Abstract
Loss of a whole chromosome 5 or a deletion of the long arm, del(5q), is a recurring abnormality in malignant myeloid diseases. In previous studies, we delineated a commonly deleted segment of ≈4 Mb within band 5q31 that was flanked by IL9 on the proximal side and D5S166 on the distal side. We have generated a physical map of P1 (PAC), bacterial (BAC), and yeast artificial chromosome (YAC) clones of this interval. The contig consists of 108 clones (78 PACs, 2 BACs, and 28 YACs) to which 125 markers (5 genes, 11 expressed sequence tags, 12 polymorphisms, and 97 sequence-tagged sites) have been mapped. Using PAC clones for fluorescence in situ hybridization analysis of leukemia cells with a del(5q), we have narrowed the commonly deleted segment to 1–1.5 Mb between D5S479 and D5S500. To search for allele loss, we used 7 microsatellite markers within and flanking the commonly deleted segment to examine leukemia cells from 28 patients with loss of 5q, and 14 patients without cytogenetically detectable loss of 5q. In the first group of patients, we detected hemizygous deletions, consistent with the cytogenetically visible loss; no homozygous deletions were detected. No allele loss was detected in patients without abnormalities of chromosome 5, suggesting that allele loss on 5q is the result of visible chromosomal abnormalities. The development of a stable PAC contig and the identification of the smallest commonly deleted segment will facilitate the molecular cloning of a myeloid leukemia suppressor gene on 5q.
Keywords: myeloid leukemia, tumor suppressor genes, chromosomal deletions, therapy-related leukemia
Recurring chromosomal abnormalities are characteristic of human malignant diseases, particularly the leukemias and lymphomas (1–3). To date, the major emphasis of the molecular analysis of chromosomal abnormalities in tumors has involved the recurring translocations, which result in the activation of a cancer-related gene in a dominant fashion (4). In addition to the recurring translocations, the loss of genetic material has been identified in many tumor types. Loss of genetic material is the hallmark of tumor suppressor genes and may result from loss of a whole chromosome or part of a chromosome (deletion) as well as by other genetic mechanisms. The consequence of these abnormalities is the unmasking of a recessive allele on the structurally “normal” homolog (5–6).
The identification of recurring chromosomal deletions in the hematological malignant diseases suggests that as for a number of solid tumors, tumor suppressor genes may be involved in the pathogenesis of some leukemias (3, 7, 8). With respect to the myeloid disorders, recurring deletions include the del(5q), del(7q), del(9q), del(11q), del(12p), del(13q), and del(20q) (3, 7). With the exception of the del(9q), a commonly deleted segment has been identified by molecular mapping for each of these rearrangements (ref. 9; see ref. 10 for references).
The occurrence of a myelodysplastic syndrome (MDS) or acute myeloid leukemia (AML) is a late complication of cytotoxic therapy used in the treatment of both malignant and nonmalignant diseases (11, 12). Therapy-related MDS or AML (t-MDS/t-AML) typically presents ≈5 years after treatment. Frequently all three hematopoietic cell lines (erythroid, myeloid, and megakaryocytic) are involved in the myelodysplastic process. Survival times of patients with t-AML are usually short (median, 8 months) (13).
At the cytogenetic level, t-MDS/t-AML is characterized by loss of an entire chromosome 5 or 7, or a deletion of the long arm of these chromosomes [del(5q)/del(7q)]. In our recently updated series of 257 consecutive patients with t-MDS/t-AML, 242 (94%) had a clonal chromosomal abnormality (ref. 13; M.M.L.B., R.A.L., E. M. Davis, J. D. Rowley, and J. W. Vardiman, unpublished work), and 183 patients (71%) had a clonal abnormality leading to loss or deletion of chromosome 5 and/or 7. Among these 183 patients, 34 had loss of chromosome 5, 52 had a del(5q), and 23 had loss of 5q following an unbalanced translocation. Overall, 109 patients (42%) had abnormalities of chromosome 5, and 132 (51%) had abnormalities of chromosome 7. Fifty-eight patients had abnormalities of both chromosomes 5 and 7. A del(5q) was the most common structural abnormality.
In addition to t-MDS/t-AML, a −5/del(5q) has also been observed in 10% of patients with AML or MDS de novo (14–16). Many of these patients have had significant occupational exposure to potential carcinogens, suggesting that abnormalities of chromosomes 5 or 7 may be a marker of mutagen-induced leukemia. A distinct clinical disorder, termed the “5q− syndrome,” is observed in a subset of patients with MDS de novo (16–18). This disorder is characterized by refractory anemia and a relatively mild course that usually does not progress to acute leukemia. These patients typically have the del(5q) as the sole abnormality.
