Abstract
Cytochrome C is a mitochondrial protein that induces apoptosis when released into the cytosol or when added to cell-free extracts. Here we show that cells that overexpress the Bcl-2-related protein Bcl-xL fail to accumulate cytosolic cytochrome C or undergo apoptosis in response to genotoxic stress. Coimmunoprecipitation studies demonstrate that Bcl-xL associates with cytochrome C. Cytochrome C binds directly and specifically to Bcl-xL and not to the proapoptotic Bcl-xs protein. The results also demonstrate that Bcl-xs blocks binding of cytochrome C to Bcl-xL. Our findings support a role for Bcl-xL in protecting cells from apoptosis by inhibiting the availability of cytochrome C in the cytosol.
The cellular response to ionizing radiation (IR) and other genotoxic agents includes cell cycle arrest and activation of DNA repair. In the event of irreparable DNA damage, cells respond with induction of apoptosis; however, the intracellular signals that control the induction of apoptosis are unclear. Whereas p53 has been implicated in promoting apoptosis induced by IR exposure (1, 2), other studies have demonstrated that Bcl-2 and Bcl-xL inhibit IR-induced apoptosis (3–5). The induction of apoptosis by diverse stimuli, including IR, is associated with activation of aspartate-specific cysteine proteases (caspases) (6) and cleavage of poly(ADP ribose) polymerase (PARP) (7), protein kinase C δ (8), and other proteins. The finding that cleavage of PARP and protein kinase C δ is blocked by Bcl-xL has suggested that the Bcl-2-related family of anti-apoptotic proteins functions upstream to the activation of caspases (8–10). More direct evidence for involvement of caspases in apoptosis is derived from studies with the baculovirus protein p35, which directly inhibits cysteine proteases and blocks induction of apoptosis (10, 11).
Recent work has suggested that mitochondria may play a role in inducing apoptosis by releasing cytochrome C (12). Addition of cytochrome C and dATP to cytosolic preparations from growing cells activates caspases, such as CPP32 (12), responsible for cleavage of PARP and protein kinase C δ (13–15). Cytochrome C also induces DNA fragmentation in isolated nuclei incubated with cytosolic lysates (12). The finding that intact cells undergo apoptosis after release of cytochrome C into the cytosol has provided further support for an apoptotic function of this mitochondrial protein (12). Cytochrome C is localized to the intermembrane space and to the surface of the inner mitochondrial membrane (16). The demonstration that the anti-apoptotic Bcl-2 family of proteins is expressed in the mitochondrial membrane also has implicated mitochondria in the induction of apoptosis (17, 18). These findings, taken together, have suggested that Bcl-2-related proteins may regulate apoptosis by controlling cytochrome C release into the cytosol.
The present studies demonstrate that the Bcl-xL protein blocks IR-induced release of cytochrome C. Similar results have been obtained with other genotoxic agents. The results also demonstrate that cytochrome C binds directly to Bcl-xL and not to the proapoptotic short Bcl-xS form.
METHODS
Cell Culture and Reagents.
Human U-937, U-937/Bcl-xL, and U-937/p35 (10) myeloid leukemia cells were grown in RPMI 1640 medium supplemented with 10% heat-inactivated fetal bovine serum, 100 units/ml penicillin, 100 μg/ml streptomycin, and 2 mM l-glutamine. Irradiation was performed with a γ-ray source (Cesium 173, Gamma Cell 1000, Atomic Energy of Canada, Ontario) at a fixed dose rate of 13 Gy/min. Cells also were treated with 50 μM cisplatinum (CDDP) (Sigma) or with 1 mM methyl methanesulfonate (Sigma).
Isolation of the Cytosolic Fraction.
Cells were washed twice with PBS, and the pellet was suspended in 5 ml of ice cold buffer A (20 mM Hepes, pH 7.5/1.5 mM MgCl2/10 mM KCl/1 mM EDTA/1 mM EGTA/1 mM DTT/0.1 mM phenylmethylsulfonyl fluoride/10 μg/ml leupeptin, aprotinin, and pepstatin A) containing 250 mM sucrose. The cells were homogenized by douncing three times in a Dounce homogenizer with a sandpaper-polished pestle. After centrifugation for 5 min at 4°C, the supernatants were centrifuged at 105,000 × g for 30 min at 4°C. The resulting supernatant was used as the soluble cytosolic fraction.
