Abstract
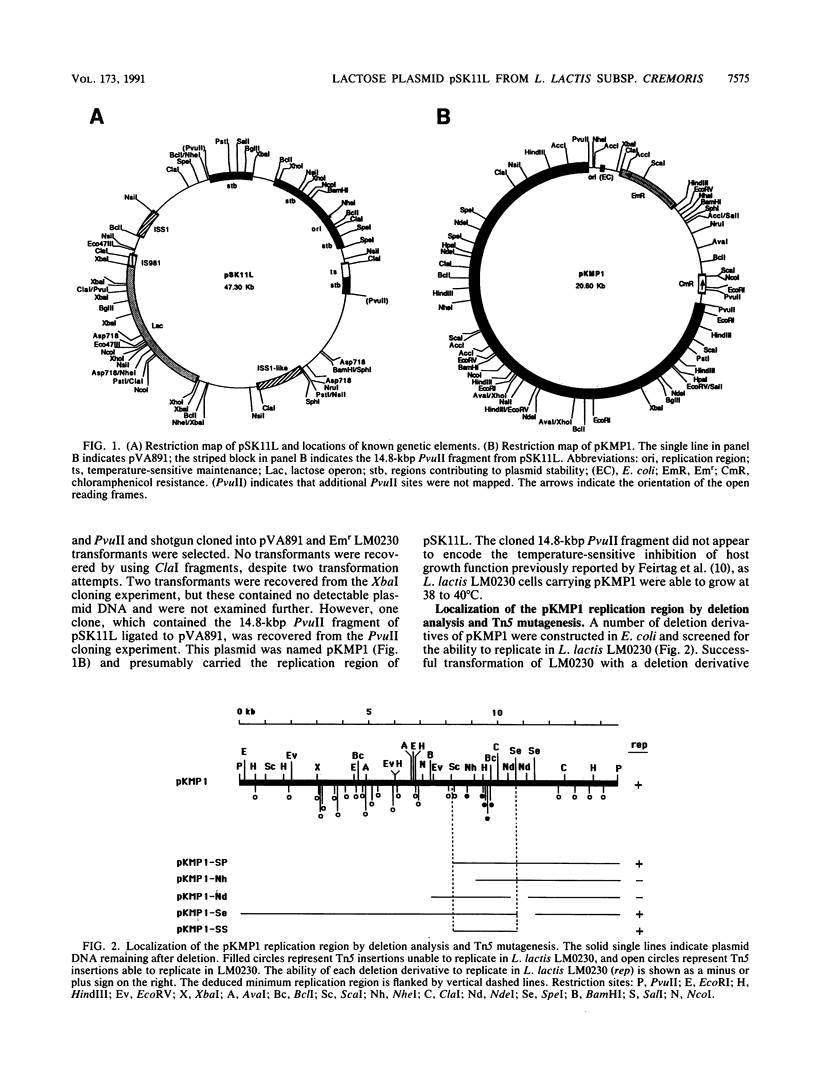
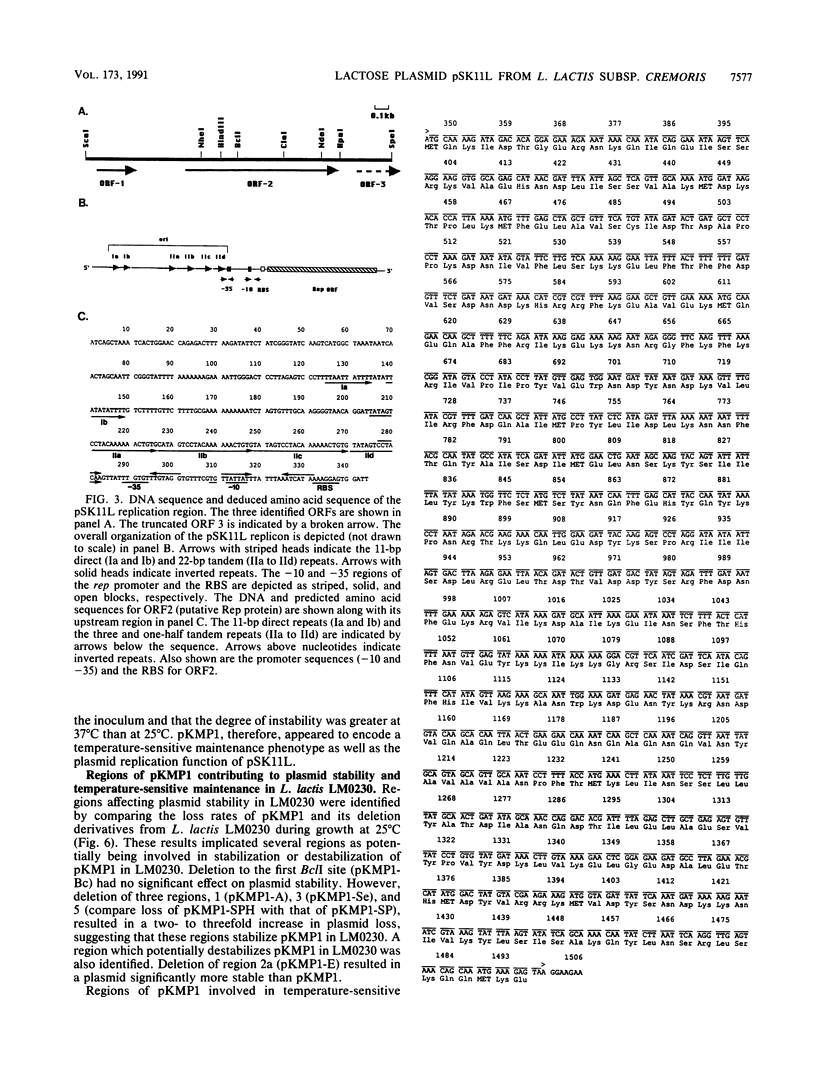
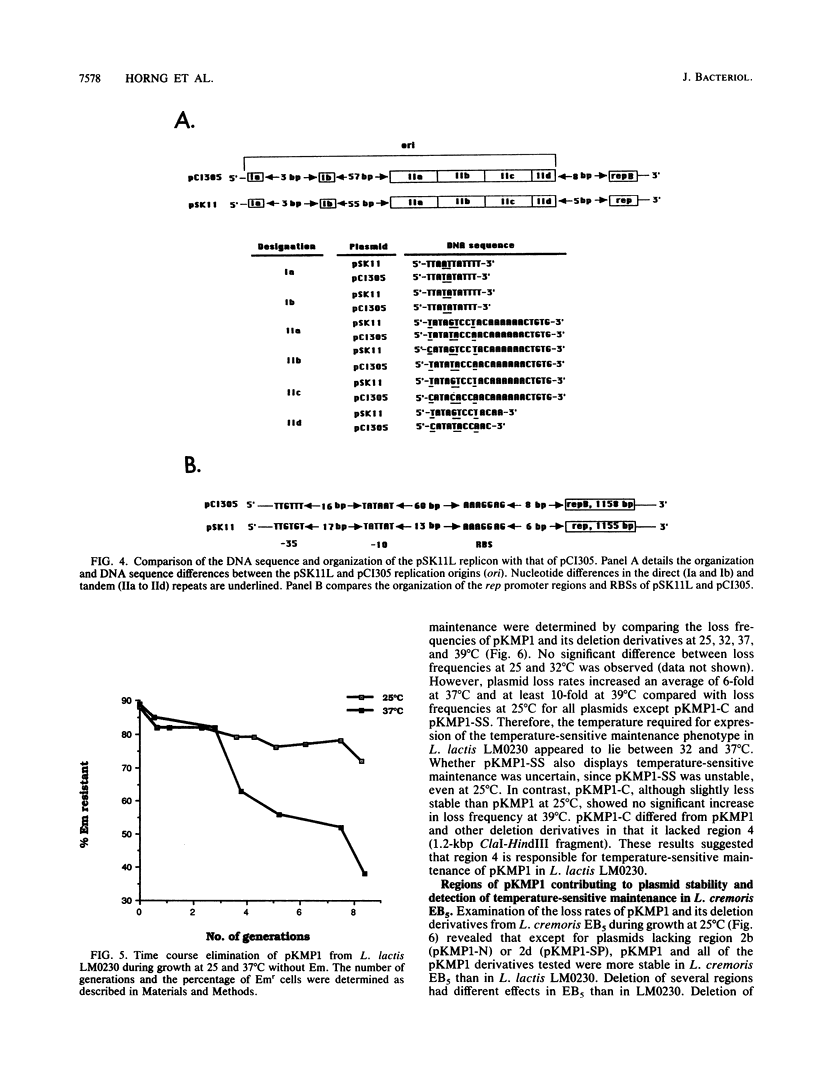
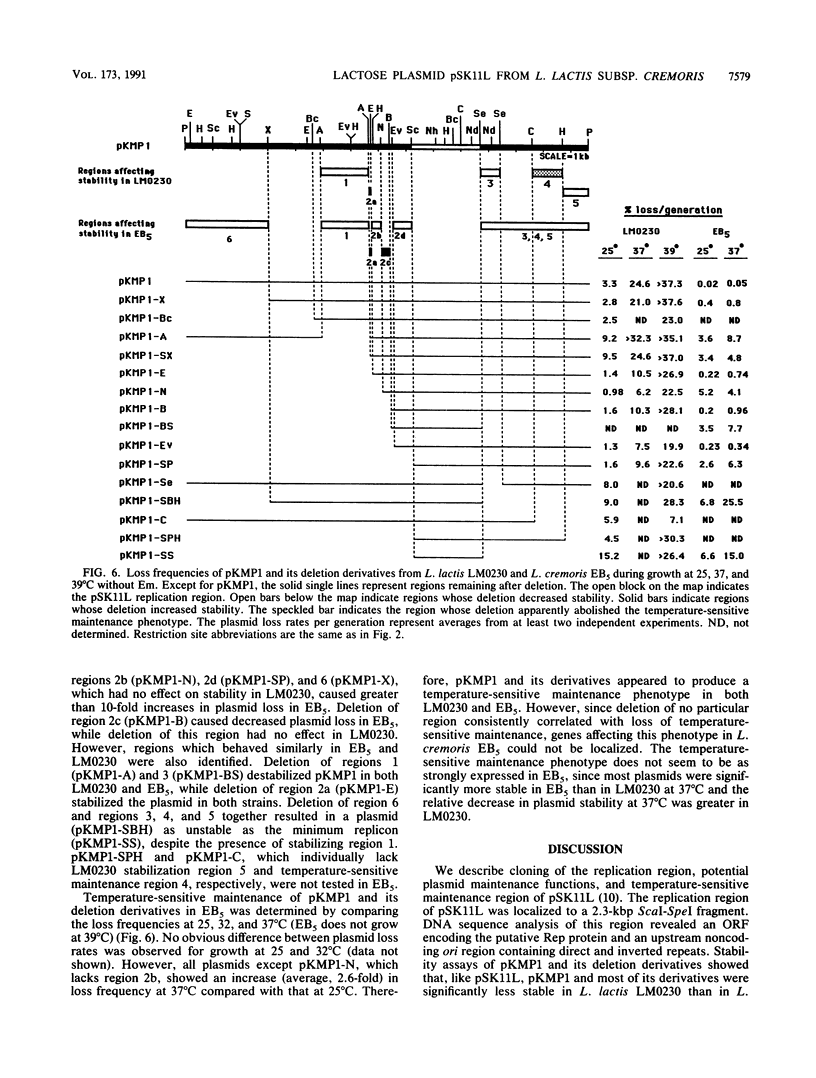
The replication region of pSK11L, the lactose plasmid of Lactococcus lactis subsp. cremoris (L. cremoris) SK11, was isolated on a 14.8-kbp PvuII fragment by shotgun cloning into an Escherichia coli vector encoding erythromycin resistance and selection for erythromycin-resistant transformants of L. lactis subsp. lactis (L. lactis) LM0230. Deletion analysis and Tn5 mutagenesis of the resulting plasmid (pKMP1) further localized the replication region to a 2.3-kbp ScaI-SpeI fragment. DNA sequence analysis of this 2.3-kbp fragment revealed a 1,155-bp open reading frame encoding the putative replication protein, Rep. The replication origin was located upstream of rep and consisted of an 11-bp imperfect direct repeat and a 22-bp sequence tandemly repeated three and one-half times. The overall organization of the pSK11L replicon was remarkably similar to that of pCI305, suggesting that pSK11L does not replicate by the rolling-circle mechanism. Like pSK11L, pKMP1 was unstable in L. lactis LM0230. Deletion analysis allowed identification of several regions which appeared to contribute to the maintenance of pKMP1 in L. lactis LM0230. pKMP1 was significantly more stable in L. cremoris EB5 than in L. lactis LM0230 at all of the temperatures compared. This stability was lost by deletion of a 3.1-kbp PvuII-XbaI fragment which had no effect on stability in L. lactis LM0230. Other regions affecting stability in L. cremoris EB5 but not in