Abstract
Bacterial superantigens (SAgs) bind to major histocompatibility complex (MHC) class II molecules and activate T cells in a Vβ-restricted fashion. We recently identified subsets of HLA-DR1 molecules that show selectivity for SAgs. Here, we extend these observations by showing that different cell lineages demonstrate distinct SAg-binding specificities although they all express HLA-DR1. Indeed, B cells bind staphylococcal enterotoxin A (SEA) and toxic shock syndrome toxin 1 (TSST-1) with high affinity while staphylococcal enterotoxin B (SEB) binding is barely detectable. In contrast, DR1-transfected HeLa cells show efficient binding of SEB, but not of SEA or TSST-1. We investigated the class II maturation events required for efficient interaction with SAgs and found that the ability of cells to bind and present the toxins can be drastically modulated by coexpression of the class II-associated invariant chain (Ii) and HLA-DM. SEA binding to DR1 molecules required coexpression of Ii, whereas TSST-1 binding was selectively enhanced by DM. Binding of SEB was affected by cell type-specific factors other than Ii or DM. The selectivity of SAgs for different MHC class II populations was minimally affected by HLA-DR intrinsic polymorphism and could not be explained by binding to alternative sites on DR molecules. Our results indicate that SAgs are sensitive to structural heterogeneity in class II molecules, which is consequent to the differential regulation of expression of antigen processing cofactors. Therefore, we speculate that Staphylococcus aureus have retained the ability to express numerous SAgs in adaptation to the micro-heterogeneity displayed by MHC class II molecules and that this may relate to their ability to infect different tissues.
Keywords: antigen processing, superantigens, enterotoxin, Staphylococcus aureus
Bacterial superantigens (SAgs) are toxins that bind to major histocompatibility complex (MHC) class II molecules outside the peptide-binding groove and trigger T cell activation by interacting with the Vβ element of the T cell receptor (for a review, see ref. 1). SAgs produced by Staphylococcus aureus constitute a family of structurally related globular proteins termed staphylococcal enterotoxins (SEA, SEB, SEC1-3, SED, SEE, SEH) and toxic shock syndrome toxin 1 (TSST-1). Their ability to induce massive secretion of pyrogenic cytokines (interleukin 1, tumor necrosis factor α) in humans is responsible for a significant number of cases of toxic shock. Apart from TSST-1, these proteins share a similar topological fold, but their MHC class II binding sites differ considerably both in their location and biochemical composition. For instance, while the C-terminal domain of SEA is involved in a coordination bond with Zn2+ and residue His-81 of the β chain of the human MHC class II molecule HLA-DR1 (2, 3), the binding of SEB and TSST-1 relies mostly on residue Lys-39 in the DRα1 domain (4–7). These differences suggest that the toxins have evolved to exert different physiological functions or alternatively, that structural constraints preclude the usage of a unique mode of binding to class II molecules.
SEB and TSST-1 share a wide binding interface on DR1 involving common residues of the α chain (5, 6) and yet they do not compete with each other for binding to cell surface HLA-DR1 (7, 8). We therefore proposed that micro-heterogeneity in the structure of cell surface MHC class II molecules leads to structurally distinct subsets of HLA-DR1 molecules displaying different specificities with regards to their interaction with SAgs (7). A role of the class II-bound peptide in modulating the affinity of SEA and TSST-1, but apparently not SEB, has been demonstrated (6, 7, 9–11). The use of soluble MHC class II constructs covalently bound to single peptides demonstrated that the kinetics of SEA interaction can greatly vary when different peptides are used (10). However, a conventional antigen presenting cell (APC) usually expresses several MHC alleles and isotypes and the peptide contents of those MHC class II molecules is heterogeneous (12). It is thus reasonable to think that the average affinity of the entire population of peptide/MHC complexes, on the surface of an APC, may not result in a significant difference in the ability of these cells to present a given SAg.
Here, we address the relevance of these findings and show that different cell lineages differ dramatically in their ability to generate MHC class II structures capable of binding SAgs. We have investigated critical events in class II assembly and maturation, such as the interaction with the invariant chain (Ii) or HLA-DM, to determine their role in the modulation of SAg binding to DR1 subsets. Ii assists the folding and targeting of MHC class II molecules to peptide-loading compartments while HLA-DM catalyzes the exchange of loosely bound Ii fragments (CLIP) and their subsequent conversion to stable MHC class II/peptide complexes (for a review, see ref. 13). We demonstrate the critical role of Ii and HLA-DM in the generation of SAg-binding subsets of MHC class II molecules and suggest that the mechanisms by which these antigen presentation cofactors affect the binding of the bacterial toxins does not simply depend on a differential display of class II-bound oligopeptides.
MATERIALS AND METHODS
Cell Lines.