Immunoprecipitation and Immunoblot Analysis.
Cell lysates were prepared as described (19). Immunoprecipitations were performed using anti-Bcl-x (Novartis, NJ) or anti-cytochrome C antibodies (20). Proteins were separated by SDS/PAGE, transferred to nitrocellulose, and probed with the indicated antibodies. The blots were developed by enhanced chemiluminescence (Amersham). Preparation of lysates and immunoblotting for PARP were carried out as described using the c-2–10 anti-PARP mAb (21).
Immunoelectron Microscopy on Frozen Cell Sections.
U-937 cells were resuspended in Tyrode buffer (pH 7.4) and fixed in 8% paraformaldehyde in phosphate buffer as described (22). The cells were then incubated with 20% poly(vinyl pyrrolidone) in 1.84 M sucrose, mounted on aluminum pins, and stored in liquid nitrogen until sectioned using the method of Stenberg et al. (23). Frozen thin sections were incubated with primary antibodies (anti-Bcl-x or anti-cytochrome C) followed by protein A or goat anti-rabbit IgG conjugated to 14 nm of gold particles. Parallel control samples included substitution of buffer or nonimmune serum for the primary antibody. After labeling, the sections were stained with poly(vinyl alcohol) and uranyl acetate. Sections were examined using transmission electron microscopy.
Generation of H6-Bcl-xLT and Bcl-xST Proteins.
Full length Bcl-xL or Bcl-xS were cloned from a cDNA library of human Raji lymphoma cells (CLONTECH) using the PCR primers: 5′ primer: 5′-GGAATTCATATGTCTCAGAGCAACCG-3′; 3′ primer: 5′-AGAATTCATTCATTTCCGACTGAAGAG-3′. After cloning into TA vector (Invitrogen), inserts were subcloned into the EcoRI site of pProEX (GIBCO/BRL) to generate His-tagged fusion constructs or pGEX-3T to generate glutathione S-transferase (GST) fusion proteins. Truncated Bcl-x proteins lacking 21 amino acids from the C terminus (Bcl-xLT or Bcl-xST) were generated by PCR cloning using the 5′ primer described above and the 3′ primer 5′-ATCGAATTCATCAGCGGTTGAAGCGTTCC-3′. All other cloning steps to generate truncated proteins were performed as described for full length proteins. GST–Bcl-xST was cleaved with thrombin to generate Bcl-xST. Folding and protein solubility were verified by circular dichroism and dynamic light scattering measurements. Fusion proteins were expressed as soluble proteins in Escherichia coli and were purified as described (24).
Far Western Analysis.
Column-purified H6-Bcl-xLT, Bcl-xST, or H6 proteins were resolved by SDS/PAGE and transferred to a nitrocellulose filter. Purified MAPK protein was used as a negative control. The filter was incubated with purified cytochrome C (bovine heart cytochrome C; Sigma) for 1 h at room temperature. The filters were then analyzed by immunoblotting with anti-cytochrome C. In separate experiments, purified cytochrome C was separated by SDS/PAGE and transferred to three nitrocellulose filters. The filters were incubated with purified, eluted H6, H6-Bcl-xLT, or Bcl-xST for 1 h at room temperature. The filters were then analyzed by immunoblotting with anti-Bcl-x.
ELISA Immunoassay.
Cytochrome C was adsorbed onto the plastic surface of a 96-well ELISA plate, followed by addition of blocking buffer (Superblock, Pierce) to reduce nonspecific binding. GST, GST–Bcl-xLT, or GST–Bcl-xST in Tris-buffered saline containing 5% BSA and 0.5% Nonidet P-40 was added to the wells. After incubation for 2 h at room temperature, the plates were washed and incubated with rabbit anti-GST, followed by washing and incubation with alkaline phosphatase-conjugated anti-rabbit antibody. The plates were developed using a fluorogenic substrate and reactivity determined by measurement of fluorescence.