L. lactis LM0230 were also identified. Stability assays conducted at various temperatures showed that pKMP1 maintenance was temperature sensitive in both L. lactis LM0230 and L. cremoris EB5, although the plasmid was more unstable in L. lactis LM0230. The region responsible for the temperature sensitivity phenotype in L. lactis LM0230 was tentatively localized to a 1.2-kbp ClaI-HindIII fragment which was distinct from the replication region of pSK11L. Our results suggest that the closely related L. lactis and L. cremoris subspecies behave differently regarding maintenance of plasmids.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson D. G., McKay L. L. Genetic and physical characterization of recombinant plasmids associated with cell aggregation and high-frequency conjugal transfer in Streptococcus lactis ML3. J Bacteriol. 1984 Jun;158(3):954–962. doi: 10.1128/jb.158.3.954-962.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson D. G., McKay L. L. Simple and rapid method for isolating large plasmid DNA from lactic streptococci. Appl Environ Microbiol. 1983 Sep;46(3):549–552. doi: 10.1128/aem.46.3.549-552.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang A. C., Cohen S. N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978 Jun;134(3):1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E. Y., Seeburg P. H. Supercoil sequencing: a fast and simple method for sequencing plasmid DNA. DNA. 1985 Apr;4(2):165–170. doi: 10.1089/dna.1985.4.165. [DOI] [PubMed] [Google Scholar]
