Abstract
The human RAD51 protein is a homologue of the bacteria RecA and yeast RAD51 proteins that are involved in homologous recombination and DNA repair. RAD51 interacts with proteins involved in recombination and also with tumor suppressor proteins p53 and breast cancer susceptibility gene 1 (BRCA1). We have used the yeast two-hybrid system to clone murine cDNA sequences that encode two RAD51-associated molecules, RAB22 and RAB163. RAB163 encodes the C-terminal portion of mouse BRCA2, the homologue of the second breast cancer susceptibility gene protein in humans, demonstrating an in vitro association between RAD51 and BRCA2. RAB22 is a novel gene product that also interacts with RAD51 in vitro. To detect RAD51 interactions in vivo, we developed a transient nuclear focus assay that was used to demonstrate a complete colocalization of RAB22 with RAD51 in large nuclear foci.
The RAD51 gene was originally identified in yeast as encoding a member of the RAD52 epistasis group (RAD50–RAD57) of proteins involved in DNA recombination and repair. RAD51 mutants are deficient in the repair of DNA damage caused by ionizing radiation, genetic recombination, and recombinational repair of the DNA lesions (1, 2). The RAD51 gene is well-conserved among eukaryotes (3), and the RAD51 protein is significantly homologous to the Escherichia coli RecA protein. Recent biochemical studies demonstrated that both yeast and human RAD51 proteins had the ability to promote ATP-dependent homologous pairing and strand-transfer reactions in vitro (2, 4, 5); but the precise function(s) of the mammalian RAD51 protein in vivo is not entirely clear. Targeted disruption of the mouse RAD51 gene resulted in an early embryonic lethal phenotype and also demonstrated that RAD51 is essential for cell proliferation (6, 7). Although targeted disruption can be a useful approach for understanding the overall function of genes in vivo, the early embryonic lethal phenotype of the RAD51 knock-out did not provide the information needed to elucidate specific functions of RAD51. Therefore, we have undertaken the strategy of cloning RAD51-associated molecules to investigate their functional role in the nuclear organization of RAD51.
Yeast RAD51 belongs to the RAD52 epistasis group whose members are functionally associated. Yeast RAD52 and RAD55 directly associate with RAD51 (8), and human RAD52 also associates with human RAD51 (9). RecA activity is essential for the SOS response, a bacterial DNA damage control pathway that is dependent on gene transcription (10); notably, RAD51 is a member of the RNA polymerase II complex (10, 11). Defects in repair molecules such as RAD51 and associated molecules could perturb genomic integrity and eventually lead to tumorigenesis. Such a model has been proposed for mutations in mismatch repair molecules in hereditary nonpolyposis colon cancer (12). Recently, RAD51 was reported to interact with the breast cancer susceptibility gene 1 (BRCA1) duplicated (10) and the p53 tumor suppressor gene product (13). Although it is not yet clear how biological function is dependent on the interactions between these nuclear proteins, these results will certainly contribute to understanding the functional connections between RAD51 and tumor suppressor genes.
Here we report the molecular cloning of cDNA sequences that encode two other proteins that can associate with RAD51. RAB163 encodes the C-terminal end of mouse breast cancer susceptibility gene 2 (BRCA2) which is another breast cancer susceptible gene product. RAB22 is a novel gene product that colocalizes in nuclear foci with RAD51. We also describe a new assay system for detecting direct interactions in vivo between RAD51 and associated molecules.
MATERIALS AND METHODS
Plasmid Constructs.
Human RAD51 (14) eukaryotic expression construct, pRc/CMV-RAD51, was generated by A.S. (unpublished data). The LexA fusion construct, pLexRAD51, for yeast-two hybrid screening was prepared from pRc/CMV-RAD51, namely, blunt NcoI–BamHI fragment which had full RAD51 cDNA sequence was recloned into the blunt EcoRI–BamHI site of the pEG202 LexA fusion plasmid (15). The other constructs were made by recloning cDNA fragments that had appropriate enzyme sites generated by polymerase chain reaction (PCR). A glutathione S-transferase (GST)-fused RAD51 construct, pGST–RAD51, was made by direct cloning of PCR-amplified BamHI fragments into the pGEX-2TK (Pharmacia). pRSET A vector (Invitrogen) was used for construction of in vitro transcription and translation products. pRSET–RAD51 and pRSET–XRCC4 (16) were generated by direct cloning of PCR-amplified BamHI fragments of each cDNA into the vector. PCR-amplified BamHI–EcoRI fragment of RAB22 which contained the whole cDNA was recloned into pRSET BamHI–EcoRI site. To make pRSET–RAB163, PCR-amplified BamHI fragment from JG4-5–RAB163, which was cloned by yeast-two hybrid system and contained the C-terminal cDNA of the mBRCA2, was recloned into the pRSET BamHI site. PCR-amplified SalI–ApaI fragment of RAB22 was constructed into XhoI–ApaI site of the green fluorescent protein (GFP) expression vector generously provided by F. Mckeon (17) to generate pGFP-RAB22. The blue flourescent protein (BFP) expression vector, pQB150, was purchased from Quantum Biotechnologies (Quebec, Canada). Partially digested NheI fragments that were amplified from RAD51 cDNA were recloned into the NheI site of this vector to generate the pBFP–RAD51 expression construct.
