing phylogenetic trees with the alignments of the conserved 7TMR domains using the neighbor-joining, least square and maximum-likelihood methods (Figure 2). These trees showed that the 7TMR-DISMs were divided into two major clusters that corresponded to forms fused to either 7TMR-DISMED1 or 7TMR-DISMED2 at their N-termini. A small number of proteins in either group lacked a distinct extracellular globular domain, suggesting secondary loss of these domains. Phyletic pattern of the 7TMR-DISMs is patchy: two relatively closely related bacteria may differ in having or lacking a gene for such a receptor, whereas two distantly related bacteria may possess closely related receptors. The phylogenetic tree shows that forms from distantly related bacteria occasionally group together, with statistically significant support for their grouping (Rell BP>80%, for 10000 boot strap replicates). For example, one such well-supported cluster (Fig. 2) contains proteins from phylogenetically distant bacteria, such as, GC Gram positive bacteria, spirochaetes and β-proteobacteria. This suggests a dynamic history for the 7TMR-DISM genes, which as in the case of many other bacterial signaling proteins, is likely to have involved lateral transfer between distantly related taxa and sporadic gene loss.
However, the most striking pattern observed in the evolutionary tree of the 7TMR-DISMs was the presence of multiple well-supported clusters (Rell BP~70–100%) comprised entirely or largely of proteins from a single organism (Figure 2). These lineage specific expansions [55] accounted for almost all of the 7TMR-DISMs in organisms such as, Desulfitobacterium, Leptospira and Cytophaga, which contained multiple copies of these 7TM domains. Furthermore, some diversity was observed in the intracellular domains associated with the 7TM domains from the proteins belonging to these lineage specific clusters (Figure 2). This suggested that the 7TMR-DISM evolved through lineage specific expansion of the 7TM domain in various bacterial lineages, accompanied by some domain shuffling in their C-terminal intracellular modules. The lineage specific expansion of these 7TMRs is reminiscent of the evolution of the eukaryotic 7TMRs such as the odorant receptors of vertebrates and chemoreceptors of nematodes [55]. The predicted role, for at least a subset of these receptors, in carbohydrate sensing, is consistent with their expansion in Cytophaga hutchinsonii that is known to actively metabolize polysaccharides [56]. The expansion in Leptospira implies that this spirochete too may respond to diverse carbohydrates. Alternatively, it could also utilize its numerous 7TMR-DISMs to recognize different carbohydrates on host cell surfaces to regulate its motility.
7TMR-HD: a novel family of bacterial receptors with a HD hydrolase domain
Another family of potential bacterial 7TMRs that was recovered by our receptor search procedure was typified by slr0104 from Synechocystis. This family of receptors is present in cyanobacteria, spirochetes like Leptospira and Treponema, most low GC Gram positive bacteria, some proteobacteria, like Magnetospirillum and Geobacter, Chloroflexus, Fusobacterium and Chlamydia pneumoniae (Figure 6). All members of this family are characterized by the presence of an intracellular HD hydrolase domain [57] C-terminal to the 7TM domain. Accordingly, we named these receptors as 7TMR-HD (for 7TM receptors with intracellular HD domains). The majority of 7TMR-HD proteins also contain a distinct domain N-terminal to the 7TM domain that is predicted to localize to the extracellular or periplasmic compartment, based on the presence of a signal peptide. Analysis of the predicted topology and searches against a position specific score matrix library of membrane-spanning domains with RPS-BLAST, suggests that the 7TM domains of the 7TMR-HDs are distant relatives of other eukaryotic and bacterial 7M receptors (e-values ~.01–.07). However, the 7TMR-HDs possess several distinctive features in their 7TM domain that clearly demarcate them from all other membrane proteins. These include, positively charged patches in the intracellular loops between helix 1–2 and helix 5–6, a glycine in helix 7, and conserved glutamate and alcoholic residues in the C-terminal cytoplasmic tail (Figure 6).
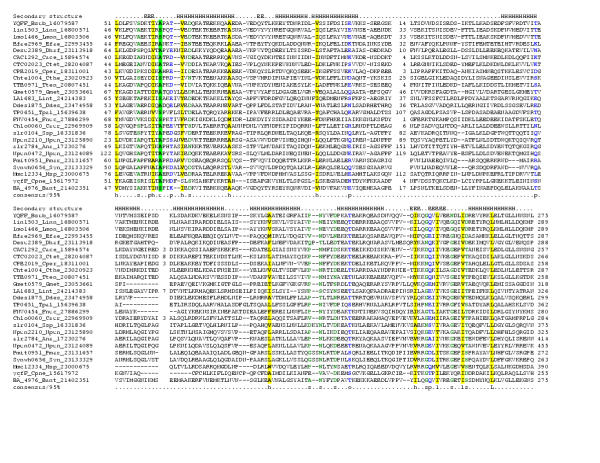
Figure 6.
Multiple sequence alignment of the 7TM domains of the 7TMR-HD family.Multiple sequence alignment the 7TM domains of the 7TMR-HD family was constructed was constructed as detailed in the legend to Figure 1. The 95% consensus follows the same convention as in Figure 1 and 2 and also shows the following classes alcohol (o: ST, Blue), the tiny subclass of small (u; GAS, Green shading) and an 'E' shows the completely conserved amino acid in that group. The species abbreviations are as shown in Table 1.
In terms of domain architecture, the 7TMR-HDs are most similar to the 7TMR-DISMs in possessing a large N-terminal extracellular domain and an intracellular signaling domain. This architectural pattern suggests that the extracellular domain (7TMR-HDED, for 7TMR-HD Extracellular Domain) is most likely to function as a sensor domain that transmits the signal via the 7TM domain to the intracellular catalytic domain. Sequence profile searches with the extracellular domain did not establish any relationship with previously known globular domains. A multiple alignment of the 7TMR-HDEDs shows that it is highly enriched in polar residues and is predicted to assume a predominantly α-helical fold with several amphipathic helical hairpins (Figure 7).
Figure 7.
Multiple sequence alignment of the 7TMR-HDEDs.Multiple sequence alignment was constructed as detailed in the legend to Figure 1. The species abbreviations are as shown in Table 1.