RESULTS AND DISCUSSION
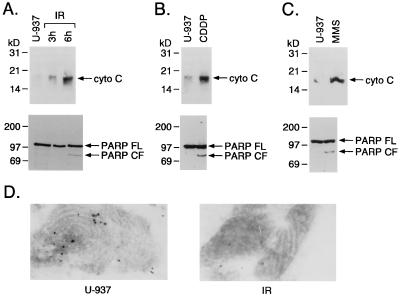
U-937 cells were exposed to 20 Gy IR to determine whether cytosolic cytochrome C levels change with induction of apoptosis. Cytochrome C levels were increased in the cytosol at 3 and 6 h after irradiation (Fig. 1A). These cells also responded to IR with (i) cleavage of PARP into characteristic apoptotic fragments (7, 25) (Fig. 1A) and (ii) induction of internucleosomal DNA fragmentation (5). Similar results were obtained with other inducers of apoptosis, including CDDP (Fig. 1B) and methyl methanesulfonate (Fig. 1C). Electron microscopic studies performed with anti-cytochrome C antibodies demonstrated ImmunoGold labeling of mitochondria from control cells whereas mitochondrial cytochrome C was depleted after exposure to IR (Fig. 1D). These results support a mechanism in which release of mitochondrial cytochrome C into the cytoplasm occurs in concert with induction of apoptosis.
Figure 1.
Cytosolic cytochrome C levels and cleavage of PARP in response to DNA-damaging agents. (A) U-937 cells were treated with 20 Gy of IR and harvested at the indicated times. (B and C) Cells were treated with 50 μM CDDP or 1 mM methyl methanesulfonate for 6 h. Cytosolic proteins were separated by 12.5% SDS/PAGE and analyzed by immunoblotting with anti-cytochrome C. Total cell lysates also were analyzed by immunoblotting with anti-PARP. No detectable reactivity was observed with a control preimmune rabbit serum. FL, full length; CF, cleaved fragment; MMS, methyl methanesulfonate. (D) Transmission electron micrographs of U-937 cells. U-937 cells were fixed in 8% paraformaldehyde and prepared for frozen thin-section immunocytochemistry. Sections were incubated with anti-cytochrome C antibodies followed by 14 nm of protein A gold. Mitochondria are shown. Left, control. Right, IR-treated cells at 20 Gy for 3 h.
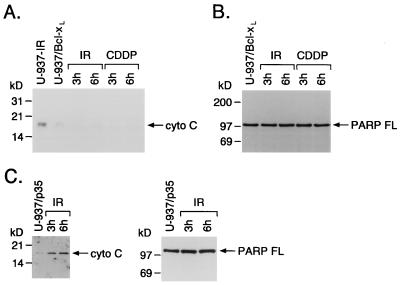
U-937 cells that stably overexpress Bcl-xL (U-937/Bcl-xL) are resistant to radiation-induced internucleosomal DNA fragmentation (5). Bcl-xL expression also prevented accumulation of cytosolic cytochrome C in response to IR or CDDP treatment (Fig. 2A). Moreover, IR-treated U-937/Bcl-xL cells failed to exhibit PARP cleavage (Fig. 2B). The finding that Bcl-xL blocks accumulation of cytosolic cytochrome C and cleavage of PARP raised the possibility that cytochrome C may function upstream of the activation of cysteine proteases. To address this issue, we irradiated U-937 cells that stably overexpress the cysteine protease inhibitor p35 (11, 26). Overexpression of p35 had no detectable effect on accumulation of cytosolic cytochrome C but blocked cleavage of PARP (Fig. 2C) and internucleosomal DNA fragmentation (data not shown). These findings suggest that cytochrome C release from mitochondria in response to apoptotic stimuli is upstream to activation of cysteine proteases.
Figure 2.
