Abstract
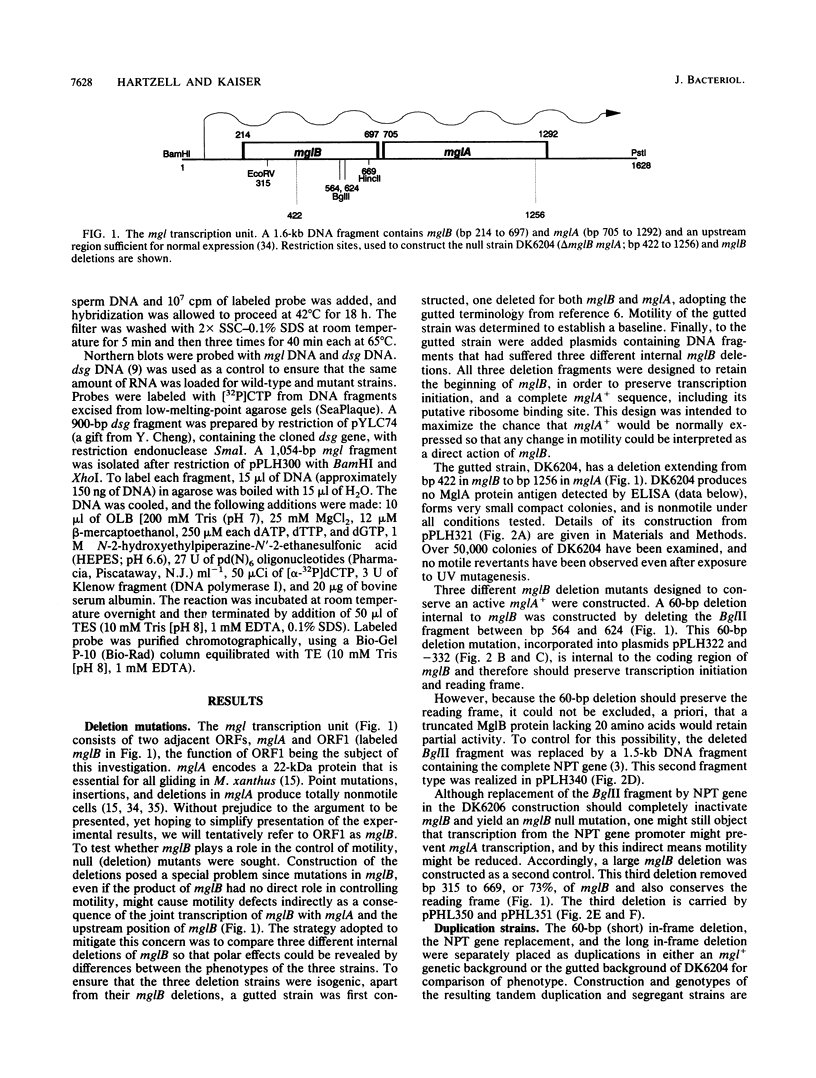
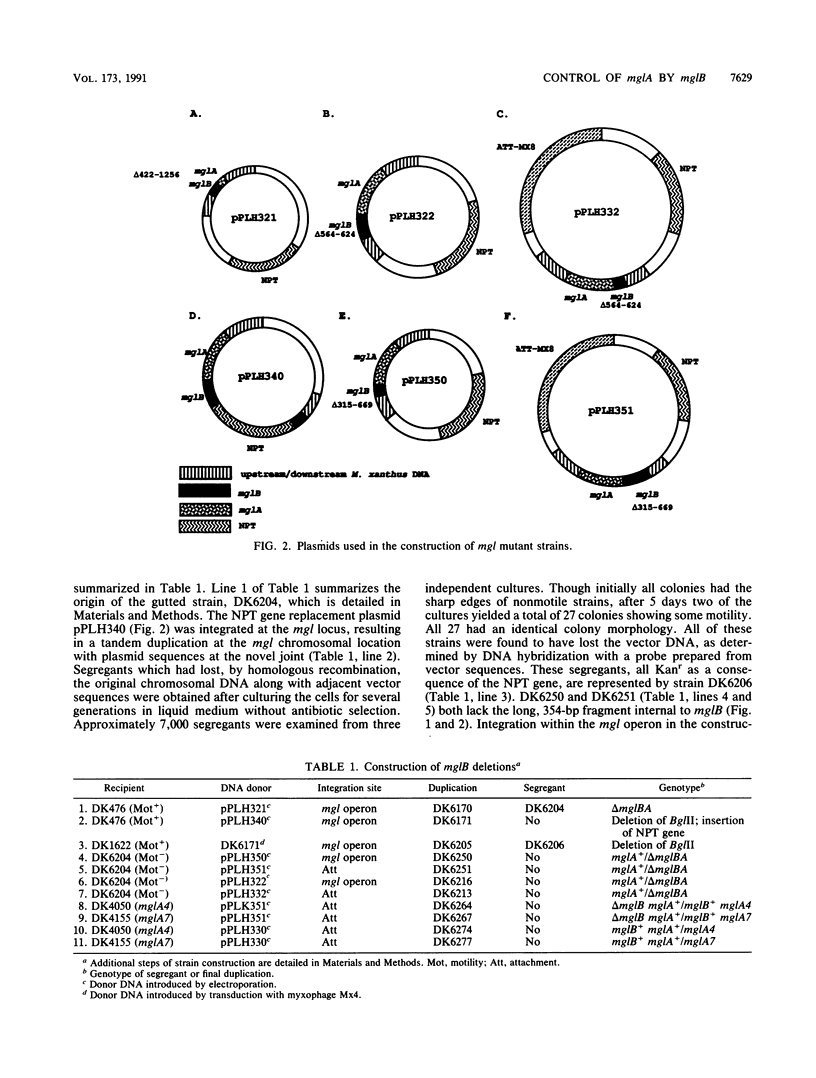
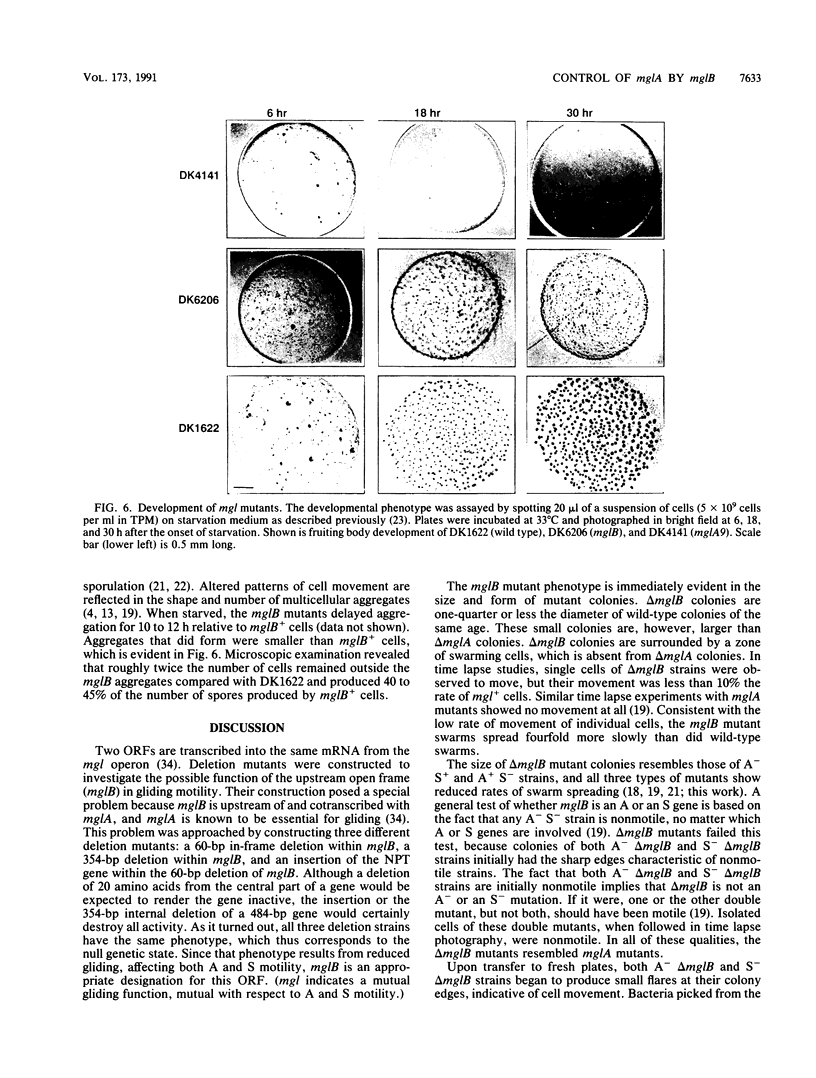
The mgl operon contains two open reading frames (ORFs) which are transcribed together. A collection of nonmotile mutants helped to define the downstream ORF as the mglA gene. Single mutations at the mglA locus completely abolish motility. A series of deletion mutations was constructed to determine the role of the upstream ORF (now called mglB). A strain carrying a deletion in mglB and with an intact mglA produces small colonies. The cells are motile, but their rate of swarm spreading is reduced. Measurements of cell movement showed that mglB mutant cells advanced, on average, less than 0.1 cell length in 5 min. The mglB+ cells advanced an average of 1.3 cell lengths in the same time. Extracts of delta mglB cells contain 15 to 20% as much of the 22-kDa MglA protein as do mglB+ cells, as measured in Western immunoblots and enzyme-linked immunosorbent assays. However, the amount of mgl transcript is the same in the delta mglB mutants as in the mglB+ strain. Heterozygous partial diploids mglB/mglA with the wild-type alleles in trans have normal motility, demonstrating that the largest of the mglB deletions is not polar on mglA. Like other motility defects, a delta mglB mutation alters fruiting body development and sporulation. The mglB mutants delayed aggregation, produced small immature fruiting bodies, and sporulated at 45 to 50% wild-type levels. All aspects of the mglB mutant phenotype are explained by the reduced levels of mglA protein and the assumption that it limits the amount of gliding.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arnold J. W., Shimkets L. J. Cell surface properties correlated with cohesion in Myxococcus xanthus. J Bacteriol. 1988 Dec;170(12):5771–5777. doi: 10.1128/jb.170.12.5771-5777.