Abstract
JAK3 is a protein tyrosine kinase that specifically associates with the common γ chain (γc), a shared subunit of receptors for interleukin (IL) 2, 4, 7, 9, and 15. Patients deficient in either JAK3 or γc presented with virtually identical forms of severe combined immunodeficiency (SCID), underscoring the importance of the JAK3–γc interaction. Despite the key roles of JAK3 and γc in lymphocytic development and function, the molecular basis of this interaction remains poorly understood. In this study, we have characterized the regions of JAK3 involved in γc association. By developing a number of chimeric JAK3–JAK2 constructs, we show that the binding specificity to γc can be conferred to JAK2 by transferring the N-terminal domains of JAK3. Moreover, those JAK3–JAK2 chimeras capable of binding γc were also capable of reconstituting IL-2 signaling as measured by inducible phosphorylation of the chimeric JAK3–JAK2 protein, JAK1, the IL-2 receptor β chain, and signal transducer and activator of transcription 5A. Subsequent deletion analyses of JAK3 have identified the N-terminal JH7-6 domains as a minimal region sufficient for γc association. Furthermore, expression of the mutant containing only the JH7-6 domains effectively competed with full-length JAK3 for binding to γc. We conclude that the JH7-6 domains of JAK3 are necessary and sufficient for γc association. These studies offer clues toward a broader understanding of JAK-mediated cytokine signaling and may provide a target for the development of novel therapeutic modalities in immunologically mediated diseases.
Cytokines are critical regulators for both the differentiation and proliferation of many cells, particularly those of the immune system (1). Cytokines such as interleukin 2 (IL-2) initiate their actions by ligand-induced oligomerization of cytokine receptors (2, 3). Although these receptors lack intrinsic kinase activity, they associate with and activate cytoplasmic protein tyrosine kinases that then phosphorylate downstream signaling molecules such as the signal transducers and activators of transcription (STATs) (4).
The Janus family of kinases (JAK1, JAK2, JAK3, and TYK2) is a structurally distinct class of protein tyrosine kinases that bind to cytokine receptors and are critical in cytokine signaling (5–10). JAK3, unique in its predominant expression in hematopoietic cells (11–14), specifically associates with the common γ chain (γc) (15, 16), a shared component of receptors for IL-2, -4, -7, -9, and -15 (17, 18). The importance of the JAK3–γc interaction in lymphocyte function was underscored by findings that human mutations of either γc or JAK3 resulted in virtually identical forms of severe combined immunodeficiency (19–21). Moreover, mice made deficient in JAK3 by gene targeting have a profound block in B cell development and T cell function similar to the immunodeficiency phenotype found in γc knockout mice (22–25). Based on these findings and the observations that most γc-using cytokines such as IL-2 are potent activators of the immune response (1, 3), targeting JAK3 itself or JAK3–γc interactions pharmacologically may provide novel immunosuppressive agents that might be useful in the treatment of autoimmunity, leukemia, and prevention of allograft rejection.
Recent studies have identified the membrane-proximal region of γc as essential for JAK3 association and activation (26, 27). This region contains two loosely conserved motifs (termed Box1 and Box2) that are also important for JAK binding and cytokine signaling by other cytokine receptors (28–30). However, the regions of JAK3 required for γc binding have not been studied. JAK3 and other JAKs have been divided into seven structural domains termed JAK homology (JH) domains 7-1 based on the homologies among the family members (31). Except for the carboxyl JH1 domain that contains the tyrosine kinase catalytic domain, little is known about the function of the other JH domains. Though it is assumed that some of these domains are involved in receptor association, the exact region or regions responsible have not been characterized. Several reports have indicated that JAK mutants lacking the JH1 kinase domain can function as a dominant negative allele, suggesting that the JH1 kinase domain is not necessary for receptor association (32–34). This is also supported by other more direct studies (30, 35, 36). In one report pertaining to the interaction of JAK2 with granulocyte–macrophage-colony stimulating factor receptor (GM-CSFR), a bacterially expressed glutathione S-transferase–JAK2 fusion protein comprising the JH7-6 domains of JAK2 was shown to be capable of binding to glutathione S-transferase–GM-CSFR βc by Far Western blotting (36). However, in another report on the interaction of JAK2 with the growth hormone receptor (GHR), the JH7-3 domains of JAK2 were all shown to be necessary for GHR binding; any further deletion of this segment abolished receptor association (30). These studies presented an apparent contradiction in the binding behavior of JAK2 and did not address a functional analysis of JAK receptor association. In light of the sequence similarity among the JAKs, studies of JAK3 and γc may provide insights in understanding JAK2 as well as other JAK-receptor interactions and how these specificities are achieved.
Given the importance of the JAK3–γc interaction in mediating lymphoid cell growth/differentiation and its potential utility as a target for immunosuppressive drugs, we sought to determine whether the ability of JAK3 to specifically bind γc principally resided in a discrete segment or whether diverse segments additively contributed to the interaction. Our data indicate that the N-terminal JH7-6 domains of JAK3 represent a necessary and sufficient region for γc association. These findings offer important advances in the understanding of the JAK receptor binding behavior and provide models for the future development of immunosuppressive agents designed to disrupt this interaction.
MATERIALS AND METHODS
Reagents and Antibodies.
Human IL-2 was obtained through C. Reynolds (National Cancer Institute, Frederick, MD). Antibodies were obtained as follows: 7G7, anti-IL-2R (receptor) α chain (anti-Tac) mAb (D. Nelson, National Cancer Institute, Bethesda); 561, anti-IL-2Rβ mAb (P. Sondel, University of Wisconsin, Madison); and ErdA, anti-IL-2Rβ (W. Leonard, National Heart, Lung, and Blood Institute, Bethesda). The following antibodies were purchased: rabbit anti-IL-2Rγc (Santa Cruz Biotechnology); JAK1 mAb (Transduction Laboratories, Lexington, KY); 4G10 mAb and rabbit anti-mouse JAK2 (Upstate Biotechnology, Lake Placid, NY). Rabbit antisera against the C and N termini of human JAK3 were raised using peptides corresponding to residues 1104–1124 (11) and 2–22, respectively. Rabbit anti-STAT5A was generated against residues 774–793 of murine STAT5A.