Selection of RAD51-Associated Molecules.
Yeast two-hybrid selection was performed in EGY48 [MATa trp1 ura3 his3 LEU2::pLexAop6-LEU2] into which pJK103 and pLexRAD51 had been introduced by transformation (15). This strain was maintained under selection for the URA3 and HIS3 makers and was transformed with a mouse thymus cDNA library made by H.-L.C. (unpublished data). A total of 1 million primary yeast transformants were selected on the Ura− His− Trp− glucose plates and the pooled yeasts (5 million) were plated on Ura− His− Trp− Leu− galactose plates. Three days later, colonies were picked and streaked on Ura− His− Trp− 5-bromo-4-chloro-3-indolyl β-d-galactoside (X-Gal) glucose and Ura− His− Trp− X-Gal galactose plates. Plasmids were isolated from colonies that grew on Leu− plates and turned blue on X-Gal. Library-derived cDNA clones were selected and expanded in KC8 cells. cDNAs were analyzed first by restriction digestion using HaeIII or AluI enzymes and sorted into classes depending on their restriction map pattern. At least one representative cDNA from each class was partially sequenced. The full-length cDNA clone for RAB22 was constructed with two cDNA clones; one, RAB22-5, from the mouse thymus cDNA library (Stratagene) that had a deletion at its 5′ end and the other, RAB22-5′-6, from a marathon mouse testis cDNA library (Clontech) that contained the intact 5′ end (see Fig. 1b). The cloning was performed as per manufacturer’s specifications. For cloning RAB22-5 we used cDNA fragments of JG4-5–RAB22, that was originally identified through the yeast two-hybrid assay, as probes to screen the phage cDNA library, For cloning RAB22-5′-6 we used a gene specific primer (RAB22-1R, 5′-tcccctgcaggtttcttctgc-3′) and a cDNA adapter primer (AP1; Clontech) to amplify the 5′ end of the RAB22 cDNA by PCR. Full-length RAB22 cDNA was made by replacing the 5′ end of the XhoI–XhoI fragment of RAB22-5 with that of RAB22-5′-6.
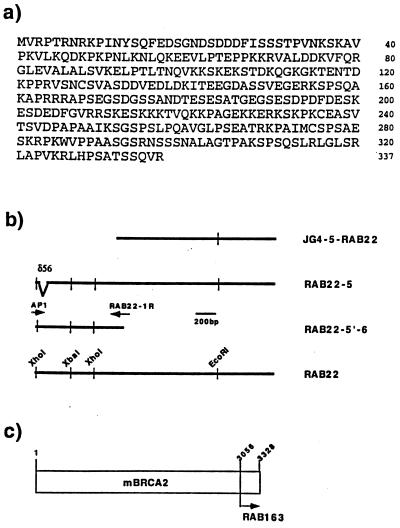
Figure 1.
RAD51-associated cDNA clones isolated by the yeast two-hybrid assay. (a) Predicted amino acid sequence of full-length RAB22, encoding 337 amino acids. No significant homology was observed to other sequences in GenBank. The GenBank accession number of the cDNA sequence is U93583. (b) Four representative cDNA fragments of RAB22 are shown as bold lines with the clone name listed on the right side. JG4-5–RAB22 was the original clone isolated by the yeast-two hybrid screening. RAB22-5 was cloned from a mouse thymus library using a cDNA probe prepared from JG4-5–RAB22, but contains a 56-nucleotide deletion (δ56) in the coding region. RAB22-5′-6 was fragment amplified from a mouse testis library by two primers (arrows), AP1 and RAB22-1R, and did not contain a deletion. RAB22 is the full-length cDNA clone constructed from RAB22-5 and RAB22-5′-6. The thin vertical lines are restriction enzyme sites as noted. (c) Relative position of the RAB163 peptide to the mouse BRCA2 (mBRCA2) peptide was shown by amino acid number and arrow. The full-length cDNA sequence of mBRCA2 has been reported (26).
Northern Blot Analysis.
The JG4-5–RAB22 cDNA, which was isolated by the yeast-two hybrid system and encoded the C-terminal 128 amino acids of the RAB22 peptide, was labeled with [α-32P]dCTP and used to probe ≈10 μg of total RNA from the various tissues indicated.
Interspecific Mouse Backcross Mapping.