HeLa (a human epithelial cell line) and DAP-3 (a murine fibroblastic cell line) are both negative for endogenous MHC class II expression (data not shown; ref. 14). HeLa DR1 and DAP DR1 are cell lines stably transfected using Ca3(PO4)2 coprecipitation with DRαDRβ1*0101 (7). DAP DR1 are transfected with human Ii (15), and the expression was confirmed by Western blot analysis using an anti-CLIP antibody (16). This polyclonal anti-CLIP antiserum also reacts with full-length Ii (gift from P. Cresswell, Yale University). HeLa DRαK39E cells are HeLa stably transfected with DRαK39E cDNA (17) and DRβ1*0101. The latter were selected on G418 1 mg/ml (GIBCO) and sorted with anti-DR antibody 50D6. HeLa DR1/Ii cells were generated by cotransfection of HeLa DR1 with the p33/35 human Ii cDNA (gift from E. O.Long, National Institutes of Health) cloned in RSV.7hygro (18) and selected in hygromycin (InterScience, Markham, Canada) (346 units/ml). The Ii transfectants were cloned and screened by intracellular immunostaining in PBS, 0.01% saponin, and Western blotting for expression using the PIN-1 Ii mAb. Two different clones were compared in toxin binding and yielded similar results. Cells were grown in DMEM supplemented with 5% calf serum and the selection agent. The human Epstein–Barr virus (EBV)-transformed B cell line LG2 (DR1/DR1) was provided by L. J. Stern (Massachusetts Institute of Technology). Raji (DR3/DRw10, DRw52) and 45.1 (DR1 hemizygous) are also EBV-transformed B cell lines. The cell lines T2DR3 and T2DR3DM+ (P. Cresswell, Yale University) were described (19). T2 DR1 was kindly provided by S. Demotz (Université de Lausanne). T2 DR1, T2 DR3, T2 DR3DM, Raji, 45.1, and LG2 were grown in RPMI 1640 medium (GIBCO) supplemented with 5% fetal calf serum (FCS) and 2-mercaptoethanol.
Human Peripheral Blood Mononuclear Cell (PBMC) Purification.
Human PBMCs were enriched in class II+ cells and purified as follows. Briefly, peripheral blood was diluted in PBS and underlayered with Ficoll-Hypaque (Pharmacia). After centrifugation, the interface was collected and diluted with an equal volume of PBS. Cells were washed and resuspended at 5 × 106 cells/ml in PBS. T cells were then removed by incubation in 1:1 (vol/vol) ratio FCS (GIBCO) preabsorbed with sheep erythrocyte and 2-aminoethylisothiouronium bromide (Sigma)-treated sheep erythrocyte (1:20) for 10 min at 37°C. The cells were then incubated 45 min−1⋅hr at 4°C, spun down, and resuspended in RPMI 1640 medium/10% FCS. The suspension was layered on top of a Ficoll gradient and centrifuged. The interface was collected, diluted, and washed in PBS.
Binding Assays.
125I-Radiolabeled toxins were obtained by incubating twenty μg of SEA, SEB or TSST-1 (Toxin Technology, Sarasota, FL) for 10 min using 0.5 μg iodogen (Pierce) and 250 μCi 125-I (Amersham; 1 Ci = 37 GBq). The reaction was stopped with PBS/0.05% NaN3. The proteins were fractionated from free iodide on Sephadex G-25 (Pharmacia) beads columns blocked with 1% BSA. SDS/PAGE separation of the labeled fraction confirmed that the toxin of interest was responsible for >95% of the total specific activity. The binding assay was performed as described (7, 20). Unless indicated otherwise, 106 cells were incubated in 200 μl for 4 h, with 100 ng of radiolabeled SEA, SEB, or TSST-1 in DMEM (2% FCS/0.05% NaN3) in the absence or presence of a 10-fold excess of unlabeled competitor. Affinity measurements and competition curves were performed by adding various amounts of cold toxin to 100 ng of radiolabeled toxin. Calculations of affinities were performed as described (7).
125I-Cell Surface Labeling and Immunoprecipitation.
For cell surface labeling with 125Iodine, cells were harvested in PBS/5 mM EDTA and washed extensively in Goding’s PBS (31). Approximately 107 cells/labeling were labeled with 1 mCi Na125I (Amersham) using lactoperoxidase/H2O2 and washed again in Goding’s PBS. Immunoprecipitation of 125I-labeled surface HLA-DR were performed as described in ref. 21. For immunoprecipitation in LG2, cells were washed four times in PBS and lysed in 500 μl lysis buffer (20 mM Tris pH 7.5/150 mM NaCl/1% Triton X-100 or Nonidet P-40). MHC class II molecules were immunoprecipitated using SEA or TSST-1 coupled to CNBr-activated Sepharose beads (Pharmacia) for 4 h in presence of proteases inhibitors [1 mM phenylmethylsulfonyl fluoride/10 μg/ml aprotinin/10 μg/ml leupeptin (Sigma-Aldrich)]. Cell surface immunoprecipitations were performed by incubating biotinylated SEA and TSST-1 fragment for 4 h at 37°C in medium containing 0,1% NaN3. The cells were then washed eight times, lysed, and incubated for 1 h using streptavidin-coupled magnetic beads. The immunoprecipitations were washed at least five times in lysis buffer, and beads were resuspended in Laemmli sample buffer containing 2% SDS.
Two-Dimensional SDS/PAGE Analysis.
