Abstract
The retinoblastoma gene family consists of the tumor suppressor protein pRB and its two relatives p107 and p130. These proteins have been implicated in the regulation of cell cycle progression, in part, through inactivation of members of the E2F transcription factor family. Overexpression of pRB, p107, or p130 leads to growth arrest in the G1 phase of the cell cycle, and this arrest is abolished by complex formation with the adenovirus E1A, human papilloma virus E7, or simian virus 40 T oncoproteins. Inactivation of pRB by gross structural alterations or point mutations in the RB-1 gene has been described in a variety of human tumors, including retinoblastomas, osteosarcomas, and small cell lung carcinomas. Despite the structural and functional similarity between pRB, p107, and p130, alterations in the latter two proteins have not been identified in human tumors. We have screened a panel of 17 small cell lung carcinoma cell lines for the presence of functional p107 and p130 by evaluating their ability to form complexes with E1A in vitro. In the GLC2 small cell lung carcinoma cells no p130 protein was detected. The loss of the p130 protein is the result of a single point mutation within a splice acceptor sequence in the GLC2 genomic DNA. This mutation eliminates exon 2, leading to an in-frame stop codon, and no detectable protein is produced. These data are, to our knowledge, the first to describe the loss of p130 as a consequence of a genetic alteration, suggesting that not only pRB but also the other members of the family may contribute to tumorigenesis, providing a rationale for the observation that the DNA tumor viruses selectively target all the members of the retinoblastoma protein family.
The retinoblastoma gene, RB-1, was identified through genetic analysis of retinoblastomas and osteosarcomas (1). The subsequent cloning of the gene and the analysis of its gene product, pRB, have demonstrated that the RB-1 gene is mutated in a variety of human tumors, including retinoblastomas, osteosarcomas, small cell lung carcinomas (SCLCs), and bladder carcinomas. Furthermore, pRB is inactivated in all cervical carcinomas analyzed so far, either by direct binding of the human papilloma virus oncoprotein E7 or by structural alterations of the RB-1 gene (2, 3). pRB is thought to regulate the progression through the G1 phase of the cell cycle by interacting with members of the E2F family of transcription factors (1, 4). Inactivation of pRB leads to constitutively active E2F family members, and loss of its ability to inhibit transcription of E2F-dependent promoters. Consistent with this model is that overexpression of E2F family members can lead to S-phase induction and transformation of rodent cells (5–7).
The G1-regulating feature of pRB is dependent on amino acids required for interaction with the E2F family members as well as with the adenovirus E1A, simian virus 40 T, and human papilloma virus E7 oncoproteins (1, 4). These amino acids, often referred to as the “pocket region,” are conserved in two additional E1A-binding proteins, p107 and p130 (8–11). The p107 and p130 pocket regions allow these proteins to bind to E2F transcription factors, E1A, T, and E7. Furthermore, p107 and p130 can, like pRB, when overexpressed inhibit G1 progression and E2F activity in mammalian cells (12, 13). The structural and functional similarities between the RB-family members suggest that they all are potential tumor suppressors. However, so far no tumors have been identified with loss of function of the p107 or p130 proteins.
Although the proteins are structurally and functionally similar, several observations suggest that members of the RB family of proteins have distinct roles in regulating the mammalian cell cycle: (i) The p107 and p130 proteins bind only a subset of the members in the E2F transcription factor family, E2F-4 and E2F-5, whereas pRB is capable of binding all members of the family (refs. 14–16; K.H., unpublished results). (ii) p107 and p130 can stably associate with cyclin A or cyclin E/CDK2 kinases, whereas pRB only transiently associates with cyclin-dependent kinases (CDKs) (4, 10, 17, 18). (iii) Overexpression studies have demonstrated the ability of p107 and p130 to suppress the growth of certain cell lines that are refractory to pRB overexpression (refs. 12 and 13; K.H., unpublished results). (iv) p107 and p130 cannot functionally substitute for pRB in RB −/− tumors or in RB-null mice (1). Furthermore, the observation that p130 associates with E2F in G0 and early G1 phase cells, while p107 binds E2F in late G1 and S phase cells (19), also supports the idea that these two molecules may have distinct roles in regulating the mammalian cell cycle. (v) Finally, data have been reported that in the T98G human glioblastoma cell line overexpression of p130 leads to cell cycle arrest in G1, whereas p107 and pRB have no effect on growth (13). The putative distinct roles of the RB-family members suggest that loss of function of more than one of these genes will confer a growth advantage to the cell, and may also provide an explanation for why the DNA tumor viruses target all the identified members of the RB family.
In this study we have examined the expression of p107 and p130 in a series of human SCLC cell lines, previously analyzed for their pRB status (20). One of these cell lines was found not to express any functional p130 protein. The loss of p130 is the consequence of a point mutation in the splice acceptor site of the first intron in the genomic DNA, resulting in the production of an RNA molecule without exon 2, and an in-frame stop codon in the p130 open reading frame.
MATERIALS AND METHODS
Cell Lines.
Seventeen SCLC cell lines established from 14 patients by three different laboratories were examined. All cell lines were grown in medium as specified below containing 10% heat-inactivated fetal calf serum at 37°C. Seven cell lines established at Dartmouth Medical School (Lebanon, NH) [DMS53, DMS79, DMS92, DMS114, DMS153, DMS273, DMS406, and DMS456) (21, 22)] were cultured in Waymouth medium (GIBCO). Seven cell lines established at Groningen Lung Cancer Center (Groningen, The Netherlands) [GLC2, GLC3, GLC14, GLC16, GLC19, GLC26, and GLC28 (23, 24)] were cultured in RPMI medium 1640 (GIBCO), and two cell lines established in one of our laboratories [CPH54A and CPH54B] (25) were grown in Eagle’s minimum essential medium (EMEM; GIBCO). The cells were passaged twice a week. Primary human foreskin fibroblasts (HFF, a gift from O. Pettengill, Dartmouth Medical School) were grown in Hanks’ basal medium Eagle (BME; GIBCO). The human embryonic kidney cell line 293 (containing E1A and E1B) (26) and the human myeloid leukemia cell line ML-1 (27) were cultured in Dulbecco’s modified Eagle’s medium (DMEM; GIBCO). All cell lines were routinely checked for, and found to be free of, mycoplasma infection.
