Abstract
Ribonucleotide reductase (RNR) catalyzes the rate limiting step in the de novo synthesis of deoxyribonucleotides by directly reducing ribonucleotides to the corresponding deoxyribonucleotides. To keep balanced pools of deoxyribonucleotides, all nonviral RNRs studied so far are allosterically regulated. Most eukaryotes contain a class I RNR, which is a heterodimer of two nonidentical subunits called proteins R1 and R2. We have isolated cDNAs encoding the R1 and R2 proteins from Trypanosoma brucei. The amino acid sequence identities with the mouse R1 and R2 subunits are 58% and 63%, respectively. Recombinant active trypanosome R1 and R2 proteins were expressed in Escherichia coli and purified. The R2 protein contains an iron–tyrosyl free radical center verified by EPR spectroscopy and iron analyses. Measurement of cytidine 5′-diphosphate reduction by the trypanosome RNR in the presence of various allosteric effectors showed that the activity is highest with dTTP, dGTP, or dATP and considerably lower with ATP. The effect of dGTP is either activating (alone) or inhibitory (in the presence of ATP). Filter binding studies indicated that there are two classes of allosteric effector binding sites that bind ATP or dATP (low-affinity dATP site) and ATP, dATP, dGTP, or dTTP (high-affinity dATP site), respectively. Therefore, the structural organization of the allosteric sites is very similar to the mammalian RNRs, whereas the allosteric regulation of cytidine 5′-diphosphate reduction is unique. Hopefully, this difference can be used to target the trypanosome RNR for therapeutic purposes.
Sleeping sickness is a major health problem in a predominant part of sub-Saharan Africa. The disease, caused by a unicellular eukaryote called Trypanosoma brucei (1), is usually fatal if left untreated, and ≈25,000 new cases are diagnosed each year. However, since only a small fraction of the population in high risk areas is under surveillance, the World Health Organization has estimated the real figure to be ≈300,000.‡ The current chemotherapy of sleeping sickness suffers from various limitations. Many of the compounds used are highly toxic and there is also a widespread drug resistance among the trypanosomes (2, 3).
Ribonucleotide reductase (RNR) catalyzes the reduction of ribonucleotides to deoxyribonucleotides. There are three different classes of RNRs (4). The substrate, which is a nucleoside diphosphate for the class I enzymes, is reduced through a radical mechanism. Class I RNR is a heterodimeric enzyme of α2β2 type. The large R1 subunit binds substrates and allosteric effectors, while the small R2 subunit contains a tyrosyl radical (5), generated by a binuclear ferric iron center. The tyrosyl radical in the R2 protein is linked to the active site in the R1 subunit through a hydrogen-bonded long-range electron transport chain (6–10). Class II RNR generates a radical from 5′-deoxyadenosylcobalamin (11, 12) and class III RNR, which only works under anaerobic conditions, contains a stable glycyl radical (13). Class I and to some extent class III RNRs are inhibited by hydroxyurea, which acts as a specific radical scavenger. Since hydroxyurea inhibits the growth of T. brucei under aerobic conditions (14), the parasite presumably contains a class I enzyme. With the exception of Euglena gracilis (15) and Pithomyces chartarum (16), which show a 5′-deoxyadenosylcobalamin-dependent activity of ribonucleotide reduction, all eukaryotes studied so far have class I RNRs. The class I enzymes are further divided into two subclasses, which show low sequence homology but still are functionally similar. Eukaryotes, viruses, and some γ-purple bacteria (Enterobacteriaceae and Haemophilus) contain class Ia RNRs (encoded in Escherichia coli by the nrdAB genes), whereas class Ib (17, 18) (encoded in E. coli by the nrdEF genes) are found only among prokaryotes.
Except those from the Herpesviridae family (19, 20), all RNRs studied are allosterically regulated. Among the class Ia enzymes, this regulation is only studied in detail for the E. coli (21, 22), the bacteriophage T4 (23, 24), the calf thymus (25, 26), and the mouse RNRs (27–29). The mammalian enzyme has two classes of effector binding sites. The activity site binds dATP or ATP and the specificity site binds dATP, ATP, dTTP, or dGTP. The specificity site determines which substrate will be reduced. ATP or dATP stimulate reduction of cytidine 5′-diphosphate (CDP) and UDP, dTTP stimulates reduction of GDP, and dGTP gives reduction of ADP. The activity site determines the overall activity of the enzyme. With ATP bound, there is a further stimulation of the activity already determined by the specificity site. With dATP bound to the activity site, the enzyme is inactive. The affinity for dATP at the activity site is lower than at the specificity site, making dATP a positive effector for CDP reduction at low concentrations (≈2 μM) (27) and a negative effector at higher concentrations. The E. coli class Ia enzyme has the same structural organization of allosteric sites and an allosteric regulation that resembles the mammalian RNR.
Many different class I RNR inhibitors have been developed. These include radical scavengers, iron chelators, inactivators of R1 sulfhydryl groups, and nucleoside analogues (30). However, the most specific inhibitors seem to be peptides that mimic the C terminus of the R2 subunit. These peptides prevent the formation of the active R1/R2 heterodimer, since the R2 C terminus is necessary for binding to the R1 subunit (31). The specificity of peptide inhibitors was first demonstrated for a nonapeptide that inhibited the herpes simplex virus (HSV) RNR without affecting the activity of the mammalian enzyme (32, 33). This observation led to the development of potent peptidomimetic inhibitors of HSV RNR with antiviral activity in vivo (34). The inhibitory effect of R2 C-terminal peptides has also been demonstrated for mammalian (35, 36) and E. coli class Ia (37) RNRs.