Plasmid Construction and Site-Directed Mutagenesis.
The following plasmids were obtained: JAK1 and JAK2 cDNAs (J. Ihle, St. Jude Children’s Research Hospital, Memphis, TN); chimeric IL-2R ααβ (Tacβ) and αγγ (Tacγc) (W. Leonard); Tacζ (T cell receptor ζ chain) (R. Klausner, National Cancer Institute, Bethesda). The kinase dead mutant J3(K855A) was created with the Transformer Site-Directed Mutagenesis kit (CLONTECH) using oligonucleotides designed to change the codons for lysine (AAA) to alanine (GCT) at aa position 855. Similarly, the plasmids J3(X1) and J3(H3) were generated by introducing an XhoI site and an HindIII site into nucleotides 1200–1205 and 1652–1657 of JAK3 cDNA, respectively. Chimera J3(7-3)J2(2-1) (see Fig. 2A) was constructed using the PCR-generated JAK2 HindIII–XbaI fragment and the pBluescript–JAK2-derived JAK2 XbaI fragment to replace the sequences coding for the JH2 and the JH1 regions of J3(H3). Similarly, the chimera J3(7-4)J2(4-1) was generated by replacing the sequences encoding the JH4-1 regions of J3(X1) with the JAK2 XhoI fragment from pBluescript–JAK2. Construction of the chimeras J3(N)J2(7-1) and J3(7)J2(6-1) was performed using the PCR-based gene SOEing strategy (37). The C-terminal deletion mutants J3(MunI), J3(StuI), J3(AccI), J3(SacI), and J3(NcoI) (see Fig. 4A) were constructed by digesting wild-type (wt) JAK3 cDNA (15) with corresponding restriction enzyme; the Myc-tagged fragments with a stop codon inserted at 3′ end were then cloned into pME18s (DNAX). For J3(ΔJH1), an EcoRI site was introduced at nucleotide 2535; the EcoRI fragment of JAK3 was then subcloned into pME18s. The N-terminal JH7 and the JH7-5 deletion mutants were generated by replacing the JAK3 sequence encoding aa 1–370 with PCR fragments containing an ATG start codon at the 5′ end and encoding aa 123–370 and 284–370, respectively (see Fig. 4A). All the constructs were verified by restriction enzyme digestion and/or automatic DNA sequencing using Applied Biosystems PRISM Dye Terminator Cycle Sequencing kit (Perkin–Elmer).
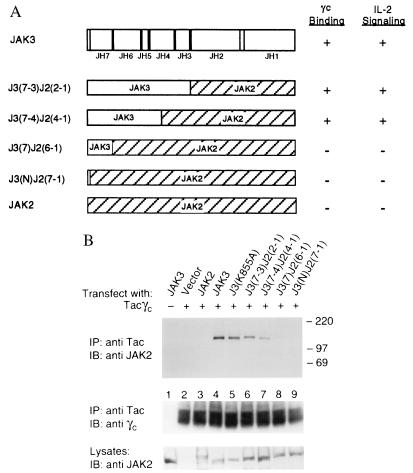
Figure 2.
The binding specificity of JAK3 to γc can be conferred to JAK2 by transferring the N-terminal domains. (A) A schematic representation of the J3J2 chimeras. J3(7-3)J2(2-1) contains the N-terminal JH7-3 domains from JAK3 (aa 1–519) fused to the C-terminal JH2-1 domains of JAK2 (aa 543–1130). J3(7-4)J2(4-1) contains the N-terminal JH7 through a portion of JH4 domains of JAK3 (aa 1–370) fused to the C-terminal JH4-1 domains of JAK2 (aa 395–1130). J3(7)J2(6-1) contains the N-terminal JH7 domain of JAK3 (aa 1–124) fused to the C-terminal JH6-1 domains of JAK2 (aa 141–1130). J3(N)J2(7-1) contains the extreme N terminus (aa 1–40) of JAK3 fused to the JH7-1 domains of JAK2 (aa 55–1130). (B) Association of J3J2 chimeras with the cytoplasmic domain of γc. COS cells were transfected with 3 μg of the indicated cDNAs. These cell lysates were immunoprecipitated with anti-Tac and blotted with anti-γc (Middle) and anti-JAK2 that crossreacts with JAK3 (Top). Cell lysates were analyzed for expression of the transfected JAK constructs by immunoblotting with anti-JAK2 (Bottom).
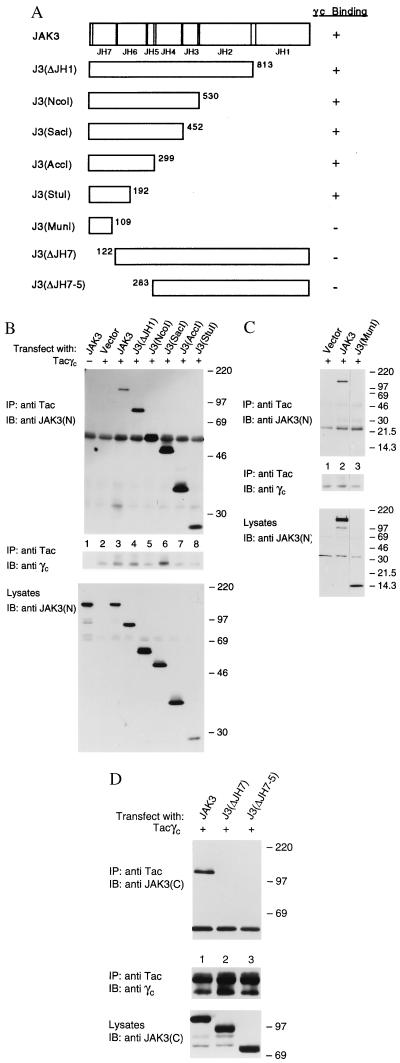
Figure 4.