Bcl-xL blocks accumulation of cytosolic cytochrome C. (A and B) U-937/Bcl-xL cells were treated with 20 Gy of IR and harvested at the indicated times. Cells also were treated with 50 μM CDDP for 6 h. IR-treated U-937 cells were used as a positive control. Cytosolic proteins were isolated, separated by SDS/PAGE and analyzed by immunoblotting with anti-cytochrome C. Total cell lysates were analyzed by immunoblotting with anti-PARP. (C) U-937 cells stably expressing p35 were treated with 20 Gy of IR and harvested at the indicated times. Cytosolic cytochrome C levels and cleavage of PARP were analyzed as described above. FL, full length.
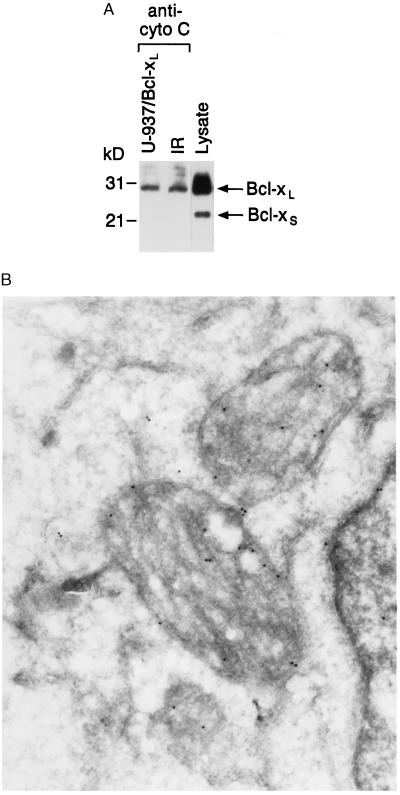
Bcl-xL blocks the increase of cytosolic cytochrome C, so we performed coimmunoprecipitation studies to determine whether these proteins form a specific complex in cells. Lysates from control and irradiated U-937/Bcl-xL cells were subjected to immunoprecipitation with anti-cytochrome C. Analysis of the immunoprecipitates with anti-Bcl-x demonstrated reactivity with Bcl-xL (but not Bcl-xS) that was independent of IR exposure (Fig. 3A). The demonstration that Bcl-xL associates with cytochrome C is in concert with the finding that Bcl-x protein is detectable within mitochondria (Fig. 3B).
Figure 3.
Bcl-xL binds directly to cytochrome C in vivo. (A) U-937/Bcl-xL cells were treated with 20 Gy of IR and harvested at 6 h. Total cell lysates were subjected to immunoprecipitation with anti-cytochrome C, and the protein precipitates were analyzed by immunoblotting with anti-Bcl-x. As a control, cell lysate was directly analyzed by immunoblotting with anti-Bcl-x to identify the Bcl-xL and Bcl-xS forms. anti-cyto C, anti-cytochrome C. (B) U-937 cells were fixed in 8% paraformaldehyde and prepared for frozen thin-section immunocytochemistry. Sections were incubated with anti-Bcl-x antibodies followed by goat anti-rabbit IgG conjugated to 14 nm of gold particles.
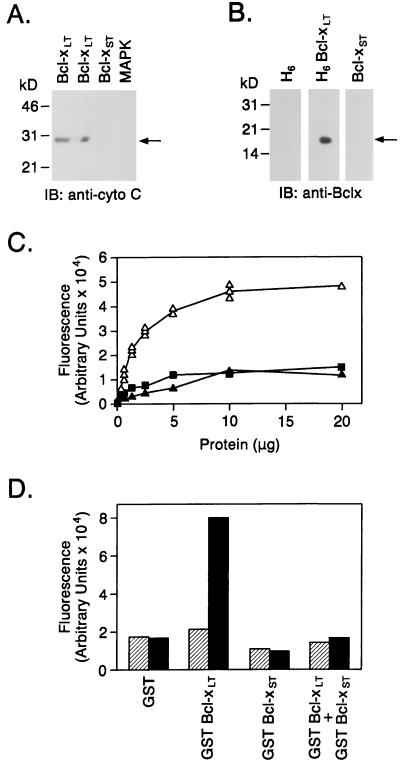
To further demonstrate that Bcl-xL interacts directly and specifically with cytochrome C, recombinant hexa-His(H6)–Bcl-xLT was resolved by gel electrophoresis, transferred to a nitrocellulose filter, and renatured in aqueous buffer. After incubation with purified cytochrome C, the filter was washed and probed with anti-cytochrome C. Reactivity with anti-cytochrome C at the position corresponding to Bcl-xL demonstrated a direct interaction between cytochrome C and Bcl-xL (Fig. 4A). By contrast, there was no detectable binding of cytochrome C to Bcl-xST or MAPK (Fig. 4A). The absence of reactivity with Bcl-xS or MAPK indicated that binding of cytochrome C to Bcl-xL is specific. Purified cytochrome C also was resolved by electrophoresis and transferred to nitrocellulose filters, which were then incubated with purified H6, H6-Bcl-xLT, or Bcl-xST proteins and analyzed by immunoblotting with anti-Bcl-x. Only the blot incubated with H6-Bcl-xLT showed reactivity with cytochrome C (Fig. 4B). Similar results were obtained