Immunoprecipitated samples were eluted in Laemmli sample buffer for >1 h. Samples were run on a 7.5 or 10% SDS/PAGE tube gels. The tracks were then soaked in sample buffer, heated at 95°C for 15 min, and run in the second dimension on SDS/PAGE slab gels. Gels were exposed for 1–2 weeks using X-Omat/AR autoradiogram films (Kodak). [See Busch et al. (21) for more details.]
RESULTS
Expression of DR1 Molecules at the Cell Surface Is Not Sufficient for Efficient Binding of SAgs.
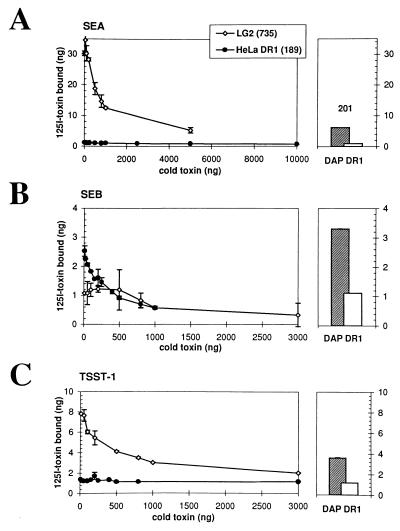
We compared binding of SEA, SEB, and TSST-1 to DR1-transfected HeLa cells, which do not express Ii and HLA-DM constitutively (14), to their binding to LG2 cells (DR1 homozygous) which express most of the molecular requirements for antigen processing and presentation. Barely detectable specific binding of SEA or TSST-1 was observed on HeLa DR1 cells (Fig. 1A and C), in the presence of 100 ng 125I-toxin (18.5 nM and 22.7 nM, respectively). In sharp contrast, HeLa cells transfected with DR1 bound strongly 100 ng (17.6 nM) of SEB (Fig. 1B). The dissociation constant (Kd) of this interaction calculated for DR1 expressed on HeLa cells is 1.1 × 10−7 M, very similar to the affinity previously reported on DAP DR1 transfectants (7, 8, 20). The binding site of SEB on DR1, as characterized both by mutagenesis (7) and x-ray diffraction studies (5), is conserved in HeLa DR1 as mutation DRαK39E (Lys to Glu) completely abrogates binding (data not shown).
Figure 1.
SEA, SEB, and TSST-1 binding to HeLa DR1 and LG2. Specific and saturable binding is demonstrated when increasing amounts of cold toxin successfully displace bound 125I-labeled SEA (A), SEB (B), or TSST-1 (C) as described (7). HLA-DR expression was analyzed by cytofluorometry using the anti-DR antibody L-243. The numbers indicate the mean of fluorescence for HLA-DR expression on all three cell types. (Right) The hatched bars represent the binding of the same amount (100 ng) of SEA (A), SEB (B), or TSST-1 (C) on DAP DR1 cells as a control. Open bars represent the binding of 100 ng of toxin in the presence of a 10-fold excess (1 μg) of cold toxin. Standard variations for each point (duplicates) is represented by the error bars. Because we do not detect binding of SEB on LG2, we approximate the dissociation constant (Kd) of the interaction to be ≥10−5 M.
We then compared the binding of all three toxins on LG2, an EBV-transformed DR1-homozygous B cell line. These cells bind SEA and TSST-1 well (Fig. 1 A and C), with affinities comparable to values previously published for DR1 on other cell lines (Kd = 4 × 10−8 M and 2 × 10−7 M, respectively) (7, 20). However, we observed very poor binding of SEB (Fig. 1B) to LG2 despite the fact that these cells express high levels of MHC class II molecules (Kd ≥ 10−5 M). Accordingly, this striking difference in the ability of LG2 and HeLa to bind toxins has dramatic consequences on T cell activation triggered by those APCs. A 1,000-fold decrease in the ability to present SEB was observed when LG2 was compared with HeLa DR1 even though the latter fail to express the appropriate costimulatory molecules (unpublished data). Conversely, HeLa DR1 required 100,000-fold more SEA to stimulate an SEA-responsive T cell hybridoma (data not shown). From these results, we conclude that cells can be deficient in the requirements to bind and present bacterial SAgs with high affinity although they express adequate levels of MHC class II molecules. Moreover, the affinities measured for SEA or TSST-1 between DAP and LG2, or for SEB between DAP DR1 and HeLa DR1, did not differ significantly.
SEA and TSST-1 Bind Well to HLA-DM and Ii-Expressing Class II-Positive B Cells.
Two distinct mechanisms could explain the selective binding of SAgs between HeLa DR1 and LG2 cells. Because class II-bound peptides can modulate SAg binding to class II molecules (6, 7, 10, 11), it is possible that individual specific, class II-bound peptides are required and not endogenously expressed in these cells. Alternatively, the differential expression of the molecular elements required for functional antigen processing, such as HLA-DM or Ii, leads to the generation of distinct MHC class II structures to which SAgs cannot bind.
