Abstract
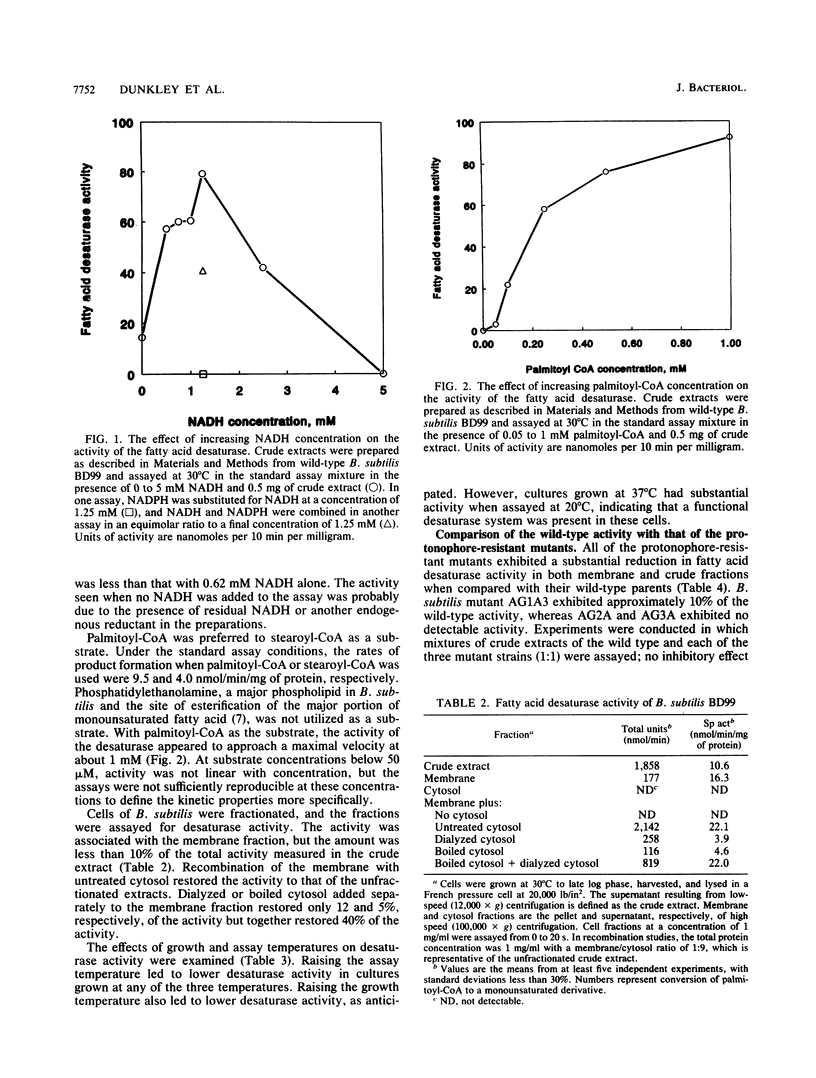
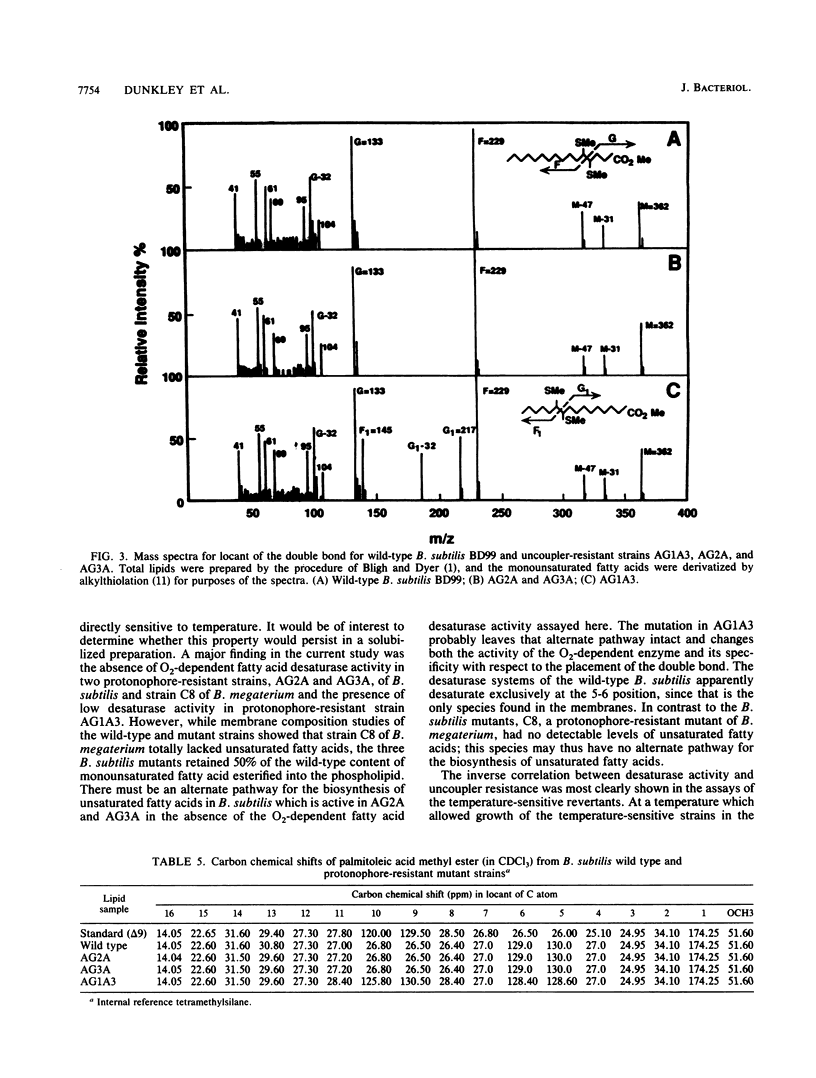
The fatty acid desaturase activity in cell extracts of Bacillus subtilis was characterized and found to be O2 dependent, NADH dependent, and cyanide sensitive. In cell fractionation studies, only 10% of the desaturase activity was recovered in the membrane fraction; the addition of cytosolic factors, which by themselves were devoid of activity, restored membrane activity to the level found in the unfractionated cell extracts. NADH was preferred over NADPH as an electron donor, and palmitoyl-coenzyme A was used preferentially over stearoyl-coenzyme A as the straight-chain fatty acid substrate. An increase in desaturase activity was observed when either the growth or the assay temperature was lowered from 37 to 20 degrees C, although the assay temperature appeared to be the more important parameter. Three protonophore-resistant mutants of B. subtilis and a comparable mutant of Bacillus megaterium had been found to possess reduced levels of unsaturated fatty acids in their membrane phospholipids; their protonophore resistance was abolished when grown in the presence of an unsaturated fatty acid supplement. All of these strains were found to be either significantly deficient in or totally lacking desaturase activity in comparison with their wild-type parent strains. Full, protonophore-sensitive revertants of the mutants had levels of desaturase activity comparable to those of the wild-type. Temperature-sensitive revertants of two of the mutants, which grew at 32 degrees C but not at 26 degrees C in the presence of protonophore, exhibited desaturase activity comparable to that of the wild-type at 26 degrees C but lacked activity at 32 degrees C. These results indicate that the biochemical basis for protonophore resistance in these Bacillus mutants is a fatty acid desaturase deficiency.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- BLIGH E. G., DYER W. J. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959 Aug;37(8):911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- Decker S. J., Lang D. R. Membrane bioenergetic parameters in uncoupler-resistant mutants of Bacillus megaterium. J Biol Chem. 1978 Oct 10;253(19):6738–6743. [PubMed] [Google Scholar]
- Decker S. J., Lang D. R. Mutants of Bacillus megaterium resistant to uncouplers of oxidative phosphorylation. J Biol Chem. 1977 Sep 10;252(17):5936–5938. [PubMed] [Google Scholar]