We and others have proposed that 5q contains a tumor suppressor gene. In previous studies, we delineated a segment of 5q, within band q31, that was deleted in all patients examined; this commonly deleted segment was ≈4 Mb in size, and was flanked by IL9 (proximal) and D5S166 (distal) (9). We have prepared a P1 (PAC), bacterial (BAC), and yeast artificial chromosome (YAC) contig of this interval. By using fluorescence in situ hybridization (FISH) of PAC clones from the contig to examine myeloid leukemia cells with a del(5q), we have narrowed the commonly deleted segment to 1–1.5 Mb. In addition, we have eliminated two known genes within this interval (EGR1 and CDC25C) as candidate tumor suppressor genes by single-strand conformation polymorphism analysis (P.W.W., A.S., N.Z., J.D.E., R.A.L., and M.M.L.B., unpublished results).
MATERIALS AND METHODS
Patients.
The diagnosis and subclassification of MDS or AML were based on morphological and cytochemical studies of peripheral blood smears, bone marrow aspirates, and biopsy specimens obtained prior to therapy, according to the French–American–British Cooperative Group criteria. Cytogenetic analysis was performed with a trypsin-Giemsa banding technique on bone marrow cells obtained at the time of diagnosis. Chromosomal abnormalities were described according to An International System for Human Cytogenetic Nomenclature (19).
YAC and PAC Contig.
The YAC libraries (CEPH A and B YAC DNA pools; Research Genetics, Huntsville, AL) developed by the Centre D’Etude du Polymorphisme Humaine (CEPH), and the PAC library (Genome Systems, St. Louis) were screened for sequence-tagged sites (STSs), expressed sequence tags (ESTs), and short tandem repeat polymorphisms (STRPs) while following the manufacturer’s recommended procedures with minor modifications. A genomic BAC library (Research Genetics) was screened for several markers to obtain redundant coverage or if clones could not be found in the YAC and PAC libraries. PCR conditions were optimized for each marker using 50 ng placental DNA as template to give a single, distinct amplified product. For library screening, PCR was carried out in a 30-μl volume containing 0.25–0.5 μM of each primer, 400 mM dNTP, 10 mM Tris⋅HCl (pH 8.3), 50 mM KCl, 2.0 mM MgCl2, and 1 unit of Taq polymerase with an initial denaturation step (94°C for 1 min), followed by 30–40 cycles of denaturation at 94°C for 40 sec, annealing for 1 min, and elongation at 72°C for 1 min. PCR products were separated by electrophoresis on 2% agarose gels and visualized by ethidium bromide staining. In addition, the databases of the CEPH-Généthon physical map of the human genome (ftp.cephb.fr.;http://www.cephb.fr/bio/infoclone.html), the Human Physical Mapping Project at the Whitehead Institute/MIT Center for Genome Research (http://www-genome.wi.mit.edu/), and the Lawrence Berkeley National Laboratory (http://www-hgc.lbl.gov/inf/ftp-message.html) (Berkeley, CA) were searched for YAC or BAC clones containing markers from this region.
PAC, BAC, and YAC End Cloning.
Bacteria containing PACs and BACs were cultured following the manufacturer’s recommended procedures, and DNA was prepared using the Qiagen Plasmid Midi Kit (Qiagen, Chatsworth, CA). Clones were sequenced using primers from the vector arms. Yeast cells containing YACs were grown in AHC media (inoculated with a single colony plated on an AHC plate), and DNA was prepared as described (20). The ends of the human DNA insert in the YACs were isolated by Alu-vector PCR as described previously (21). The PCR products were cloned into the HincII site of pGEM-4Z or -3Z (Promega) and sequenced. To extend the map, PCR primers were designed from the unique end sequences of clones and were used for the next round of library screening.
Estimating the Insert Sizes of PAC, BAC, and YAC Clones.
Pulse-field gel electrophoresis (PFGE) was used to determine the size of the insert in the genomic clones. YAC DNA was prepared in plugs as described (20); 1–3 μg of PAC or BAC DNA in solution was loaded in a 1% agarose gel. PFGE was performed using a Bio-Rad CHEF Mapper using an electrophoresis profile to separate DNA in the range of 50–500 kb (PAC clones) or 100–1,500 kb (YAC clones).
DNA Sequencing.
DNA sequencing was performed using the Sequenase Kit (Version 2.0, United States Biochemical) or the AmpliCycle Sequencing Kit (Roche Diagnostics). Automated fluorescent DNA sequencing was performed using the ABI Prism Dye Terminator Cycle Sequencing Kit (Perkin–Elmer), and the reactions were analyzed using an ABI Prism 377 DNA Sequencer (Perkin–Elmer).
Fluorescence in Situ Hybridization.