Antibodies.
The monoclonal antibody to p130, Z83, and the rabbit polyclonal antisera to p107 (10) were the kind gift of P. Whyte (McMaster University, Hamilton, ON, Canada). Two mouse monoclonal antibodies to pRB were used: PMG245 obtained from Pharmigen, and XZ77 (28). The E1A monoclonal antibody M73 has previously been described (29).
Glutathione S-Transferase (GST)-Coupled Proteins.
GST, GSTE1A12S, and GSTE1A-(1–139) (30) were purified from Escherichia coli as previously described (31).
In Vitro Binding Assay, Immunoprecipitation, and Western Blotting.
Exponentially growing cells were washed twice in phosphate-buffered saline (PBS) and lysed in E1A lysis buffer (50 mM Hepes, pH 7.0/250 mM NaCl/0.1% Nonidet P-40) (32), containing 1 mM EDTA, 1 mM dithiothreitol, 100 μg/ml phenylmethylsulfonyl fluoride (PMSF), 1 μg/ml leupeptin, and 1 μg/ml aprotinin. Protein concentrations were determined by the Bradford method (Bio-Rad). For in vitro binding experiments 500 μg of cell lysate was incubated with 5 μg of purified GST, GSTE1A12S, or GSTE1A-(1–139) for 2 h on ice. The GST proteins and any associated proteins were then recovered by using glutathione-agarose (Sigma), washed three times with E1A lysis buffer, separated on SDS/8% polyacrylamide gels, and transferred to nitrocellulose by using standard techniques (33). Transferred proteins were detected by using the antibodies indicated in the figures and developed by the ECL system (Amersham). As a positive control for E1A-associated proteins, 100 μg of 293 cell lysate was immunoprecipitated with M73 as described (32), and immunoprecipitated E1A and associated proteins were processed for Western blotting as described above. For detection of proteins in cell lysates, a 50-μg sample of the total cell lysate was separated on SDS/8% polyacrylamide gels and processed for Western blotting.
RNA Extraction, Electrophoresis, and Blotting.
Exponentially growing cells were harvested and centrifuged at 1,100 × g for 5 min. The cell pellet was frozen immediately in liquid nitrogen and stored at −70°C until analysis. Poly(A)+ RNA was extracted directly from the frozen (−70°C) cell pellets by the guanidinium thiocyanate method followed by two sequential purifications by oligo(dT)-cellulose chromatography using a commercial kit (QuickPrep mRNA Purification Kit, Pharmacia). The concentration of RNA was determined by spectrophotometry, and the RNA was stored in ethanol at −70°C, in aliquots of 3–5 μg. RNA was dissolved in sample buffer [50% (vol/vol) formamide/2.2 M formaldehyde/20 mM 3-morpholinopropanesulfonic acid (Mops)/5 mM sodium acetate/1 mM EDTA/2% Ficoll 400/0.25% bromophenol blue) containing 0.033 mg/ml ethidium bromide and electrophoresed under denaturing conditions in 1% agarose gels containing 2.2 M formaldehyde, together with 5 mg of RNA size marker (RNA ladder, GIBCO/BRL). Transfer to charged nylon membranes (GeneScreenPlus, NEN/DuPont) was done in 10× saline/sodium citrate (SSC; 1× SSC = 150 mM sodium chloride/15 mM sodium citrate, pH 7.0). Northern blots were prehybridized for at least 3 h and hybridized for 18 h at 42°C in a buffer containing 50% formamide, 1% sodium dodecyl sulfate, 1 M NaCl, 5% dextran sulfate, and 100 mg/ml denatured salmon testes DNA. Maximal washing stringency was 65°C in 2× SSC/1% sodium dodecyl sulfate. The Northern blots were exposed to x-ray films (Amersham) at −80°C with an intensifying screen for 1–7 days. The blots were hybridized with full-length p130 cDNA (10) or a BamHI/DraIII fragment of the p107 cDNA (12) and rehybridized with the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) probe, which was used as an internal standard to compare the amounts of poly(A)+ RNA transferred to the membranes.
DNA Extraction, Electrophoresis, and Blotting.
DNA was extracted with phenol and chloroform by standard procedures (34). Digestion with restriction endonucleases PvuI, PstI, EcoRI, BsmI, and the methylation-sensitive HinPI was performed as recommended by the supplier (GIBCO/BRL, Life Technologies, or New England Biolabs). DNA (10 μg per lane) was electrophoresed in 0.8% agarose gels and transferred to charged nylon membranes (GeneScreenPlus, NEN/DuPont). Prehybridization, hybridization, and washing were as recommended by the supplier: washing stringency was 2× SSC/1% sodium dodecyl sulfate twice for 30 min at 60°C. Membranes were exposed to x-ray film (Amersham) at −80°C with an intensifying screen for 7 days. Southern blots were probed with full-length 32P-labeled p130 cDNA.
Cloning of p130 cDNA and Genomic Fragments.