The JAK3 N terminus is necessary and sufficient for γc binding. (A) A schematic representation of the JAK3 C-terminal and N-terminal deletion mutants. The numbers indicate the last (C-terminal) or the second (N-terminal) aa residue in each mutant with respect to the original JAK3 sequence. (B-D) COS cells were transfected with 5 μg of the indicated cDNAs. Lysates were immunoprecipitated with anti-Tac and blotted with anti-JAK3(N) (B and C Top) or anti-JAK3 C-terminal [anti-JAK3(C)] (D Top) and anti-γc (Middle). Expression levels of various mutants were analyzed by immunoblotting with anti-JAK3(N) (B and C Bottom) or anti-JAK3(C) (D Bottom).
Cells, Transfections, Immunoprecipitation, and Immunoblotting.
COS7 and NIH 3T3αβγ cells (38) cells were maintained as described (15, 38). COS7 cells were transiently transfected using DEAE-dextran method and were harvested 2 days later as previously described (15). For generation of stable transfectants, wt JAK3 or JAK2–JAK3 (J3J2) chimeras cloned in pCDNA3 (Invitrogen) were transfected into NIH 3T3αβγ cells, using DOTAP (Boehringer Mannheim). Forty-eight hours later, cells were selected in DMEM containing 1 mg/ml G418 (GIBCO). Resistant clones were analyzed for JAK3 or J3J2 chimera expression and IL-2 responsiveness by immunoblotting. For cell stimulation, NIH 3T3αβγ cells (10–15 × 106) were washed, serum-starved for 4 h, and then stimulated with 1,000 units/ml IL-2 for 15 min at 37°C prior to lysis. Immunoprecipitation and immunoblotting were performed as previously described (10).
RESULTS
The J3J2 Chimeras Bind γc and Mediate IL-2-Dependent Signaling.
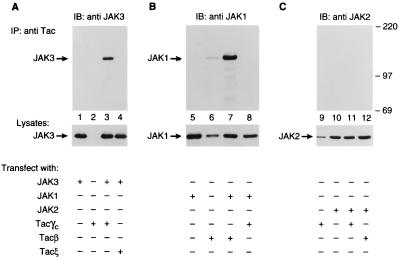
JAK3 has been previously shown to associate with γc and is only activated upon stimulation with γc binding cytokines (12, 15, 16, 39). We first confirmed the specific interaction between γc and JAK3 using a chimeric receptor containing the cytoplasmic γc fused to the extracellular domain of the IL-2Rα chain (Tacγc). COS cells were transfected with Tacγc either alone or together with JAK3 (Fig. 1A), JAK1 (Fig. 1B), or JAK2 (Fig. 1C). Anti-Tac immunoprecipitates from transfected cells were blotted with antibodies against JAK1, JAK2, or JAK3. As shown in Fig. 1, anti-Tac coprecipitated JAK3 from COS cells expressing both the Tacγc and JAK3 (lane 3) but not Tacγc alone (lane 2). This interaction is specific for Tacγc since JAK3 failed to coprecipitate with a control chimeric protein Tacξ (lane 4) and was not immunoprecipitated nonspecifically by anti-Tac (lane 1). Moreover, γc selectively interacted with JAK3 since JAK1, JAK2, or TYK2 did not coprecipitate with Tacγc (lanes 8 and 11, and data not shown, respectively) despite the fact that JAK1 was capable of associating with Tacβ as previously described (lane 7) (15). The lack of association between JAK2 and γc is also illustrated below (Fig. 2B, lane 3). A faint JAK1 band was detected in lane 6 when Tacβ was transfected alone due to endogenous expression of JAK1 in COS cells. These data demonstrate that JAK3 specifically associates with the cytoplasmic domain of γc. This specific interaction was also confirmed by in vitro studies using purified glutathione S-transferase-γc fusion protein and baculovirus expressed JAK3 (L.K., M.C., J.J.O., and P.C., unpublished work).
Figure 1.
JAK3 specifically associates with the cytoplasmic domain of γc. COS cells were transfected by the DEAE–dextran method with 5 μg of the indicated cDNAs. Cell lysates were immunoprecipitated (IP) with anti-Tac mAb and immunoblotted (IB) with antisera to JAK3 (lanes 1–4), JAK1 (lanes 5–8), or JAK2 (lanes 9–12). The levels of expression of JAK1, JAK2, or JAK3 were assessed by immunoblotting with corresponding antibodies (Bottom).
Having demonstrated that JAK3 but not other JAKs specifically associates with γc, we next investigated the region of JAK3 conferring γc-binding specificity by generating a series of chimeric JAK kinases. By using chimeric constructs in these analyses, discrete changes in JAK3 can be made without perturbing its autokinase activity or dramatically changing its tertiary structure. In light of the homology between JAK3 and JAK2, we generated chimeras by replacing the C terminus of JAK3 with that of JAK2, which does not interact with γc (Figs. 1, lane 11 and 2B, lane 3). Four chimeras were constructed as illustrated in Fig. 2A. The cDNAs of these constructs were transfected into COS cells either alone or together with Tacγc. Their ability to associate with the cytoplasmic domain of γc was analyzed by immunoprecipitation with anti-Tac followed by immunoblotting with anti-JAK2, which also recognizes JAK3 (Fig. 2B Top). The expression levels of the various JAKs (Fig. 2B Bottom) and of Tacγc (Fig. 2B Middle) did not significantly vary among the transfected cells. Consistent with our earlier findings (Fig. 1C), JAK2 did not bind to the cytoplasmic domain of γc (Fig. 2B, lane 3). Interestingly, a JAK3 mutant lacking kinase activity, J3(K855A), bound efficiently to γc compared with wt JAK3 (lane 5 vs. 4), suggesting that tyrosine kinase activity is not required for γc binding. Among the four chimeras generated, J3(7-4)J2(4-1) and J3(7-3)J2(2-1) were also shown to interact with γc (lanes 6 and 7), although to a lesser extent compared with wt JAK3. In contrast, chimeras J3(7)J2(6-1) and J3(N)J2(7-1) did not bind (lanes 8 and 9). As expected, all chimeras retained autokinase activity comparable to wt JAK3 and JAK2 (data not shown). These results indicate that the N-terminal JH7-4 domains of JAK3 (aa 1–370) confer specific binding to γc, whereas transferring the JH7 domain alone onto JAK2 is not sufficient in conferring γc-interacting ability.