- Dunkley E. A., Jr, Clejan S., Guffanti A. A., Krulwich T. A. Large decreases in membrane phosphatidylethanolamine and diphosphatidylglycerol upon mutation to duramycin resistance do not change the protonophore resistance of Bacillus subtilis. Biochim Biophys Acta. 1988 Aug 4;943(1):13–18. doi: 10.1016/0005-2736(88)90341-0. [DOI] [PubMed] [Google Scholar]
- Dunkley E. A., Jr, Guffanti A. A., Clejan S., Krulwich T. A. Facultative alkaliphiles lack fatty acid desaturase activity and lose the ability to grow at near-neutral pH when supplemented with an unsaturated fatty acid. J Bacteriol. 1991 Feb;173(3):1331–1334. doi: 10.1128/jb.173.3.1331-1334.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enoch H. G., Strittmatter P. Role of tyrosyl and arginyl residues in rat liver microsomal stearylcoenzyme A desaturase. Biochemistry. 1978 Nov 14;17(23):4927–4932. doi: 10.1021/bi00616a011. [DOI] [PubMed] [Google Scholar]
- FULCO A. J., BLOCH K. COFACTOR REQUIREMENTS FOR THE FORMATION OF DELTA-9-UNSATURATED FATTY ACIDS IN MYCOBACTERIUM PHLEI. J Biol Chem. 1964 Apr;239:993–997. [PubMed] [Google Scholar]
- Ferrante G., Kates M. Pathways for desaturation of oleoyl chains in Candida lipolytica. Can J Biochem Cell Biol. 1983 Nov;61(11):1191–1196. doi: 10.1139/o83-153. [DOI] [PubMed] [Google Scholar]
- Fulco A. J. Fatty acid metabolism in bacteria. Prog Lipid Res. 1983;22(2):133–160. doi: 10.1016/0163-7827(83)90005-x. [DOI] [PubMed] [Google Scholar]
- Fulco A. J. The biosynthesis of unsaturated fatty acids by bacilli. I. Temperature induction of the desaturation reaction. J Biol Chem. 1969 Feb 10;244(3):889–895. [PubMed] [Google Scholar]
- Guffanti A. A., Clejan S., Falk L. H., Hicks D. B., Krulwich T. A. Isolation and characterization of uncoupler-resistant mutants of Bacillus subtilis. J Bacteriol. 1987 Oct;169(10):4469–4478. doi: 10.1128/jb.169.10.4469-4478.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krulwich T. A., Quirk P. G., Guffanti A. A. Uncoupler-resistant mutants of bacteria. Microbiol Rev. 1990 Mar;54(1):52–65. doi: 10.1128/mr.54.1.52-65.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kshirsagar S. S., Nair P. M. Isolation and properties of a particulate fraction for the desaturation of palmitic acid from Alcaligenes faecalis. Biochim Biophys Acta. 1979 Sep 28;574(3):369–378. doi: 10.1016/0005-2760(79)90233-9. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Nagai J., Bloch K. Enzymatic desaturation of stearyl acyl carrier protein. J Biol Chem. 1968 Sep 10;243(17):4626–4633. [PubMed] [Google Scholar]