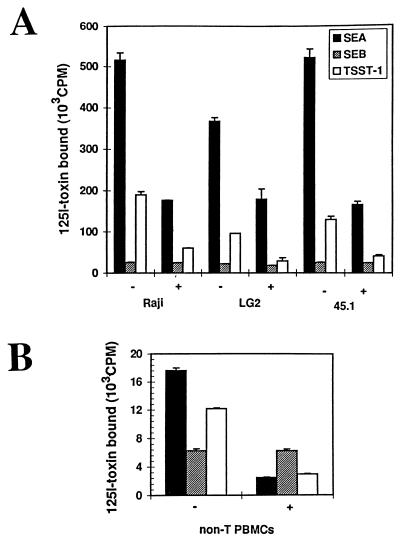
We tested EBV-transformed B cell lines expressing class II molecules constitutively, as well as peripheral blood class II-expressing mononuclear cells (PBMCs) for their ability to bind SAgs (Fig. 2). We found that SEA and TSST-1 bind efficiently to MHC class II expressed in other B cells whereas SEB binding was poor (Fig. 2 A and B). The binding of SEA, SEB, and TSST-1 on 45.1 (DR1), another B lymphoma-derived tumor cell line, was comparable to LG2. PBMCs isolated from a DR1 homozygous individual bound SEA and TSST-1 well. However, these cells bind SEB poorly (Fig. 2B). This result shows that the LG2 phenotype can be generalized to primary cells. Finally, Raji cells which express another set of DR alleles (DR3/DRw10, DRw52) nevertheless gave the same pattern of SAg reactivity as LG2 (Fig. 2A), despite the different spectrum of peptides displayed on this DR allele (22). This observation suggests that differences in the array of peptides due to HLA-DR polymorphism do not alter the ability of B cell lines to bind the toxins. Clearly then, the dramatic differences in SAg reactivity for the same DR allele expressed in different cell lines (see above) pointed to additional factors influencing SAg binding.
Figure 2.
Binding of SEA, SEB, and TSST-1 on DR-expressing cells lines and purified non-T PBMCs from a DR1 homozygous individual. (A) Specific binding of 125I-SEA, -SEB, or -TSST-1 with (+) or without (−) a 10-fold excess of cold competitor. (B) Binding of the toxins to class II-expressing peripheral mononuclear cells (5 × 106) from a DR1 homozygous individual with (+) or without (−) a 10-fold excess of cold toxin. In all these experiments the binding was compared (using the same toxin preparation and comparable amounts) to DAP DR1 cells to confirm the functional integrity of the toxins (data not shown). Standard variations for each point (duplicates) is represented in the error bars.
Ii Expression Affects the Binding of SEA and TSST-1 on MHC Class II Molecules.
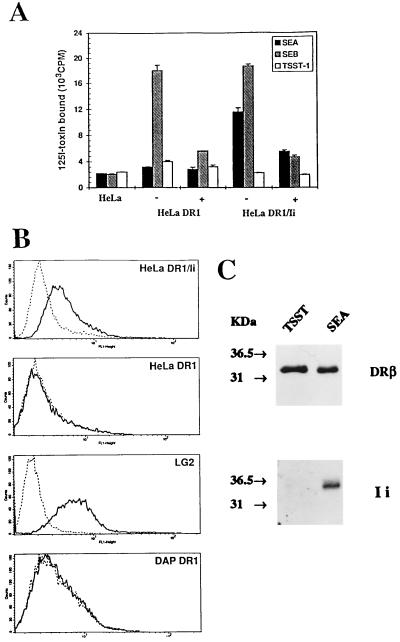
We specifically addressed the role of cellular factors involved in the generation of different SAg-binding subsets. We first transfected the p33–35 Ii cDNA in HeLa DR1 cells. Fig. 3A shows that expression of Ii in HeLa-DR1 dramatically restores high affinity binding of SEA. This binding is dependent on the presence of Zn2+ in solution suggesting that it is mediated through the β81 histidine residue (data not shown), as previously described for DAP DR1 and LG2 cells (2, 3). On the other hand, expression of Ii did not restore the binding of TSST-1 (Fig. 3A). Ii could possibly affect the binding of SAgs by either affecting the nature of the class II ligands (21) or directly through its expression at the cell surface as previously suggested for TSST-1 (23). As indicated in Fig. 3B, Ii is expressed at the cell surface of HeLa DR1/Ii cells. Indeed, the p35 form of Ii is bound to a majority of cell surface DR1 in these cells (data not shown and see below).
Figure 3.
Effect of the Ii on the binding of SEA, SEB, and TSST-1 on HeLa DR1 cells. (A) Specific binding of 125I-SEA, -SEB, or -TSST-1 on HeLa transfectants in presence (+) or absence (−) of a 10-fold excess of cold competitor. (B) Surface staining confirm the expression of Ii at the surface of HeLa DR1/Ii or LG2, but not DAP or HeLa DR1. The control antibody (dotted line) is fluorescein isothiocyanate-coupled goat anti-mouse Ig. Cell surface Ii expression (solid line) was determined using the anti-Ii antibody BU45 (which recognizes the extracellular C-terminal end of Ii). (C) SEA, but not TSST-1, bind to DR1/Ii complexes. The cell surface immunoprecipitation was carried out by incubating intact LG2 cells with biotinylated SEA or TSST-1. After washing, cells were lysed and incubated with streptavidin-coupled magnetic beads. (Upper) Samples blotted using XD5, an anti-DRβ antibody, and the same samples (Lower) were blotted with an anti-CLIP antibody (obtained from Peter Cresswell, Yale University).