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck E., Ludwig G., Auerswald E. A., Reiss B., Schaller H. Nucleotide sequence and exact localization of the neomycin phosphotransferase gene from transposon Tn5. Gene. 1982 Oct;19(3):327–336. doi: 10.1016/0378-1119(82)90023-3. [DOI] [PubMed] [Google Scholar]
- Blackhart B. D., Zusman D. R. "Frizzy" genes of Myxococcus xanthus are involved in control of frequency of reversal of gliding motility. Proc Natl Acad Sci U S A. 1985 Dec;82(24):8767–8770. doi: 10.1073/pnas.82.24.8767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackshear P. J. Systems for polyacrylamide gel electrophoresis. Methods Enzymol. 1984;104:237–255. doi: 10.1016/s0076-6879(84)04093-3. [DOI] [PubMed] [Google Scholar]
- Block S. M., Berg H. C. Successive incorporation of force-generating units in the bacterial rotary motor. 1984 May 31-Jun 6Nature. 309(5967):470–472. doi: 10.1038/309470a0. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Burchard R. P. Growth of surface colonies of the gliding bacterium Myxococcus xanthus. Arch Microbiol. 1974 Mar 7;96(3):247–254. doi: 10.1007/BF00590180. [DOI] [PubMed] [Google Scholar]
- Cheng Y., Kaiser D. dsg, a gene required for cell-cell interaction early in Myxococcus development. J Bacteriol. 1989 Jul;171(7):3719–3726. doi: 10.1128/jb.171.7.3719-3726.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis T. N., Urdea M. S., Masiarz F. R., Thorner J. Isolation of the yeast calmodulin gene: calmodulin is an essential protein. Cell. 1986 Nov 7;47(3):423–431. doi: 10.1016/0092-8674(86)90599-4. [DOI] [PubMed] [Google Scholar]
- Dworkin M., Keller K. H., Weisberg D. Experimental observations consistent with a surface tension model of gliding motility of Myxococcus xanthus. J Bacteriol. 1983 Sep;155(3):1367–1371. doi: 10.1128/jb.155.3.1367-1371.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisselsoder J., Campos J. M., Zusman D. R. Physical characterization of bacteriophage MX4, a generalized transducing phage for Myxococcus xanthus. J Mol Biol. 1978 Feb 25;119(2):179–189. doi: 10.1016/0022-2836(78)90432-1. [DOI] [PubMed] [Google Scholar]