Interspecific backcross progeny were generated by mating (C57BL/6J × Mus spretus)F1 females and C57BL-6J males as described (18). A total of 205 N2 mice were used to map the Rab22 and Rab163 (Brca2) loci (see text for details). DNA isolation, restriction enzyme digestion, agarose gel electrophoresis, Southern blot transfer, and hybridization were performed essentially as described (19). All blots were prepared with Zetabind nylon membrane (AMF Cuno). Probes, which were specific for each locus, were labeled with [α-32P]dCTP using a nick translation labeling kit (Boehringer Mannheim); washing was done to a final stringency of 0.8–1.0× SSCP (0.12 M NaCl2/0.015 M Na citrate/0.01 M KPO4)/0.1% SDS at 65°C. The Rab22 probe, an ≈0.8-kb EcoRI fragment of mouse cDNA, JG4-5–RAB22, detected a major fragment of 2.6 kb in C57BL/6J (B) DNA and a major fragment of 17.0 kb in M. spretus (S) DNA following digestion with XbaI. The Rab163 (Brca2) probe, an ≈1.2-kb EcoRI/XhoI fragment of mouse cDNA, detected TaqI fragments of 1.0 and 0.7 kb (B) and 1.3 and 1.0 kb (S). The presence or absence of M. spretus-specific fragments was followed in backcross mice.
A description of the probes and restriction fragment length polymorphisms (RFLPs) for the loci linked to the RAB22 and RAB163 loci in the interspecific backcross has been reported. These include Pdgfa, Pms2, and Flt3, chromosome 5 (20, 21) and Cd4, Fgf6, and Grin2b, chromosome 6 (22, 23). Recombination distances were calculated using map manager, Version 2.6.5. Gene order was determined by minimizing the number of recombination events required to explain the allele distribution patterns.
In Vitro Affinity Pull Down Assay.
E. coli DH5α was transformed with pGEX–RAD51 or pGEX–2TK vector, and GST fusion proteins were extracted as described by the manufacturer (Pharmacia). The supernatant was mixed with glutathione-Sepharose 4B beads (Pharmacia), and washed several times with 1× NETN buffer (0.5% Nonidet P-40/20 mM Tris, pH 8/100 mM NaCl/1 mM EDTA/2 mM phenylmethylsulfonyl fluoride/protease inhibitor cocktail). 35S-labeled RAD51, RAB22, RAB163/mBRCA2, XRCC4, and luciferase were synthesized in vitro from pRSET vectors containing those sequences, using TnT coupled reticulocyte lysate systems (Promega). 35S-labeled proteins were bound to the immobilized GST proteins. After washing several times, immobilized proteins were separated by 10% SDS/PAGE. The gel was dried and exposed to x-ray film.
Transient Nuclear Focus (TrNF) Assay.
Approximately 1 μg of the pGFP–RAB22 and pRc/CMV–RAD51 or pBFP–RAD51 were transiently transfected into Chinese hamster ovary (CHO-K1) cells on the coverslips by the calcium phosphate precipitation method (17). Cells on the coverslips were fixed in 3% formaldehyde in PBS, permeabilized with 0.2% Triton X-100 PBS, and incubated with rabbit polyclonal anti-human RAD51 (1:200 dilution). Rhodamine-conjugated anti-rabbit antibody (BioSource International, Camarillo, CA; 1:2,000 dilution) was used as the second antibody. Coverslips were mounted on glass slides in 90% glycerol, 20 mM Tris⋅HCl (pH 9.35) and 1 μg/ml 4′,6-diamidine-2′-phenylindole dihydrochloride (DAPI; Boehringer Mannheim), and examined on a Zeiss Axioplan 2 fluorescent microscope equipped with a ×63, NA 1.4 objective. Images of single optical sections were captured individually using appropriate filters and a grayscale charge-coupled device (CCD) camera (Photometrics, Tucson, AZ) then merged, pseudocolored, and analyzed for colocalization in iplab spectrum version 3.1 with multiprobe extension (Signal Analytics, Vienna, VA).
RESULTS
Isolation of RAD51, RAB22, and RAB163.
To identify molecules that associate with RAD51, we used the yeast two-hybrid assay (15). A yeast strain expressing LexA–RAD51 “bait” was transformed with a mouse thymus library to generate one million independent transformants. Cells containing candidate RAD51-associated molecules were identified by their ability to grow in the absence of leucine on galactose plates. Of the 200 colonies that passed this first selection, 27 contained plasmids that induced transcription in a galactose-dependent manner from both the LEU2 and LacZ reporter constructs. The 27 plasmids were sorted into eight groups based on restriction digest pattern, and their nucleotide sequences were determined. Twelve of 27 plasmids were identical, and named RAB22. None of the sequences in the database showed clear similarity to RAB22. One clone from 27 plasmids was mouse RAD51, which was anticipated; because the RAD51 protein has been shown to associate with itself to make nuclear filaments (24). Another unique clone was RAB163. Of note, this clone encodes the mouse homologue of human BRCA2 (25, 26).
RAB22 Sequence.