To confirm that TSST-1 cannot bind to DR/Ii complexes, we performed immunoprecipitation experiments on LG2 cells, which bind SEA and TSST-1 with high affinity (Fig. 1). The experiment was performed on those cells because Ii is present on the surface of LG2 cells (Fig. 3B), although only a minority of cell surface DR1 molecules are complexed to Ii (24). Whereas SEA could interact with DR/Ii complexes from the surface of LG2 cells, Ii could not be detected from DR1 molecules associated with TSST-1 (Fig. 3C). Also, we could easily immunoprecipitate DR/Ii complexes from the surface of HeLa DR1/Ii cells with biotinylated SEA and streptavidin-coupled beads, but not with TSST-1 (data not shown). Of note, the DAP DR1 cells used were previously transfected with Ii (15) and showed low levels of Ii expression by Western blotting (data not shown). However, differences in trafficking or recycling of DR1/Ii results in the absence of detectable expression on their surface (Fig. 3B), which is sufficient to allow efficient binding of TSST-1 (Fig. 1). As indicated by the LG2 and DAP DR1 cells, SEA and TSST-1 bind with high affinity to MHC class II molecules that have folded in the presence of Ii. However, lack of TSST-1 binding to HeLa DR1/Ii cells allow us to conclude that surface expression of Ii impairs the binding of TSST-1. Finally, the high affinity binding of SEB remained unaffected by expression or absence of Ii in all cells analyzed.
Efficient Binding of TSST-1 to Cell Surface HLA-DR1 Is Dependent on HLA-DM Expression.
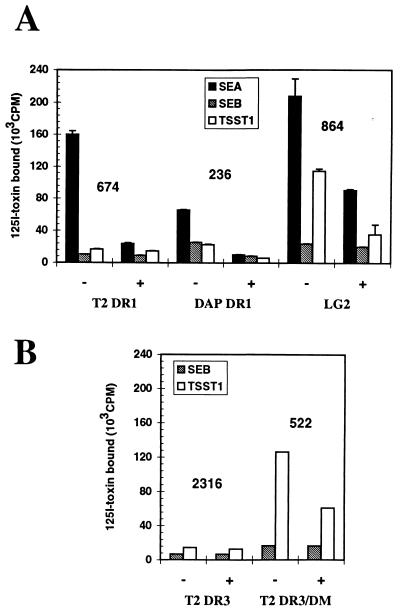
With the knowledge that HLA-DM can modulate the interaction of Ii with HLA-DR and following our demonstration that cell surface Ii prevents TSST-1 binding, we tested the binding of SEA, SEB, and TSST-1 to DR1 expressed in T2 cells that lack functional HLA-DM. DR molecules at the surface of these cells largely remain bound to Ii fragments (CLIP) (19). Efficient binding of SEA was detected on T2 DR1 cells (Fig. 4A). However, binding of TSST-1 could not be detected on this cell line, confirming the inability of this toxin to interact with DR1 molecules bound to Ii or CLIP, and suggesting a role for DM in generating TSST-1-binding DR molecules. Therefore, we used T2 cells transfected with both HLA-DR and DM to confirm its effect on the binding of TSST-1. Cells expressing DR3 were used because SDS-unstable CLIP/DR3 complexes are long-lived at the cell surface, allowing their accumulation prior to displacement by serum peptides (16). The binding of TSST-1 is dramatically restored on T2 DR3 expressing HLA-DM molecules even though these cells express lower levels of DR compared with the DM-negative cells (Fig. 4B). This result shows that removal of CLIP peptides in Ii-positive cells is required to bind TSST-1. Binding of TSST-1 on T2 cells was indeed augmented by an exogenous addition of class II-restricted peptides (9). The observation that DAP DR1 displays a significant fraction of peptide/MHC complexes (SDS-stable) at the cell surface explains the binding of TSST-1 on these cells despite the lack of HLA-DM (unpublished data). Lastly, it is also interesting to note that DM expression in T2 cells had little effect on SEB binding (Fig. 4) and is in agreement with the poor binding of SEB on B cell lines. Therefore, the difference in the affinity of SEB for DR on fibroblasts versus B cells cannot be ascribed to differential HLA-DM expression.
Figure 4.
Binding of SAgs to DM+ and DM-T2 cells. (A) Comparison of the binding of SEA, SEB, and TSST-1 on T2 DR1 cells, LG2, or DAP DR1 cells. (B) SEB and TSST-1 binding was compared in T2 DR3 cells with or without HLA-DM expression. Efficient binding of all three toxins is confirmed on DAP DR1 and LG2 cells as a control. The numbers represent the mean of fluorescence in cytofluorometry (FACScan; Becton Dickenson) for each cell line as detected using the L243 anti-DR antibody.
Impaired Binding of SEA and TSST-1 on Ii-Negative Cells Is Related to the Presence of Polypeptides Occupying the Class II Groove.