- Hartzell P., Kaiser D. Function of MglA, a 22-kilodalton protein essential for gliding in Myxococcus xanthus. J Bacteriol. 1991 Dec;173(23):7615–7624. doi: 10.1128/jb.173.23.7615-7624.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heumann H. G., Kühlwein H. Effects of drugs that influence eucaryotic motile processes on motility of Cystobacter fuscus (Myxobacterales). Z Allg Mikrobiol. 1979;19(8):547–552. doi: 10.1002/jobm.3630190804. [DOI] [PubMed] [Google Scholar]
- Hodgkin J., Kaiser D. Cell-to-cell stimulation of movement in nonmotile mutants of Myxococcus. Proc Natl Acad Sci U S A. 1977 Jul;74(7):2938–2942. doi: 10.1073/pnas.74.7.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KUHLWEIN H. Weitere Untersuchungen an Myxobakterien. Arch Mikrobiol. 1953;19(4):365–371. [PubMed] [Google Scholar]
- Kim S. K., Kaiser D. Cell motility is required for the transmission of C-factor, an intercellular signal that coordinates fruiting body morphogenesis of Myxococcus xanthus. Genes Dev. 1990 Jun;4(6):896–904. doi: 10.1101/gad.4.6.896. [DOI] [PubMed] [Google Scholar]
- Kroos L., Hartzell P., Stephens K., Kaiser D. A link between cell movement and gene expression argues that motility is required for cell-cell signaling during fruiting body development. Genes Dev. 1988 Dec;2(12A):1677–1685. doi: 10.1101/gad.2.12a.1677. [DOI] [PubMed] [Google Scholar]
- Kroos L., Kuspa A., Kaiser D. A global analysis of developmentally regulated genes in Myxococcus xanthus. Dev Biol. 1986 Sep;117(1):252–266. doi: 10.1016/0012-1606(86)90368-4. [DOI] [PubMed] [Google Scholar]