Because the first identified clone of RAB22 (JG4-5–RAB22) contained only the 3′ end of the cDNA, a mouse thymus library was then screened to obtain a full-length cDNA clone. The nucleotide sequence of the three longest clones was determined, but minor differences in their 5′ ends and an ambiguous open reading frame suggested either alternative splicing or cloning artifacts. We then screened the mouse testis marathon-cDNA library with the 5′-rapid amplification of cDNA ends (RACE) method to generate additional 5′ end clones. Three new clones were isolated, RAB22-5′-6 was the longest while RAB22-5′-3 and RAB22-4 were identical and overlapped with RAB22-5′-6 (Fig. 1b). The new sequence contained a potential start codon proceeded by consensus Kozak sequences and revealed that the previously identified clone RAB22–5 had a 56-nucleotide deletion (δ56) just downstream of the authentic initial codon (Fig. 1b). To make full-length RAB22 cDNA we replaced the 5′ end of the XhoI–XhoI fragment of RAB22-5 with that of RAB22-6. The complete RAB22 cDNA encoded 337 amino acids, and there was no highly homologous proteins identified in the GenBank (including yeast) database (Fig. 1a). The first two-thirds of the protein showed an abundance of charged amino acids and last one-third was hydrophobic.
Expression Pattern of the RAB22.
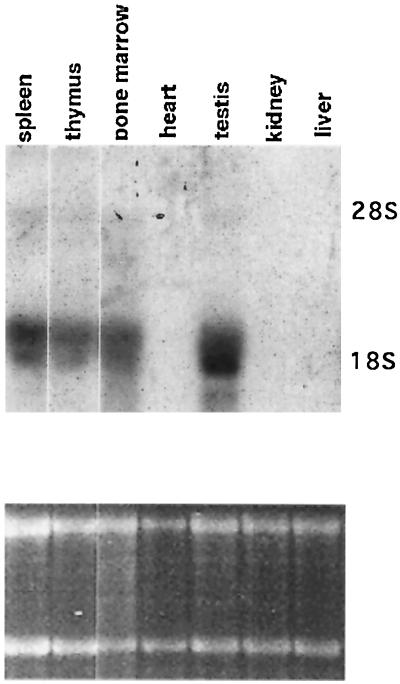
The expression pattern of the RAB22 in various tissues was determined by Northern blot analysis (Fig. 2). The RAB22 probe identified a transcript of the predicted size (2 kb) in RNA from spleen, thymus, bone marrow and, most abundantly, in testis; while no hybridizing transcripts were detected in RNA from heart, kidney, and liver (Fig. 2 Upper). This tissue distribution of RAB22 expression appears essentially identical to that of RAD51 (14), BRCA1 (27), and BRCA2 (25), suggesting the possibility of a functional correlation among these molecules.
Figure 2.
Analysis of RAB22 expression. (Upper) Total RNA from multiple mouse tissues was probed with the full-length RAB22 cDNA. (Lower) Relative amount of total RNA in each lane is demonstrated by etidium bromide staining of 28S and 18S rRNA.
Chromosome Mapping of RAB22 and RAB163/mBRCA2 Genes.
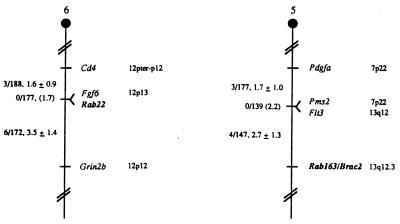
The murine chromosomal location of Rab22 and Rab163/Brca2 was determined using an interspecific backcross mapping panel derived from crosses of [C57BL/6J × (M. spretus)F1 × C57BL/6J] mice. This mapping panel has been typed for over 2,300 loci that are well dispersed among all the autosomes as well as the X chromosome (18). cDNA fragments specific for each of the loci were used as probes in Southern blot hybridization analysis of C57BL/6J and M. spretus genomic DNAs that were separately digested with several different restriction enzymes to identify informative RFLPs useful for gene mapping (see Materials and Methods). The strain distribution pattern of each RFLP in the interspecific backcross was then determined by following the presence or absence of RFLPs specific for M. spretus in backcross mice. The mapping results (Fig. 3) assigned the two loci to two different mouse autosomes: Rab22 mapped to the distal region of mouse chromosome 6, 1.6 centimorgan (cM) distal of Cd4 and 3.5 cM proximal of Grin2b; RAB163/Brca2 to the distal region of mouse chromosome 5, 2.7 cM distal to Flt3 and Pms2.
Figure 3.
Chromosomal locations of the two RAB loci in the mouse genome. RAB loci were mapped by interspecific backcross analysis. The number of recombinant N2 animals is presented over the total number of N2 animals typed to the left of the chromsome maps between each pair of loci. The recombination frequencies, expressed as genetic distance in centimorgans (±1 SE) are also shown. The upper 95% confidence limit of the recombination distance is given in parentheses when no recombinants were found between loci. Gene order was determined by minimizing the number of recombination events required to explain the allele distribution patterns. The positions of loci on human chromosomes, where known, are shown to the right of the chromosome maps. References for the map positions of most human loci can be obtained from the Genome Data Base, a computerized database of human linkage information maintained by The William H. Welch Medical Library of The Johns Hopkins University (Baltimore).
Direct Interaction of RAB22 and RAB163/mBRCA2 with RAD51 in Vitro.