Human metaphase cells were prepared from phytohemagglutinin-stimulated lymphocytes from healthy individuals, or from bone marrow cells from patients with MDS or AML. FISH was performed as described previously (10). Labeled PAC probes were prepared by nick-translation using Bio-16-dUTP (Enzo Diagnostics), digoxigenin-11-dUTP (Boehringer Mannheim), or directly labeled nucleotides (Vysis, Downers Grove, IL). Labeled YAC probes were prepared by sequence-independent amplification or Alu-PCR using biotin-labeled or directly labeled nucleotides (Spectrum-Orange-dUTP, Vysis) (22, 23). Chromosomes were identified by staining with 4,6-diamidino-2-phenylindole-dihydrochloride. For the analysis of leukemia samples, 10–15 metaphase cells with a del(5q) were scored by two independent observers; in a few instances, only 5–9 metaphase cells were available for analysis.
Extended chromatin fibers were prepared using the alkaline treatment method of Fidlerova et al. (24). For estimating distances between markers or clones, length measurements for each segment of the fluorescent signal or gap between signals were determined in pixels using NIH image 1.57, and were normalized to the signal length of a probe of known size. An estimate of the gap between probes was generated by averaging the relative lengths from 30 images.
Microsatellite PCR.
Ten nanograms of genomic DNA from leukemia cells was amplified independently with each microsatellite primer pair, along with normal control DNA. Primer sequences for the 7 markers analyzed are available from the Genome Data Base (GDB; Johns Hopkins University, Baltimore, MD) and the Center for Genome Research at the Whitehead Institute. Reactions were carried out in 10 μl containing 10 mM Tris⋅HCl (pH 8.3), 50 mM KCl, 2.0 mM MgCl2, 200 μM dNTPs, 0.3 units of Taq polymerase, and 4 pmol of each primer;  of one of the primers was end-labeled with [γ-32P]ATP (6,000 Ci/mmol; 1 Ci = 37 GBq; Amersham) using T4 polynucleotide kinase (New England Biolabs). PCR conditions consisted of an initial denaturation at 94°C for 4 min, followed by 28–30 cycles of denaturation at 94°C for 40 s, annealing at an optimized temperature between 55 and 65°C, for 1 min, and an extension at 72°C for 1 min, with a final extension at 72°C for 7 min. The amplified products were separated in 5% denaturing polyacrylamide gels (1× TBE at 1,400 V) for several hours; the gels were dried and autoradiographed overnight.
of one of the primers was end-labeled with [γ-32P]ATP (6,000 Ci/mmol; 1 Ci = 37 GBq; Amersham) using T4 polynucleotide kinase (New England Biolabs). PCR conditions consisted of an initial denaturation at 94°C for 4 min, followed by 28–30 cycles of denaturation at 94°C for 40 s, annealing at an optimized temperature between 55 and 65°C, for 1 min, and an extension at 72°C for 1 min, with a final extension at 72°C for 7 min. The amplified products were separated in 5% denaturing polyacrylamide gels (1× TBE at 1,400 V) for several hours; the gels were dried and autoradiographed overnight.
RESULTS
Construction of a PAC, BAC, and YAC Genomic Contig.
In previous studies, we identified a small segment of 5q31 that was deleted in all patients with MDS or AML and a del(5q) (9). This commonly deleted segment was ≈4 Mb and was flanked by IL9 (proximal) and D5S166 (distal). To generate a YAC contig of this interval, we screened the CEPH A and B libraries using 10 STRPs (GATA-P6551, D5S393, D5S89, D5S399, D5S479, AFM350YB1, D5S414, D5S500, D5S476, and D5S166) that were mapped to 5q31 by genetic linkage analysis, and 7 STS markers for three genes (IL9, EGR1, and CDC25C), 3 anonymous sequences (D5S1697, WI-9259, and WI-9261), and 1 EST (WI-8192). Positive YACs were subjected to end-cloning to identify unique human fragments. Of 120 YACs identified by the initial screening, 41 YAC clones were confirmed to be positive using the original STS or STRP primer pair.
The preparation of a YAC contig proved be an exceedingly difficult task, in large part due to the apparent instability of this region of the genome. Of the 41 YACs, ≈90% were chimeric and mapped by FISH to 5q31 as well as to one or more other chromosomal sites. Moreover, a number of them had internal deletions, as revealed by FISH of the YACs to extended chromatin fibers. PCR analysis also suggested that the YACs were chimeric, because YACs with large inserts (800 kb–1.5 Mb) were found to contain only one or a few STS markers from 5q31. Finally, there were gaps in the contig that could not be filled by rescreening the CEPH libraries (A and B), as these sequences apparently were not represented in these libraries.