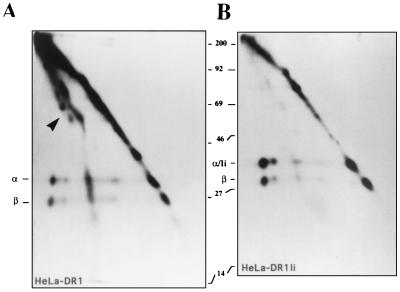
A recent study demonstrated that large polypeptides bound to cell surface MHC class II molecules, including DR1, in the absence of Ii expression (21). We compared 125I-labeled MHC class II molecules immunoprecipitated from the cell surface of HeLa DR1 (Fig. 5A) vs. HeLa DR1/Ii (Fig. 5A) and ran the complexes on two-dimensional SDS/PAGE electrophoresis. The first dimension (horizontal) is run under nonboiled conditions. The sample is then boiled to dissociate the MHC class II α/β heterodimeric complexes before it is run in the second dimension (vertical). As previously described, a significant fraction of the class II α/β formed SDS-stable complexes with various polypeptides (Fig. 5A) in the absence of Ii expression (21). However, when Ii was coexpressed, these polypeptides were largely absent from MHC class II molecules (Fig. 5B). We also observed in both Ii-positive and Ii-negative HeLa DR1 cells an appreciable proportion of dissociated (SDS-unstable) DRα/β heterodimers migrating on the diagonal at the expected molecular weight for the individual α and β chains. Finally, in B cells such as LG2, the great majority of cell surface MHC class II molecules are SDS-resistant and bound to conventional peptides and migrate at a molecular weight of 55 kDa (C55) in the nondenaturing dimension (data not shown). Thus, one possible explanation for SEA binding by HeLa-DR1Ii, but not HeLa-DR1 cells, is that SEA is unable to bind class II molecules bound to intact polypeptides (other than Ii). Accordingly, the low levels of Ii expression in DAP DR1 cells appeared to be sufficient to prevent most, but not all, of the DR heterodimers from associating with large polypeptides (unpublished data and ref. 21). The number of MHC class II molecules able to bind SEA and TSST-1 is indeed reduced in DAP DR1 (7).
Figure 5.
Comparison of the polypeptides bound to DR molecules immunoprecipitated from Ii+ or Ii- HeLa DR1 cells. DR1 molecules from HeLa DR1 (A) or HeLa DR1/Ii (B) cells were immunoprecipitated using the anti-class II antibody ISCR3 (which binds specifically to intact αβ heterodimers) and run on a two-dimensional SDS/PAGE without boiling in the first dimension (left to right), and boiled in the second dimension (top to bottom). The α and β chain of the DR dimer are easily separated in the second dimension in boiled/reducing conditions. The arrow in A points to a heterogeneous array of polypeptides, which form 1:1 complexes with DR1 dimers in the absence of Ii. The polypeptide/DR1 complexes are shifted to the left of the diagonal because of their SDS stability (A). Ii comigrates with the DR α chain in the second dimension in B. Cells were labeled and immunoprecipitated as described in ref. 21.
DISCUSSION
Since the first reports suggesting that MHC class II-bound peptides have a role in the fine tuning of SAg cell surface binding (6, 7), various groups have provided interesting results supporting this concept (9–11). These studies revealed at most a 10-fold difference in affinity of SAgs for peptide-loaded murine class II molecules that cannot fully explain the total lack of binding that we report here on the different cell types. Additionally, SEA seems to bind to the majority of cell surface HLA-DR1 molecules on B cells (J.T., P.M.L., and R.P.S., unpublished data), suggesting that the peptides that are not permissive to SAg binding represent a minority of class II-bound antigens and cannot account for the total lack of binding to certain cell types. In the experiments presented here, we show that cells lack the requirements to bind SAgs efficiently although they express adequate levels of MHC class II molecules. Moreover, those studies illustrate that specific antigen presentation cofactors can drastically modulate the binding of the toxins to MHC class II molecules independently of the HLA-haplotype.
Our results show that expression of Ii in HeLa-DR1 cells is sufficient for SEA binding and that removal of Ii or CLIP is critical for binding of TSST-1. Apparently, differences in the peptide profile due to polymorphism of DR molecules, as well as those due to species- or lineage-specific peptides, did not greatly influence SAg binding. Rather, the profound effects of antigen processing/presentation cofactors on SAg binding raise the intriguing possibility that qualitatively different sets of class II groove ligands, loaded in the presence or absence of specific cofactors, confer a differential specificity for SAgs. Since the expression of HLA-DM and Ii is required to bind SEA and TSST-1 with high affinity, it is therefore not surprising to find that cells expressing fully mature peptide/MHC complexes at the cell surface bind both toxins with the highest efficiency. For example, the inability to detect SEA binding Ii-negative HeLa cells appears to be due to the fact that DR1 molecules in these cells are associated with intact polypeptides, rather than short peptides. Such polypeptides have previously been shown to bind in the antigen-binding groove (21), and their protruding ends may sterically block regions of the DR molecule that are important for SEA binding, such as β81His. Since the binding site of TSST-1 spans the peptide-binding groove, a similar mechanism could be put forward to explain the low binding of this toxin to HeLa DR1 cells. Preliminary biochemistry analysis shows that TSST-1 is also affected by class II-bound polypeptides (P.M.L., J.T., and R.P.S., unpublished data).