Our original intentions were to investigate the possibility of using the peptide approach to specifically inhibit T. brucei proliferation without affecting the host. Therefore, we have cloned, expressed, purified, and characterized both subunits of RNR from a procyclic T. brucei cDNA library. However, the C terminus of the T. brucei R2 protein was very similar to the mammalian R2 protein, which may make it difficult to develop specific peptidomimetics against the parasite. Indeed, a mouse R2 C-terminal heptapeptide (identical in human) did inhibit the trypanosome RNR equally as well as the mouse enzyme. Instead, we found that the allosteric regulation of CDP reduction is quite different for the trypanosome RNR compared with the mammalian enzyme. Therefore, new antitrypanosome drugs may be developed, targeting the allosteric properties of the trypanosome RNR.
MATERIALS AND METHODS
PCR.
DNA from Trypanosoma congolense was a generous gift from David Steenkamp (Department of Chemical Pathology, Medical School, Cape Town, South Africa). PCR primers for both the R1 and the R2 genes were derived from eukaryotic consensus amino acid sequences. These were APMPTA and KTGMYYLRT for the R1 gene and VEGIFF and KTNFFEK for the R2 gene. All the primers were made by a 392 DNA/RNA synthesizer from Applied Biosystems. The degenerate primers are 5′-TACCGGAATTCAGTCGACAAGCNCCNATGCCNACNGC-3′ and 5′-CACGTCGACGAATTCTGNCKNARRTARTACATNCCNGTYTT-3′. Sites for SalI and EcoRI used in the subcloning are underlined. After a hot start at 97°C for 5 min without enzyme, T. congolense DNA (20 ng) was amplified for 32 cycles. Each cycle includes denaturation at 94°C for 30 sec, annealing at 42°C for 1 min, and elongation at 72°C for 2 min. The amplification was performed with 40 pmol of each primer, 1 mM MgCl2, and 2.5 units of Taq DNA polymerase from GIBCO/BRL in their recommended buffer. The single PCR product was digested with SalI and EcoRI and subcloned into pUC18. In a similar way, a T. congolense R2 gene fragment was amplified using the primers 5′-ACCGGAATTCAGTCGACAAGTNGARGGNATHTTYTT-3′ and 5-′GCAGGTCGACGAATTCTAYTTYTCRAARAARTTNGTYTT-3′. The amplification was run for 30 cycles at 94°C for 30 sec, 39°C for 1 min, and 72°C for 2 min. Another 2.5 units of fresh enzyme was added and the reaction was run for 15 more cycles. The single PCR product was digested with EcoRI and SalI. Because it happened to contain an internal EcoRI site, the digestion gave fragments of 260 bp and 170 bp. The larger fragment was subcloned into pUC18. The subcloned R1 and R2 gene fragments were sequenced from both directions using the dideoxyribonucleotide method.
Library Screening.
The cloned fragment of the R1 gene was excised, labeled with [32P]dCTP, and successfully used to screen a procyclic Trypanosoma brucei brucei 427 (TbGARP 16) λgt22 cDNA library (a generous gift from Isabel Roditi, Allgemeine Mikrobiologie der Universität, Bern, Switzerland). The R2 gene fragment was first used to screen a T. congolense IL3000 genomic λgt11 DNA library (a generous gift from Noel Murphy, International Laboratory for Research on Animal Diseases, Nairobi), since it gave a too high background on the T. brucei library. After isolating plaques containing the T. congolense R2 gene, the T. brucei library was successfully rescreened with another piece of the R2 gene. The R1 and R2 cDNAs were excised from the λ vector with NotI and SalI and subcloned into a modified pUC18, containing these sites in the polylinker. The plasmids containing the T. brucei R1 and R2 cDNAs were called pUCT1 and pUCT2, respectively. Both strands of the R1 and R2 cDNAs were sequenced using the dideoxyribonucleotide method.
Cloning of the T. brucei R1 cDNA into an Expression Vector.
A double-stranded oligonucleotide was synthesized, containing the pET3a T7 promoter sequence between the unique XbaI site and the start codon (38). This sequence was followed by the first 15 coding nucleotides of the T. brucei R1 cDNA ending in a 3′ HindIII overhang. A 5′ AatII overhang was included upstream from the XbaI site to allow cloning into the unique AatII and HindIII sites of pUCT1 (see above). The resulting plasmid was called pUCMT1. A DNA fragment containing the T7 promotor and the T. brucei R1 cDNA was excised from the pUCMT1 plasmid with XbaI and EcoRI. EcoRI cuts downstream from the stop codon of the R1 cDNA in pUCMT1. The excised DNA fragment was inserted between the unique XbaI (T7 promoter) and EcoRI sites of pETR2-Cla/R1 (8), a pET3a derivative containing the mouse R2 cDNA. EcoRI cuts 39 nucleotides upstream from the stop codon in the mouse R2 cDNA. The part of the resulting plasmid that was originally derived from the synthesized oligonucleotides was verified by dideoxyribonucleotide sequencing. The final plasmid, called pETT1, was transfected into the E. coli strain BL21(DE3)pLysS (38).