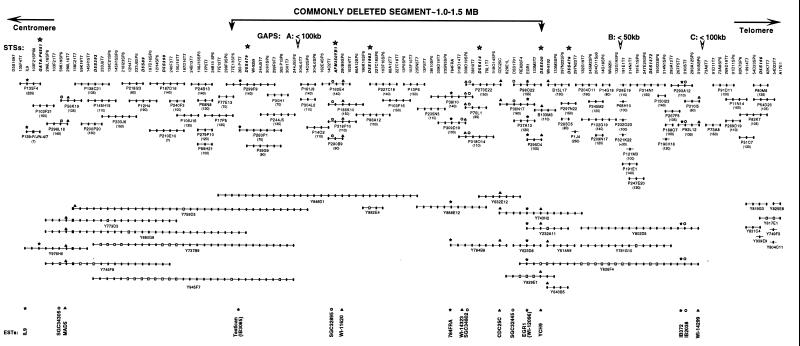
To minimize the problem of instability, we converted the contig to PAC clones by screening the PAC library using the 17 markers described above (Fig. 1). To walk between PAC clones, sequencing with vector arm primers was performed to obtain sequences from the insert adjacent to the vector arms, which were then used to rescreen the PAC library. The PAC and BAC contig shown in Fig. 1 contains 78 PAC clones and 2 BACs, to which 125 markers were uniquely localized. The markers include 12 polymorphic STSs (described above), 97 nonpolymorphic STSs, 5 known genes (IL9, MAD5, EGR1, CDC25C, and Testican), and 11 ESTs. The localization of all clones to 5q31 was confirmed by FISH. In addition, FISH to extended DNA fibers was used to confirm the order of PAC clones and, in some cases, to identify clones that overlap. Although it was possible to deduce STS order, the absolute physical size of the contig is not known precisely, as the extent of the overlapping sequences between adjacent clones has not been determined. The presence of YACs that are chimeric or contain deletions can also confound the estimation of the physical size of the contig.
Figure 1.
STS content map of the genomic contig of 5q31. PAC, BAC, and YAC clones are prefaced by a P, B, or Y, respectively. The clones are presented as lines; their size reflects the number of STSs contained in each clone. Physical size in kilobases is given in braces. Solid circles indicate positive STSs. Open squares indicate that a clone was found to be negative or ambiguous for a given STS. STS markers are listed at the top of the map; those indicated by an asterisk represent microsatellite markers used for allele loss studies in leukemia cells. Gaps in the contig are indicated by an open arrow at the top of the map, and the estimated sizes of the gaps in kilobases are listed. The location of known genes and ESTs are shown at the bottom of the map.
There is currently one small gap in the PAC contig on the centromeric side, and two gaps on the telomeric side (Fig. 1); only one of these is not covered by YAC clones (i.e., the gap between clones P92L12 and P91D11 at the telomeric end of the contig). We used dual-color FISH analysis of released DNA fibers (24) to estimate the sizes of these gaps: gap A, P244J5–P161J9, <100 kb; B, P132G19–P68H10, <50 kb; and C, P92L12–P91D11, <100 kb.
Identification of Transcribed Sequences in the Physical Map.
The availability of a PAC contig that spans the commonly deleted segment in myeloid leukemias provides an opportunity to begin the search for a myeloid tumor suppressor gene. Five known genes were placed in the contig: IL9 encodes a T cell growth factor (25), MAD5 encodes a member of the MAD family (26, 27), Testican encodes a progenitor of a unique heparan/chondroitin-sulfate-bearing peptide present in human seminal fluid, EGR1 encodes a transcription factor (28), and CDC25C encodes a phosphatase that regulates the G2–M transition (29).
Through the GenBank EST database (dbEST) and the integrated gene resource (30), we identified 13 ESTs in the interval between IL9 and D5S166, and have localized these ESTs within our contig (Fig. 1). Nine ESTs are within our 1–1.5 Mb commonly deleted segment described below (IB3085, SGC-32895, WI-11620, 784FRA, WI-14323, SGC-34802, SGC-32445, WI-12096, and YCH9). EST IB3085 has high homology to Testican, and EST WI-12096 is highly homologous to EGR1.
Molecular Delineation of the Commonly Deleted Segment at 5q31.
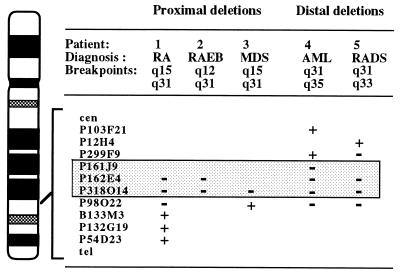
To refine the commonly deleted segment at 5q31 in myeloid leukemias, we performed FISH of PAC clones within the contig to metaphase cells from five patients with myeloid diseases characterized by a del(5q) who had either a proximal (two patients) or distal breakpoint (three patients) within 5q31. The results revealed that PAC P299F9 and probes centromeric of this PAC are retained in some patients and, thus, are proximal to the commonly deleted segment. PAC P98O22 and clones telomeric to this marker are distal to the commonly deleted segment and are retained in some patients (Figs. 2 and 3). In all cases, hybridization of the PAC clones was observed on the normal chromosome 5 homolog at 5q31, suggesting that submicroscopic deletions had not occurred. We have complete coverage of this interval in the combined YAC and PAC contig. A simple addition of the insert sizes in the PACs spanning this interval (P299F9, P244J5, P161J9, P14O2, P318P10, P86A12, P103F16, P13P6, P235N5, P309D19, P273E22, P36N17, and P98O22) totals 1.7 Mb. This is a considerable overestimate of the actual size of the region, as there is substantial overlap among the PAC clones. This estimate also includes the flanking PACs (P299F9 and P98O22), which are 140 kb and 135 kb, respectively.