The 5′ end of the p130 cDNA was isolated from 1 μg of GLC2, GLC3, and DMS92 poly(A)+ RNA by reverse transcription followed by PCR. For the PCR the following primers were used: p130.1, 5′-GACGGATCCGCCATGCCGTCGGGAGGTGACCAG-3′ (nucleotides 70–90 of the p130 gene); and p130.2, 5′-GACGGATCCGTTCAGACACCTTGAGAGAG-3′ (nucleotides 1180–1160). The PCR products were cloned into BamHI-cut pBSK (−) (Stratagene), and inserts were sequenced using standard techniques (34). To identify mutations in the genomic DNA, 200 ng of genomic DNA prepared from GLC2 or GLC3 cells was amplified with exon 1 and exon 2 specific primers: p130.10, 5′-CTAGGATCCACAGCTACCGCAGCATGAG-3′ (nucleotides 272–290); and p130.11, 5′-CTAGGATCCTTGCTTACAGTTGGAACAG-3′ (nucleotides 380–362). The resulting PCR products were cloned and sequenced as described above. The genomic mutation identified in the GLC2 cell line was confirmed by amplifying a 200-bp fragment of the p130 gene containing the mutation using primer p130.11 and an intron-specific primer, p130.14: 5′-TACGGATCCGGGTCATCATTGAAACTAAG-3′. Several clones from independent PCRs were sequenced to confirm the detected mutation.
DNA Sequencing.
Double-stranded DNA sequencing was performed using Sequenase 2.0 (Amersham) according to the supplier’s procedure.
Fluorescence in Situ Hybridization (FISH) Analysis.
Metaphase chromosomes were prepared from GLC2 by treatment of exponentially growing cells with Colcemid at 1:100 in standard medium (KaryoMAX Colcemid, GIBCO/BRL) for 2½ h. The cells were fixed in 3:1 (vol/vol) methanol/acetic acid and kept at −20°C. The slides were washed for 60 min in 2× SSC (37°C), dehydrated, and denatured in 70% (vol/vol) formamide/2× SSC (2 min). A p130 genomic probe was obtained commercially from screening a P1 bacteriophage library (Genome Systems, St. Louis) by the use of p130-specific primers, p.130.11 and p.130.14, which revealed two positive clones, 13347 and 13348. The P1 constructs were subsequently transfected into a Cre-containing E. coli host. A 16.5-kb circular plasmid containing a genomic insert of 75–100 kb was recovered by a standard miniprep procedure (34). The P1 DNA was labeled with biotinylated dUTP (GIBCO/BRL) by nick translation. Hybridization was performed at 37°C for 1 day. Detection was carried out at room temperature with fluorescein isothiocyanate (FITC)-conjugated avidin (Vector Laboratories), and the signals were amplified with one additional layer of biotinylated goat anti-avidin, followed by an avidin-FITC layer. Chromosomes were stained with 4′,6-diamidino-2-phenylindole (DAPI; 50 ng/μg), rinsed in PBS, and mounted in propidium iodide/antifade. The slides were examined in a Leica MD epifluorescence microscope, equipped with a Ludl filter wheel with a standard Pinkel filter set, and a Xillix Microimager charge-coupled device camera. Images were obtained with the Xquips imaging program and printed on a Codonics NP1600 printer. As a control for the hybridization, the two p130-positive P1 probes were hybridized to normal lymphocytic chromosomes, and both hybridized to chromosome 16q12–13.
Microsatellite Analysis of GLC2.
Nine chromosome 16 microsatellite loci (with cytogenetic and genetic location) were tested for homo-/heterozygosity on an Applied Biosystems Prism (Perkin–Elmer) as recommended by the manufacturer: D16S2622 (pter), D16S406 (p13.3), D16S497 (p13.13), D16S2619 (p13), D16S403 (p12.3), D16S771 (q11.2–q13), D16S1624 (q22.1), and D16S753 and D16S422 (q24.2).
RESULTS
Analysis of p107 and p130 in SCLC Cells.
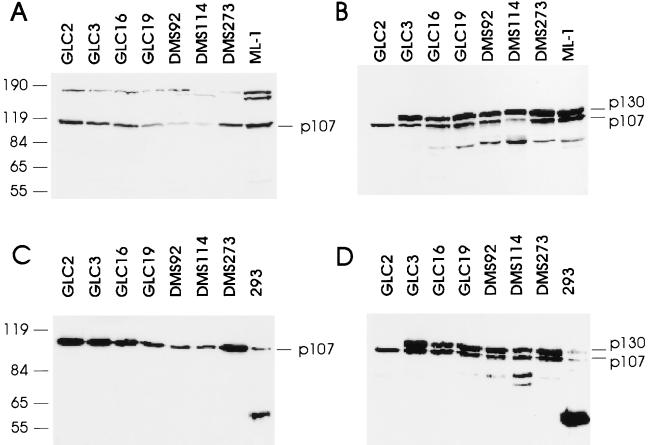
We have previously analyzed a panel of SCLC cell lines for the expression of functional pRB (20). To extend this analysis to the other known members of the RB family, we tested for the presence of functional p107 and p130 proteins by in vitro complex formation with the adenovirus E1A protein. In this assay, cells are lysed under mild detergent conditions and incubated with bacterially purified GST or GSTE1A containing the domains of E1A sufficient for interaction with the RB-family members (35, 36). After precipitation of the GST proteins and associated proteins, Western blotting was performed with antibodies specific for pRB, p107, or p130. No RB-family protein was found in precipitates from lysates incubated with the GST protein alone (data not shown). To know if mutated RB-family proteins were expressed, Western blotting was also performed on “native” cell lysates prepared from the different cell lines. The summary of our analysis is shown in Table 1, and examples of our assay are shown in Fig. 1. Of the 17 SCLCs analyzed in this assay, all were found to express wild-type p107, and all but one expressed a wild-type p130 protein. The GLC2 cell line did not express a functional p130 protein (Fig. 1D), and there was no evidence of expression of a mutated protein (Fig. 1B). Since the p130 monoclonal antibody used to probe the filter shown in Fig. 1 B and D recognizes an epitope in the pocket region of p130, and since an antibody that recognizes the C terminus of p130 did not detect any aberrantly migrating p130 protein (data not shown), our data suggest that the GLC2 cell line does not express a p130 protein containing more than the N terminus.