To gain further insight into the functions of the 7TMR-HDs we used the contextual information available in the form of their gene neighborhoods (Figure 8A). In Gram-positive bacteria, Fusobacterium, Treponema, Leptospira, Geobacter and Thermotoga, the 7TMR-HD gene is always associated with the conserved PhoH-YbeY-diacylglycerol kinase gene neighborhood, and is specifically located adjacent to the YbeY and PhoH genes. The PhoH-YbeY-diacylglycerol kinase gene neighborhood [58] is defined by several conserved genes (Figure 8A), which often occur adjacent to each other or the gene for the well-studied bacterial membrane-associated enzyme diacylglycerol kinase (DgkA) [59,60]. The PhoH gene encodes a member of the helicase-like superclass of the P-loop NTPase fold, and has been linked to the response to phosphate starvation in E. coli [58]. The third major member of this neighborhood encodes a highly conserved, uncharacterized protein typified by E. coli YbeY. An alignment of the YbeY orthologs (unpublished data VA & LA) indicates that it possesses 3 conserved histidines that are arranged in a manner very similar to the Zincin-like metallopeptidases [61,62]. The occurrence of YbeY next to the DgkA gene, and the fusions of the YbeY protein with the DgkA protein, suggest a function for it in the same pathway as DgkA, namely lipid-head-group metabolism. This function is also supported by the fusion of the YbeY protein to the acyl carrier protein synthase domain in Plasmodium. These observations, taken together with the predicted metalloprotease-like active site of the YbeY proteins, suggest that YbeY is most likely to function as an endogenous lecithinase (phospholipase C) in lipid metabolism to generate diacylglycerol, the substrate for DgkA, from phosphatidylcholine. This would imply that the 7TMR-HDs, which show a strong association with the PhoH-YbeY-diacylglycerol kinase gene neighborhood, are likely to function as a regulator of this pathway of lipid metabolism.
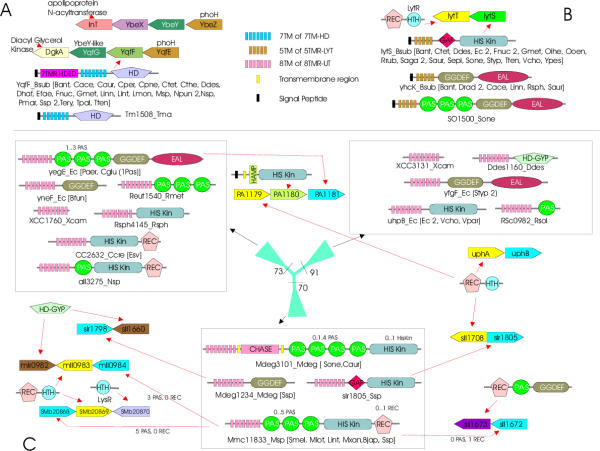
Figure 8.
Gene neighborhoods of 7TMR-HD, 5TMR-LYTs and the Phylogenetic tree, domain architectures and gene neighborhoods of the 8TMR-UT.A) The domain architectures found in 7TMR-HD proteins and the PhoH-YbeY gene neighborhoods are shown. The upper panel shows the PhoH-YbeY neighborhood in proteobacteria (E. coli DgkA-YqfG-YqfF-YqfE operon), while the lower one shows the neighborhood typical of bacteria with the 7TMR-HD proteins. The organisms, possessing a particular domain architecture, are indicated by abbreviations in brackets). YbeY or its ortholog YqfG is the predicted lecithinase with a metal binding active site. YbeX is a Cystathionine beta-synthase domain (CBS) containing protein.B) The domain architectures found in 5TMR-LYT containing proteins and the conserved operon LytT-LytS found in Bacillus and gram-positive bacterial genomes are shown. C) Phylogenetic relationships of the 8TMR-UT domain containing proteins along with their domain architectures are shown. The RELL bootstrap values for the major branches are shown at their base. The thickness of a given branch is approximately proportional to the number of proteins contained within it. Domain architectures of the proteins in each branch of the tree are shown in boxes pointed to by the black arrows. The gene neighborhood data of some of the genes encoding 7TMR-DISM containing proteins are shown. The red arrow points to the architecture of protein encoded by a particular gene in a depicted neighborhood. Domain abbreviations are as shown in Figure 2 and HD, hydrolase of the HD superfamily; HD-GYP – cyclic diaguanylate phosphodiesterases of the HD-GYP variety; GAF – domain found in cGMP- specific phosphodiesterases, Adenylyl cyclases and Escherichia coli FhlA. Species abbreviations are as shown in Table 1
It is possible that the highly polar 7TMR-HDED may sense particular changes to ion concentrations, and regulate the YbeY-DgkA-dependent lipid metabolism pathway in order to regulate membrane properties. The nature of the intracellular signal transmitted by the HD hydrolase domain of the 7TMR-HDs is unclear. This HD domain is very distinct from the HD-GYP variety, which acts as a cyclic diaguanylate phosphodiesterase. Contextual analysis also provides no evidence for any association with GGDEF proteins, thus ruling out a role in cDGMP signaling [39]. Likewise, contextual information does not provide any evidence for association with cyclic NMP signaling even though HD domains are known to act as phosphodiesterases in this signaling pathway. These observations imply that the HD hydrolase domain of the 7TMR-HDs may have a distinct function of its own. One possibility is suggested by the contextual association of the 7TMR-HD genes with the genes for the P-loop protein PhoH (Figure 8A). These two proteins could potentially constitute a kinase-phosphoesterase couple that regulates YbeY-DgkA-dependent lipid metabolism. In Chlamydia pneumoniae 7TMR-HD gene occurs in the neighborhood of genes for several uncharacterized membrane proteins with no detectable homologs in other bacterial lineages. Hence, it is likely that in C. pneumoniae the 7TMR-HD has acquired a distinct function, which may be related to the expression of these pathogen-specific surface proteins.