Figure 2.
Diagram of the banding pattern of chromosome 5 illustrating the results of FISH analysis of five patients with a del(5q) involving either a proximal (two patients) or distal (three patients) breakpoint in 5q31. The PAC clones are shown on the right of the chromosome. Hybridization results for the deleted homologs are indicated by a “+” (positive signal) or “−” (no signal) sign. The genomic segment between P299F9 and P98O22 (shaded box) was deleted in each patient examined. RA, refractory anemia; RAEB, RA with excess blasts; RADS, RA with dysplasia. Patients 1 and 4 were also examined in a previous report (patients 5 and 9, respectively, in ref. 9).
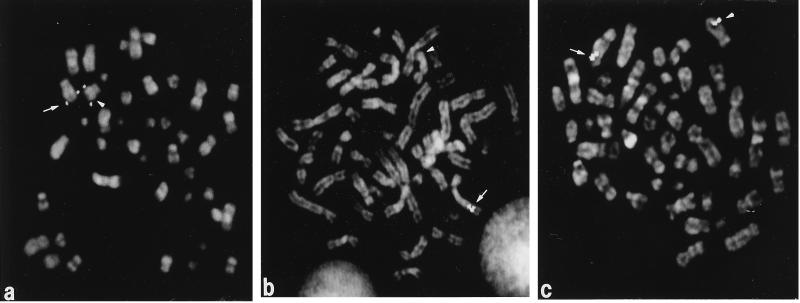
Figure 3.
PAC clones within the contig were hybridized to metaphase cells from patients with a del(5q). (a) PAC P299F9 hybridized to both the normal (arrow) and deleted (arrowhead) homologs in leukemia cells with a del(5)(q31q35) (patient 4 in Fig. 2). Thus, it is proximal to the commonly deleted segment. (b) Clone P318O14 is deleted on the rearranged homolog (arrowhead) in leukemia cells with a del(5)(q15q31) (patient 1 in Fig. 2). (c) Clone P98O22 is retained on the deleted (arrowhead) and normal (arrow) homologs in leukemia cells with a del(5)(q15q31) (patient 3 in Fig. 2), and is distal to the commonly deleted segment.
Analysis of Allele Loss.
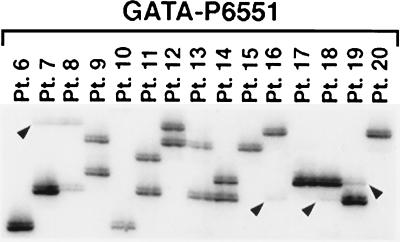
To determine whether submicroscopic deletions of sequences within the 1–1.5 Mb commonly deleted segment occur in myeloid leukemia cells, we performed allele-loss studies using seven STRPs within the contig. Five markers are within the commonly deleted segment (D5S479, AFM350YB1, D5S1983, D5S414, and D5S500), and two markers flank this region on the proximal (GATA-P6551/D5S816) and distal (D5S476) sides (these microsatellite markers are identified with an asterisk in Fig. 1). DNA was extracted from leukemia cells from 28 patients with t-AML or AML de novo characterized by loss of chromosome 5 or a del(5q). All of the patients with a del(5q) had large deletions that included 5q31; thus, analysis of microsatellite markers was not used to define the commonly deleted segment. In each patient, we identified hemizygous loss of the informative markers, consistent with the cytogenetically detectable loss of 5q (Fig. 4). No homozygous deletions were detected.
Figure 4.
Allele-loss studies of DNA from myeloid leukemia cells using the GATA-P6551 (D5S816) STRP centromeric to the commonly deleted segment. Hemizygous loss of alleles is indicated with arrowheads. Patients 6–8, 10, 13, and 15–20 had cytogenetic abnormalities leading to loss of 5q, whereas patients 9, 11, 12, and 14 had no visible loss of chromosomal material from 5q. Single bands are visible for patients 6, 10, 15, 17, and 20; thus, we cannot distinguish between homozygosity and complete loss of one allele.
