Abstract
A strain of Bacillus species which produced an enzyme named glutaryl 7-ACA acylase which converts 7 beta-(4-carboxybutanamido)cephalosporanic acid (glutaryl 7-ACA) to 7-amino cephalosporanic acid (7-ACA) was isolated from soil. The gene for the glutaryl 7-ACA acylase was cloned with pHSG298 in Escherichia coli JM109, and the nucleotide sequence was determined by the M13 dideoxy chain termination method. The DNA sequence revealed only one large open reading frame composed of 1,902 bp corresponding to 634 amino acid residues. The deduced amino acid sequence contained a potential signal sequence in its amino-terminal region. Expression of the gene for glutaryl 7-ACA acylase was performed in both E. coli and Bacillus subtilis. The enzyme preparations purified from either recombinant strain of E. coli or B. subtilis were shown to be identical with each other as regards the profile of sodium dodecyl sulfate-polyacrylamide gel electrophoresis and were composed of a single peptide with the molecular size of 70 kDa. Determination of the amino-terminal sequence of the two enzyme preparations revealed that both amino-terminal sequences (the first nine amino acids) were identical and completely coincided with residues 28 to 36 of the open reading frame. Extracellular excretion of the enzyme was observed in a recombinant strain of B. subtilis.
Full text
PDF