Other bacterial receptors with evolutionarily mobile membrane-associated sensory domains
Most of the above-identified bacterial 7TMRs contain additional N-terminal domains that are likely to play a crucial role in recognition of an extracellular signal. We were also interested in identifying novel families of membrane-associated receptors that do not contain any extracellular N-terminal domains, but primarily utilize their multi-TM domains for sensory purposes. While there are numerous prokaryotic signaling proteins with TM domains, a widely utilized sensory TM domain is likely to exhibit the following characteristics: 1) distinctive sequence or structural features that clearly distinguish them from generic multi-TM proteins and previously characterized transporters. 2) Evolutionary mobility, which means that the same conserved multi-TM domain could be associated with different types of intracellular signaling domains. These criteria are supported by the precedence offered by the domain architectures of previously identified membrane-associated bacterial receptors, such as those of the MHYT family [63]. Analysis of the clusters of conserved Multi-TM domains recovered in our receptor search procedure identified two widespread groups of conserved membrane-associated domains that were combined in different proteins with different types of intracellular signaling domains, but lacked any other extracellular domains.
The first group of these domains is typified by the Bacillus proteins, LytS and YhcK, which share a conserved membrane-spanning domain with 5 TM helices (Figure 8B and 9). In LytS-type proteins the 5TM domains are combined with C-terminal intracellular GAF and histidine kinase domains, while in the YhcK-type proteins the 5TM domain is combined with intracellular GGDEF (diguanylate cyclase catalytic) domains. Occasionally, some members of the latter group, such as SO1500 from Shewanella, are also combined with additional intracellular EAL (cyclic diguanylate phosphodiesterase) and PAS domains (Figure 8B). We named this family of conserved 5TM domains the 5TMR-LYT family (for 5 transmembrane receptors of the LytS-YhcK type). 5TMR-LYTs are widely distributed in bacteria, with multiple members in Gram-positive bacteria, various proteobacteria, Fusobacteria, and Deinococcus (Table 1). The presence of a strongly predicted signal peptide in all members of this family suggests that it adopts a topology analogous to the classic 7TMRs: the N-terminus of the first helix is extracellular (or periplasmic), while the C-terminal tail with the fused signaling domain is intracellular (Figure 9). The membrane-spanning domain of the 5TMR-LYT family is distinguished from other membrane spanning domains by the presence of certain distinctive sequence features. These include the presence of a characteristic NXR motif in the loop between helix-1 and 2, multiple small residues, like glycine and proline, in the middle of helix-2, and a small residue (typically glycine) in the midst of the 5th helix. These small residues in the middle of the TM helices are likely to distort them, and this conformation may be critical to accommodate a ligand, or provide flexibility for transmission of a signal.
Figure 9.
Multiple sequence alignment of the 5TM domains of the 5TMR-LYT family.Multiple sequence alignment the 5TM domains of the 5TMR-LYT family was constructed as detailed in the legend to Figure 1. The two subgroups 1) LytS and 2) YhcK are shown on the right. The species abbreviations are as shown in Table 1.
In several bacterial lineages, such as the Gram-positive bacteria and Vibrionaceae, the 5TMR-LYTs of the LytS variety is encoded by a gene that occurs in the same operon as a gene for a LytR type transcription factor (Figure 8B). This suggests that they transmit a signal via a LytR protein to regulate transcription. In Gram-positive bacteria, LytS and LytR affect the composition of the cell wall by regulating murein hydrolases, and disruption of the LytS gene results in increased autolysis [64,65]. This suggests that the membrane-spanning domain of the 5TMR-LYT family may act as a receptor for derivatives of murein. Some of the 5TMR-LYTs of the YchK variety occur in gene neighborhoods containing genes encoding various signaling proteins with GAF [25], histidine kinase and receiver modules. Hence, the YchK-like proteins could be part of a distinct signaling complex that could relay signals, which may be dependent on cell wall composition.
A second family of evolutionarily mobile TM domains that was recovered in our search procedure is typified by the conserved N-terminal TM domain, which is found in the E. coli sugar response histidine kinase UhpB (Figure 8C and 10). In addition to orthologs of the UhpB protein, this conserved TM domain was recovered in diverse signaling proteins that are particularly widespread in proteobacteria. Sporadic occurrences of this domain were also encountered in signaling proteins from actinomycetes like Corynebacterium glutamicum, cyanobacteria, spirochetes like Leptospira interrogans, flexibacteria like Chloroflexus, and the Ectocarpus siliculosus virus (Table 1). A search for signal peptides using the SignalP program [66,67] with bacterial signal peptide models did not yield strong signal predictions for these proteins. TM helix prediction with an alignment of this conserved TM domain using the PHDhtm program [37] suggested the presence of 8 membrane-spanning helices. Further, helix prediction, individually for all members of this family, with the TOPRED, TMHMM2.0 and TMPRED programs [34-36] also suggested the presence of 8 membrane-spanning helices on an average. These algorithms also predicted a topology with an intracellular N and C terminus for this TM domain, which is compatible with the C-terminal signaling domains occurring immediately after the last predicted membrane spanning helix in most instances. Accordingly, we refer to this domain as the 8TMR-UT domain (for 8 trans membrane UhpB type domain).
Figure 10.
Multiple sequence alignment of the 8TMR-UT family.Multiple sequence alignment the 8TMR was constructed as detailed in the legend to Figure 1. The species abbreviations are as shown in Table 1.
8TMR-UTs are approximately 290–300 residues in length and are characterized by several distinctive features that differentiate them from all other TM regions (Figure 10). These include, an aromatic position followed by a proline in the second helix, a pair of small residues typically glycine in the 5th helix, and a charged patch just C-terminal to the last helix. The conserved prolines and glycines within the predicted helices suggest that they may possess conformational distortions that could be critical for signal sensing and transduction. The clearest functional clues for the 8TMR-UT domain comes from the E. coli UhpB protein, whose C-terminal intracellular histidine kinase transfers a phosphate to the receiver domain of the transcription factor UhpA. Currently available experimental evidence suggests that the 8TMR-UT domain of UhpB interacts with the transporter UhpC, which binds glucose 6-phosphate [68,69]. When UhpC binds glucose 6-phosphate it appears to transmit a signal via the 8TMR-UT domain of the UhpB protein to activate its kinase domain. Operons related to the Uhp operon are seen in number of bacteria, suggesting that a similar signal relay system is widely employed in sugar sensing by bacteria.