Cloning of the T. brucei R2 cDNA into an Expression Vector.
A double-stranded oligonucleotide was synthesized, containing a 5′ NcoI overhang and the first 60 coding nucleotides of the T. brucei R2 cDNA. The R2 encoding part was ended in a unique BseRI site. The BseRI site was followed by a SacI site and a 3′ XmaI overhang. The double-stranded oligonucleotide was inserted into an NcoI (start codon) and XmaI-digested pETH2, a pET3d derivative containing the HSV type 1 (HSV-1) R2 gene (39). The resulting plasmid was called pETHT2. The rest of the T. brucei R2 sequence was excised with BseRI and SacI from pUCT2 (see above). SacI cuts downstream from the stop codon in the T. brucei R2 cDNA in pUCT2. The excised fragment was inserted into pETHT2, digested with BseRI and SacI. SacI cuts downstream from the stop codon in the HSV-1 R2 sequence. The part of the resulting plasmid that was originally derived from the synthesized oligonucleotides was verified by dideoxyribonucleotide sequencing. The final plasmid, called pETT2, was transfected into BL21(DE3) (38).
Expression and Purification of Recombinant Trypanosome R1 and R2 Proteins.
The R1 protein was expressed and purified as described earlier (40). Bacteria containing the R2 protein were disintegrated in a French press and centrifuged at 45,000 rpm for 1 h (Beckman L-90 ultracentrifuge, Ti70 rotor). The supernatant was treated with streptomycin sulfate (2.5% wt/vol) and then ammonium sulfate. The material precipitating between 40% and 45% ammonium sulfate saturation (0.229–0.262 g/ml at 0°C) was dissolved and desalted on a Sephadex G-25 column and then purified on a DE52 column. A gradient from 0–200 mM KCl (buffered with 10 mM potassium phosphate, pH 7.0) was used. R2 protein, eluting between 100 and 130 mM KCl, was desalted on a Sephadex G-25 column equilibrated with 50 mM Tris⋅HCl (pH 7.6). Other details in the purification scheme are the same as for the recombinant mouse R2 protein (39). The purity of the R1 and R2 proteins was estimated by scanning a Coomassie brilliant blue stained SDS/polyacrylamide gel using a laser densitometer (Pharmacia). Protein concentrations were assessed using the extinction coefficients at 280 nm determined for calf thymus protein R1, E1%1 cm = 12.0 (26), and at 280–310 nm for recombinant mouse R2 protein, E1%1 cm = 13.8 (39).
EPR Spectroscopy.
First derivative EPR spectra were recorded with a Bruker ESP-300 X-band spectrometer equipped with an Oxford ISR9 liquid helium cryostat. A sample of 0.16 ml of R2 protein (2 mg/ml) was used for determination of microwave power saturation behavior (41) at 3.6K, 10K, and 29K. Similar results were obtained using the double integral of the EPR signal or its amplitude.
Filter Binding Assay to Measure Nucleotide Binding to R1.
[8-3H]dATP, [8-3H]dGTP, and [methyl-3H]dTTP were purchased from Amersham. They were diluted to specific activities of 250–350 cpm/pmol in solutions of unlabeled nucleotides (Pharmacia), adjusted to pH 7, and stored at −20°C. Protein R1 (5–30 μg) was incubated for 5 min in 50 mM Tris⋅HCl, 0.1 M KCl, 10 mM DTT, 6.4 mM MgCl2, and a labeled nucleotide in concentrations ranging from 0.2–4 × KD. The amount of bound and free nucleotide was determined by scintillation counting of aliquots of the solution before and after centrifugation through a membrane (42).
CDP Reducing Activity of T. brucei RNR.
The assay was essentially made as described (43). When testing the influence of the different allosteric effectors, the R2 protein was always in excess (77 pmol) over the R1 protein (22 pmol). The MgCl2 concentration was kept at 6.4 mM except when the total concentration of NTP + dNTP was more than 2 mM. After that point, 2 mol of MgCl2 were added per mol of extra nucleoside triphosphate (43). The CDP concentration was always kept at 0.5 mM. The nucleoside triphosphates were either ultrapure dNTP or rNTP sets from Pharmacia or standard nucleotides from the same company. No significant difference could be detected between the two sources of nucleotides. Tritiated CDP was purchased from Amersham and diluted with unlabeled CDP from Sigma to a specific activity of 20 cpm/μmol.
RESULTS
Cloning and Sequencing of T. brucei R1 and R2 cDNAs.
DNA fragments encoding a part of the R1 and R2 proteins were isolated by PCR from genomic T. congolense DNA, using primers to highly conserved regions of other eukaryotic R1 and R2 proteins. The entire coding region of the R1 cDNA was obtained from a procyclic T. brucei cDNA library, using the isolated T. congolense fragment as a probe. The R1 cDNA (GenBank accession no. U80910; 3,540 bp) contained an open reading frame of 2,514 bp, which when translated gives a predicted protein of 838 amino acids with an estimated molecular weight of 94,630. The entire coding region of the R2 cDNA was isolated by first screening a T. congolense genomic DNA library with the R2 PCR fragment and then using another part of the T. congolense R2 gene to obtain an R2 clone from the T. brucei cDNA library. The R2 cDNA (GenBank accession no. U80911; 1,695 bp) contained an open reading frame of 1,011 bp, which when translated gives a predicted protein of 337 amino acids with an estimated molecular weight of 39,013.