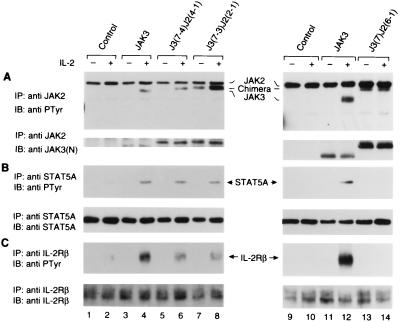
Previous studies have shown that the JAK3–γc interaction is essential for IL-2-induced downstream signaling events such as the activation of Janus kinases (JAK1 and JAK3), the phosphorylation of receptor subunits (IL-2Rβ and γc) and the subsequent activation of STATs (STAT3 and STAT5) (10, 39–42). These events, therefore, represent suitable functional readouts of the ability of the J3J2 chimeras in associating with γc. By using a JAK3-negative NIH 3T3-derived cell line expressing endogenous JAK1 and reconstituted IL-2R complex (3T3αβγ) (38), we assessed whether our chimeras could functionally couple to γc and mediate IL-2-dependent signaling. 3T3αβγ cells have previously been shown to respond to IL-2 in the presence of ectopically expressed JAK3 (16). We therefore generated stable transfectants of J3J2 chimeras in these cells and evaluated their responses to IL-2 stimulation by measuring the presence or absence of inducible phosphorylation. Specifically, stable transfectants of JAK3 or J3J2 chimeras were stimulated with IL-2, lysed, immunoprecipitated with JAK2 antiserum (that crossreacts with JAK3), and subjected to immunoblotting analysis with an anti-phosphotyrosine antibody (4G10). In spite of its basal level of phosphorylation, chimera J3(7-3)J2(2-1) demonstrated IL-2-inducible phosphorylation (Fig. 3A, lane 7 vs. lane 8) similar to wt JAK3 (lane 3 vs. lane 4 and lane 11 vs. lane 12) and the other γc-binding chimera J3(7-4)J2(4-1) (lane 5 vs. lane 6). In contrast, the constitutively phosphorylated J3(7)J2(6-1) chimera that did not bind γc was not further phosphorylated upon IL-2 treatment (lane 13 vs. lane 14) nor was endogenous JAK2 inducibly phosphorylated despite its abundant expression in these cells (lanes 1–14, top band and data not shown). The lack of inducible phosphorylation of chimera J3(7)J2(6-1) was confirmed in other clones (data not shown). Thus, the IL-2-inducible phosphorylation of J3J2 chimeras correlated well with the ability of these chimeras to interact with γc.
Figure 3.
Reconstitution of IL-2-dependent signaling in IL-2R-expressing fibroblast cells requires physical coupling of J3J2 chimeras to γc. Serum-starved cells (1.5 × 107 cells) were left unstimulated or stimulated with IL-2 (1,000 units/ml) as indicated for 15 min; cell lysates were immunoprecipitated with anti-JAK2 (A), anti-STAT5A (B), or anti-IL-2Rβ (561) (C). Tyrosine phosphorylation was assayed by blotting with anti-phosphotyrosine (anti-PTyr) (Top). Membranes were reprobed with the anti-JAK3 N-terminal [anti-JAK3(N)] (A), anti-STAT5A (B), or anti-IL-2Rβ (ErdA) (C), to confirm equal loading (Bottom).
We next assessed IL-2-mediated phosphorylation of other downstream signaling molecules in the presence of J3J2 chimeras. As shown in Fig. 3B, in the absence of JAK3, very low levels of STAT5 phosphorylation were detected (lanes 1 and 2); however, this IL-2-dependent phosphorylation was dramatically increased upon ectopic expression of wt JAK3 (lane 3 vs. lane 4) and the chimeric JAKs that bound γc [J3(7-4)J2(4-1], lane 5 vs. lane 6 and J3(7-3)J2(2-1), lane 7 vs. lane 8). In contrast, the chimera that bound γc poorly [J3(7)J2(6-1)] did not demonstrate IL-2-dependent STAT5 phosphorylation (lane 13 vs. lane 14). We also analyzed IL-2Rβ chain phosphorylation in these reconstituted cells. Consistent with the data provided in Fig. 3B, chimera J3(7)J2(6-1) did not allow for inducible phosphorylation (Fig. 3C, lane 13 vs. lane 14). In contrast, chimeras J3(7-4)J2(4-1) and J3(7-3)J2(2-1) permitted IL-2-dependent IL-2Rβ chain phosphorylation (lane 5 vs. lane 6 and lane 7 vs. lane 8) compared with untransfected cells in which minimal phosphorylation occurred (lane 2). Interestingly though, the level of IL-2Rβ chain phosphorylation by either chimera was less than that seen in wt JAK3 expressing cells (lanes 6 and 8 vs. lane 4). As expected, the IL-2 inducible phosphorylation of JAK1 also correlated with the ability of the JAK chimeras to bind γc (data not shown). These data support the notion that physical coupling of JAK3 to γc is essential for activation of IL-2 induced downstream signaling events. Furthermore, the ability of JAK3 to bind γc can be conferred onto JAK2 by the N-terminal JH7-5 and a portion of the JH4 domains of JAK3, strongly supporting our previous conclusions that these domains of JAK3 are important for the specific association with γc.