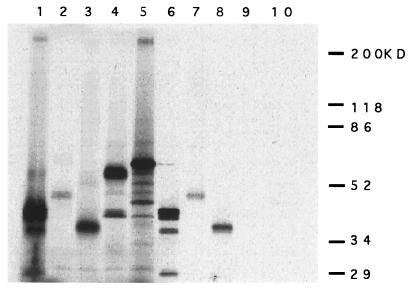
The yeast two-hybrid system is a very useful technique for cloning associated molecules, but it can be prone to artifacts. To verify the interaction we performed a GST-affinity pull down assay. RAD51–GST fusion protein and GST protein were synthesized and immobilized on glutathione-Sepharose beads to test for their ability to bind to 35S-labeled RAD51, RAB22, and RAB163/mBRCA2. A 35S-labeled XRCC4 and luciferase was used as a control. In agreement with the results from the yeast two-hybrid system RAD51, RAB22, and RAB163/mBRCA2 were specifically retained on beads coupled to GST–RAD51 (Fig. 4, lanes 1–3 and 6–8), while none of them were retained on beads coupled with GST alone (data not shown). RAD51 is known to make nuclear polymers (24), so the RAD51–RAD51 homologous interaction served as a positive control (lane 6). As negative controls, one of the V(D)J recombination related molecules XRCC4 (16) and luciferase were not retained on beads coupled to GST–RAD51 (Fig. 4, lanes 4, 5, 9, and 10).
Figure 4.
Specific association between RAD51 and RAB22 or RAB163 in vitro by GST-affinity pull-down assay. 35S-labeled RAD51 (≈40 kDa, lane 1), RAB22 (≈45 kDa, lane 2), RAB163/mBRCA2 (≈35 kDa, lane 3), XRCC4 (≈53 kDa, lane 4), and luciferase (≈60 kDa, lane 5) were generated in vitro and visualized following SDS/PAGE. For these lanes, 2.5 μl of in vitro-translated product was separated on the gel. Each in vitro-translated protein (15 μl of reaction) also was incubated with GST–RAD51 fusion protein that was synthesized in bacteria and immobilized on glutathione-Sepharose beads (40 μl). After washing with 1× NETN buffer several times, the beads were suspended in 25 μl of 2× sample buffer (2% SDS/10% glycerol/0.01% Bromophenol Blue), boiled, and spun to pellet the beads. Then 5 μl of each supernatant was assayed by SDS/PAGE; RAD51 (lane 6), RAB22 (lane 7), RAB163/mBRCA2 (lane 8), XRCC4 (lane 9), and luciferase (lane 10).
Direct Evidence of RAB22–RAD51 Interaction in Vivo.
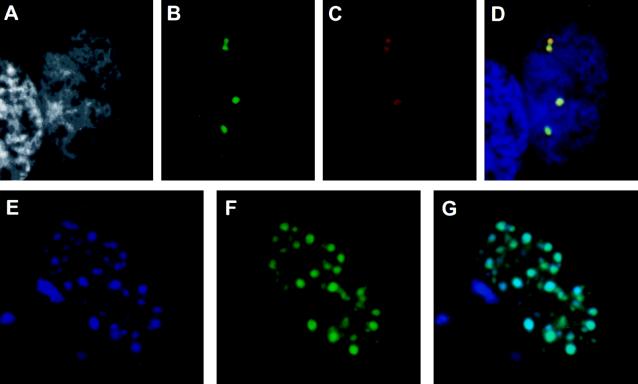
To confirm the RAB22–RAD51 interaction in vivo, we developed a simple assay system, called the TrNF assay. Previously, the RAD51 protein was shown to organize as discrete foci in the interphase nucleus (28). We therefore hypothesized that associated molecules may colocalize with RAD51 in nuclear foci in cells that have been cotransfected with RAD51 and the newly isolated clone. To detect colocalization by fluorescence microscopy, a GFP tag sequence was introduced in the 5′ end of the RAB22 cDNA (pGFP–RAB22). The RAD51 expression construct (pRc/CMV–RAD51) and RAB22 expression construct (pGFP–RAB22) were cotransfected into CHO-K1 cells. The cells were fixed, stained with anti-RAD51 antibody, and examined by fluorescence microscopy (Fig. 5 A–D). Transfection-positive cells showed several large nuclear foci (Fig. 5 B and C), although the size and number of the foci was variable. GFP–RAB22 (Fig. 5B, green foci) and RAD51 (Fig. 5C, red foci) clearly colocalized in the same nuclear foci (Fig. 5D, yellow). GFP vector-alone transfectants showed a diffuse staining pattern in both nucleus and cytoplasm with the absence of foci (data not shown). The same assay was then performed for direct detection of both proteins by inserting a BFP tag sequence in the 3′ end of RAD51 cDNA (pBFP–RAD51) and cotransfecting with pGFP–RAB22. The BFP–RAD51 (Fig. 5E, blue foci) and the GFP–RAB22 (Fig. 5F, green foci) colocalized within multiple nuclear foci (Fig. 5G, aqua foci), demonstrating that the colocalization was not the result of an artifact from antibody detection.
Figure 5.