Phylogenetic analysis of this family suggests that the 8TMR-UT proteins are divided into 3 major groups (Fig. 8C). Proteins belonging to each of these sub-divisions often show a sporadic phyletic pattern, and often 8TMR-UT domains from distantly related organisms group closely together in the tree (Fig. 8C). These observations suggest a dynamic evolutionary history with gene loss and lateral transfer as in the case of the 7TM-DISMs. Likewise, the 8TMR-UT domains also appear to have extensively combined with a range of intracellular domains in various bacteria (Fig. 8C). These combinations often include linkages with GGDEF and HD-GYP domains, which are cyclic diguanylate generating and degrading enzymes respectively, histidine kinase and receiver domains, and PAS domains. In some cases the 8TMR-UT is combined with extracellular CHASE domains [23,70] (Fig. 8C), which suggests that it may transmit the conformational changes arising from the interactions of ligands with the CHASE domain, to intracellular signaling domains. These observations suggest that the 8TMR-UT domain might, in general interact with other membrane-associated proteins, and act a switch that senses conformational changes in them to transmit signals. Examination of the gene neighborhoods of the 8TMR-UTs showed that they often co-occurred with genes predicted to encode molecules with HTH, HAMP, PAS, GGDEF, HD-GYP, histidine kinase and receiver domains (Fig. 8C). These predicted operon organizations suggest that the 8TMR-UTs might functionally interact with other signaling molecules to form complex signaling networks that might help in sensing a wide diversity of stimuli [9].
Conclusions
The presence of a relatively small set of well-understood signaling domains that are combined with a variety of other accessory domains allowed us to set up a sieve for new receptors in prokaryotes. As a result, we were able to identify two distinct families of 7TM receptors, namely 7TMR-DISM and 7TMR-HD that are unique to bacteria. The discovery of these new 7TMRs in diverse bacteria suggests that they may be more widely utilized in prokaryotes than has been previously suspected. Importantly, the domain architectures of the 7TMR-DISMs suggest that they are likely to activate a variety of intracellular signaling cascades including adenylyl cyclases and kinases. This suggests that these bacterial 7TMRs are functional analogues of the eukaryotic receptors, and could serve as models for the non-G protein linked pathways downstream of the eukaryotic receptors. Most members of the 7TMR-DISM family are fused to one of two extracellular domains at their N-termini. Both these domains are predicted to adopt all-β fold with a jellyroll topology similar to the discoidin-type sugar binding domains. One of them, 7TMR-DISMED1 can be unified with the carbohydrate binding domains of β-galactosidases and β-glucoronidases. Accordingly, the 7TMR-DISM family is predicted to function as receptors for carbohydrates and related molecules.
Based on the contextual information from gene neighborhoods, the 7TMR-HD proteins are predicted to act as receptors that regulate the highly conserved glycerolipid metabolism pathway in response to stimuli sensed by their extracellular domains. The architectures of most of the 7TMR-HD and 7TMR-DISM proteins are reminiscent of the animal metabotropic glutamate and taste receptors. These animal receptors contain an extracellular periplasmic solute-binding domain that is typical of several bacterial signaling proteins [54]. This architecture, along with the limited phyletic pattern of these 7TMR (only found in animals), could imply that they were acquired from an as yet unknown prokaryotic source. In more general terms, the eukaryotic 7TMRs are thus far restricted in their phyletic spread to a few crown group lineages (animals, slime molds fungi and plants) and appear to have proliferated principally through lineage specific expansions from a few founders. The herein-reported discovery of new representatives of 7TMRs in bacteria suggests that they are ancient and widespread in the bacterial lineage. Hence, it is possible that a subset of the crown group eukaryotes may have ultimately acquired the founding members of their 7TMR families from a prokaryotic source through lateral transfer.
We also detected two evolutionarily mobile membrane-spanning domains, namely 5TM-LYT and 8TMR-UT that are associated with several different types of intracellular domains. These conserved TM domains, unlike members of the 7TMR-DISM and 7TMR-HD families, are not associated with large globular extracellular domains. We propose that one of these families, the 5TMR family, is likely to function as receptors for murein or its components. 8TMR-UT in contrast may sense conformational changes in other membrane-associated proteins and relay these signals to the intracellular signaling domains.
Identification of these receptors suggests new paradigms in bacterial signal transduction, and could also provide models for the functions of an important class of eukaryotic proteins. Experimental investigation of these proteins, particularly those from pathogenic bacteria, such as Bacillus anthracis, Leptospira and Cytophaga, are likely to be of interest in understanding microbial physiology and pathogenesis.
Methods
The non-redundant (NR) database of protein sequences (National Center for Biotechnology Information, NIH, Bethesda, Date: April 1, 2003) was searched using the BLASTP program [71]. The searches were supplemented with a new round of searches at the time of revision of the manuscript (June 20, 2003). All completely sequenced and assembled microbial genomes that were submitted to the NCBI GenBank database as of April 2003, including 16 species of archaea and 96 species of bacteria were included used in this analysis. A complete list of these genomes and the predicted proteomes in fasta format can be downloaded from: http://www.ncbi.nlm.nih.gov/PMGifs/Genomes/micr.html
Additional sequences, from microbial genomes that have been sequenced but not completely assembled and submitted to the GenBank database were also used in this analysis. A list of these prokaryotic genomes, from which sequences have been deposited in GenBank can be accessed from the following URL: http://www.ncbi.nlm.nih.gov/PMGifs/Genomes/eub_u.html
Profile searches were conducted using the PSI-BLAST program with either a single sequence or an alignment used as the query, with a default profile inclusion expectation (E) value threshold of 0.01 (unless specified otherwise), and was iterated until convergence [71,72]. For all searches involving membrane-spanning domains we used a statistical correction for compositional bias to reduce false positives due the general hydrophobicity of these proteins. Multiple alignments were constructed using the T_Coffee program [73], followed by manual correction based on the PSI-BLAST results. Signal peptides were predicted using the SIGNALP program [66,67]http://www.cbs.dtu.dk/services/SignalP-2.0/. Multiple alignments of the N-terminal regions of proteins were used additionally to verify the presence of a conserved signal peptide, and only those signal peptides that were conserved across orthologous groups of proteins were considered as true positives. Transmembrane regions were predicted in individual proteins using the TMPRED, TMHMM2.0 and TOPRED1.0 program with default parameters [34-36]. For TOPRED1.0, the organism parameter was set to "prokaryote" [34]http://bioweb.pasteur.fr/seqanal/interfaces/toppred.html. Additionally, the multiple alignments were used to predict TM regions with the PHDhtm program [37]. The library of profiles for membrane proteins was prepared by extracting all membrane protein alignments from the PFAM database http://www.sanger.ac.uk/Software/Pfam/index.shtml) and updating them by adding new members from the NR database. These updated alignments were then used to make HMMs with the HMMER package [74] or PSSM with PSI-BLAST. All large-scale sequence analysis procedures were carried out using the SEALS package http://www.ncbi.nlm.nih.gov/CBBresearch/Walker/SEALS/index.html.