by an ELISA in which purified cytochrome C adsorbed to wells was incubated with GST, GST–Bcl-xLT, or GST–Bcl-xST. After development with a fluorogenic substrate, only incubations containing GST–Bcl-xL produced a fluorescent product (Fig. 4C). Our results supported binding of cytochrome C to Bcl-xL but not Bcl-xS, so we asked whether Bcl-xS, which can form a complex with Bcl-xL, blocks the interaction between cytochrome C and Bcl-xL. The results demonstrate that addition of excess Bcl-xS inhibits binding of cytochrome C to Bcl-xL (Fig. 4D). These findings with recombinant proteins confirm that the binding between cytochrome C and Bcl-xL found by coimmunoprecipitation of cellular lysates is due to a specific interaction that can be blocked by Bcl-xS.
Figure 4.
(A) Column-purified H6-Bcl-xL (lanes: 1, 5 μg; 2, 2.5 μg) and Bcl-xST (lane 3, 5 μg) proteins were resolved by SDS/PAGE and transferred to a nitrocellulose filter. Purified MAPK protein was used as a negative control (lane 4). The filter was incubated with purified cytochrome C (bovine heart cytochrome C) for 1 h at room temperature. The filters then were analyzed by immunoblotting with anti-cytochrome C. (B) Purified cytochrome C was separated by SDS/PAGE and transferred to three nitrocellulose filters. The filters were incubated with purified, eluted H6, H6-Bcl-xLT. or Bcl-xST for 1 h at room temperature. The filters then were analyzed by immunoblotting with anti-Bcl-x. (C) Cytochrome C was adsorbed onto the plastic surface of a 96-well ELISA plate. Various concentrations of GST (▴), GST–Bcl-xLT (▵), or GST–Bcl-xST (▪) were added to the wells. Binding of Bcl-x to cytochrome C was measured by ELISA immunoassay. (D) Cytochrome C adsorbed to plates was incubated with 1 μg/ml GST, 1 μg/ml GST–Bcl-xLT, 5 μg/ml GST–Bcl-xST, or a mixture of 1 μg/ml GST–Bcl-xLT and 5 μg/ml GST–Bcl-xST. Binding of Bcl-x was assessed by ELISA. Hatched bars represent controls with no cytochrome C. Solid bars represent assays performed in the presence of cytochrome C.
Bcl-xL inhibits apoptosis induced by diverse stimuli although the basis for this effect has been unclear (5, 27–29). Recent studies have shown that cytochrome C is released from mitochondria during the induction of apoptosis (12). Our studies demonstrate that Bcl-xL blocks the increase in cytosolic cytochrome C accumulation associated with induction of apoptosis by IR and genotoxic drugs. Moreover, our results show that the interaction is specific for Bcl-xL and not the proapoptotic Bcl-xS protein. Bcl-xL is predominantly localized to the mitochondrial membrane (18). The crystal structure of Bcl-xL supports an arrangement of α-helices that is found in the membrane translocation domains of bacterial toxins (30, 31). Bcl-xL domains that extend across the mitochondrial membrane may contribute to the interaction with cytochrome C. Taken together, our findings support a model in which release of cytochrome C from mitochondria during induction of apoptosis can be prevented by Bcl-xL.