The observation that the TSST-1-binding site overlaps with Ii confirms previous results in which a cytoplasmic-tail truncated version of Ii, expressed at the cell surface, affected the presentation of this toxin (23). Herein, the CLIP fragments of Ii also seem to interfere with the binding of TSST-1, although the structure of the SDS-unstable CLIP/DR3 complex is structurally and serologically similar to stable HA/DR1 complexes (25, 26). Whether the interference with TSST-1 binding is due to the peptide side chains of CLIP or due to the SDS-instability induced by this peptide will be the subject of future reports (P.M.L., et al, unpublished data). Nevertheless, these observations clearly demonstrate the potential of SAgs to serve as molecular probes to characterize the fine structural features of MHC class II antigens.
The contrasting phenotypes observed among different DR1-expressing cells, together with other studies on the biochemistry of MHC class II molecules, confirm that expression of the latter, at the cell surface, can be very heterogeneous. The physical inability of SAgs to interact with MHC class II of different conformations may explain why S. aureus has maintained such a diversified arsenal of toxins, all sharing the ability to bind MHC class II molecules and target T cell receptors in a seemingly redundant fashion. Since we show here that bacterial SAgs are sensitive to different MHC class II structures generated in absence of HLA-DM or Ii, it is possible that MHC class II-positive cells which discordantly express these cofactors may bind and present various toxins differentially (27). Indeed, early classification of the different strains of S. aureus suggested that TSST-1 expression is over-represented in strains targeting the blood compartment, as compared with SEB whose expression is more restricted to strains invading the gastrointestinal tract (28). Accordingly, our results suggest that TSST-1, in contrast to SEB, would bind and be presented much more efficiently by fully functional APCs as opposed to cells physiologically expressing low levels of Ii and DM (29, 30). An intestinal pathogen could possibly gain a selective advantage by expressing a given SAg rather than another and this heterogeneity may well be functionally exploited by bacteria encoding these SAgs. Hence, our results bring up very exciting questions about the function of the individual SAgs in the context of an infectious process and merit further investigation into this apparent pathogen tropism.
The factors responsible for the difference in SEB binding observed here between B cells and nonhematopoetic class II-expressing cells appear to be independent of variations in class II-ligands imposed by Ii or DM. These results are in agreement with reports that fail to demonstrate a role for the peptide in the modulation of SEB binding (10, 11) and x-ray diffraction studies on SEB/DR1 which show that SEB binds at a distance from the peptide groove (5). The observation that fully folded MHC/peptide complexes on B cells yet do not bind SEB well supports the notion that SAgs are not merely sensitive to perturbations in class II structure, but rather specifically designed to bind to cells expressing different MHC class II structures.
Acknowledgments
We are grateful to Peter Cresswell and Liza Denzin for providing T2DR3 and T2DR3DM cell lines, Peter Cresswell for the anti-CLIP antibody, Eric Long for the p33/p35 Ii cDNA, Larry Stern for the LG2 cell line, and Stephane Demotz for the T2 DR1 cell line. We thank Jean-Pierre Fortin and Helen McGrath for technical assistance, P. A. Cazenave (Institut Pasteur, Paris) for his kind hospitality in his laboratory, and our colleagues for helpful discussions and reading of the manuscript. P.M.L. is funded by the Medical Research Council of Canada. This work was supported by a National Cancer Institute of Canada grant (NCIC-007273).
ABBREVIATIONS
- APC
antigen presenting cell
- Ii
invariant chain
- MHC
major histocompatibility complex
- SEA and SEB
staphylococcal enterotoxin A and B
- SAg
superantigen
- TSST-1
toxic shock syndrome toxin 1
- EBV
Epstein–Barr virus
- PBMC
peripheral blood mononuclear cell
- FCS
fetal calf serum
References
- 1.Scherer M T, Ignatowicz L, Winslow G M, Kappler J W, Marrack P. Annu Rev Cell Biol. 1993;9:101–128. doi: 10.1146/annurev.cb.09.110193.000533. [DOI] [PubMed] [Google Scholar]