Structural manipulations were carried out using the Swiss-PDB viewer program [75] and the ribbon diagrams were constructed with MOLSCRIPT [76]. Searches of the PDB database with query structures was conducted using the DALI program [77]. Protein secondary structure was predicted using a multiple alignment as the input for the PHD program [78]. Homology modeling was carried out using the Swiss-PDB viewer, version 3.7 to align a target sequence with template structures. This alignment was then provided as input to the SWISS-MODEL server [75] to generate a homology model using the PROMODII program. This model was then energy-minimized using the GROMOS96 routine of the SPDBV.
Similarity based clustering of proteins was carried out using the BLASTCLUST program ftp://ftp.ncbi.nih.gov/blast/documents/blastclust.txt). Phylogenetic analysis was carried out using the maximum-likelihood, neighbor-joining and least squares methods [79,80]. Briefly, this process involved the construction of a least squares tree using the FITCH program or a neighbor joining tree using the NEIGHBOR program (both from the Phylip package) [81], followed by local rearrangement using the Protml program of the Molphy package [80] to arrive at the maximum likelihood (ML) tree. The statistical significance of various nodes of this ML tree was assessed using the relative estimate of logarithmic likelihood bootstrap (Protml RELL-BP), with 10,000 replicates. Text versions of all alignments reported in this study can be downloaded from: ftp://ftp.ncbi.nih.gov/pub/aravind
Authors' contributions
Author 1 (VA) contributed to the discovery process, preparation of most of the figures. Author 2 (LA) conceived the study and contributed to the discovery process, preparation of one of the figures and the manuscript. All authors read and approved the final manuscript.
Contributor Information
Vivek Anantharaman, Email: ananthar@ncbi.nlm.nih.gov.
L Aravind, Email: aravind@ncbi.nlm.nih.gov.
References
- Alberts B, Bray D, Lewis J, Raff M, Roberts K, Watson JD. In Molecular Biology of the Cell. Garland Publishing, Inc; 1999. [Google Scholar]
- Lodish H, Berk A, Zipursky SL, Matsudaira P, Baltimore D, Darnell J, Zipursky L. In Molecular Cell Biology. WH Freeman & Co; 1999. [Google Scholar]
- Koretke KK, Lupas AN, Warren PV, Rosenberg M, Brown JR. Evolution of two-component signal transduction. Mol Biol Evol. 2000;17:1956–1970. doi: 10.1093/oxfordjournals.molbev.a026297. [DOI] [PubMed] [Google Scholar]
- Robinson VL, Buckler DR, Stock AM. A tale of two components: a novel kinase and a regulatory switch. Nat Struct Biol. 2000;7:626–633. doi: 10.1038/77915. [DOI] [PubMed] [Google Scholar]
- Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu Rev Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- Blume-Jensen P, Hunter T. Oncogenic kinase signalling. Nature. 2001;411:355–365. doi: 10.1038/35077225. [DOI] [PubMed] [Google Scholar]
- Manning G, Plowman GD, Hunter T, Sudarsanam S. Evolution of protein kinase signaling from yeast to man. Trends Biochem Sci. 2002;27:514–520. doi: 10.1016/S0968-0004(02)02179-5. [DOI] [PubMed] [Google Scholar]
- Bourret RB, Stock AM. Molecular information processing: lessons from bacterial chemotaxis. J Biol Chem. 2002;277:9625–9628. doi: 10.1074/jbc.R100066200. [DOI] [PubMed] [Google Scholar]
- Bray D. Genomics. Molecular prodigality. Science. 2003;299:1189–1190. doi: 10.1126/science.1080010. [DOI] [PubMed] [Google Scholar]
- Lu ZL, Saldanha JW, Hulme EC. Seven-transmembrane receptors: crystals clarify. Trends Pharmacol Sci. 2002;23:140–146. doi: 10.1016/S0165-6147(00)01973-8. [DOI] [PubMed] [Google Scholar]
- Pierce KL, Premont RT, Lefkowitz RJ. Seven-transmembrane receptors. Nat Rev Mol Cell Biol. 2002;3:639–650. doi: 10.1038/nrm908. [DOI] [PubMed] [Google Scholar]
- Mombaerts P. Seven-transmembrane proteins as odorant and chemosensory receptors. Science. 1999;286:707–711. doi: 10.1126/science.286.5440.707. [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann N, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, Gregory S, Hubbard T, Humphray S, Hunt A, Jones M, Lloyd C, McMurray A, Matthews L, Mercer S, Milne S, Mullikin JC, Mungall A, Plumb R, Ross M, Shownkeen R, Sims S, Waterston RH, Wilson RK, Hillier LW, McPherson JD, Marra MA, Mardis ER, Fulton LA, Chinwalla AT, Pepin KH, Gish WR, Chissoe SL, Wendl MC, Delehaunty KD, Miner TL, Delehaunty A, Kramer JB, Cook LL, Fulton RS, Johnson DL, Minx PJ, Clifton SW, Hawkins T, Branscomb E, Predki P, Richardson P, Wenning S, Slezak T, Doggett N, Cheng JF, Olsen A, Lucas S, Elkin C, Uberbacher E, Frazier M, Gibbs RA, Muzny DM, Scherer SE, Bouck JB, Sodergren EJ, Worley KC, Rives CM, Gorrell JH, Metzker ML, Naylor SL, Kucherlapati RS, Nelson DL, Weinstock GM, Sakaki Y, Fujiyama A, Hattori M, Yada T, Toyoda A, Itoh T, Kawagoe C, Watanabe H, Totoki Y, Taylor T, Weissenbach J, Heilig R, Saurin W, Artiguenave F, Brottier P, Bruls T, Pelletier E, Robert C, Wincker P, Smith DR, Doucette-Stamm L, Rubenfield M, Weinstock K, Lee HM, Dubois J, Rosenthal A, Platzer M, Nyakatura