Abstract
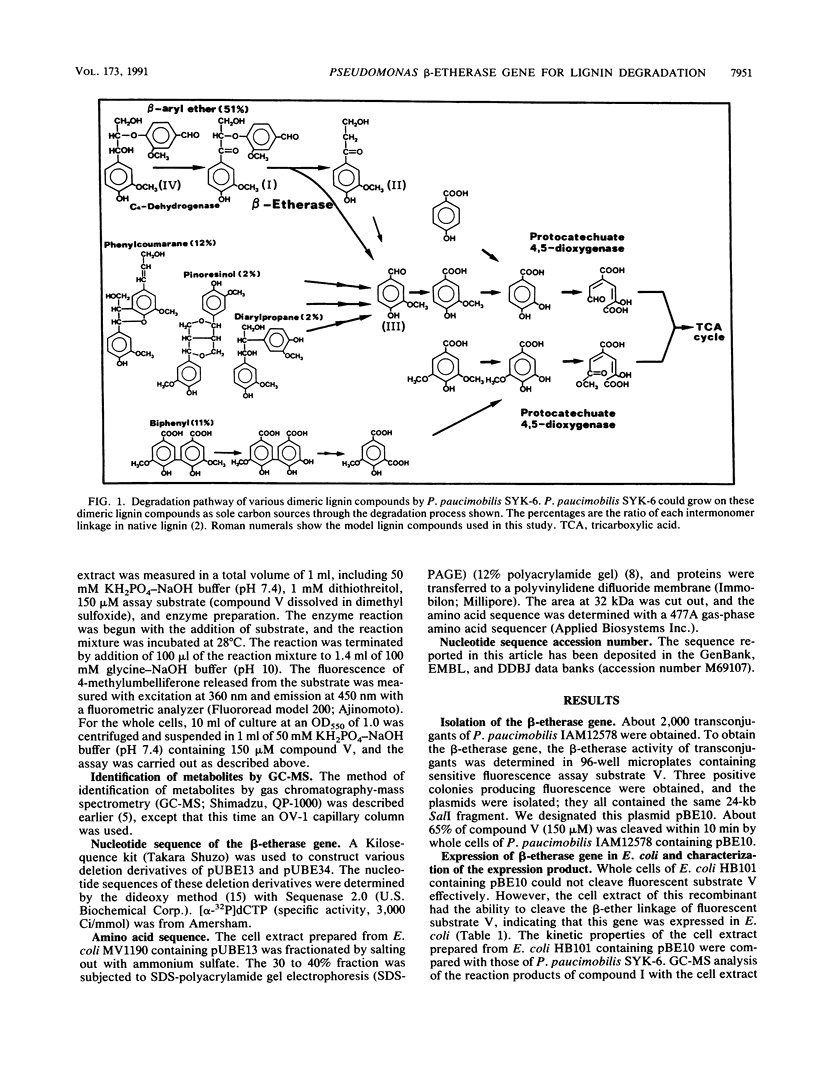
We isolated Pseudomonas paucimobilis SYK-6, which was able to degrade various dimeric lignin compounds (Y. Katayama, S. Nishikawa, M. Nakamura, K. Yano, M. Yamasaki, N. Morohoshi, and T. Haraguchi, Mokuzai Gakkaishi 33:77-79, 1987). This metabolic process is a distinct characteristic of this bacterium, which is equipped with an enzymatic modification system for various dimeric lignin compounds involved in the tricarboxylic acid cycle. Cleavage of the beta-aryl ether linkage is essential in this process, because this linkage is the most abundant (approximately 50%) in lignin. Here, we report the isolation and characterization of the beta-etherase gene, which contains an open reading frame of 843 bp and which we call ligE. This gene was expressed in Escherichia coli, and the enzyme had the same kinetic properties as the P. paucimobilis SYK-6 enzyme.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ditta G., Stanfield S., Corbin D., Helinski D. R. Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7347–7351. doi: 10.1073/pnas.77.12.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