- 2.Herman A, Labrecque N, Thibodeau J, Marrack P, Kappler J W, Sékaly R-P. Proc Natl Acad Sci USA. 1991;88:9954–9958. doi: 10.1073/pnas.88.22.9954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hudson K R, Tiedemann R E, Urban R G, Lowe S C, Strominger J L, Fraser J D. J Exp Med. 1995;182:711–720. doi: 10.1084/jem.182.3.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thibodeau J, Labrecque N, Denis F, Huber B T, Sékaly R-P. J Exp Med. 1994;179:1029–1034. doi: 10.1084/jem.179.3.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jardetzky T S, Brown J H, Gorga J C, Stern L J, Urban R G, Chi Y-I, Stauffacher C, Strominger J L, Wiley D C. Nature (London) 1994;368:711–718. doi: 10.1038/368711a0. [DOI] [PubMed] [Google Scholar]
- 6.Kim J, Urban R G, Strominger J L, Wiley D C. Science. 1994;266:1870–1874. doi: 10.1126/science.7997880. [DOI] [PubMed] [Google Scholar]
- 7.Thibodeau J, Cloutier I, Lavoie P M, Labrecque N, Mourad W, Jardetzky T, Sékaly R-P. Science. 1994;266:1874–1878. doi: 10.1126/science.7997881. [DOI] [PubMed] [Google Scholar]
- 8.Chintagumpala M M, Mollick J A, Rich R R. J Immunol. 1991;147:3876–3881. [PubMed] [Google Scholar]
- 9.Von Bonin A, Ehrlich S, Malcherek G, Fleischer B. Eur J Immunol. 1995;25:2894–2898. doi: 10.1002/eji.1830251028. [DOI] [PubMed] [Google Scholar]
- 10.Kozono H, Parker D, White J, Marrack P, Kappler J. Immunity. 1995;3:187–196. doi: 10.1016/1074-7613(95)90088-8. [DOI] [PubMed] [Google Scholar]
- 11.Wen R, Cole G A, Surman S, Blackman M A, Woodland D L. J Exp Med. 1996;183:1083–1092. doi: 10.1084/jem.183.3.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chicz R M, Urban R G, Lane W S, Gorga J C, Stern L J, Vignali D A A, Strominger J L. Nature (London) 1992;358:764–768. doi: 10.1038/358764a0. [DOI] [PubMed] [Google Scholar]
- 13.Busch R, Mellins E D. Curr Opin Immunol. 1996;8:51–58. doi: 10.1016/s0952-7915(96)80105-1. [DOI] [PubMed] [Google Scholar]
- 14.Chang C-H, Flavell R A. J Exp Med. 1995;181:765–767. doi: 10.1084/jem.181.2.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jacobson S, Sékaly R-P, Jacobson C L, McFarland H E, Long E O. J Virol. 1989;63:1756–1762. doi: 10.1128/jvi.63.4.1756-1762.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Avva R R, Cresswell P. Immunity. 1994;1:763–774. doi: 10.1016/s1074-7613(94)80018-9. [DOI] [PubMed] [Google Scholar]
- 17.Thibodeau J, Dohlsten M, Cloutier I, Lavoie P M, Björk P, Michel F, Leveille C, Mourad W, Kalland T, Sékaly R-P. J Immunol. 1997;158:3698–3704. [PubMed] [Google Scholar]
- 18.Long E O, Rosen-Bronson S, Kard D R, Malnati M, Sékaly R-P, Jaraquemada D. Hum Immunol. 1991;31:229–235. doi: 10.1016/0198-8859(91)90092-n. [DOI] [PubMed] [Google Scholar]
- 19.Denzin L K, Robbins N F, Carboy-Newcomb C, Cresswell P. Immunity. 1994;1:595–606. doi: 10.1016/1074-7613(94)90049-3. [DOI] [PubMed] [Google Scholar]
- 20.Mollick J A, Chintagumpala M, Cook R G, Rich R R. J Immunol. 1991;146:463–468. [PubMed] [Google Scholar]
- 21.Busch R, Cloutier I, Sékaly R-P, Hammerling G J. EMBO J. 1996;15:418–428. [PMC free article] [PubMed] [Google Scholar]
- 22.Sidney J, Oseroff C, Southwood S, Wall M, Ishioka G, Koning F, Sette A. J Immunol. 1992;149:2634–2640. [PubMed] [Google Scholar]
- 23.Karp D R, Jenkins R N, Long E O. Proc Natl Acad Sci USA. 1992;89:9657–9661. doi: 10.1073/pnas.89.20.9657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gorga J C, Horejsia V, Johnson D R, Raghupathy R, Strominger J L. J Biol Chem. 1987;262:16087–16094. [PubMed] [Google Scholar]
- 25.Ghosh P, Amaya M, Mellins E, Wiley D C. Nature (London) 1995;378:457–462. doi: 10.1038/378457a0. [DOI] [PubMed] [Google Scholar]
- 26.Mellins E, Smith L, Arp B, Cotner T, Celis E, Pious D. Nature (London) 1990;343:71–74. doi: 10.1038/343071a0. [DOI] [PubMed] [Google Scholar]
- 27.Lavoie P M, Sékaly R P, Thibodeau J, Denis F. In: Superantigens: Molecular Biology, Immunology, and Relevance to Human Disease. Huber B T, Leung D Y M, Schlievert P M, editors. New York: Dekker; 1997. pp. 61–84. [Google Scholar]
- 28.Bonventre P F, Weckbach L, Harth G, Haidaris C. Rev Infect Dis. 1989;11:S90–S95. doi: 10.1093/clinids/11.supplement_1.s90. [DOI] [PubMed] [Google Scholar]
- 29.Neib U, Reske K. Int Immunol. 1994;6:61–71. doi: 10.1093/intimm/6.1.61. [DOI] [PubMed] [Google Scholar]
- 30.Schneider F J, Opel B, Ballhausan W, Henkes W, Steinlein P, Reske K. Eur J Immunol. 1987;17:1235–1242. doi: 10.1002/eji.1830170904. [DOI] [PubMed] [Google Scholar]
- 31.Goding J W. J Immunol. 1980;154:1048–1056. [Google Scholar]