Sequence Comparison of the T. brucei R2 Protein with R2 Proteins from Other Species.
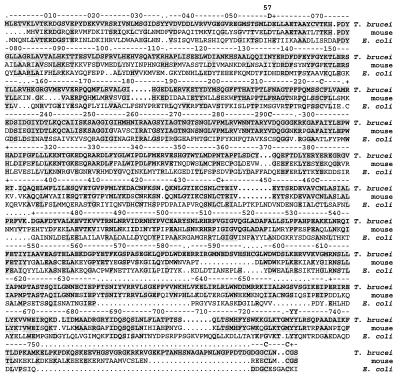
The T. brucei R2 protein shows 63% amino acid sequence identity to the mouse R2 protein (44) and 17% identity to the E. coli R2 protein (45) (Fig. 1). The iron ligands (Asp-84, Glu-115, His-118, Glu-204, Glu-238, His-241) (6, 46), the tyrosyl free radical (Tyr-122) (5, 6, 46), the residues in the hydrophobic pocket surrounding the radical (Phe-208, Phe-212, Ile-234) (6, 46), and the residues that are proposed to participate in the long-range electron transport pathway (Trp-48, Asp-237, Glu-350, Tyr-356) (6, 8, 9, 46), are all conserved (E. coli class Ia numbering is used throughout the text unless stated otherwise). A striking difference between the mouse and the T. brucei R2 proteins is that the mouse R2 protein is larger due to 53 extra residues in the N terminus. This is probably a highly flexible part of the protein, since it cannot be detected in the crystal structure (47). Furthermore, it does not seem to be essential for catalytic activity (39). The C-terminal seven residues, which were shown to be essential for binding between the R1 and R2 proteins in the mouse RNR (35, 36), show high similarity between the trypanosome and mouse R2 proteins. The only difference is that Thr-385 (mouse R2 numbering) is replaced by a serine in the T. brucei R2 protein.
Figure 1.
Comparison between the T. brucei, the mouse (44), and the E. coli (45) class Ia R2 proteins. The ruler follows the conventionally used numbering of the E. coli R2 protein (45). Breaks in alignments are indicated with dots; breaks in the E. coli R2 protein sequence are indicated as dots in the ruler as well. Unless written out, every tenth residue is indicated as a cross in the ruler. Shaded residues are identical to the T. brucei R2 protein amino acid sequence. Residues mentioned in the text are indicated as the one-letter abbreviation. These are the iron ligands (D-84, E-115, H-118, E-204, E-238, H-241), the tyrosyl radical (Y-122), the residues in the hydrophobic pocket surrounding the radical (F-208, F-212, I-234), and the electron transport pathway residues (W-48, D-237, E-350, Y-356). E. coli R2 protein amino acid numbering is used. The part of the C terminus in the mouse R2 protein important for binding to the R1 subunit is indicated with five-point stars.
Sequence Comparison of T. brucei. R1 Protein with Other Cloned R1 Proteins.
The T. brucei R1 protein shows 58% amino acid sequence identity to the mouse R1 protein (48) and 22% identity to the E. coli R1 protein (45) (Fig. 2). The active site cysteines (Cys-225, Cys-439, Cys-462) (7, 49, 50), the proposed electron transport pathway (Tyr-730, Tyr-731) (7, 10), and the two cysteines that transfer electrons between thioredoxin or glutaredoxin and the active site sulfhydryl groups (Cys-754, Cys-759) (7, 49, 50) are conserved in the trypanosome R1 subunit.
Figure 2.
Alignment of the T. brucei R1 protein with the mouse (48) and E. coli (45) class Ia R1 proteins. The ruler follows the conventionally used numbering of the E. coli R1 protein (45). Breaks in the alignments are indicated with dots; breaks in the E. coli R1 sequence are indicated as dots in the ruler as well. Unless written out, every tenth residue is indicated as a cross in the ruler. Shaded residues are identical to the T. brucei R1 protein amino acid sequence. Residues mentioned in the text are indicated as the one-letter abbreviation. These are the active site cysteines (C-225, C-439, C-462), the residues in the electron transport pathway (Y-730, Y731), the cysteines that shuttle electrons from thioredoxin or glutaredoxin (C-754, C-759), and the allosteric effector binding-site residues (D-57, C-292). Mouse R1 protein amino acid numbering is used for the allosteric activity site residue (D-57). Otherwise, E. coli R1 protein amino acid numbering is used.
Expression and Purification.
R1 and R2 cDNAs were cloned into pET vectors, expressed in E. coli, and purified. The purification schemes are very similar to the ones used for mouse R1 (40) and R2 (39). The final yields were 2 mg R1 protein per liter of culture in Terrific broth and 3 mg R2 protein per liter of culture in Luria broth. The R1 protein had an estimated purity of 80–90% and the R2 protein a purity of 85–90%.
EPR Signal.