Table 1.
Expression of RB-family members in SCLC cells
| Cell line | pRB | p107 | p130 |
|---|---|---|---|
| GLC2 | − | + | − |
| GLC3 | + | + | + |
| GLC14 | − | + | + |
| GLC16 | − | + | + |
| GLC19 | − | + | + |
| GLC26 | − | + | + |
| GLC28 | − | + | + |
| DMS53 | + | + | + |
| DMS79 | − | + | + |
| DMS92 | + | + | + |
| DMS114 | + | + | + |
| DMS153 | − | + | + |
| DMS273 | + | + | + |
| DMS406 | − | + | + |
| DMS456 | + | + | + |
| CPH54A | + | + | + |
| CPH54B | + | + | + |
Expression of p107 and p130 was evaluated by Western blotting and binding to GSTE1A. The data for pRB expression have previously been published by Rygård et al. (20), except for GLC2, GLC26, GLC28, and DMS406, which were tested by Western blotting and binding to GSTE1A. A + indicates that the wild-type protein is expressed; a − indicates no expression or that a mutated protein is expressed. The GLC14, GLC16, and GLC19 cell lines are derived from the same patient. CPH54A and CPH54B are derived from a different single patient.
Figure 1.
Loss of p130 expression in the GLC2 cell line. (A) Fifty micrograms of cell lysate from the indicated cell lines was separated on an SDS/8% polyacrylamide gel and processed for Western blotting. The ML-1 cell line is a control for the migration of wild-type p107. This blot was probed with a polyclonal rabbit antibody raised against p107. (B) Fifty micrograms of cell lysate was processed as described above. The blot was probed with a monoclonal antibody to p130, Z83, that also recognizes p107, and two related proteins (or breakdown products) of 80 and 90 kDa. (C) Five hundred micrograms of cell lysate was incubated with 5 μg of GSTE1A-(1–139) for 2 h on ice, and GSTE1A-(1–139) and associated proteins were precipitated and processed for Western blotting. E1A and associated proteins from 293 cells precipitated with the monoclonal antibody M73 served as a control for the migration of p107 and p130. The blot was probed with the rabbit polyclonal antibody to p107. (D) As in C; however, the blot was probed with Z83. At the left are positions of molecular mass markers, indicated in kDa.
No Gross Structural Alterations of the p130 Gene Are Found in GLC2 Cells, and an mRNA of Apparently Normal Size Is Expressed.
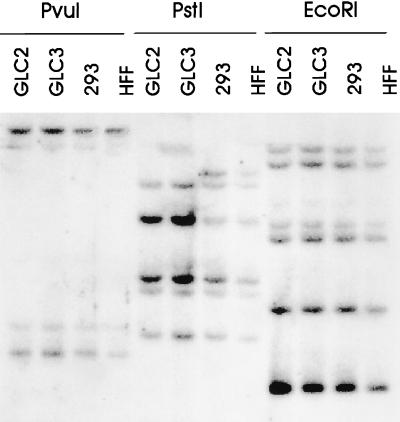
To investigate the mechanism leading to the loss of p130 expression, Southern blotting of genomic DNA prepared from GLC2 cells was performed. As controls for the restriction pattern seen for GLC2 cells, genomic DNA from GLC3, HFF, and 293 cells was used (Fig. 2). The restriction pattern of p130 genomic DNA digested with PvuI, PstI, or EcoRI for the GLC2 cells was identical to the pattern seen for the GLC3 cells when probed with the full-length p130 cDNA. This pattern is almost identical to the one observed for 293 cells and HFFs. DNA digested with PstI led to an extra p130-specific fragment in the latter two cell lines. The reason for this extra fragment is not known. The lack of p130 in GLC2 cells is therefore not due to gross structural alterations of the p130 gene.
Figure 2.
The structural organization of the p130 gene appears normal in GLC2 cells. Ten micrograms of genomic DNA isolated from the indicated cell lines was digested with PvuI, PstI, or EcoRI, and fragments were separated on an 0.8% agarose gel and processed for Southern blotting. The blot was probed with 32P-labeled full-length p130 cDNA.
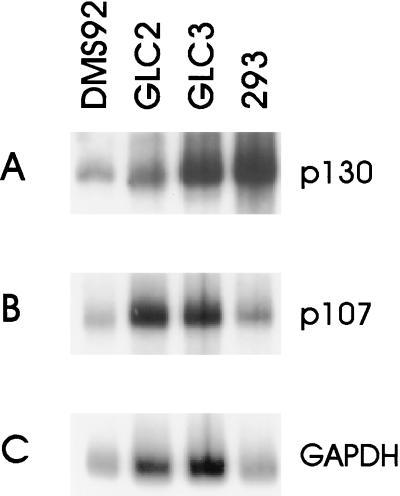
To investigate whether the GLC2 cell line expresses the p130 mRNA, Northern blots were prepared. As shown in Fig. 3, GLC2 cells contain an apparently normal-sized p130 mRNA expressed at levels comparable to the p130-positive cell lines GLC3, DMS92, and 293. The lack of p130 in GLC2 cells is therefore not due to promoter mutations, gross alterations of the genomic structure, or the abundance of expressed mRNA, but more likely is caused by a subtle mutation in the p130 gene.