To determine whether submicroscopic deletions of 5q occurred in patients who did not have loss of 5q at the cytogenetic level, we examined leukemia cells from 14 patients with t-MDS/t-AML who did not have cytogenetic abnormalities of chromosome 5, but who had a disease that was clinically and morphologically identical to that of t-AML patients with loss of 5q. Of these, eight patients had −7/del(7q), four patients had other chromosomal abnormalities, and two patients had a normal karyotype. No allele loss was detected in these patients (Fig. 4). These results suggest that loss of alleles on 5q is the result of visible chromosomal abnormalities. Submicroscopic deletions, if they occur, may be rare events.
DISCUSSION
Several myeloid disorders with different clinical features may be associated with a del(5q). Although the breakpoints and extent of the deletions are variable, the deletions in the Refractory Anemia 5q- Syndrome, t-AML, and AML de novo appear to be relatively similar, with a proximal breakpoint commonly in q13–q15 and a distal breakpoint within region q3, at bands q31–35 (9, 13, 17, 18). In previous studies, we identified a region of 5q31 flanked by IL9 (proximal) and D5S166 (distal) that was deleted in each patient with MDS or AML (9). We have generated a physical map of this region; the sex-averaged length is ≈3 cM, and the maximum physical length is ≈4 Mb. Overall, the order of the markers based on genetic or physical methods is in close agreement, and markers that were tightly linked genetically could be ordered on the physical map. By molecular mapping of the deletions of chromosome 5 in myeloid disorders, we have narrowed the commonly deleted segment of 5q31 to a 1–1.5 Mb region between the D5S479 and D5S500 loci, which are contained in PACs P299F9 and P98O22, respectively. Our studies have focused on defining the commonly deleted segment in patients with MDS or AML, rather than the 5q- Syndrome, because only 1 out of 20 patients with a breakpoint in 5q31 examined (ref. 9 and this study) had the 5q- Syndrome (patient 1 in Fig. 2).
Fairman et al. (31) have generated a YAC contig spanning IL9 and EGR1, which is estimated to be <2.4 Mb. The order of the loci was cen-IL9-(D5S525-D5S558-D5S89-D5S526-D5S393)-D5S399-D5S396-D5S414-EGR1-tel. These investigators also noted instability of the YACs examined from this region of the genome. Molecular analysis of a patient with a complex translocation, t(5;18;17) (q31;p11;q11), revealed allele loss of D5S399 and D5S396. Similar analysis of leukemia cells with a del(5q) revealed that one patient had retained IL9 and D5S396 but lost D5S399, suggesting that there is a narrow region of loss between IL9 and D5S396 (31). However, the interpretation of these findings is difficult in light of recent maps that position D5S396 proximal to IL9, rather than distal (this manuscript, and the YAC contig prepared by the Whitehead Institute/MIT Center for Genome Research).
Willman et al. (32) identified a more centromeric-deleted region encompassing the IRF1 gene using gene dosage and FISH analysis in 11 patients with a del(5q). The IRF1 protein possesses growth-inhibitory and antioncogenic activities and, thus, is a candidate tumor suppressor. Of note is that their more recent analysis of IRF1 mRNA and protein in leukemia cells with hemizygous loss of one IRF1 allele on the deleted homolog revealed significant exon skipping of IRF1 transcripts from the remaining allele, resulting in a loss of IRF1 protein in leukemia cells. Such functional loss of IRF1 could contribute to the phenotype of the disease in patients with a del(5q) (C. Willman, personal communication). We and others have found that IRF1 is proximal to, and excluded from, the commonly deleted segment of 5q; thus, the role of this gene remains unclear (9, 33).
Boultwood et al. (34) have examined three patients with the 5q- Syndrome and small deletions extending from q31 to q33 by using gene dosage and in situ hybridization analysis (Boultwood, J., Jaju, R. J., Fidler, C., Sheridan, H., Oliver, F., Walker, H., Parker, N., Jabs, E. W., McPherson, J., Muller, U., Wainscoat, J. S., and Kearney, L., unpublished work). The commonly deleted segment was an ≈3 Mb region between ADRB2 and NKSF1. This region is distal to that identified in our studies and those of Fairman et al. (31), suggesting that there is likely to be more than one region and, hence, gene involved in the pathogenesis of myeloid disorders associated with abnormalities of chromosome 5. Whether all cases of the 5q- Syndrome involve a gene in this distal region and whether this gene plays a role in the pathogenesis of AML or other subtypes of MDS are unknown.
With respect to candidate tumor suppressor genes located within the commonly deleted segment identified in our studies, we have placed two known genes, EGR1 and CDC25C, and nine ESTs within this interval. EGR1 encodes a DNA-binding zinc finger protein that is essential for the differentiation of monocytic cells; thus, this gene is a particularly good candidate for a tumor suppressor gene in myeloid leukemias (28). CDC25C encodes a phosphatase that is required for entry of the cell into mitosis via dephosphorylation and activation of CDC2 kinase activity in CDC2/cyclin B complexes (29). To evaluate the role of EGR1 and CDC25C in leukemogenesis, we examined myeloid leukemia cells with −5 or del(5q) for mutations of EGR1 or CDC25C using single-strand conformation polymorphism analysis. No mutations were detected; thus, EGR1 and CDC25C are unlikely to be involved in the pathogenesis of myeloid leukemias characterized by abnormalities of chromosome 5 (P.W.W. et al., unpublished results).