Acknowledgments
We thank Dr. Lawrence Prochaska for providing antibody to cytochrome C. We also thank Andrew Place, Eileen McMillan-Ward, and Zeinab Moustafa for technical assistance. This investigation was supported by Public Health Service Grants CA55241 and CA66996 awarded by the National Cancer Institute, Department of Health and Human Services.
ABBREVIATIONS
- IR
ionizing radiation
- PARP
poly(ADP ribose) polymerase
- CDDP
cisplatinum
- GST
glutathione S-transferase
References
- 1.Lowe S W, Schmitt E M, Smith S W, Osborne B A, Jacks T. Nature (London) 1993;362:847–849. doi: 10.1038/362847a0. [DOI] [PubMed] [Google Scholar]
- 2.Clarke A R, Purdie C A, Harrison D J, Morris R G, Bird C C, Hooper M L, Wylie A H. Nature (London) 1993;362:849–852. doi: 10.1038/362849a0. [DOI] [PubMed] [Google Scholar]
- 3.Sentman C L, Shutter J R, Hockenbery D, Kanagawa O, Korsmeyer S J. Cell. 1991;67:879–888. doi: 10.1016/0092-8674(91)90361-2. [DOI] [PubMed] [Google Scholar]
- 4.Strasser A, Harris A W, Cory S. Cell. 1991;67:889–899. doi: 10.1016/0092-8674(91)90362-3. [DOI] [PubMed] [Google Scholar]
- 5.Datta R, Manome Y, Taneja N, Boise L H, Weichselbaum R R, Thompson C B, Slapak C A, Kufe D W. Cell Growth Differ. 1995;6:363–370. [PubMed] [Google Scholar]
- 6.Martin S J, Green D R. Cell. 1995;82:349–352. doi: 10.1016/0092-8674(95)90422-0. [DOI] [PubMed] [Google Scholar]
- 7.Kaufmann S H, Desnoyers S, Ottaviano Y, Davidson N E, Poirier G G. Cancer Res. 1993;53:3976–3985. [PubMed] [Google Scholar]
- 8.Emoto Y, Manome G, Meinhardt G, Kisaki H, Kharbanda S, Robertson M, Ghayur T, Wong W W, Kamen R, Weichselbaum R, Kufe D. EMBO J. 1995;14:6148–6156. doi: 10.1002/j.1460-2075.1995.tb00305.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chinnaiyan A M, Orth K, O’Rourke K, Duan H, Poirier G G, Dixit V M. J Biol Chem. 1996;271:4573–4576. doi: 10.1074/jbc.271.9.4573. [DOI] [PubMed] [Google Scholar]
- 10.Datta R, Kojima H, Banach D, Bump N J, Talanian R V, Alnemri E S, Weichselbaum R R, Wong W W, Kufe D W. J Biol Chem. 1997;272:1965–1969. doi: 10.1074/jbc.272.3.1965. [DOI] [PubMed] [Google Scholar]
- 11.Bump N J, Hackett M, Hugunin M, Seshagiri S, Brady K, Chen P, Ferenz C, Franklin S, Ghayur T, Li P, Licari P, Mankovich J, Shi L, Greenberg A H, Miller L K, Wong W W. Science. 1995;269:1885–1888. doi: 10.1126/science.7569933. [DOI] [PubMed] [Google Scholar]
- 12.Liu X, Kim C N, Yang J, Jemmerson R, Wang X. Cell. 1996;83:147–157. doi: 10.1016/s0092-8674(00)80085-9. [DOI] [PubMed] [Google Scholar]
- 13.Tewari M, Quan L T, O’Rourke K, Desnoyers S, Zeng Z, Beidler D R, Poirier G G, Salvesen G S, Dixit V M. Cell. 1995;81:801–809. doi: 10.1016/0092-8674(95)90541-3. [DOI] [PubMed] [Google Scholar]