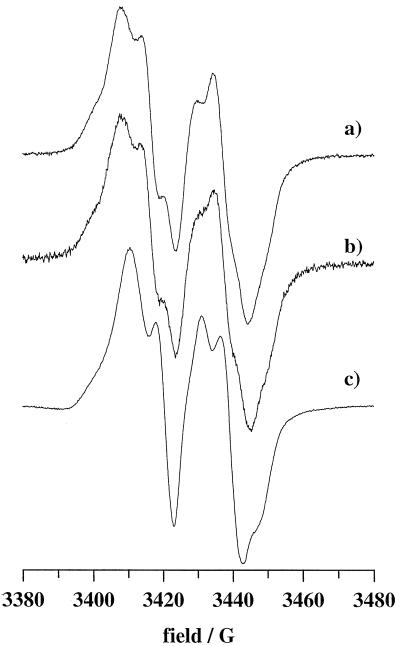
An EPR signal was recorded at 14K for the purified trypanosome R2 protein. The lineshape of this signal is very similar to that seen in mouse but different from the one in E. coli (Fig. 3). This proves the presence of a tyrosyl radical also in the trypanosome RNR and indicates a similar environment for the radicals in the R2 proteins of T. brucei and mouse. The protein had not been reconstituted with iron prior to the EPR measurement. Still, it contained 0.28 tyrosyl radical per dimeric R2 protein which was estimated from the double integral of the EPR signal compared with a standard sample of 1 mM CuClO4. A colorimetric ferroin assay (51) showed that the same sample contained 1.4 iron atoms per R2 dimer indicating loss of radical during the purification of the R2 protein.
Figure 3.
First derivative EPR spectra of the stable tyrosyl free radical in recombinantly expressed R2 protein from mouse at 10K (a), T. brucei at 14K (b), and E. coli at 4K (c). The concentrations of R2 protein dimer were 20 μM (24 μM free radical), 25 μM (7 μM free radical), and 350 μM (370 μM free radical), respectively. All spectra were recorded at 9.62 GHz (X-band) under nonsaturating microwave power conditions, 3G modulation amplitude, and 100 kHz modulation frequency.
The microwave power saturation behavior of the trypanosome R2 protein, evaluated as the microwave power at half saturation P1/2 (41), is similar but not identical to that in mouse. At 3.6K, 10K, and 29K the P1/2 values were 21 μW, 42 μW, and 13 mW for the trypanosome R2 protein and 29 μW, 680 μW, and 3 mW for the mouse R2 protein, respectively. The difference at 3.6K is within the error range and therefore the iron center should be in its diamagnetic groundstate in T. brucei comparable of that in the mouse R2 protein (52). The differences seen between the two proteins at 10K and 29K are larger than the experimental uncertainty, indicating a species-specific magnetic interaction between the radical and the iron center at higher temperatures (53).
Enzyme Activity.
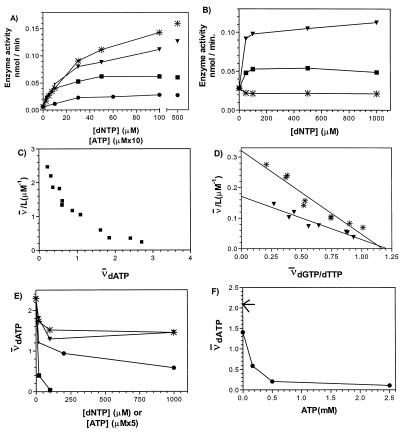
CDP reduction was first measured in the presence of 3 mM ATP as a positive effector. The assay was linear with time for at least 30 min at 37°C . The specific activities were 120 units/mg for the T. brucei R2 protein and 20 units/mg for the T. brucei R1 protein. The corresponding specific activities for the mouse RNR are 120–140 units/mg for the R1 protein (40) and 280 units/mg for the R2 protein (39). The low specific activity of the trypanosome RNR compared with the mammalian enzyme urged us to investigate if ATP really is the optimal allosteric effector for CDP reduction. To our surprise, we found that dATP, dGTP, and dTTP all induced higher activities than ATP (Fig. 4A). This is in strong contrast to the mammalian enzyme, where these three deoxyribonucleotides are inhibitory for CDP reduction.
Figure 4.
Effects of nucleoside triphosphates on CDP reduction catalyzed by the trypanosome RNR (A and B) and binding of nucleoside triphosphate effectors to the T. brucei R1 protein (C–F). (A) Enzyme activity in the presence of dGTP (∗), dTTP (▾), dATP (▪), or ATP (•). Note that the scale on the abscissa is different for ATP compared with the other effectors. (B) Enzyme activity in the presence of a fixed concentration of ATP (1 mM) and varying concentrations of dTTP (▾), dATP (▪), or dGTP (∗). (C) Scatchard plots of dATP binding at 4°C. A similarly shaped curve with lower affinity was obtained at 25°C (not shown). (D) Scatchard plots of dGTP (∗ and dTTP (▾) binding at 25°C. The dissociation constants (KD) as determined from the Scatchard plots were 3.7 and 7.1 μM, respectively. The corresponding values at 4°C were 2.0 μM (dGTP) and 3.7 μM (dTTP). L is the ligand concentration in micromolar and ν̄ is the average number of ligand molecules bound per R1 protein dimer (2 × 95 kDa). (E) Binding of 6.4 μM tritiated dATP to trypanosome R1 protein at 4°C in the presence of increasing concentrations of unlabeled dGTP (∗), dTTP (▾), ATP (•), or dATP (▪). Note that the scale on the abscissa is different for ATP compared with the dNTPs. (F) Binding of tritiated 6.4 μM dATP to trypanosome R1 protein at 4°C in the presence of increasing concentrations of unlabeled ATP and a constant amount of unlabeled dGTP (400 μM). Arrow indicates the amount of tritiated dATP bound to the R1 protein when neither cold ATP nor dGTP is added.