Figure 3.
Normal-sized mRNA for p130 is present in GLC2 cells. The RNA from the indicated cell lines was processed for Northern blotting, and the blots were probed with p130 cDNA (A), p107 cDNA (B), or glyceraldehyde-3-phosphate dehydrogenase (GAPDH) cDNA (C).
Identification of a Splice Acceptor Site Mutation in the p130 Genomic DNA.
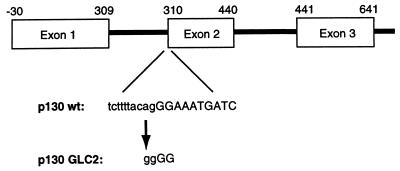
To define the nature of the mutation(s) leading to loss of p130, mRNA prepared from GLC2 cells was reverse transcribed and used for PCR amplification. Primers specific for the 5′-coding region of the p130 gene were designed, since it was anticipated (on the basis of the results described above) that the mutation(s) leading to loss of p130 was residing in this part of the molecule. The amplification of the p130 cDNA from GLC2 cells gave rise to a fragment of around 1 kb, slightly shorter than the expected 1.1 kb observed in a control reaction performed with cDNA prepared from GLC3 cells (data not shown). When the resulting cDNAs were subcloned and used for coupled in vitro transcription/translation reaction, the p130 cDNA amplified from GLC3 gave rise to a peptide of around 45 kDa, whereas no peptide was synthesized from the p130 cDNA amplified from GLC2 cells (data not shown). By sequencing the GLC2 p130 cDNA, a deletion of nucleotides 310–440 was found. This deletion leads to an in-frame stop codon 86 amino acids downstream of the initiation codon. Eight clones from two independent PCRs all contained the same deletion, suggesting that no wild-type allele of p130 is expressed and that the PCR did not lead to introduction of mutations in the gene.
While this work was in progress the genomic organization of the p130 gene was published (37). Exon 2 of the p130 gene contains nucleotides 310–440 of the p130 cDNA, and the mRNA produced in GLC2 cells therefore does not contain exon 2. To identify the cause of the exon skipping, primers specific for exon 2 and exon 1 of the p130 gene were used to amplify genomic DNA from GLC2 and GLC3 cells. Sequencing of the amplified product revealed a single point mutation in the GLC2 genomic DNA compared with the GLC3 cell line and the published genomic sequence (Fig. 4). This point mutation changes a canonical splice-acceptor site ACAGGGAT to ACGGGGAT, presumably leading to skipping of exon 2 in the genesis of the mRNA for p130. Since we amplified only GLC2 genomic DNA with this point mutation and found only p130 mRNA without exon 2, we assumed that the mutation is hemi- or homozygous in the cell line. To investigate the possible reason for the lack of a wild-type p130 transcript in GLC2 cells, we examined all the SCLC cell lines for altered methylation pattern in the CpG island at the 5′ end of the p130 gene. This region fulfills the criteria for an island, having a C+G content over 70%. Numerous restriction enzymes are methylation sensitive, including HinPI, for which there are six recognition sites in the 5′-end region of p130 (37). Southern analysis of DNA from the SCLC cell lines digested with HinPI and BsmI showed that the restriction pattern was identical for all cell lines, including GLC2 (data not shown). We therefore concluded that the loss of the wild-type p130 transcript in GLC2 is not caused by silencing of transcription due to DNA hypermethylation.
Figure 4.
Exon–intron structure of the p130 gene with the indicated splice acceptor mutation in the p130 gene present in GLC2 cells. Numbers above the exons indicate the nucleotide numbers in the wild-type p130 cDNA.
Cytogenetics and FISH Analysis of GLC2.
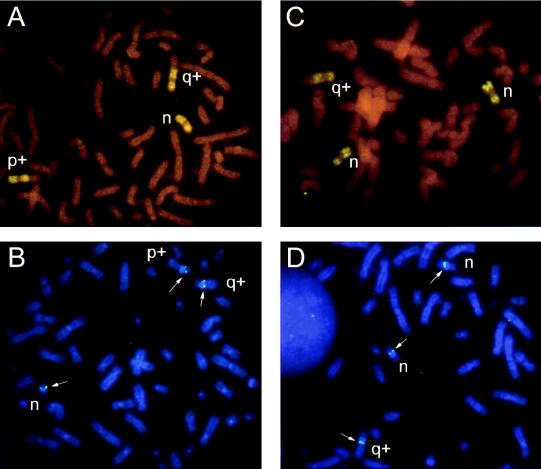
The GLC2 cell line has a hypotriploid chromosome number (53–60, n = 20), with three copies of chromosome 16 as shown by chromosome painting (Fig. 5 A and C). In 25% of the cells (n = 5) two of these copies were normal, whereas one copy contained extra material on the long arm of unknown origin (q+) (Fig. 5C). In a majority of cells (n = 15) only one chromosome 16 was normal with the same 16q+ chromosome as seen above, and in addition there was a rearranged chromosome 16 with extra non-16 material on the short arm (p+) (Fig. 5A). FISH analysis with both of the P1 probes revealed specific signals on all of these chromosomes corresponding to 16q12–13—i.e., three copies of the p130 genomic region in all examined cells (Fig. 5 B and D). Microsatellite analysis of the GLC2 cell line demonstrated that the cell line is homozygous for all nine examined chromosome 16 loci (data not shown). Therefore we conclude that the GLC2 cells do not contain a wild-type allele of the p130 gene, but three copies of the mutant allele.
Figure 5.