- Resnick M. A., Mortimer R. K. Unsaturated fatty acid mutants of Saccharomyces cerevisiae. J Bacteriol. 1966 Sep;92(3):597–600. doi: 10.1128/jb.92.3.597-600.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville C., Browse J. Plant lipids: metabolism, mutants, and membranes. Science. 1991 Apr 5;252(5002):80–87. doi: 10.1126/science.252.5002.80. [DOI] [PubMed] [Google Scholar]
- Spizizen J. TRANSFORMATION OF BIOCHEMICALLY DEFICIENT STRAINS OF BACILLUS SUBTILIS BY DEOXYRIBONUCLEATE. Proc Natl Acad Sci U S A. 1958 Oct 15;44(10):1072–1078. doi: 10.1073/pnas.44.10.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strittmatter P., Enoch H. G. Purification of stearyl-CoA desaturase from liver. Methods Enzymol. 1978;52:188–193. doi: 10.1016/s0076-6879(78)52020-x. [DOI] [PubMed] [Google Scholar]
- Strittmatter P., Thiede M. A., Hackett C. S., Ozols J. Bacterial synthesis of active rat stearyl-CoA desaturase lacking the 26-residue amino-terminal amino acid sequence. J Biol Chem. 1988 Feb 15;263(5):2532–2535. [PubMed] [Google Scholar]
- Stukey J. E., McDonough V. M., Martin C. E. Isolation and characterization of OLE1, a gene affecting fatty acid desaturation from Saccharomyces cerevisiae. J Biol Chem. 1989 Oct 5;264(28):16537–16544. [PubMed] [Google Scholar]
- Thiede M. A., Ozols J., Strittmatter P. Construction and sequence of cDNA for rat liver stearyl coenzyme A desaturase. J Biol Chem. 1986 Oct 5;261(28):13230–13235. [PubMed] [Google Scholar]
- Thiede M. A., Strittmatter P. The induction and characterization of rat liver stearyl-CoA desaturase mRNA. J Biol Chem. 1985 Nov 25;260(27):14459–14463. [PubMed] [Google Scholar]
- Wada H., Gombos Z., Murata N. Enhancement of chilling tolerance of a cyanobacterium by genetic manipulation of fatty acid desaturation. Nature. 1990 Sep 13;347(6289):200–203. doi: 10.1038/347200a0. [DOI] [PubMed] [Google Scholar]
- Wada H., Murata N. Temperature-Induced Changes in the Fatty Acid Composition of the Cyanobacterium, Synechocystis PCC6803. Plant Physiol. 1990 Apr;92(4):1062–1069. doi: 10.1104/pp.92.4.1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Mendoza D., Klages Ulrich A., Cronan J. E., Jr Thermal regulation of membrane fluidity in Escherichia coli. Effects of overproduction of beta-ketoacyl-acyl carrier protein synthase I. J Biol Chem. 1983 Feb 25;258(4):2098–2101. [PubMed] [Google Scholar]