Characterization of Allosteric Effector Binding Sites.
In the mammalian and the E. coli class Ia RNRs, dATP inhibits enzyme activity for all substrates when bound to the activity site (low-affinity dATP site), whereas it stimulates CDP reduction when bound to the specificity site (high-affinity dATP site). Since dATP even at high concentrations activated CDP reduction by the trypanosome RNR, we wanted to determine if this enzyme really contained two classes of allosteric effector binding sites. Fig. 4 C–F presents results from filter binding assays using trypanosome R1 protein and labeled nucleotides. From the Scatchard plots, it appears that the trypanosome RNR also contains at least two classes of binding sites showing different affinities for dATP (Fig. 4C) and one class of dGTP/dTTP binding sites (Fig. 4D). The amount of nucleotide bound per R1 dimer was somewhat low, possibly due to impurities (10–20%) or instability of the R1 protein. However, we interpret our data in such a way that there are four sites for dATP and two sites for dGTP or dTTP per R1 dimer. As compared with the recombinantly expressed E. coli R1 protein (42), the affinities for the nucleotides are considerably lower for the trypanosome R1 protein and the temperature dependence of the dissociation constants less pronounced (Fig. 4D).
To further characterize the effector binding sites, we measured the ability of unlabeled nucleotides to compete with 6.4 μM tritiated dATP for binding to the trypanosome R1 protein (Fig. 4E). ATP could compete out 75% of the label, whereas dTTP or dGTP competed out less than 50% of the label. This supports the presence of two different classes of dATP binding sites in the R1 protein, where dTTP/dGTP competes with only one class, while ATP competes with both. The model is further strengthened by the finding that in the presence of 400 μM unlabeled dGTP, ATP could compete out most of the labeled dATP bound to the R1 protein (Fig. 4F). Finally, to verify that both dTTP and ATP can bind to the dGTP binding sites, an experiment was performed where 400 μM dTTP chased 95% and 5 mM ATP chased 35% of the labeled dGTP (3.5 μM) bound to the R1 protein.
Functional Interaction Between the Two Classes of Allosteric Effector Binding Sites.
The filter binding assay indicated that the trypanosome R1 protein indeed has two classes of allosteric effector binding sites. Still, dATP did not inhibit the trypanosome RNR catalyzed CDP reduction and therefore we wanted to investigate the function of the low-affinity dATP binding site. The problem was addressed by measuring CDP reduction in the presence of ATP or dATP at a fixed concentration high enough to saturate all allosteric sites. Then, increasing concentrations of competing nucleotides were added. Because dGTP and dTTP compete for only one class of allosteric sites, the plateau of the activity curve can be interpreted as the CDP reduction obtained when the low-affinity dATP effector binding site is occupied with ATP (Fig. 4B) or dATP (data not shown) and the specificity site (high-affinity dATP effector binding site) is occupied by dGTP or dTTP. The ATP concentration was chosen to be 1 mM, since at this concentration the activity curve with ATP alone reaches a plateau (Fig. 4A). The concentration of dATP was chosen to be 100 μM, which should easily saturate all allosteric sites as evaluated from the Scatchard plot at 25°C (data not shown). Using this approach, we found that the combination of dGTP and ATP was slightly inhibitory compared with ATP alone (Fig. 4B). This is in contrast to the stimulatory effect when dGTP was used by itself (Fig. 4A). While dGTP acted differently whether ATP was present or not, dTTP in combination with ATP gave a plateau value very similar to the one with dTTP alone (Fig. 4A).
When dATP was combined with dGTP or dTTP no obvious combination effect was observed, since the curves reached very similar plateau values (data not shown), as with dGTP or dTTP alone (Fig. 4A). As dATP was shown to bind to the same allosteric sites as ATP in the filter binding assay, it is reasonable to assume that the ratio of [dATP]/[ATP] will determine whether dGTP is a positive or negative effector for CDP reduction.
The ability of dCTP to function as an allosteric effector was also tested. However, it had no effect on CDP reduction regardless of whether ATP was present or not (data not shown).
Peptide Inhibition and Lack of Cross Reactivity with the Host RNR.
To evaluate the importance for subunit interaction of the Thr to Ser exchange in the C terminus of the trypanosome R2 protein, we determined the IC50 of a heptapeptide corresponding to the mouse R2 C terminus for the mouse and the T. brucei RNRs. The IC50 of the peptide was 70 μM for both the mouse and the T. brucei RNR using equimolar concentrations of each subunit (0.44 μM). The identical IC50 values raised the question of whether the mouse R1 protein could form an active complex with the trypanosome R2 protein and vice versa. T. brucei has for a long time been regarded as a strictly extracellular parasite (2). However, intracellular forms have recently been found in the choroid plexus, which is where the invasion of the central nervous system begins (54). When tested, interspecies mixtures of R1 and R2 proteins were totally inactive in contrast to the positive controls where proteins from the same species were mixed (data not shown).