- 14.Nicholson D W, Ali A, Thornberry N A, Vaillancourt J P, Ding C K, Gallant M, Gareau Y, Griffin P R, Labelle M, Lazebnik Y A, Munday N A, Raju S M, Smulson M E, Yamin T-T, Yu V L, Miller D K. Nature (London) 1995;376:37–43. doi: 10.1038/376037a0. [DOI] [PubMed] [Google Scholar]
- 15.Ghayur T, Hugunin M, Talanian R V, Ratnofsky S, Quinlan C, Emoto Y, Pandey P, Datta R, Kharbanda S, Allen H, Kamen R, Wong W, Kufe D. J Exp Med. 1996;184:2399–2404. doi: 10.1084/jem.184.6.2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gonzales D H, Neupert W. J Bioenerg Biomembr. 1990;22:753–768. doi: 10.1007/BF00786929. [DOI] [PubMed] [Google Scholar]
- 17.Monaghan P, Robertson D, Amos T A S, Dyer M J S, Mason D Y, Greaves M F. J Histochem Cytochem. 1992;40:1819–1825. doi: 10.1177/40.12.1453000. [DOI] [PubMed] [Google Scholar]
- 18.Gonsalez-Garcia M, R, P-B, Ding L, Duan L, Boise L H, Thompson C B, Nunuz G. Development. 1994;120:3033–3042. doi: 10.1242/dev.120.10.3033. [DOI] [PubMed] [Google Scholar]
- 19.Kharbanda S, Yuan Z M, Rubin E, Weichselbaum R, Kufe D. J Biol Chem. 1994;269:20739–20743. [PubMed] [Google Scholar]
- 20.Kirken R A, Lincoln A J, Fink P S, Prochaska L J. Protein Expression Purif. 1995;6:707–715. doi: 10.1006/prep.1995.1093. [DOI] [PubMed] [Google Scholar]
- 21.Desnoyers S, Shah G M, Brochu G, Poirier G G. Ann Biochem. 1994;218:470–473. doi: 10.1006/abio.1994.1212. [DOI] [PubMed] [Google Scholar]
- 22.Israels S J, Gerrard J M, Jacques Y V, McNicol A, Cham B, Nishibori M, Bainton D F. Blood. 1992;80:143–152. [PubMed] [Google Scholar]
- 23.Stenberg P E, Shuman M A, Levine S P, Bainton D F. Histochem J. 1984;16:983–1001. doi: 10.1007/BF01003853. [DOI] [PubMed] [Google Scholar]
- 24.Polayes D, Hughes A J., Jr Focus. 1994;16:81. [Google Scholar]
- 25.Lazebnik Y A, Kaufmann S H, Desnoyers S, Poirier G G, Earnshaw W C. Nature (London) 1994;371:346–347. doi: 10.1038/371346a0. [DOI] [PubMed] [Google Scholar]
- 26.Datta R, Banach D, Kojima H, Talanian R V, Alnemri E S, Wong W W, Kufe D. Blood. 1996;88:1936–1943. [PubMed] [Google Scholar]
- 27.Boise L H, Gonzaiz-Garcia M, Postema C E, Ding L, Lindsten T, Turka L A, Mao X, Nunez G, Thompson C B. Cell. 1993;74:597–608. doi: 10.1016/0092-8674(93)90508-n. [DOI] [PubMed] [Google Scholar]
- 28.Gottschalk A R, Boise L H, Thompson C B, Quintans J. Proc Natl Acad Sci USA. 1994;91:7350–7354. doi: 10.1073/pnas.91.15.7350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shimizu S, Eguchi Y, Kosaka H, Kamlike W, Matsuda H, Tsujimoto Y. Nature (London) 1995;374:811–813. doi: 10.1038/374811a0. [DOI] [PubMed] [Google Scholar]
- 30.Muchmore S W, Sattler M, Liang H, Meadows R P, Harlan J E, Yoon H S, Nettesheim D, Chang B S, Thompson C B, Wong S-L, Ng S-C, Fesik S W. Nature (London) 1996;381:335–341. doi: 10.1038/381335a0. [DOI] [PubMed] [Google Scholar]
- 31.Parker M W, Pattus F. Trends Biochem Sci. 1993;18:391–395. doi: 10.1016/0968-0004(93)90096-6. [DOI] [PubMed] [Google Scholar]