Colocalization of RAD51 and RAB22 in discrete nuclear foci by TrNF assay. (A–D) CHO-K1 cells were transiently transfected with pGFP–RAB22 and pRc/CMV–RAD51 constructs and subsequently stained with anti-RAD51 antiserum and a rhodamine-conjugated secondary antibody. A representative nucleus of a transfection-positive cell is shown. (A) DAPI nuclear counterstain in grayscale. (B) pGFP–RAB22 localization, green foci. (C) pRc/CMV–RAD51 localization by antibody detection, red foci. No foci were observed with preimmune serum (data not shown). (D) Merged image of A–C. Where green and red signals overlap, a yellow signal is observed, indicating colocalization of RAB22 and RAD51. DNA counterstain is in blue. (E–G) CHO-K1 cells were transiently cotransfected with pGFP–RAB22 and pBFP–RAD51 constructs and directly analyzed for nuclear foci. (E) pBFP–RAD51 localization, blue foci. (F) pGFP–RAB22 localization, green foci. (G) In merged image of E and F, aqua foci indicate colocalization of blue and green signals.
The same assay was performed to detect a possible in vivo association of RAB163/mBRCA2 with RAD51. A construct containing a RAB163 cDNA fragment, which encoded only 273 amino acids of the mBRCA2 C terminus (1/12th of full-length mBRCA2 peptide; 3,328 amino acids) (Fig. 1c) (26) was cloned into the pGFP vector and cotransfected with pRc/CMV–RAD51 or pBFP–RAD51 into CHO-K1 cells. GFP–RAB163 did not show specific localization and was diffusively stained in both nucleus and cytoplasm (data not shown). In these cotransfected nuclei, RAD51 showed the same absence of nuclear foci formation as GFP–RAB163 (data not shown), suggesting that RAD51 nuclear foci formation might be altered by the associated molecules.
DISCUSSION
We have identified two mouse genes, RAB22 and RAB163, whose products interact with the human RAD51 protein in the yeast two-hybrid system. RAB163 is the mouse homologue of human BRCA2 which has been linked to hereditary breast cancer (25, 26). Recently, it was reported that RAD51 colocalized in nuclear foci with BRCA1, the other breast cancer-associated gene product, but a direct association between RAD51 and BRCA1 in vitro was not detected (10). In this report, we describe a direct association between RAD51 and mBRCA2 in vitro, but we were unable to detect an in vivo association due to the lack of foci formation in the cotransfected cells. The latter result does not rule out the possibility of nuclear foci formation with a full-length BRCA2 protein; this will be testable when a full length cDNA or specific antibody is available. Perhaps RAD51 associates directly with BRCA2, which itself or with other molecule(s) binds to BRCA1, creating a large complex of associated molecules. The formation of such complexes to form nuclear foci may depend on the presence of certain molecules within the complex, and each molecule may have a distinct role in the assembly or disassembly of the nuclear foci. This model of interdependent interactions between RAD51-associated molecules can be tested by the TrNF assay with various cotransfection combinations of RAD51-associated molecules.
In support of a functional relationship among RAD51, BRCA1, and BRCA2, BRCA2 is coordinately regulated with BRCA1 during proliferation and differentiation (29). Both BRCA1 and BRCA2 mRNAs are up-regulated at the G1/S boundary (29), completely coincident with RAD51 expression (30). Such interactions may have an important role in tumorigenesis as many of the BRCA2 mutations in patients are frameshift mutations which make BRCA2 products truncated at the C terminus. Since the RAB163 clone of mBRCA2 encodes only the C-terminal 273 amino acids (1/12 of whole product), and interacts with RAD51 in vitro, perhaps truncated BRCA2 products in these patients lack the ability to interact with RAD51.
The novel RAB22 protein that we identified based on interaction with RAD51 also has an expression pattern similar to that of human RAD51 and BRCA2, with high level expression in testis, thymus, spleen, and bone marrow (refs. 14 and 25; Fig. 2). RAB22 appears to be a novel protein with no sequence in GenBank showing significant homology. The N-terminal two-thirds of this protein is hydrophilic and C-terminal third is hydrophobic. This hydrophobic C-terminal region contained 128 amino acids and was sufficient for an in vitro interaction with RAD51. A typical nuclear localization signal was not observed, but one candidate sequence was 215-KKKTVQKK-222, and transient transfection experiments demonstrated that RAB22 is transported into the nucleus (Fig. 5). The RAB22 protein is serine rich (15% overall), especially the S/TP sequence which appeared six times, suggesting the possibility of cell cycle regulated phosphorylation in several sites (31). Mouse RAB22 mapped to chromosome 6; but no gene that has been correlated with breast/ovarian cancer has been reported in that region.
Evidence for direct association between RAB22 and RAD51 in vivo was obtained by the TrNF assay (Fig. 5). When RAB22 was cotransfected with RAD51, the number of nuclei with large multiple foci was dramatically increased compared with transfection with RAB22 or RAD51 alone which showed more diffuse staining (data not shown). Native human RAD51 and human BRCA1 were also reported to show diffuse distribution in addition to nuclear foci, and it was proposed that RAD51 and BRCA1 might be assembled to form nuclear foci in S phase to participate in a specific function (10, 28). These data, together with the coordinate expression pattern of RAD51, BRCA1, and BRCA2, suggest potential cell cycle regulation of in vivo complex formation of these three molecules and RAB22.