The N-Terminal JH7-6 Domains of JAK3 Are Necessary and Sufficient for Association with γc.
Having established that the N-terminal portion of JAK3 is necessary for γc association, we next attempted to define a minimal segment that was necessary and sufficient for γc binding by generating a series of C-terminal and N-terminal deletion mutants of JAK3 (Fig. 4A). cDNAs corresponding to various JAK3 mutants were cotransfected with Tacγc into COS cells. The association between Tacγc and the JAK3 mutants was then assessed by immunoprecipitation with anti-Tac and subsequent immunoblotting with antibodies against the N or C terminus of JAK3. Consistent with a previous report that JAK3 lacking the JH1 domain can dominantly inhibit wt JAK3 function (32), deletion of the JH1 domain or both the JH1 and JH2 domains had no effect on the ability of JAK3 to associate with γc (Fig. 4B, lanes 4 and 5). However, in contrast to a report suggesting that the entire JH7 through JH3 domains were required for JAK2–GHR interaction (30), we observed that the JH5-3 domains were not required for JAK3–γc interaction (lanes 6–8). The J3(SacI), J3(AccI), and J3(StuI) mutants, all containing the JH7-6 domains, efficiently coprecipitated with the chimeric γc receptor (lanes 6–8, respectively). However, the J3(MunI) mutant containing further deletions of this region failed to bind Tacγc (Fig. 4C, lane 3). This suggested that the JH7-6 domains represent a minimal region required for JAK3 association with γc. This conclusion was further supported by the results generated using N-terminal deletion mutants lacking either the JH7 or the JH7-5 domains (Fig. 4A). These constructs did not coprecipitate with Tacγc (Fig. 4D, lanes 2 and 3), indicating that the N-terminal JH7 domain was especially important for binding to γc. However, this segment was not sufficient since the J3J2 chimera containing the JH7 domain of JAK3 did not bind (Fig. 2B, lane 8). Therefore, the association of JAK3 with γc involves more than just the JH7 domain. In light of our findings regarding the C-terminal deletion mutants, these data point to the N-terminal JH7-6 domains as a necessary and sufficient region for γc association.
The N-Terminal JH7-6 Domains of JAK3 Compete with wt JAK3 for γc Binding.
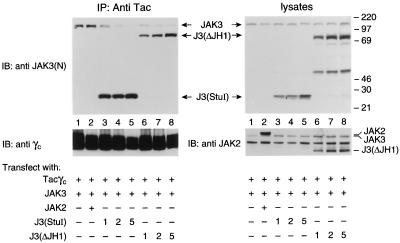
If the segment containing the JH7-6 domains was the major γc interactive region, the mutant J3(StuI) comprising the JH7-6 domains should bind γc as well as full-length JAK3. To test this hypothesis, COS cells were transfected with Tacγc, JAK3, and increasing amounts of the J3(StuI) or J3(ΔJH1) mutants. Extracts were then immunoprecipitated with anti-Tac and immunoblotted with anti-JAK3 N terminal. As shown in Fig. 5, expression of J3(StuI) (Left, lanes 3–5) and J3(ΔJH1) (lanes 6–8) greatly reduced the amount of full-length JAK3 coprecipitating with Tacγc without reducing the levels of expression of JAK3 (Right Upper) or Tacγc (Left Lower) in these cells. In contrast, overexpression of JAK2 failed to generate a similar effect (Left, lane 2), indicating that J3(StuI) specifically competes with wt JAK3 for γc binding. Lysates were blotted with anti-JAK3 and anti-JAK2 (that crossreacts with JAK3) (Fig. 5 Right) demonstrating that the levels of expression of JAK2 were comparable to that of the JAK3 constructs. These data further support that the N-terminal JH7-6 domains of JAK3 are necessary and sufficient for γc association and that the major site of interaction with γc resides in this segment.
Figure 5.
The JAK3 N terminus competes with full-length JAK3 for binding to γc. COS cells were cotransfected with JAK3 (0.5 μg) and Tacγc (0.5 μg) together with the vector pME18s (5 μg) (lane 1), JAK2 (5 μg) (lane 2), or 1, 2, or 5 μg of J3(StuI) (lanes 3–5, respectively) or J3(ΔJH1) (lanes 6–8, respectively). These cell lysates were immunoprecipitated with anti-Tac and then blotted with anti-JAK3(N) (Left Upper) or anti-γc (Left Lower). The expression levels of various JAK3 mutants and JAK2 were analyzed by immunoblotting with anti-JAK3(N) (Right Upper) and anti-JAK2 (Right Lower), which immunoblots both JAK2 and JAK3. The unspecified middle band in all lanes shown on the anti-JAK2 blot is consistently recognized by this antibody as described by the manufacturer.
DISCUSSION
In this study, we have characterized the regions of JAK3 required for interacting with γc. Our data indicate that the N-terminal JH7-6 domains (aa 1–192) of JAK3 comprise a minimal region necessary and sufficient for γc association. This was demonstrated by the ability of chimeric forms of JAK3 to reconstitute IL-2-dependent signaling in a fibroblast cell line expressing the IL-2 receptor complex, by coimmunoprecipitation with the cytoplasmic domain of γc, and finally by the ability of this region to compete with full-length JAK3 for binding to γc. Though previous studies have indicated that the N-terminal domains of JAKs are important for association with their cognate receptors (30, 35, 36), the minimal binding domain has not been defined nor has the conversion of receptor specificity from one JAK member to another been achieved. This study therefore provides, to the best of our knowledge, the most precise characterization of JAKs to date in terms of their interactions with cytokine receptors.