FISH analysis of cell line GLC2. (A) Chromosome 16 paint showing the normal (n), and the two rearranged 16s (q+, p+) observed in 75% of the cells. (B) FISH signals (arrows) at 16q12–13 on all three chromosomes 16 obtained with the p130 gene. (C) Chromosome 16 paint showing the two normal (n) and the single rearranged (q+) chromosome 16 observed in 25% of the cells. (D) FISH signals (arrows) of p130 at 16q12–13 on all three chromosomes 16 obtained with the p130 gene.
DISCUSSION
The data presented in this paper are, to our knowledge, the first describing a loss of function of a member of the RB family of proteins other than pRB. A mutation converts a canonical splice-acceptor sequence, 5′-TCTTTTACAGGG-3′ present in the wild-type p130 allele into the sequence 5′-TCTTTTACGGGG-3′. This point mutation leads to synthesis of an mRNA for p130 lacking exon 2, resulting in an in-frame stop codon 86 amino acids after translation initiation, and therefore absence of a functional p130 protein. Since the GLC2 cells express only the mutated version of p130, we investigated a possible mechanism for the lack of expression of the wild-type allele. FISH analyses showed the presence of three copies of the p130 gene (Fig. 5 B and D) without any visible difference in the signal strength between the three chromosomes. These data confirmed the localization of p130 to 16q12–13 (9, 10) and demonstrated that the lack of the wild-type p130 allele is not due to gross deletions within the gene. However, the detection of complete homozygosity for nine different microsatellite loci, each with a >70% a priori chance of revealing heterozygosity (38), strongly supports the interpretation that the three copies of the p130 gene are duplications of the mutant allele. These data also suggest that the point mutation of p130 has been an early event, where subsequent hits have involved complete loss of chromosome 16 carrying the wild-type p130, followed by chromosome duplication of the copy containing the mutated p130 allele and secondary chromosomal rearrangements. This sequence of genetic alterations resembles what has previously been shown for the RB-1 gene (39). Although we were not able to obtain primary clinical material from the patient from whom the GLC2 cell line was derived, and therefore were not able to address whether the p130 mutation was present in the patient, we have been able to confirm that only the mutant allele of p130 is present in early passage GLC2 cells (passage 7–8). Since we found only the mutant allele, our data strongly suggest that the mutation was indeed present in the patient, and that it did not arise during in vitro culture of the GLC2 cell line.
Interestingly, 80–90% of SCLCs do not express a functional pRB (see, for instance, ref. 40), and the GLC2 cell line, in which we found the p130 gene mutated, does not either. p130 has been shown to associate with members of the E2F family in quiescent cells and early G1 (15, 19). In fact, p130 appears to be the major partner of E2F in G0 and early G1, and it has therefore been suggested that it might have a specific role in regulating E2F activity in quiescent/differentiated cells and in early G1. Indeed, other experiments have demonstrated a correlation between the abundance of p130–E2F complexes and terminal differentiation of skeletal muscle cells (41, 42) and growth arrest induced by transforming growth factor β1 (43). Since the DNA tumor virus oncoproteins selectively target all the members of the RB family and p130 is the most abundant pRB-related protein associated with the E2Fs in differentiated cells, it is expected that loss of p130 will confer a growth advantage to the cell. However, recent data suggest that loss of p130 is not sufficient to deregulate the cell cycle: under normal physiological conditions p130 associates only with E2F-4 and E2F-5, and not E2F-1, E2F-2, and E2F-3 (4), strongly suggesting that p130 regulates the activity of only these two members of the E2F family. Moreover, it was recently demonstrated that overexpression of E2F-4 or E2F-5 is not sufficient to induce S-phase progression in quiescent fibroblasts, contrary to the other E2F family members (7). Therefore, loss of p130 might not be sufficient to deregulate the orderly progression through the cell cycle, since it does not control the activity of the E2Fs that directly participate in the regulation of the S-phase genes. Accordingly, one may expect to find loss of p130 only in cancers in which the function of pRB has been compromised, since there is no selective advantage of p130 loss alone. Our model is supported by recent data demonstrating that mice unable to express the p107 or p130 proteins do not have any altered tumor disposition or development defects (44, 45). However, mice which have been mutated in several RB-family proteins have an enhanced or altered phenotype compared with the single knock-out animals (44, 45).
We have not been able to determine any specific growth alterations of the GLC2 cell line, compared with the other SCLCs. Reintroduction of the wild-type p130 gene into the GLC2 carcinoma resulted in the isolation of few stable transfected colonies, and none of them expressed p130; however, a similar phenomenon was observed when expression plasmids for pRB or p107 were introduced into GLC2 cells (data not shown). This result is in agreement with previous data demonstrating that pRB inhibits the growth of cell lines lacking pRB (46–48), and overexpression of p130 or p107 leads to G1 arrest even in cells containing the wild-type protein (12, 13).
Loss of p130 was found in only 1 of 17 SCLCs analyzed, suggesting that inactivation of p130 might not be a very frequent event in this type of cancer. The p130 gene has been localized to 16q12–13 (9, 10, 49), a chromosomal region that is frequently lost in several types of cancers, including Wilms tumor (50, 51) and ovarian carcinomas (52). The development of an assay that uses bacterially produced E1A protein, combined with in vitro complex formation and Western blotting, allows for the rapid screening of cell lines and primary tumor material for mutations in the RB family of proteins. Our data should initiate a second round of very extensive search for p130 mutations based on this E1A assay, and this search might provide the information of how frequent p130 is mutated in human cancer.
Acknowledgments
We gratefully acknowledge the technical assistance of Jette Røhrmann. We thank Hetty Timmer-Boscha for early passage GLC2 cells and Peter Whyte for antibodies. We thank Giulio Draetta for critical reading of the manuscript and Pier Giuseppe Pelicci for discussions. This work was supported by grants from the Associazione Italiana per la Ricerca sul Cancro, the Danish Cancer Society, the Danish Medical Research Council, the Leo Nielsen Foundation, the Danish Cancer Research Foundation, the Einar Willumsen Memorial Foundation, and the Danish Research Academy.