Another candidate tumor suppressor gene, MAD5, is localized proximal to our commonly deleted segment. MAD family genes encode proteins of ≈450 amino acids with highly conserved N- and C-terminal domains, and provide the link in signal transduction from the cytoplasm to the nucleus in response to TGF-β growth factors (27). Two human MAD homologs at 18q21.1 (DPC4 and JV18–1) are candidate tumor suppressor genes in pancreatic and colon carcinoma, respectively (26). Our preliminary analysis of DNA from myeloid leukemia cells revealed no mutations in the highly conserved C-terminal region of MAD5 (N.Z., J.D.E., R.A.L., and M.M.L.B., unpublished results). In conclusion, our studies delineate a 1- to 1.5-Mb region that must be searched to isolate a putative myeloid leukemia suppressor gene, and they provide the necessary cloned DNA for more detailed physical mapping and gene isolation.
Acknowledgments
We thank Elizabeth M. Davis, Anthony A. Fernald, Steven Minaglia, and Cynthia Klestinec as well as the technologists in the Hematology/Oncology Cytogenetics Laboratory for expert technical assistance, and Marjorie Isaacson for data management. We thank Drs. Kevin Shannon and Janet D. Rowley for helpful discussions. This work was supported by Public Health Service Grant P01 CA40046 (M.M.L. and R.A.L.).
ABBREVIATIONS
- AML
acute myeloid leukemia
- MDS
myelodysplastic syndrome
- t-MDS/t-AML
therapy-related MDS or AML
- PAC
P1 artificial chromosome
- BAC
bacterial artificial chromosome
- YAC
yeast artificial chromosome
- FISH
fluorescence in situ hybridization
- STS
sequence-tagged site
- EST
expressed sequence tag
- STRP
short tandem repeat polymorphism
References
- 1.Le Beau M M, Rowley J D. In: Hematology. Beutler E, Erslev A J, Lichtman M A, Coller B S, Kipps T J, editors. New York: McGraw–Hill; 1995. pp. 98–106. [Google Scholar]
- 2.Mitelman F. Catalog of Chromosome Aberrations in Cancer. 5th Ed. New York: Wiley; 1994. [Google Scholar]
- 3.Mitelman F, Kaneko Y, Berger R. In: Human Gene Mapping 1993. Cuticchia A J, Pearson P L, editors. Baltimore, MD: Johns Hopkins Univ. Press; 1994. pp. 773–812. [Google Scholar]
- 4.Rabbitts T H. Nature (London) 1994;372:143–149. doi: 10.1038/372143a0. [DOI] [PubMed] [Google Scholar]
- 5.Knudson A G. Proc Natl Acad Sci USA. 1993;90:10914–10921. doi: 10.1073/pnas.90.23.10914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levine A J. Annu Rev Biochem. 1993;62:623–651. doi: 10.1146/annurev.bi.62.070193.003203. [DOI] [PubMed] [Google Scholar]
- 7.Johansson B, Mertens F, Mitelman F. Genes Chromosomes Cancer. 1993;8:205–218. doi: 10.1002/gcc.2870080402. [DOI] [PubMed] [Google Scholar]
- 8.Pedersen-Bjergaard J, Rowley J D. Blood. 1994;83:2780–2786. [PubMed] [Google Scholar]
- 9.Le Beau M M, Espinosa R, III, Neuman W L, Stock W, Roulston D, Larson R A, Keinanen M, Westbrook C A. Proc Natl Acad Sci USA. 1993;90:5484–5488. doi: 10.1073/pnas.90.12.5484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Le Beau M M, Espinosa R, III, Davis E M, Eisenbart J D, Larson R A, Green E D. Blood. 1996;88:1930–1935. [PubMed] [Google Scholar]
- 11.Thirman M, Larson R A. Hematol Oncol Clin North America. 1996;10:293–320. doi: 10.1016/s0889-8588(05)70340-3. [DOI] [PubMed] [Google Scholar]
- 12.Smith M A, McCaffrey R P, Karp J E. J Natl Cancer Inst. 1996;88:407–418. doi: 10.1093/jnci/88.7.407. [DOI] [PubMed] [Google Scholar]
- 13.Le Beau M M, Albain K S, Larson R A, Vardiman J W, Davis E M, Blough R R, Golomb H M, Rowley J D. J Clin Oncol. 1986;4:325–345. doi: 10.1200/JCO.1986.4.3.325. [DOI] [PubMed] [Google Scholar]