G, Taudien S, Rump A, Yang H, Yu J, Wang J, Huang G, Gu J, Hood L, Rowen L, Madan A, Qin S, Davis RW, Federspiel NA, Abola AP, Proctor MJ, Myers RM, Schmutz J, Dickson M, Grimwood J, Cox DR, Olson MV, Kaul R, Shimizu N, Kawasaki K, Minoshima S, Evans GA, Athanasiou M, Schultz R, Roe BA, Chen F, Pan H, Ramser J, Lehrach H, Reinhardt R, McCombie WR, de la Bastide M, Dedhia N, Blocker H, Hornischer K, Nordsiek G, Agarwala R, Aravind L, Bailey JA, Bateman A, Batzoglou S, Birney E, Bork P, Brown DG, Burge CB, Cerutti L, Chen HC, Church D, Clamp M, Copley RR, Doerks T, Eddy SR, Eichler EE, Furey TS, Galagan J, Gilbert JG, Harmon C, Hayashizaki Y, Haussler D, Hermjakob H, Hokamp K, Jang W, Johnson LS, Jones TA, Kasif S, Kaspryzk A, Kennedy S, Kent WJ, Kitts P, Koonin EV, Korf I, Kulp D, Lancet D, Lowe TM, McLysaght A, Mikkelsen T, Moran JV, Mulder N, Pollara VJ, Ponting CP, Schuler G, Schultz J, Slater G, Smit AF, Stupka E, Szustakowski J, Thierry-Mieg D, Thierry-Mieg J, Wagner L, Wallis J, Wheeler R, Williams A, Wolf YI, Wolfe KH, Yang SP, Yeh RF, Collins F, Guyer MS, Peterson J, Felsenfeld A, Wetterstrand KA, Patrinos A, Morgan MJ, Szustakowki J, de Jong P, Catanese JJ, Osoegawa K, Shizuya H, Choi S, Chen YJ. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Jones AM. G-protein-coupled signaling in Arabidopsis. Curr Opin Plant Biol. 2002;5:402–407. doi: 10.1016/S1369-5266(02)00288-1. [DOI] [PubMed] [Google Scholar]
- Lanyi JK, Luecke H. Bacteriorhodopsin. Curr Opin Struct Biol. 2001;11:415–419. doi: 10.1016/S0959-440X(00)00226-8. [DOI] [PubMed] [Google Scholar]
- Spudich JL, Luecke H. Sensory rhodopsin II: functional insights from structure. Curr Opin Struct Biol. 2002;12:540–546. doi: 10.1016/S0959-440X(02)00359-7. [DOI] [PubMed] [Google Scholar]
- Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Le Trong I, Teller DC, Okada T, Stenkamp RE, Yamamoto M, Miyano M, et al. Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- Teller DC, Okada T, Behnke CA, Palczewski K, Stenkamp RE. Advances in determination of a high-resolution three-dimensional structure of rhodopsin, a model of G-protein-coupled receptors (GPCRs) Biochemistry. 2001;40:7761–7772. doi: 10.1021/bi0155091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beja O, Aravind L, Koonin EV, Suzuki MT, Hadd A, Nguyen LP, Jovanovich SB, Gates CM, Feldman RA, Spudich JL, Spudich EN, DeLong EF, et al. Bacterial rhodopsin: evidence for a new type of phototrophy in the sea. Science. 2000;289:1902–1906. doi: 10.1126/science.289.5486.1902. [DOI] [PubMed] [Google Scholar]
- Jung KH, Trivedi VD, Spudich JL. Demonstration of a sensory rhodopsin in eubacteria. Mol Microbiol. 2003;47:1513–1522. doi: 10.1046/j.1365-2958.2003.03395.x. [DOI] [PubMed] [Google Scholar]
- Bieszke JA, Spudich EN, Scott KL, Borkovich KA, Spudich JL. A eukaryotic protein, NOP-1, binds retinal to form an archaeal rhodopsin-like photochemically reactive pigment. Biochemistry. 1999;38:14138–14145. doi: 10.1021/bi9916170. [DOI] [PubMed] [Google Scholar]
- Anantharaman V, Koonin EV, Aravind L. Regulatory potential, phyletic distribution and evolution of ancient, intracellular small-molecule-binding domains. J Mol Biol. 2001;307:1271–1292. doi: 10.1006/jmbi.2001.4508. [DOI] [PubMed] [Google Scholar]
- Anantharaman V, Aravind L. The CHASE domain: a predicted ligand-binding module in plant cytokinin receptors and other eukaryotic and bacterial receptors. Trends Biochem Sci. 2001;26:579–582. doi: 10.1016/S0968-0004(01)01968-5. [DOI] [PubMed] [Google Scholar]
- Anantharaman V, Aravind L. Cache – a signaling domain common to animal Ca 2+ channel subunits and a class of prokaryotic chemotaxis receptors. Trends Biochem Sci. 2000;25:535–537. doi: 10.1016/S0968-0004(00)01672-8. [DOI] [PubMed] [Google Scholar]
- Aravind L, Ponting CP. The GAF domain: an evolutionary link between diverse phototransducing proteins. Trends Biochem Sci. 1997;22:458–459. doi: 10.1016/S0968-0004(97)01148-1. [DOI] [PubMed] [Google Scholar]
- Ponting CP, Aravind L. PAS: a multifunctional domain family comes to light. Curr Biol. 1997;7:R674–R677. doi: 10.1016/s0960-9822(06)00352-6. [DOI] [PubMed] [Google Scholar]
- Taylor BL, Zhulin IB. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol Mol Biol Rev. 1999;63:479–506. doi: 10.1128/mmbr.63.2.479-506.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer LM, Anantharaman V, Aravind L. Ancient conserved domains shared by animal soluble guanylyl cyclases and bacterial signaling proteins. BMC Genomics. 2003;4:5. doi: 10.1186/1471-2164-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Aravind L, Grishin NV, Rogozin IB, Koonin EV. A DNA repair system specific for thermophilic Archaea and bacteria predicted by genomic context analysis. Nucleic Acids Res. 2002;30:482–496. doi: 10.1093/nar/30.2.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf YI, Rogozin IB, Kondrashov AS, Koonin EV. Genome alignment, evolution of prokaryotic genome organization, and prediction of gene function using genomic context. Genome Res. 2001;11:356–372. doi: 10.1101/gr.GR-1619R. [DOI] [PubMed] [Google Scholar]