To determine the ability of two proteins to interact in vivo, we developed a simple assay for nuclear foci formation in cotransfected cells. This TrNF assay is based on the organization of RAD51 protein into foci in S phase (28, 32) and directly tests the ability of candidate associating molecule(s) to colocalize with RAD51 in these foci. This assay system is very simple and can be performed in the absence of an antibody to either protein if separate GFP and BFP constructs are designed. We used this assay to demonstrate a clear colocalization between RAD51 and RAB22 in large nuclear foci in cotransfected cells. The functional role of such large complexes between RAD51 and RAB22 remains to be determined. Since the organization of endogenous RAD51 into foci is S phase-specific and RAD51 exhibits DNA binding and strand transferase activity, several groups have suggested some S phase-specific function. RAD51 complexes may include sites of the sister chromatid exchange or the recombinational repair of double-strand DNA breaks (28, 32).
In mammalian somatic cells, it is thought that there are two recombination pathways, a homologous recombination that occurs during the S and G2 phase and a nonhomologous recombination that occurs in the G1 phase (7). Because RAD51 mRNA is highly expressed in lymphoid tissues, RAD51 has been speculated to be involved in V(D)J recombination and/or class switch recombination, lymphocyte-specific recombination events. However, because V(D)J recombination is a nonhomologous recombination process specific to the G0 and G1 phase, it is unlikely that RAD51 is involved in V(D)J recombination (7, 33). Recently, it was found that RAD51 was expressed and localized in B cells carrying out class switch recombination (34); further circumstantial evidence suggests that the class switch recombination might occur in S phase and correlate with DNA replication (35). Further identification of proteins that specifically interact or colocalize with RAD51 may facilitate analyses of the potential involvement of RAD51 in class switch recombination.
Acknowledgments
We thank Mary Barnstead for excellent technical assistance. This research was supported in part by National Institutes of Health Grants AI315714 and CA42335 and by the National Cancer Institute, Department of Health and Human Services, under contract with ABL. F.W.A. and M.L. are supported by the Howard Hughes Medical Institute.
ABBREVIATIONS
- BRCA1
breast cancer susceptibility gene 1
- BRCA2
breast cancer susceptibility gene 2
- TrNF
transient nuclear focus
- GFP
green flourescent protein
- BFP
blue flourescent protein
- GST
glutathione S-transferase
- RFLP
restriction fragment length polymorphism
- DAPI
4′,6-diamidine-2′-phenylindole dihydrochloride
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. U93583).
References
- 1.Shinohara A, Ogawa H, Ogawa T. Cell. 1992;69:457–470. doi: 10.1016/0092-8674(92)90447-k. [DOI] [PubMed] [Google Scholar]
- 2.Edelmann W, Kucherlapati R. Proc Natl Acad Sci USA. 1996;93:6225–6227. doi: 10.1073/pnas.93.13.6225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Terasawa M, Shinohara A, Hotta Y, Ogawa H, Ogawa T. Genes Dev. 1995;9:925–934. doi: 10.1101/gad.9.8.925. [DOI] [PubMed] [Google Scholar]
- 4.Sung P, Robberson D L. Cell. 1995;82:453–461. doi: 10.1016/0092-8674(95)90434-4. [DOI] [PubMed] [Google Scholar]
- 5.Baumann P, Benson F E, West S C. Cell. 1996;87:757–766. doi: 10.1016/s0092-8674(00)81394-x. [DOI] [PubMed] [Google Scholar]
- 6.Tsuzuki T, Fujii Y, Sakumi K, Tominaga Y, Nakao K, Sekiguchi M, Matsushiro A, Yoshimura Y, Morita T. Proc Natl Acad Sci USA. 1996;93:6236–6240. doi: 10.1073/pnas.93.13.6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lim D-S, Hasty P. Mol Cell Biol. 1996;16:7133–7143. doi: 10.1128/mcb.16.12.7133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson R D, Symington L S. Mol Cell Biol. 1995;15:4843–4850. doi: 10.1128/mcb.15.9.4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shen Z, Cloud K G, Chen D J, Park M S. J Biol Chem. 1996;271:148–152. doi: 10.1074/jbc.271.1.148. [DOI] [PubMed] [Google Scholar]
- 10.Scully R, Chen J, Plug A, Xiao Y, Weaver D, Feunteun J, Ashley T, Livingston D M. Cell. 1997;88:265–275. doi: 10.1016/s0092-8674(00)81847-4. [DOI] [PubMed] [Google Scholar]