- 14.Fourth International Workshop on Chromosomes in Leukemia 1982. Cancer Genet Cytogenet. 1984;11:249–360. [PubMed] [Google Scholar]
- 15.Samuels B L, Larson R A, Le Beau M M, Daly K M, Bitter M A, Vardiman J W, Barker C M, Rowley J D, Golomb H M. Leukemia. 1988;2:79–83. [PubMed] [Google Scholar]
- 16.Nimer S D, Golde D W. Blood. 1987;70:1705–1712. [PubMed] [Google Scholar]
- 17.Van den Berghe H, Vermaelen C, Mecucci C, Barbieri D, Tricot G. Cancer Genet Cytogenet. 1985;17:189–255. doi: 10.1016/0165-4608(85)90016-0. [DOI] [PubMed] [Google Scholar]
- 18.Boultwood J, Lewis S, Wainscoat J S. Blood. 1994;84:3253–3260. [PubMed] [Google Scholar]
- 19.Mitelman F. ISCN (1995): An International System for Human Cytogenetic Nomenclature. Basel: Karger; 1995. [Google Scholar]
- 20.Foote S, Munroe D J, Segre J A. In: Current Protocols in Human Genetics. Boyle A L, editor. New York: Wiley; 1994. , Chapter 5, pp. 5.0.1–5.10.23. [Google Scholar]
- 21.Stoffel M, Le Beau M M, Espinosa R, III, Bohlander S F, Le Paslier D, Cohen D, Xiang K-S, Cox N J, Fajans S S, Bell G I. Proc Natl Acad Sci USA. 1996;93:3937–3941. doi: 10.1073/pnas.93.9.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bohlander S K, Espinosa R, III, Fernald A A, Rowley J D, Le Beau M M, Diaz M O. Cytogenet Cell Genet. 1994;65:108–110. doi: 10.1159/000133612. [DOI] [PubMed] [Google Scholar]
- 23.Lengauer C, Green E D, Cremer T. Genomics. 1992;13:826–828. doi: 10.1016/0888-7543(92)90160-t. [DOI] [PubMed] [Google Scholar]
- 24.Fidlerova H, Senger G, Kost M, Sanseau P, Sheer D. Cytogenet Cell Genet. 1994;65:203–205. doi: 10.1159/000133632. [DOI] [PubMed] [Google Scholar]
- 25.McPherson J D, Le Beau M M. In: Human Gene Mapping 1993. Cuticchia A J, Pearson P L, editors. Baltimore, MD: Johns Hopkins Univ. Press; 1994. pp. 280–301. [Google Scholar]
- 26.Riggins G J, Thiagalingam S, Rozenblum E, Weinstein C L, Kern S E, Hamilton S R, Willson J K V, Markowitz S D, Kinzler K W, Vogelstein B. Nat Genet. 1996;13:347–349. doi: 10.1038/ng0796-347. [DOI] [PubMed] [Google Scholar]
- 27.Liu F, Hata A, Baker J C, Doody J, Carcamo J, Harland R M, Massague J. Nature (London) 1996;381:620–623. doi: 10.1038/381620a0. [DOI] [PubMed] [Google Scholar]
- 28.Nguyen H Q, Hoffman-Liebermann B, Liebermann D. Cell. 1993;72:197–209. doi: 10.1016/0092-8674(93)90660-i. [DOI] [PubMed] [Google Scholar]
- 29.Sadhu K, Reed S I, Richardson H, Russell P. Proc Natl Acad Sci USA. 1990;87:5139–5143. doi: 10.1073/pnas.87.13.5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schuler G D, Boguski M S, Stewart E A, Stein L D, Gyapay G, et al. Science. 1996;274:540–546. [PubMed] [Google Scholar]
- 31.Fairman J, Chumakov I, Chinault A C, Nowell P C, Nagarajan L. Proc Natl Acad Sci USA. 1995;92:7406–7410. doi: 10.1073/pnas.92.16.7406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Willman C L, Sever C E, Pallavicini M G, Harada H, Tanaka N, Slovak M L, Yamamoto H, Harada K, Meeker T C, List A F, Taniguchi T. Science. 1993;259:968–971. doi: 10.1126/science.8438156. [DOI] [PubMed] [Google Scholar]
- 33.Boultwood J, Fidler C, Lewis S, MacCarthy A, Sheridan H, Kelly S, Oscier D, Buckle V J, Wainscoat J S. Blood. 1993;82:2611–2616. [PubMed] [Google Scholar]
- 34.Boultwood J, Fidler C, Lewis S, Kelly S, Sheridan H, Littlewood T J, Buckle V J, Wainscoat J S. Genomics. 1994;19:425–432. doi: 10.1006/geno.1994.1090. [DOI] [PubMed] [Google Scholar]