- Aravind L. Guilt by association: contextual information in genome analysis. Genome Res. 2000;10:1074–1077. doi: 10.1101/gr.10.8.1074. [DOI] [PubMed] [Google Scholar]
- Huynen M, Snel B, Lathe W, 3rd, Bork P. Predicting protein function by genomic context: quantitative evaluation and qualitative inferences. Genome Res. 2000;10:1204–1210. doi: 10.1101/gr.10.8.1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer AA, Wolf YI, Ponting CP, Koonin EV, Aravind L, Altschul SF. IMPALA: matching a protein sequence against a collection of PSI-BLAST-constructed position-specific score matrices. Bioinformatics. 1999;15:1000–1011. doi: 10.1093/bioinformatics/15.12.1000. [DOI] [PubMed] [Google Scholar]
- Claros MG, von Heijne G. TopPred II: an improved software for membrane protein structure predictions. Comput Appl Biosci. 1994;10:685–686. doi: 10.1093/bioinformatics/10.6.685. [DOI] [PubMed] [Google Scholar]
- Hofmann K, Stoffel W. TMbase – A database of membrane spanning proteins segments. Biol Chem Hoppe-Seyler. 1993;374:166. [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Rost B, Fariselli P, Casadio R. Topology prediction for helical transmembrane proteins at 86% accuracy. Protein Sci. 1996;5:1704–1718. doi: 10.1002/pro.5560050824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer AA, Aravind L, Madden TL, Shavirin S, Spouge JL, Wolf YI, Koonin EV, Altschul SF. Improving the accuracy of PSI-BLAST protein database searches with composition-based statistics and other refinements. Nucleic Acids Res. 2001;29:2994–3005. doi: 10.1093/nar/29.14.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin MY, Natale DA, Aravind L, Koonin EV. A specialized version of the HD hydrolase domain implicated in signal transduction. J Mol Microbiol Biotechnol. 1999;1:303–305. [PMC free article] [PubMed] [Google Scholar]
- Bork P, Brown NP, Hegyi H, Schultz J. The protein phosphatase 2C (PP2C) superfamily: detection of bacterial homologues. Protein Sci. 1996;5:1421–1425. doi: 10.1002/pro.5560050720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L, Ponting CP. The cytoplasmic helical linker domain of receptor histidine kinase and methyl-accepting proteins is common to many prokaryotic signalling proteins. FEMS Microbiol Lett. 1999;176:111–116. doi: 10.1016/S0378-1097(99)00197-4. [DOI] [PubMed] [Google Scholar]
- Kobe B, Kajava AV. When protein folding is simplified to protein coiling: the continuum of solenoid protein structures. Trends Biochem Sci. 2000;25:509–515. doi: 10.1016/S0968-0004(00)01667-4. [DOI] [PubMed] [Google Scholar]
- Pei J, Grishin NV. GGDEF domain is homologous to adenylyl cyclase. Proteins. 2001;42:210–216. doi: 10.1002/1097-0134(20010201)42:2<210::AID-PROT80>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Egan SM. Growing repertoire of AraC/XylS activators. J Bacteriol. 2002;184:5529–5532. doi: 10.1128/JB.184.20.5529-5532.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itou H, Tanaka I. The OmpR-family of proteins: insight into the tertiary structure and functions of two-component regulator proteins. J Biochem (Tokyo) 2001;129:343–350. doi: 10.1093/oxfordjournals.jbchem.a002863. [DOI] [PubMed] [Google Scholar]
- Kenney LJ. Structure/function relationships in OmpR and other winged-helix transcription factors. Curr Opin Microbiol. 2002;5:135–141. doi: 10.1016/S1369-5274(02)00310-7. [DOI] [PubMed] [Google Scholar]
- el Hassouni M, Henrissat B, Chippaux M, Barras F. Nucleotide sequences of the arb genes, which control beta-glucoside utilization in Erwinia chrysanthemi: comparison with the Escherichia coli bgl operon and evidence for a new beta-glycohydrolase family including enzymes from eubacteria, archeabacteria, and humans. J Bacteriol. 1992;174:765–777. doi: 10.1128/jb.174.3.765-777.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies G, Henrissat B. Structures and mechanisms of glycosyl hydrolases. Structure. 1995;3:853–859. doi: 10.1016/S0969-2126(01)00220-9. [DOI] [PubMed] [Google Scholar]
- Jacobson RH, Zhang XJ, DuBose RF, Matthews BW. Three-dimensional structure of beta-galactosidase from E. coli. Nature. 1994;369:761–766. doi: 10.1038/369761a0. [DOI] [PubMed] [Google Scholar]
- Baumgartner S, Hofmann K, Chiquet-Ehrismann R, Bucher P. The discoidin domain family revisited: new members from prokaryotes and a homology-based fold prediction. Protein Sci. 1998;7:1626–1631. doi: 10.1002/pro.5560070717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bork P, Doolittle RF. Drosophila kelch motif is derived from a common enzyme fold. J Mol Biol. 1994;236:1277–1282. doi: 10.1016/0022-2836(94)90056-6. [DOI] [PubMed] [Google Scholar]
- Lichtarge O, Sowa ME, Philippi A. Evolutionary traces of functional surfaces along G protein signaling pathway. Methods Enzymol. 2002;344:536–556. doi: 10.1016/s0076-6879(02)44739-8. [DOI] [PubMed] [Google Scholar]
- Madabushi S, Yao H, Marsh M, Kristensen DM, Philippi A, Sowa ME, Lichtarge O. Structural clusters of evolutionary trace residues are statistically significant and common in proteins. J Mol Biol. 2002;316:139–154. doi: 10.1006/jmbi.2001.5327. [DOI] [PubMed] [Google Scholar]