Previous studies on a JAK3 mutant lacking the JH1 kinase domain was shown to dominantly inhibit the function of wt JAK3 in BAF/3-derived cells, suggesting that the kinase domain is not necessary for receptor association (32). Other reports on JAK1 and JAK2 have also confirmed this observation (33, 34). Our data support these conclusions; however, it is plausible that the JAK kinase domain may still be playing a dynamic role in stabilizing or facilitating JAK receptor binding by affecting the stoichiometry or affinity of JAK recruitment to its receptor following cytokine-dependent activation.
A number of reports have identified the conserved membrane-proximal region (termed Box1 and Box2) of cytokine receptors as being necessary for JAK association (28–30). A comparable region in γc has also been documented to be important for JAK3 binding (26, 27). Based on this and the strong sequence homology among the JAK family members, it is likely that a conserved region of the JAKs mediates binding to the membrane-proximal region. However, the JAKs must also be able to bind different receptors selectively. While some receptors are rather indiscriminate (e.g., gp130 interacts with TYK2, JAK1, and JAK2) (43), our findings confirm that the interaction of JAK3 and γc is rather specific. Moreover, our data provide insights in understanding the molecular mechanisms that account for this specificity. The N-terminal JH7-6 domains of the JAKs contain both loosely conserved and highly variable sequences (11). The model predicted by our data would suggest that some well-conserved motifs in the JH7-6 domains allow for generalized binding ability to Box1/Box2 regions within the cytokine receptor, while the neighboring less conserved sequences govern binding specificity (i.e., JAK3 to γc).
Our results demonstrate the strong correlation between the functional and physical coupling of the JAK chimeras with γc and support the contention that the JAK3–γc interaction is important for IL-2 signaling. Wild-type JAK3, as well as chimeras J3(7-4)J2(4-1) and J3(7-3)J2(2-1) that bound to γc, restored IL-2-inducible phosphorylation of the chimeric JAK, JAK1, the IL-2Rβ chain, and STAT5A in 3T3αβγ cells (Figs. 2B and 3). In contrast, the chimera J3(7)J2(6-1) that failed to associate with γc also failed to respond to IL-2 stimulation (Figs. 2B and 3). It is unclear why chimeras J3(7-3)J2(2-1) and J3(7)J2(6-1) demonstrated basal levels of tyrosine phosphorylation similar to endogenous JAK2, though one can imagine subtle conformational changes or altered interactions with other proteins such as phosphatases as likely causes. While the levels of STAT5A phosphorylation in cells expressing the chimeras were similar to that of wt JAK3, we observed a decrease in IL-2Rβ chain phosphorylation that directly correlated with a decrease in γc binding by the chimeras (Figs. 2B and 3C). This suggests that changes in the binding affinity or functional behavior may result as a consequence of replacing the JAK3 JH4-1 domains with JAK2. Future efforts toward elucidating the functional aspects of all the JH domains may help in explaining this phenomenon.
Our findings offer clues in reconciling the conflicting reports on the interaction between JAK2 and its cognate receptors. In one report, the entire JH7-3 domains of JAK2 were all shown to be necessary in GHR association (30). In another, the N-terminal region comprising the JH7-6 domains was said to be sufficient for GM-CSFR binding (36). In light of our data, a number of possible explanations may account for this discrepancy. First, while the JH5-3 domains of JAK2 are not absolutely required for association with GM-CSFR, they may in fact be playing a role that affects receptor binding affinity for one receptor type over another. Our data suggest that this may be a possibility (Figs. 2B and 3C). However, inherent differences between the JAK2–GHR interaction and the JAK2–GM-CSFR association governing binding behavior may exist that limit the extent of interpreting studies from one JAK receptor interaction to another. More importantly, the discrepancies may also be explained by the constructs used. In the study by Tanner et al. (30), a series of JAK2–Fyn chimeric constructs were used in which the C terminus of JAK2 was replaced by the Fyn kinase domain. It is thus possible that conformational alterations in the mutant caused by fusion with the Fyn kinase domain interfered with GHR association. In comparison, our studies on JAK3 suggest that the JH7-6 domains are in fact necessary and sufficient for receptor binding. However, we cannot rule out a potential role of the JH5-3 domains in mediating receptor binding affinity. As demonstrated by our results, a decreased association with γc was observed in both chimeras [J3(7-4)J2(4-1) and J3(7-3)J2(2-1)] (Fig. 2B) compared with wt JAK3. In addition, our use of the J3J2 chimeras has allowed us to further assess the functional implications of these binding studies, thus offering more insight to the receptor binding behavior of the JAKs.
In summary, our data indicate that the N-terminal JH7-6 domains are necessary and sufficient for association with γc. Although the exact sequences within this region have yet to be determined, our findings have delineated a restricted domain that is essential for JAK3–γc interaction. Given the similarity among all members of the JAK family, these data can serve as a model in addressing broader questions regarding the specific and general interactions between the JAK kinases and their cytokine receptors. In light of the critical role of cytokines in regulating many cellular processes including malignant transformation (44–46), targeting these interactions is an attractive possibility. Moreover, the specific consequences of mutations in γc and JAK3 make this interaction a particularly compelling target.
Acknowledgments
We thank Drs. J. Ihle, R. Klausner, W. Leonard, D. Nelson, and P. Sondel for reagents, and Drs. J. Johnston and C. Sudarshan for insightful suggestions.