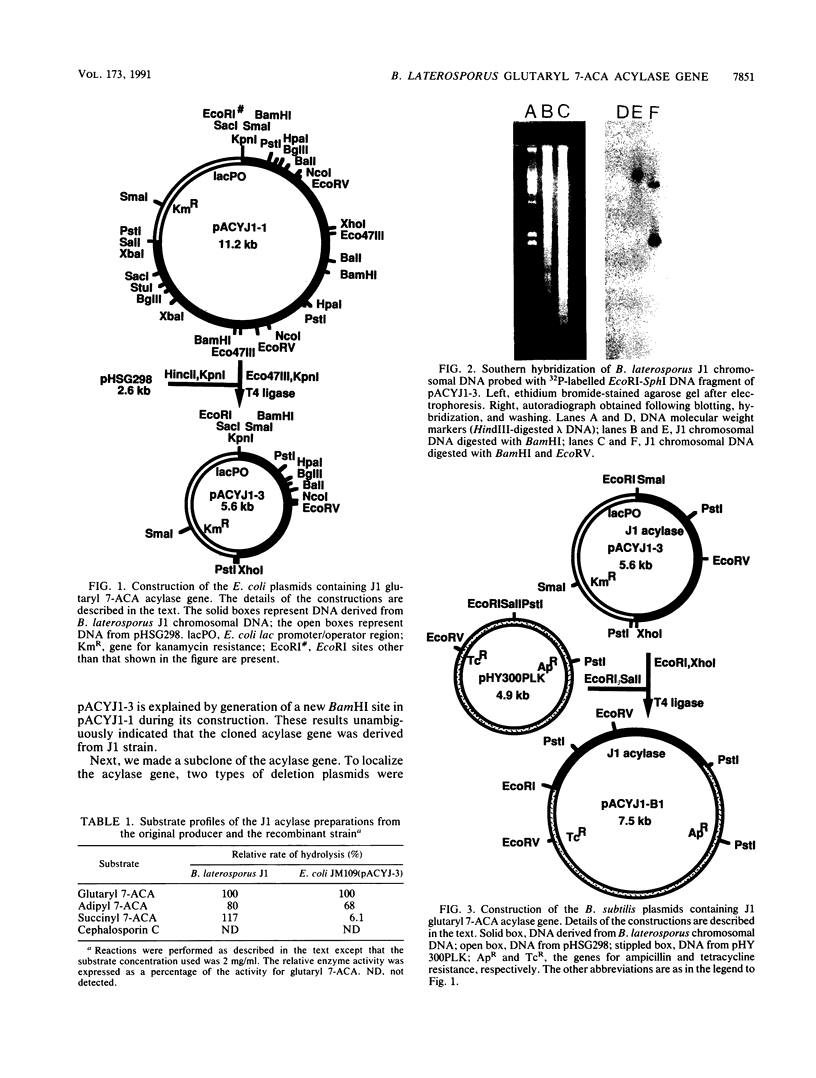

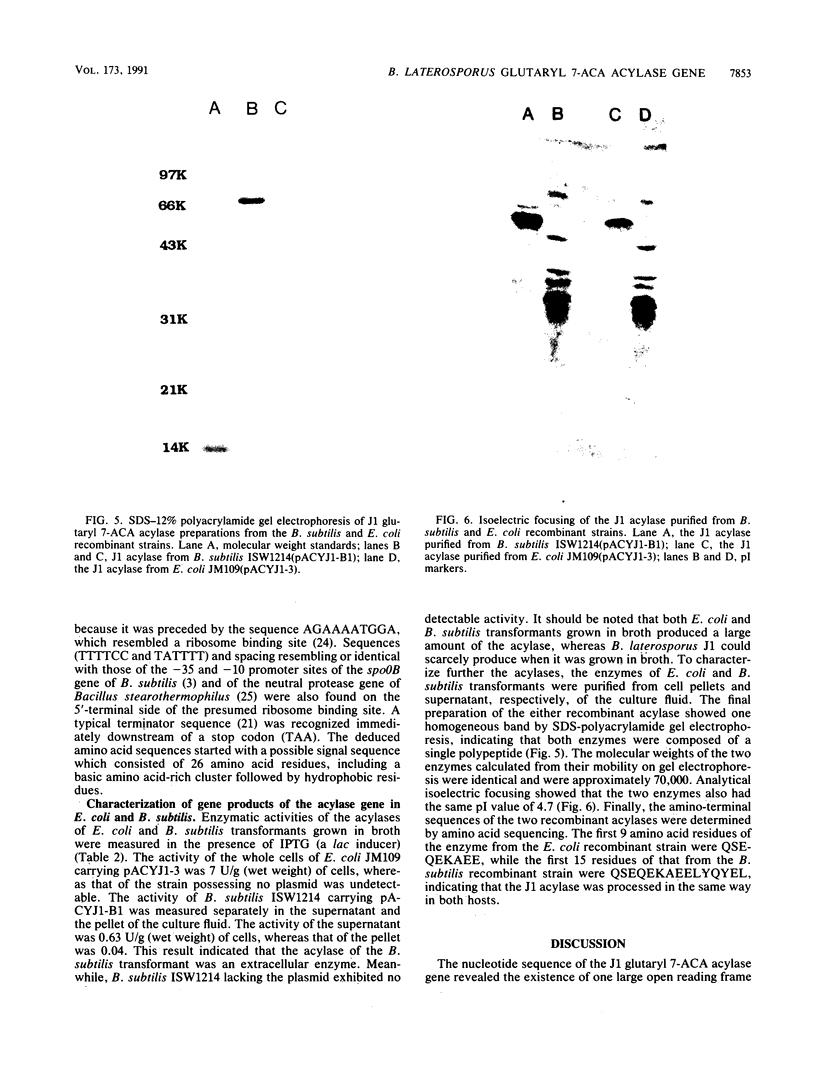


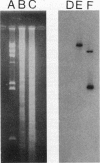
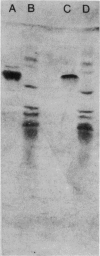
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvier J., Stragier P., Bonamy C., Szulmajster J. Nucleotide sequence of the spo0B gene of Bacillus subtilis and regulation of its expression. Proc Natl Acad Sci U S A. 1984 Nov;81(22):7012–7016. doi: 10.1073/pnas.81.22.7012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clewell D. B., Helinski D. R. Supercoiled circular DNA-protein complex in Escherichia coli: purification and induced conversion to an opern circular DNA form. Proc Natl Acad Sci U S A. 1969 Apr;62(4):1159–1166. doi: 10.1073/pnas.62.4.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gormley E. P., Cantwell B. A., Barker P. J., Gilmour R. S., McConnell D. J. Secretion and processing of the Bacillus subtilis endo-beta-1,3-1,4-glucanase in Escherichia coli. Mol Microbiol. 1988 Nov;2(6):813–819. doi: 10.1111/j.1365-2958.1988.tb00093.x. [DOI] [PubMed] [Google Scholar]
- Harris-Warrick R. M., Elkana Y., Ehrlich S. D., Lederberg J. Electrophoretic separation of Bacillus subtilis genes. Proc Natl Acad Sci U S A. 1975 Jun;72(6):2207–2211. doi: 10.1073/pnas.72.6.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikemura H., Takagi H., Inouye M. Requirement of pro-sequence for the production of active subtilisin E in Escherichia coli. J Biol Chem. 1987 Jun 5;262(16):7859–7864. [PubMed] [Google Scholar]
- Imanaka T., Fujii M., Aiba S. Isolation and characterization of antibiotic resistance plasmids from thermophilic bacilli and construction of deletion plasmids. J Bacteriol. 1981 Jun;146(3):1091–1097. doi: 10.1128/jb.146.3.1091-1097.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Matsuda A., Komatsu K. I. Molecular cloning and structure of the gene for 7 beta-(4-carboxybutanamido)cephalosporanic acid acylase from a Pseudomonas strain. J Bacteriol. 1985 Sep;163(3):1222–1228. doi: 10.1128/jb.163.3.1222-1228.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda A., Matsuyama K., Yamamoto K., Ichikawa S., Komatsu K. Cloning and characterization of the genes for two distinct cephalosporin acylases from a Pseudomonas strain. J Bacteriol. 1987 Dec;169(12):5815–5820. doi: 10.1128/jb.169.12.5815-5820.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi H., Katsuta Y., Hashizume T., Abe S. N., Kajiura H., Hattori H., Kamei T., Yano M. Molecular cloning of the penicillin G acylase gene from Arthrobacter viscosus. Appl Environ Microbiol. 1988 Nov;54(11):2603–2607. doi: 10.1128/aem.54.11.2603-2607.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt T. Transcription termination and the regulation of gene expression. Annu Rev Biochem. 1986;55:339–372. doi: 10.1146/annurev.bi.55.070186.002011. [DOI] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. Terminal-sequence analysis of bacterial ribosomal RNA. Correlation between the 3'-terminal-polypyrimidine sequence of 16-S RNA and translational specificity of the ribosome. Eur J Biochem. 1975 Sep 1;57(1):221–230. doi: 10.1111/j.1432-1033.1975.tb02294.x. [DOI] [PubMed] [Google Scholar]
- Takagi M., Imanaka T., Aiba S. Nucleotide sequence and promoter region for the neutral protease gene from Bacillus stearothermophilus. J Bacteriol. 1985 Sep;163(3):824–831. doi: 10.1128/jb.163.3.824-831.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshita S., Sato M., Toba M., Masahashi W., Hashimoto-Gotoh T. High-copy-number and low-copy-number plasmid vectors for lacZ alpha-complementation and chloramphenicol- or kanamycin-resistance selection. Gene. 1987;61(1):63–74. doi: 10.1016/0378-1119(87)90365-9. [DOI] [PubMed] [Google Scholar]