- O'Hara PJ, Sheppard PO, Thogersen H, Venezia D, Haldeman BA, McGrane V, Houamed KM, Thomsen C, Gilbert TL, Mulvihill ER. The ligand-binding domain in metabotropic glutamate receptors is related to bacterial periplasmic binding proteins. Neuron. 1993;11:41–52. doi: 10.1016/0896-6273(93)90269-w. [DOI] [PubMed] [Google Scholar]
- Lespinet O, Wolf YI, Koonin EV, Aravind L. The role of lineage-specific gene family expansion in the evolution of eukaryotes. Genome Res. 2002;12:1048–1059. doi: 10.1101/gr.174302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt JG. In Bergey's Manual of Systematic Bacteriology. Baltimore;Williams & Wilkins; 1989. [Google Scholar]
- Aravind L, Koonin EV. The HD domain defines a new superfamily of metal-dependent phosphohydrolases. Trends Biochem Sci. 1998;23:469–472. doi: 10.1016/S0968-0004(98)01293-6. [DOI] [PubMed] [Google Scholar]
- Kazakov AE, Vassieva O, Gelfand MS, Osterman A, Overbeek R. Bioinformatics classification and functional analysis of PhoH homologs. In Silico Biol. 2002;3:1. [PubMed] [Google Scholar]
- Smith RL, O'Toole JF, Maguire ME, Sanders CR., 2nd Membrane topology of Escherichia coli diacylglycerol kinase. J Bacteriol. 1994;176:5459–5465. doi: 10.1128/jb.176.17.5459-5465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh JP, Bell RM. Diacylglycerol kinase from Escherichia coli. Methods Enzymol. 1992;209:153–162. doi: 10.1016/0076-6879(92)09019-y. [DOI] [PubMed] [Google Scholar]
- Hooper NM. Families of zinc metalloproteases. FEBS Lett. 1994;354:1–6. doi: 10.1016/0014-5793(94)01079-X. [DOI] [PubMed] [Google Scholar]
- Bond JS, Beynon RJ. The astacin family of metalloendopeptidases. Protein Sci. 1995;4:1247–1261. doi: 10.1002/pro.5560040701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin MY, Gaidenko TA, Mulkidjanian AY, Nakano M, Price CW. MHYT, a new integral membrane sensor domain. FEMS Microbiol Lett. 2001;205:17–23. doi: 10.1016/S0378-1097(01)00424-4. [DOI] [PubMed] [Google Scholar]
- Brunskill EW, Bayles KW. Identification and molecular characterization of a putative regulatory locus that affects autolysis in Staphylococcus aureus. J Bacteriol. 1996;178:611–618. doi: 10.1128/jb.178.3.611-618.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunskill EW, Bayles KW. Identification of LytSR-regulated genes from Staphylococcus aureus. J Bacteriol. 1996;178:5810–5812. doi: 10.1128/jb.178.19.5810-5812.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen H, Engelbrecht J, Brunak S, von Heijne G. A neural network method for identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Int J Neural Syst. 1997;8:581–599. doi: 10.1142/S0129065797000537. [DOI] [PubMed] [Google Scholar]
- Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- Island MD, Kadner RJ. Interplay between the membrane-associated UhpB and UhpC regulatory proteins. J Bacteriol. 1993;175:5028–5034. doi: 10.1128/jb.175.16.5028-5034.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhamme DT, Postma PW, Crielaard W, Hellingwerf KJ. Cooperativity in signal transfer through the Uhp system of Escherichia coli. J Bacteriol. 2002;184:4205–4210. doi: 10.1128/JB.184.15.4205-4210.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mougel C, Zhulin IB. CHASE: an extracellular sensing domain common to transmembrane receptors from prokaryotes, lower eukaryotes and plants. Trends Biochem Sci. 2001;26:582–584. doi: 10.1016/S0968-0004(01)01969-7. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L, Koonin EV. Gleaning non-trivial structural, functional and evolutionary information about proteins by iterative database searches. J Mol Biol. 1999;287:1023–1040. doi: 10.1006/jmbi.1999.2653. [DOI] [PubMed] [Google Scholar]
- Notredame C, Higgins DG, Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- Bateman A, Birney E, Cerruti L, Durbin R, Etwiller L, Eddy SR, Griffiths-Jones S, Howe KL, Marshall M, Sonnhammer EL. The Pfam protein families database. Nucleic Acids Res. 2002;30:276–280. doi: 10.1093/nar/30.1.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- Kraulis PJ. Molscript. J Appl Cryst. 1991;24:946–950. doi: 10.1107/S0021889891004399. [DOI] [Google Scholar]
- Holm L, Sander C. Protein structure comparison by alignment of distance matrices. J Mol Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- Rost B, Sander C. Prediction of protein secondary structure at better than 70% accuracy. J Mol Biol. 1993;232:584–599. doi: 10.1006/jmbi.1993.1413. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Inferring phylogenies from protein sequences by parsimony, distance, and likelihood methods. Methods Enzymol. 1996;266:418–427. doi: 10.1016/s0076-6879(96)66026-1. [DOI] [PubMed] [Google Scholar]
- Hasegawa M, Kishino H, Saitou N. On the maximum likelihood method in molecular phylogenetics. J Mol Evol. 1991;32:443–445. doi: 10.1007/BF02101285. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP – Phylogeny Inference Package (Version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]