ABBREVIATIONS
- γc
common γ chain
- IL
interleukin
- STAT
signal transducer and activator of transcription
- JH
JAK homology
- GHR
growth hormone receptor
- J3J2
JAK3–JAK2
- Tac
IL-2Rα chain
- wt
wild type
- GM-CSFR
granulocyte–macrophage-colony stimulating factor receptor
- IL-2R
IL-2 receptor
References
- 1.Paul W E, Seder R A. Cell. 1994;76:241–251. doi: 10.1016/0092-8674(94)90332-8. [DOI] [PubMed] [Google Scholar]
- 2.Ihle J N, Witthuhn B A, Quelle F W, Yamamoto K, Silvennoinen O. Annu Rev Immunol. 1995;13:369–398. doi: 10.1146/annurev.iy.13.040195.002101. [DOI] [PubMed] [Google Scholar]
- 3.Taniguchi T. Science. 1995;268:251–255. doi: 10.1126/science.7716517. [DOI] [PubMed] [Google Scholar]
- 4.Schindler C, Darnell J E J. Annu Rev Biochem. 1995;64:621–651. doi: 10.1146/annurev.bi.64.070195.003201. [DOI] [PubMed] [Google Scholar]
- 5.Darnell J E J, Kerr I M, Stark G R. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 6.Johnston J A, Bacon C M, Riedy M C, O’Shea J J. J Leukocyte Biol. 1996;60:441–452. doi: 10.1002/jlb.60.4.441. [DOI] [PubMed] [Google Scholar]
- 7.Velazquez L, Fellous M, Stark G R, Pellegrini S. Cell. 1992;70:313–322. [PubMed] [Google Scholar]
- 8.Muller M, Briscoe J, Laxton C, Guschin D, Ziemiecki A, Silvennoinen O, Harpur A G, Barbieri G, Witthuhn B A, Schindler C, Pellegrini S, Wilks A F, Ihle J N, Stark G R, Kerr I M. Nature (London) 1993;366:129–135. doi: 10.1038/366129a0. [DOI] [PubMed] [Google Scholar]
- 9.Watling D, Guschin D, Muller M, Silvennoinen O, Witthuhn B A, Quelle F W, Rogers N C, Schindler C, Stark G R, Ihle J N, Kerr I M. Nature (London) 1993;366:166–170. doi: 10.1038/366166a0. [DOI] [PubMed] [Google Scholar]
- 10.Oakes S A, Candotti F, Johnston J A, Chen Y-Q, Ryan J J, Taylor N, Liu X, Hennighausen L, Notarangelo L D, Paul W E, Blaese R M, O’Shea J J. Immunity. 1996;5:605–615. doi: 10.1016/s1074-7613(00)80274-5. [DOI] [PubMed] [Google Scholar]
- 11.Kawamura M, McVicar D W, Johnston J A, Blake T B, Chen Y Q, Lal B K, Lloyd A R, Kelvin D J, Staples J E, Ortaldo J R, O’Shea J J. Proc Natl Acad Sci USA. 1994;91:6374–6378. doi: 10.1073/pnas.91.14.6374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Witthuhn B A, Silvennoinen O, Miura O, Lai K S, Cwik C, Liu E T, Ihle J N. Nature (London) 1994;370:153–157. doi: 10.1038/370153a0. [DOI] [PubMed] [Google Scholar]
- 13.Gurniak C B, Berg L J. Blood. 1996;87:3151–3160. [PubMed] [Google Scholar]
- 14.Rane S G, Reddy E P. Oncogene. 1994;9:2415–2423. [PubMed] [Google Scholar]
- 15.Russell S M, Johnston J A, Noguchi M, Kawamura M, Bacon C M, Friedmann M, Berg M, McVicar D W, Witthuhn B A, Silvennoinen O, Goldman A S, Schmalsteig F C, Ihle J N, Oshea J J, Leonard W J. Science. 1994;266:1042–1045. doi: 10.1126/science.7973658. [DOI] [PubMed] [Google Scholar]
- 16.Miyazaki T, Kawahara A, Fujii H, Nakagawa Y, Minami Y, Liu Z J, Oishi I, Silvennoinen O, Witthuhn B A, Ihle J N, Taniguchi T. Science. 1994;266:1045–1047. doi: 10.1126/science.7973659. [DOI] [PubMed] [Google Scholar]
- 17.Leonard W J, Noguchi M, Russell S M. Adv Exp Med Biol. 1994;365:225–232. doi: 10.1007/978-1-4899-0987-9_23. [DOI] [PubMed] [Google Scholar]
- 18.Giri J G, Ahdieh M, Eisenman J, Shanebeck K, Grabstein K, Kumaki S, Namen A, Park L S, Cosman D, Anderson D. EMBO J. 1994;13:2822–2830. doi: 10.1002/j.1460-2075.1994.tb06576.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Noguchi M, Yi H, Rosenblatt H M, Filipovich A H, Adelstein S, Modi W S, McBride O W, Leonard W J. Cell. 1993;73:147–157. doi: 10.1016/0092-8674(93)90167-o. [DOI] [PubMed] [Google Scholar]
- 20.Russell S M, Tayebi N, Nakajima H, Riedy M C, Roberts J L, Aman M J, Migone T S, Noguchi M, Markert M L, Buckley R H, O’Shea J J, Leonard W J. Science. 1995;270:797–800. doi: 10.1126/science.270.5237.797. [DOI] [PubMed] [Google Scholar]
- 21.Macchi P, Villa A, Gillani S, Sacco M G, Frattini A, Porta F, Ugazio A G, Johnston J A, Candotti F, O’Shea J J, Vezzoni P, Notarangelo L D. Nature (London) 1995;377:65–68. doi: 10.1038/377065a0. [DOI] [PubMed] [Google Scholar]
- 22.Nosaka T, van Deursen J M, Tripp R A, Thierfelder W E, Witthuhn B A, McMickle A P, Doherty P C, Grosveld G C, Ihle J N. Science. 1995;270:800–802. doi: 10.1126/science.270.5237.800. [DOI] [PubMed] [Google Scholar]