DISCUSSION.
The trypanosome RNR clearly belongs to the class Ia RNRs. This is evident from the amino acid sequence homology to the mouse RNR, the presence of an iron–tyrosyl free radical center and the importance of the R2 protein carboxyl-terminal end for binding to the R1 protein. However, the allosteric regulation of CDP reduction is quite different from the mammalian or E. coli class Ia RNRs. The inability of dATP to inhibit CDP reduction catalyzed by the trypanosome RNR is similar to the situation for the Herpesviridae family class Ia (19, 20), bacteriophage T4 class Ia (23, 24), Salmonella typhimurium class Ib (55), and Lactobacillus leichmannii class II (56) RNRs. The two last-named enzymes have been shown to contain only one class of allosteric site (55, 57) similar to the specificity site in the class I enzymes, whereas the Herpesviridae RNRs seem to completely lack allosteric regulation (19, 20). The bacteriophage T4 enzyme is believed to have two classes of allosteric sites (58), but dATP/ATP does not seem to influence the catalytic activity (23, 24). Conversely, we observed that dGTP plus ATP give lower activity than either effector alone in the trypanosome RNR, which indicates that the two different classes of effector binding sites communicate to determine the activity of the enzyme. This resembles the earlier observation that for the E. coli class Ia RNR, ATP plus dTTP together give a considerably lower pyrimidine nucleotide reducing activity than either effector alone (22).
The functional consequences of binding allosteric effectors to the specificity site also seem to be quite unique for the trypanosome enzyme. This RNR reduces CDP more efficiently in the presence of dTTP or dGTP than in the presence of ATP or dATP, which are the best allosteric effectors for CDP reduction in the case of the other class Ia nonviral RNRs. Regardless of class, neither dGTP nor dTTP were ever found to be the optimal effectors for CDP reduction (19–29, 55, 56, 59).
The parts of the amino acid sequence of the R1 protein responsible for the allosteric regulation are not yet fully defined. However, a G-to-A transition at codon 57, changing an Asp to an Asn, made the mouse R1 protein resistant to feedback regulation by dATP (29). This indicates that the activity site is located in the amino-terminal region of the R1 protein close to Asp-57. The aspartic acid residue is conserved in the trypanosome R1 protein (Fig. 2). The substrate specificity site was located to the region around Cys-292 by photoaffinity labeling (60). Recently, binding of dTTP close to this residue was observed in E. coli R1 protein crystals (U. Uhlin, personal communication). The amino acid residues immediately downstream from the position corresponding to Cys-292, DQGGNKRPG, are conserved in human (61), mouse (48), Caenorhabditis elegans (GenBank accession no. Z19158), Schizosaccharomyces pombe (62), and Saccharomyces cerevisiae (RNR1, GenBank accession no. U18813 and RNR3, GenBank accession no. Z38060) R1 proteins. The only eukaryotic R1 proteins known to differ in this amino acid sequence are those from T. brucei (Fig. 2) and Plasmodium falciparum (the malaria parasite) (63), which instead contain the sequence DQGGGKRKG. This sequence identity in these two otherwise unrelated protozoans is interesting from an evolutionary standpoint.
Clearly, more data are needed for a full understanding of the allosteric regulation of the trypanosome enzyme and work is in progress to study the regulation of the reduction of UDP, GDP, and ADP. Many nucleotide analogues have been developed acting as suicide substrates for RNR (30). Their efficiencies are heavily influenced by the presence of a certain allosteric effector (64). Hopefully, a combination of a suicide substrate with a specific allosteric effector may be much more harmful to the trypanosome RNR than to the host enzyme.
Acknowledgments
We would like to thank Dr. David Steenkamp, Dr. Isabel Roditi, and Dr. Noel Murphy for supplying us with trypanosome DNA and DNA libraries. This work was supported by grants from the Swedish Natural Science Research Council and the Kempe Foundation.
ABBREVIATIONS
- RNR
ribonucleotide reductase
- CDP
cytidine 5′-diphosphate
- HSV
herpes simplex virus
Footnotes
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. U80910 and U80911).