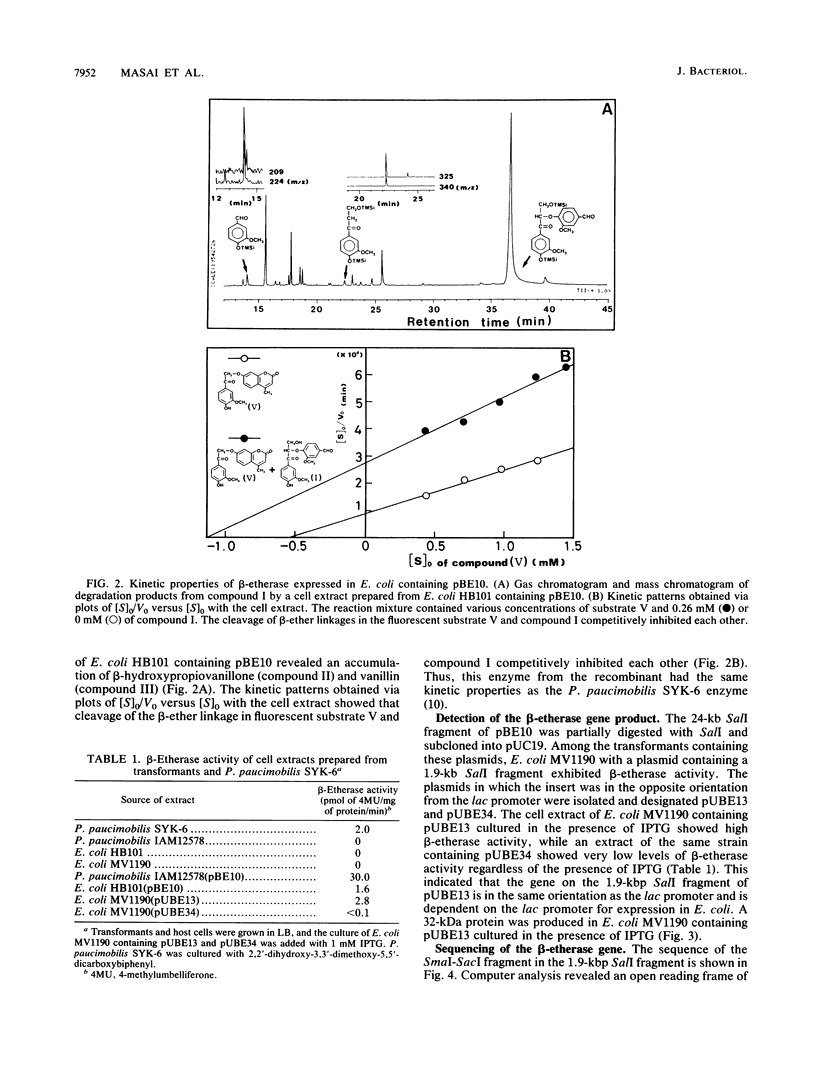
- Masai E., Katayama Y., Nishikawa S., Yamasaki M., Morohoshi N., Haraguchi T. Detection and localization of a new enzyme catalyzing the beta-aryl ether cleavage in the soil bacterium (Pseudomonas paucimobilis SYK-6). FEBS Lett. 1989 Jun 5;249(2):348–352. doi: 10.1016/0014-5793(89)80656-8. [DOI] [PubMed] [Google Scholar]
- Noda Y., Nishikawa S., Shiozuka K., Kadokura H., Nakajima H., Yoda K., Katayama Y., Morohoshi N., Haraguchi T., Yamasaki M. Molecular cloning of the protocatechuate 4,5-dioxygenase genes of Pseudomonas paucimobilis. J Bacteriol. 1990 May;172(5):2704–2709. doi: 10.1128/jb.172.5.2704-2709.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelmont J., Barrelle M., Hauteville M., Gamba D., Romdhane M., Dardas A., Beguin C. A new bacterial dehydrogenase oxidizing the lignin model compound guaiacylglycerol beta-O-4-guaiacyl ether. Biochimie. 1985 Sep;67(9):973–986. doi: 10.1016/s0300-9084(85)80292-3. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]