- Kuspa A., Kaiser D. Genes required for developmental signalling in Myxococcus xanthus: three asg loci. J Bacteriol. 1989 May;171(5):2762–2772. doi: 10.1128/jb.171.5.2762-2772.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride M. J., Weinberg R. A., Zusman D. R. "Frizzy" aggregation genes of the gliding bacterium Myxococcus xanthus show sequence similarities to the chemotaxis genes of enteric bacteria. Proc Natl Acad Sci U S A. 1989 Jan;86(2):424–428. doi: 10.1073/pnas.86.2.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCurdy H. D., Ho J., Dobson W. J. Cyclic nucleotides, cyclic nucleotide phosphodiesterase, and development in Myxococcus xanthus. Can J Microbiol. 1978 Dec;24(12):1475–1481. doi: 10.1139/m78-237. [DOI] [PubMed] [Google Scholar]
- McCurdy H. D. Studies on the taxonomy of the Myxobacterales. I. Record of Canadian isolates and survey of methods. Can J Microbiol. 1969 Dec;15(12):1453–1461. doi: 10.1139/m69-259. [DOI] [PubMed] [Google Scholar]
- O'Connor K. A., Zusman D. R. Patterns of cellular interactions during fruiting-body formation in Myxococcus xanthus. J Bacteriol. 1989 Nov;171(11):6013–6024. doi: 10.1128/jb.171.11.6013-6024.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spratt B. G., Hedge P. J., te Heesen S., Edelman A., Broome-Smith J. K. Kanamycin-resistant vectors that are analogues of plasmids pUC8, pUC9, pEMBL8 and pEMBL9. Gene. 1986;41(2-3):337–342. doi: 10.1016/0378-1119(86)90117-4. [DOI] [PubMed] [Google Scholar]
- Stephens K., Hartzell P., Kaiser D. Gliding motility in Myxococcus xanthus: mgl locus, RNA, and predicted protein products. J Bacteriol. 1989 Feb;171(2):819–830. doi: 10.1128/jb.171.2.819-830.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Womack B. J., Gilmore D. F., White D. Calcium requirement for gliding motility in myxobacteria. J Bacteriol. 1989 Nov;171(11):6093–6096. doi: 10.1128/jb.171.11.6093-6096.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]