- Efstathiou J. D., McKay L. L. Inorganic salts resistance associated with a lactose-fermenting plasmid in Streptococcus lactis. J Bacteriol. 1977 Apr;130(1):257–265. doi: 10.1128/jb.130.1.257-265.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feirtag J. M., Petzel J. P., Pasalodos E., Baldwin K. A., McKay L. L. Thermosensitive plasmid replication, temperature-sensitive host growth, and chromosomal plasmid integration conferred by Lactococcus lactis subsp. cremoris lactose plasmids in Lactococcus lactis subsp. lactis. Appl Environ Microbiol. 1991 Feb;57(2):539–548. doi: 10.1128/aem.57.2.539-548.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasson M. J. In vivo genetic systems in lactic acid bacteria. FEMS Microbiol Rev. 1990 Sep;7(1-2):43–60. doi: 10.1111/j.1574-6968.1990.tb04878.x. [DOI] [PubMed] [Google Scholar]
- Hayes F., Daly C., Fitzgerald G. F. Identification of the Minimal Replicon of Lactococcus lactis subsp. lactis UC317 Plasmid pCI305. Appl Environ Microbiol. 1990 Jan;56(1):202–209. doi: 10.1128/aem.56.1.202-209.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes F., Vos P., Fitzgerald G. F., de Vos W. M., Daly C. Molecular organization of the minimal replicon of novel, narrow-host-range, lactococcal plasmid pCI305. Plasmid. 1991 Jan;25(1):16–26. doi: 10.1016/0147-619x(91)90003-f. [DOI] [PubMed] [Google Scholar]
- Imanaka T., Ano T., Fujii M., Aiba S. Two replication determinants of an antibiotic-resistance plasmid, pTB19, from a thermophilic bacillus. J Gen Microbiol. 1984 Jun;130(6):1399–1408. doi: 10.1099/00221287-130-6-1399. [DOI] [PubMed] [Google Scholar]