- 11.Maldonado E, Shiekhattar R, Sheldon M, Cho H, Drapkin R, Rickert P, Lees E, Anderson C W, Linn S, Reinberg D. Nature (London) 1996;381:86–89. doi: 10.1038/381086a0. [DOI] [PubMed] [Google Scholar]
- 12.Kolodner R. Genes Dev. 1996;10:1433–1442. doi: 10.1101/gad.10.12.1433. [DOI] [PubMed] [Google Scholar]
- 13.Sturzbecher H-W, Donzelmann B, Henning W, Knippschild U, Buchop S. EMBO J. 1996;15:1992–2002. [PMC free article] [PubMed] [Google Scholar]
- 14.Shinohara A, Ogawa H, Matsuda Y, Ushio N, Ikeo K, Ogawa T. Nat Genet. 1993;4:239–243. doi: 10.1038/ng0793-239. [DOI] [PubMed] [Google Scholar]
- 15.Zervos A S, Gyuris J, Brent R. Cell. 1993;72:223–232. doi: 10.1016/0092-8674(93)90662-a. [DOI] [PubMed] [Google Scholar]
- 16.Li Z, Otevrel T, Gao Y, Cheng H-L, Seed B, Stamato T D, Taccioli G E, Alt F W. Cell. 1995;83:1079–1089. doi: 10.1016/0092-8674(95)90135-3. [DOI] [PubMed] [Google Scholar]
- 17.Shibasaki F, Price E R, Milan D, Mckeon F. Nature (London) 1996;382:370–373. doi: 10.1038/382370a0. [DOI] [PubMed] [Google Scholar]
- 18.Copeland N G, Jenkins N A. Trends Genet. 1991;7:113–118. doi: 10.1016/0168-9525(91)90455-y. [DOI] [PubMed] [Google Scholar]
- 19.Jenkins N A, Copeland N G, Taylor B A, Lee B K. J Virol. 1982;43:26–36. doi: 10.1128/jvi.43.1.26-36.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kolodner R D, Hall N R, Lipford J, Kane M F, Rao M R S, et al. Cold Spring Harbor Symp Quant Biol. 1994;59:331–338. doi: 10.1101/sqb.1994.059.01.037. [DOI] [PubMed] [Google Scholar]
- 21.Tsui C C, Copeland N G, Gilbert D J, Jenkins N A, Barnes C, Worley P F. J Neurosci. 1996;16:2463–2478. doi: 10.1523/JNEUROSCI.16-08-02463.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goodwin R G, Anderson D, Jerzy R, Davis T, Brannan C I, Copeland N B, Jenkins N A, Smith C A. Mol Cell Biol. 1991;11:3020–3026. doi: 10.1128/mcb.11.6.3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nagasawa M, Sakimura K, Mori K J, Bedell M A, Copeland N G, Jenkins N A, Mishina M. Mol Brain Res. 1996;36:1–11. doi: 10.1016/0169-328x(95)00225-h. [DOI] [PubMed] [Google Scholar]
- 24.Benson F E, Stasiak A, West S C. EMBO J. 1994;13:5764–5771. doi: 10.1002/j.1460-2075.1994.tb06914.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tavtigian S V, Simard J, Rommens J, Couch F, Shattuck-Eidens D, et al. Nat Genet. 1996;12:333–337. doi: 10.1038/ng0396-333. [DOI] [PubMed] [Google Scholar]
- 26.Sharan S K, Bradley A. Genomics. 1997;40:234–241. doi: 10.1006/geno.1996.4573. [DOI] [PubMed] [Google Scholar]
- 27.Miki Y, Swensen J, Shattuck-Eidens D, Futreal P A, Harshman K, et al. Science. 1994;266:66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- 28.Tashiro S, Kotomura N, Shinohara A, Tanaka K, Ueda K, Kamada N. Oncogene. 1996;12:2165–2170. [PubMed] [Google Scholar]
- 29.Rajan J V, Wang M, Marquis S T, Chodosh L A. Proc Natl Acad Sci USA. 1996;93:13078–13083. doi: 10.1073/pnas.93.23.13078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamamoto A, Taki T, Yagi H, Habu T, Yoshida K, Yoshimura Y, Yamamoto K, Matsushiro A, Nishimune Y, Morita T. Mol Gen Genet. 1996;251:1–12. doi: 10.1007/BF02174338. [DOI] [PubMed] [Google Scholar]
- 31.Songyang Z, Lu K P, Kwon Y T, Tsai L H, Filhol O, Cochet C, Brickey D A, Soderling T R, Bartleson C, Graves D J, DeMaggio A J, Hoekstra M F, Blenis J, Hunter T, Cantley L C. Mol Cell Biol. 1996;16:6486–6493. doi: 10.1128/mcb.16.11.6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haaf T, Golub E I, Reddy G, Radding C M, Ward D C. Proc Natl Acad Sci USA. 1995;92:2298–2302. doi: 10.1073/pnas.92.6.2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Z, Dordai D D, Lee J, Desiderio S. Immunity. 1996;5:575–589. doi: 10.1016/s1074-7613(00)80272-1. [DOI] [PubMed] [Google Scholar]
- 34.Li M-J, Peakman M C, Golub E I, Reddy G, Ward D C, Radding C M, Maizels N. Proc Natl Acad Sci USA. 1996;93:10222–10227. doi: 10.1073/pnas.93.19.10222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lundgren M, Strom L, Bergquist L O, Skog S, Heiden T, Stavnezer J, Severinson E. Eur J Immunol. 1995;25:2042–2051. doi: 10.1002/eji.1830250736. [DOI] [PubMed] [Google Scholar]