ABBREVIATIONS
- RB
retinoblastoma
- pRB
retinoblastoma protein
- SCLC
small cell lung carcinoma
- GST
glutathione S-transferase
- HFF
human foreskin fibroblast
- FISH
fluorescence in situ hybridization
References
- 1.Weinberg R A. Cell. 1995;81:323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 2.Scheffner M, Münger K, Byrne J C, Howley P M. Proc Natl Acad Sci USA. 1991;88:5523–5527. doi: 10.1073/pnas.88.13.5523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vousden K H. Semin Cancer Biol. 1995;6:109–116. doi: 10.1006/scbi.1995.0014. [DOI] [PubMed] [Google Scholar]
- 4.Cobrinik D. Curr Top Microbiol Immunol. 1996;208:31–60. doi: 10.1007/978-3-642-79910-5_2. [DOI] [PubMed] [Google Scholar]
- 5.Adams P D, Kaelin W G., Jr Curr Top Microbiol Immunol. 1996;208:79–93. doi: 10.1007/978-3-642-79910-5_4. [DOI] [PubMed] [Google Scholar]
- 6.Mellilo R M, Helin K, Lowy D R, Schiller J T. Mol Cell Biol. 1994;14:8241–8249. doi: 10.1128/mcb.14.12.8241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lukas J, Petersen B O, Holm K, Bartek J, Helin K. Mol Cell Biol. 1996;16:1047–1057. doi: 10.1128/mcb.16.3.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ewen M E, Xing Y, Lawrence J B, Livingston D M. Cell. 1991;66:1155–1164. doi: 10.1016/0092-8674(91)90038-z. [DOI] [PubMed] [Google Scholar]
- 9.Hannon G J, Demetrick D, Beach D. Genes Dev. 1993;7:2378–2391. doi: 10.1101/gad.7.12a.2378. [DOI] [PubMed] [Google Scholar]
- 10.Li Y, Graham C, Lacy S, Duncan A M V, Whyte P. Genes Dev. 1993;7:2366–2377. doi: 10.1101/gad.7.12a.2366. [DOI] [PubMed] [Google Scholar]
- 11.Mayol X, Graña X, Baldi A, Sang N, Hu Q, Giordano A. Oncogene. 1993;8:2561–2566. [PubMed] [Google Scholar]
- 12.Zhu L, van den Heuvel S, Helin K, Fattaey A, Ewen M, Livingston D, Dyson N, Harlow E. Genes Dev. 1993;7:1111–1125. doi: 10.1101/gad.7.7a.1111. [DOI] [PubMed] [Google Scholar]
- 13.Claudio P P, Howard C M, Baldi A, Luca A D, Fu Y, Condorelli G, Sun Y, Colburn N, Calabretta B, Giordano A. Cancer Res. 1994;54:5556–5560. [PubMed] [Google Scholar]
- 14.Dyson N, Dembski M, Fattaey A, Ngwu C, Ewen M, Helin K. J Virol. 1993;67:7641–7647. doi: 10.1128/jvi.67.12.7641-7647.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vairo G, Livingston D M, Ginsberg D. Genes Dev. 1995;9:869–881. doi: 10.1101/gad.9.7.869. [DOI] [PubMed] [Google Scholar]
- 16.Moberg K, Starz M A, Lees J A. Mol Cell Biol. 1996;16:1436–1449. doi: 10.1128/mcb.16.4.1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Faha B, Ewen M, Tsai L-H, Livingston D, Harlow E. Science. 1992;255:87–90. doi: 10.1126/science.1532458. [DOI] [PubMed] [Google Scholar]
- 18.Lees E, Faha B, Dulic V, Reed S I, Harlow E. Genes Dev. 1992;6:1874–1885. doi: 10.1101/gad.6.10.1874. [DOI] [PubMed] [Google Scholar]
- 19.Cobrinik D, Whyte P, Peeper D S, Jacks T, Weinberg R A. Genes Dev. 1993;7:2392–2404. doi: 10.1101/gad.7.12a.2392. [DOI] [PubMed] [Google Scholar]
- 20.Rygård K, Sorenson G D, Pettengill O S, Cate C C, Spang-Thomsen M. Cancer Res. 1990;50:5312–5317. [PubMed] [Google Scholar]
- 21.Pettengill O S, Sorenson G D, Wurster-Hill D, Curpey T J, Noll W W, Cate C C, Maurer L H. Cancer. 1980;45:906–918. doi: 10.1002/1097-0142(19800301)45:5<906::aid-cncr2820450513>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 22.Sorenson G D, Pettengill O S, Cate C C, DelPrete S A. In: Lung Cancer. Aisner J, editor. New York: Churchill Livingstone; 1984. pp. 203–240. [Google Scholar]
- 23.Berendsen H H, De Leij L, De Vries E G E, Mesander G, Mulder N H, De Jong B, Buys C H C M, Postmus P E, Poppema S, Sluiter H J, The H T. Cancer Res. 1988;48:6891–6899. [PubMed] [Google Scholar]
- 24.De Leij L, Postmus P E, Buys C H C M, Elema J D, Ramaekers F, Poppema S, Brouwer M, van der Veen A Y, Mesander G, The T H. Cancer Res. 1985;45:6024–6033. [PubMed] [Google Scholar]
- 25.Engelholm S A, Spang-Thomsen M, Vindeløv L L, Brünner N, Nielsen M H, Hirsch F, Hansen H H. Acta Pathol Microbiol Immunol Scand Sect A Pathol. 1986;94:325–336. doi: 10.1111/j.1699-0463.1986.tb03001.x. [DOI] [PubMed] [Google Scholar]