- 23.Thomis D C, Gurniak C B, Tivol E, Sharpe A H, Berg L J. Science. 1995;270:794–797. doi: 10.1126/science.270.5237.794. [DOI] [PubMed] [Google Scholar]
- 24.Park S Y, Saijo K, Takahashi T, Osawa M, Arase H, Hirayama N, Miyake K, Nakauchi H, Shirasawa T, Saito T. Immunity. 1995;3:771–782. doi: 10.1016/1074-7613(95)90066-7. [DOI] [PubMed] [Google Scholar]
- 25.Cao X, Shores E W, Hu-Li J, Anver M R, Kelsall B L, Russell S M, Drago J, Noguchi M, Grinberg A, Bloom E T, Paul W E, Katz S I, Love P E, Leonard W L. Immunity. 1995;2:223–238. doi: 10.1016/1074-7613(95)90047-0. [DOI] [PubMed] [Google Scholar]
- 26.Nelson B H, Lord J D, Greenberg P D. Mol Cell Biol. 1996;16:309–317. doi: 10.1128/mcb.16.1.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goldsmith M A, Lai S Y, Xu W, Amaral M C, Kuczek E S, Parent L J, Mills G B, Tarr K L, Longmore G D, Greene W C. J Biol Chem. 1995;270:21729–21737. doi: 10.1074/jbc.270.37.21729. [DOI] [PubMed] [Google Scholar]
- 28.Witthuhn B A, Quelle F W, Silvennoinen O, Yi T, Tang B, Miura O, Ihle J N. Cell. 1993;74:227–236. doi: 10.1016/0092-8674(93)90414-l. [DOI] [PubMed] [Google Scholar]
- 29.Quelle F W, Sato N, Witthuhn B A, Inhorn R C, Eder M, Miyajima A, Griffin J D, Ihle J N. Mol Cell Biol. 1994;14:4335–4341. doi: 10.1128/mcb.14.7.4335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tanner J W, Chen W, Young R L, Longmore G D, Shaw A S. J Biol Chem. 1995;270:6523–6530. doi: 10.1074/jbc.270.12.6523. [DOI] [PubMed] [Google Scholar]
- 31.Harpur A G, Andres A C, Ziemiecki A, Aston R R, Wilks A F. Oncogene. 1992;7:1347–1353. [PubMed] [Google Scholar]
- 32.Kawahara A, Minami Y, Miyazaki T, Ihle J N, Taniguchi T. Proc Natl Acad Sci USA. 1995;92:8724–8728. doi: 10.1073/pnas.92.19.8724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guschin D, Rogers N, Briscoe J, Witthuhn B, Watling D, Horn F, Pellegrini S, Yasukawa K, Heinrich P, Stark G R, Ihle J N, Kerr I M. EMBO J. 1995;14:1421–1429. doi: 10.1002/j.1460-2075.1995.tb07128.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Briscoe J, Rogers N C, Witthuhn B A, Watling D, Harpur A G, Wilks A F, Stark G R, Ihle J N, Kerr I M. EMBO J. 1996;15:799–809. [PMC free article] [PubMed] [Google Scholar]
- 35.Frank S J, Yi W, Zhao Y, Goldsmith J F, Gilliland G, Jiang J, Sakai I, Kraft A S. J Biol Chem. 1995;270:14776–14785. doi: 10.1074/jbc.270.24.14776. [DOI] [PubMed] [Google Scholar]
- 36.Zhao Y, Wagner F, Frank S J, Kraft A S. J Biol Chem. 1995;270:13814–13818. doi: 10.1074/jbc.270.23.13814. [DOI] [PubMed] [Google Scholar]
- 37.Pogulis R J, Vallejo A N, Pease L R. Methods Mol Biol. 1996;57:167–176. doi: 10.1385/0-89603-332-5:167. [DOI] [PubMed] [Google Scholar]
- 38.Minami Y, Oishi I, Liu Z J, Nakagawa S, Miyazaki T, Taniguchi T. J Immunol. 1994;152:5680–5690. [PubMed] [Google Scholar]
- 39.Johnston J A, Kawamura M, Kirken R A, Chen Y Q, Blake T B, Shibuya K, Ortaldo J R, McVicar D W, O’Shea J J. Nature (London) 1994;370:151–153. doi: 10.1038/370151a0. [DOI] [PubMed] [Google Scholar]
- 40.Lin J X, Migone T S, Tsang M, Friedmann M, Weatherbee J A, Zhou L, Yamauchi A, Bloom E T, Mietz J, John S, Leonard W J. Immunity. 1995;2:331–339. doi: 10.1016/1074-7613(95)90141-8. [DOI] [PubMed] [Google Scholar]
- 41.Hou J, Schindler U, Henzel W J, Wong S C, McKnight S L. Immunity. 1995;2:321–329. doi: 10.1016/1074-7613(95)90140-x. [DOI] [PubMed] [Google Scholar]
- 42.Fujii H, Nakagawa Y, Schindler U, Kawahara A, Mori H, Gouilleux F, Groner B, Ihle J N, Minami Y, Miyazaki T, Taniguchi T. Proc Natl Acad Sci USA. 1995;92:5482–5486. doi: 10.1073/pnas.92.12.5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stahl N, Boulton T G, Farruggella T, Ip N Y, Davis S, Witthuhn B A, Quelle F W, Silvennoinen O, Barbieri G, Pellegrini S, Ihle J N, Yancopoulos G D. Science. 1994;263:92–95. doi: 10.1126/science.8272873. [DOI] [PubMed] [Google Scholar]
- 44.Migone T S, Lin J X, Cereseto A, Mulloy J C, O’Shea J J, Franchini G, Leonard W J. Science. 1995;269:79–81. doi: 10.1126/science.7604283. [DOI] [PubMed] [Google Scholar]
- 45.Danial N N, Pernis A, Rothman P B. Science. 1995;269:1875–1877. doi: 10.1126/science.7569929. [DOI] [PubMed] [Google Scholar]
- 46.Meydan N, Grunberger T, Dadi H, Shahar M, Arpaia E, Lapidot Z, Leeder J S, Freedman M, Cohen A, Gazit A, Levitzki A, Roifman C M. Nature (London) 1996;379:645–648. doi: 10.1038/379645a0. [DOI] [PubMed] [Google Scholar]