- Lin L. S., Meyer R. J. DNA synthesis is initiated at two positions within the origin of replication of plasmid R1162. Nucleic Acids Res. 1987 Oct 26;15(20):8319–8331. doi: 10.1093/nar/15.20.8319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L. S., Meyer R. J. Directly repeated, 20-bp sequence of plasmid R1162 DNA is required for replication, expression of incompatibility, and copy-number control. Plasmid. 1986 Jan;15(1):35–47. doi: 10.1016/0147-619x(86)90012-0. [DOI] [PubMed] [Google Scholar]
- Ludwig W., Seewaldt E., Kilpper-Bälz R., Schleifer K. H., Magrum L., Woese C. R., Fox G. E., Stackebrandt E. The phylogenetic position of Streptococcus and Enterococcus. J Gen Microbiol. 1985 Mar;131(3):543–551. doi: 10.1099/00221287-131-3-543. [DOI] [PubMed] [Google Scholar]
- Macrina F. L., Evans R. P., Tobian J. A., Hartley D. L., Clewell D. B., Jones K. R. Novel shuttle plasmid vehicles for Escherichia-Streptococcus transgeneric cloning. Gene. 1983 Nov;25(1):145–150. doi: 10.1016/0378-1119(83)90176-2. [DOI] [PubMed] [Google Scholar]
- Meissner P. S., Sisk W. P., Berman M. L. Bacteriophage lambda cloning system for the construction of directional cDNA libraries. Proc Natl Acad Sci U S A. 1987 Jun;84(12):4171–4175. doi: 10.1073/pnas.84.12.4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordström K., Austin S. J. Mechanisms that contribute to the stable segregation of plasmids. Annu Rev Genet. 1989;23:37–69. doi: 10.1146/annurev.ge.23.120189.000345. [DOI] [PubMed] [Google Scholar]
- Persson C., Nordström K. Control of replication of the broad host range plasmid RSF1010: the incompatibility determinant consists of directly repeated DNA sequences. Mol Gen Genet. 1986 Apr;203(1):189–192. doi: 10.1007/BF00330402. [DOI] [PubMed] [Google Scholar]