- 26.Graham F L, Smiley J, Russell W C, Nairn R. J Gen Virol. 1977;36:59–74. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- 27.Takeda K, Minowada J, Bloch A. Cancer Res. 1982;42:5151–5158. [PubMed] [Google Scholar]
- 28.Hu Q, Bautista C, Edwards G, Defeo-Jones D, Jones R, Harlow E. Mol Cell Biol. 1991;11:5792–5799. doi: 10.1128/mcb.11.11.5792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harlow E, Franza B J, Schley C. J Virol. 1985;55:533–546. doi: 10.1128/jvi.55.3.533-546.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fattaey A R, Harlow E, Helin K. Mol Cell Biol. 1993;13:7267–7277. doi: 10.1128/mcb.13.12.7267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Helin K, Lees J A, Vidal M, Dyson N, Harlow E, Fattaey A. Cell. 1992;70:337–350. doi: 10.1016/0092-8674(92)90107-n. [DOI] [PubMed] [Google Scholar]
- 32.Harlow E, Whyte P, Franza B J, Schley C. Mol Cell Biol. 1986;6:1579–1589. doi: 10.1128/mcb.6.5.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harlow E, Lane D. Antibodies: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. [Google Scholar]
- 34.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 35.Whyte P, Williamson N M, Harlow E. Cell. 1989;56:67–75. doi: 10.1016/0092-8674(89)90984-7. [DOI] [PubMed] [Google Scholar]
- 36.Whyte P, Ruley H E, Harlow E. J Virol. 1988;62:257–265. doi: 10.1128/jvi.62.1.257-265.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baldi A, Boccia V, Claudio P P, Luca A D, Giordano A. Proc Natl Acad Sci USA. 1996;93:4629–4632. doi: 10.1073/pnas.93.10.4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dib C, Fauré S, Fizames C, Samson D, Drouot N, Vignal A, Millasseau P, Marc S, Hazan J, Seboun E, Lathrop M, Gyapay G, Morisette J, Weissenbach J. Nature (London) 1996;380:152–154. doi: 10.1038/380152a0. [DOI] [PubMed] [Google Scholar]
- 39.Cavenee W K, Dryja T P, Phillips R A, Benedict W F, Godbout R, Gallie B L, Murphree A L, Strong L C, White R L. Nature (London) 1983;305:779–784. doi: 10.1038/305779a0. [DOI] [PubMed] [Google Scholar]
- 40.Shimuzu E, Coxon A, Otterson G A, Steinberg S M, Kratzke R A, Kim Y W, Fedorko J, Oie H, Johnson B E, Mulshine J L, Minna J D, Gazdar A F, Kaye F J. Oncogene. 1994;9:2441–2448. [PubMed] [Google Scholar]
- 41.Halevy O, Novitch B G, Spicer D B, Skapek S X, Rhee J, Hannon G J, Beach D, Lassar A B. Science. 1995;267:1018–1021. doi: 10.1126/science.7863327. [DOI] [PubMed] [Google Scholar]
- 42.Kiess M, Gill R M, Hamel P A. Cell Growth Diff. 1995;6:1287–1298. [PubMed] [Google Scholar]
- 43.Herzinger T, Wolf D A, Eick D, Kind P. Oncogene. 1995;10:2079–2084. [PubMed] [Google Scholar]
- 44.Cobrinik D, Lee M-H, Hannon G, Mulligan G, Bronson R T, Dyson N, Harlow E, Beach D, Weinberg R A, Jacks T. Genes Dev. 1996;10:1633–1644. doi: 10.1101/gad.10.13.1633. [DOI] [PubMed] [Google Scholar]
- 45.Lee M-H, Williams B O, Mulligan G, Mukai S, Bronson R T, Dyson N, Harlow E, Jacks T. Genes Dev. 1996;10:1621–1632. doi: 10.1101/gad.10.13.1621. [DOI] [PubMed] [Google Scholar]
- 46.Hinds P W, Mittnacht S, Dulic V, Arnold A, Reed S I, Weinberg R A. Cell. 1992;70:993–1006. doi: 10.1016/0092-8674(92)90249-c. [DOI] [PubMed] [Google Scholar]
- 47.Qin X-Q, Chittenden T, Livingston D, Kaelin W G. Genes Dev. 1992;6:953–964. doi: 10.1101/gad.6.6.953. [DOI] [PubMed] [Google Scholar]
- 48.Goodrich D W, Wang N P, Qian Y-W, Lee E Y-H P, Lee W-H. Cell. 1991;67:293–302. doi: 10.1016/0092-8674(91)90181-w. [DOI] [PubMed] [Google Scholar]
- 49.Yeung R S, Bell D W, Testa J R, Mayol X, Baldi A, Graña X, Klinga-Levan K, Knudson A G, Giordano A. Oncogene. 1993;8:3465–3468. [PubMed] [Google Scholar]
- 50.Newsham I, Kindler-Rohrbon A, Daub D, Cavenee W. Genes Chromosomes Cancer. 1995;12:1–7. doi: 10.1002/gcc.2870120102. [DOI] [PubMed] [Google Scholar]
- 51.Austruy E, Candon S, Henry I, Gyapay G, Tournade M F, Mannens M, Callen D, Junien C, Jeanpierre C. Genes Chromosomes Cancer. 1995;14:285–294. doi: 10.1002/gcc.2870140407. [DOI] [PubMed] [Google Scholar]
- 52.Sato T, Saito H, Morita R, Koi S, Lee J H, Nakamura Y. Cancer Res. 1991;51:5118–5122. [PubMed] [Google Scholar]