- Polzin K. M., McKay L. L. Identification, DNA sequence, and distribution of IS981, a new, high-copy-number insertion sequence in lactococci. Appl Environ Microbiol. 1991 Mar;57(3):734–743. doi: 10.1128/aem.57.3.734-743.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polzin K. M., Shimizu-Kadota M. Identification of a new insertion element, similar to gram-negative IS26, on the lactose plasmid of Streptococcus lactis ML3. J Bacteriol. 1987 Dec;169(12):5481–5488. doi: 10.1128/jb.169.12.5481-5488.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts R. C., Burioni R., Helinski D. R. Genetic characterization of the stabilizing functions of a region of broad-host-range plasmid RK2. J Bacteriol. 1990 Nov;172(11):6204–6216. doi: 10.1128/jb.172.11.6204-6216.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terzaghi B. E., Sandine W. E. Improved medium for lactic streptococci and their bacteriophages. Appl Microbiol. 1975 Jun;29(6):807–813. doi: 10.1128/am.29.6.807-813.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winberg G., Hammarskjöld M. L. Isolation of DNA from agarose gels using DEAE-paper. Application to restriction site mapping of adenovirus type 16 DNA. Nucleic Acids Res. 1980 Jan 25;8(2):253–264. doi: 10.1093/nar/8.2.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruijn F. J., Lupski J. R. The use of transposon Tn5 mutagenesis in the rapid generation of correlated physical and genetic maps of DNA segments cloned into multicopy plasmids--a review. Gene. 1984 Feb;27(2):131–149. doi: 10.1016/0378-1119(84)90135-5. [DOI] [PubMed] [Google Scholar]