World Health Organisation, Division of Control of Tropical Diseases. www.who.ch/programmes/ctd/diseases/slee/sleemain.htm
References
- 1.Hajduk S L, Englund P T, Smith D H. In: Tropical and Geographical Medicine. Warren K S, Mahmoud A A F, editors. New York: McGraw–Hill; 1990. pp. 268–281. [Google Scholar]
- 2.Poltera A A. Br Med Bull. 1985;41:169–174. doi: 10.1093/oxfordjournals.bmb.a072045. [DOI] [PubMed] [Google Scholar]
- 3.Hajduk S, Adler B, Bertrand K, Fearon K, Hager K, Hancock K, Harris M, le Blanc A, Moore R, Pollard V. Am J Med Sci. 1992;303:258–270. doi: 10.1097/00000441-199204000-00011. [DOI] [PubMed] [Google Scholar]
- 4.Reichard P. Science. 1993;260:1773–1777. doi: 10.1126/science.8511586. [DOI] [PubMed] [Google Scholar]
- 5.Larsson Å, Sjöberg B M. EMBO J. 1986;5:2037–2040. doi: 10.1002/j.1460-2075.1986.tb04461.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nordlund P, Sjöberg B-M, Eklund H. Nature (London) 1990;345:593–598. doi: 10.1038/345593a0. [DOI] [PubMed] [Google Scholar]
- 7.Uhlin U, Eklund H. Nature (London) 1994;370:533–539. doi: 10.1038/370533a0. [DOI] [PubMed] [Google Scholar]
- 8.Rova U, Goodtzova K, Ingemarson R, Behravan G, Gräslund A, Thelander L. Biochemistry. 1995;34:4267–4275. doi: 10.1021/bi00013a016. [DOI] [PubMed] [Google Scholar]
- 9.Climent I, Sjöberg B-M, Huang C K. Biochemistry. 1992;31:4801–4807. doi: 10.1021/bi00135a009. [DOI] [PubMed] [Google Scholar]
- 10.Ekberg M, Sahlin M, Eriksson M, Sjöberg B-M. J Biol Chem. 1996;271:20655–20659. doi: 10.1074/jbc.271.34.20655. [DOI] [PubMed] [Google Scholar]
- 11.Tamao Y, Blakley R L. Biochemistry. 1973;12:24–34. doi: 10.1021/bi00725a005. [DOI] [PubMed] [Google Scholar]
- 12.Orme-Johnson W H, Beinert H, Blakley R L. J Biol Chem. 1974;249:2338–2343. [PubMed] [Google Scholar]
- 13.Sun X, Harder J, Krook M, Jörnvall H, Sjöberg B-M, Reichard P. Proc Natl Acad Sci USA. 1993;90:577–581. doi: 10.1073/pnas.90.2.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brun R. J Protozool. 1980;27:122–128. doi: 10.1111/j.1550-7408.1980.tb04241.x. [DOI] [PubMed] [Google Scholar]
- 15.Gleason F K, Hogenkamp H P C. J Biol Chem. 1970;245:4894–4899. [PubMed] [Google Scholar]
- 16.Stutzenberger F. J Gen Microbiol. 1973;81:501–503. doi: 10.1099/00221287-81-2-501. [DOI] [PubMed] [Google Scholar]
- 17.Jordan A, Gibert I, Barbé J. J Bacteriol. 1994;176:3420–3427. doi: 10.1128/jb.176.11.3420-3427.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jordan A, Pontis E, Atta M, Krook M, Gibert I, Barbé J, Reichard P. Proc Natl Acad Sci USA. 1994;91:12892–12896. doi: 10.1073/pnas.91.26.12892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lankinen H, Gräslund A, Thelander L. J Virol. 1982;41:893–900. doi: 10.1128/jvi.41.3.893-900.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Averett D R, Lubbers C, Elion G B, Spector T. J Biol Chem. 1983;258:9831–9838. [PubMed] [Google Scholar]
- 21.Larsson Å, Riechard P. J Biol Chem. 1966;241:2533–2549. [PubMed] [Google Scholar]
- 22.Brown N C, Reichard P. J Mol Biol. 1969;46:25–55. doi: 10.1016/0022-2836(69)90055-2. [DOI] [PubMed] [Google Scholar]
- 23.Berglund O. J Biol Chem. 1972;247:7276–7281. [PubMed] [Google Scholar]
- 24.Berglund O. J Biol Chem. 1975;250:7450–7455. [PubMed] [Google Scholar]
- 25.Eriksson S, Thelander L, Åkerman M. Biochemistry. 1979;18:2948–2952. doi: 10.1021/bi00581a005. [DOI] [PubMed] [Google Scholar]
- 26.Thelander L, Eriksson S, Åkerman M. J Biol Chem. 1980;255:7426–7432. [PubMed] [Google Scholar]
- 27.Eriksson S, Gudas L J, Clift S M, Caras I W, Ullman B, Martin D W., Jr J Biol Chem. 1981;256:10193–10197. [PubMed] [Google Scholar]
- 28.Ullman B, Gudas L J, Caras I W, Eriksson S, Weinberg G L, Wormsted M A, Martin D W., Jr J Biol Chem. 1981;256:10189–10192. [PubMed] [Google Scholar]
- 29.Caras I W, Martin D W., Jr Mol Cell Biol. 1988;8:2698–2704. doi: 10.1128/mcb.8.7.2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nocentini G. Crit Rev Oncol Hematol. 1996;22:89–126. doi: 10.1016/1040-8428(95)00187-5. [DOI] [PubMed] [Google Scholar]
- 31.Lycksell P-O, Ingemarson R, Davis R, Gräslund A, Thelander L. Biochemistry. 1994;33:2838–2842. doi: 10.1021/bi00176a013. [DOI] [PubMed] [Google Scholar]
- 32.Cohen E A, Gaudreau P, Brazeau P, Langelier Y. Nature (London) 1986;321:441–443. doi: 10.1038/321441a0. [DOI] [PubMed] [Google Scholar]
- 33.Dutia B M, Frame M C, Subak-Sharpe J H, Clark W N, Marsden H S. Nature (London) 1986;321:439–441. doi: 10.1038/321439a0. [DOI] [PubMed] [Google Scholar]
- 34.Liuzzi M, Dèziel R, Moss N, Beaulieu P, Bonneau A-M, Bosquet C, Chafouleas J G, Garneau M, Jaramillo J, Krogsrud R L, Lagacé L, McCollum R S, Nawoot S, Guindon Y. Nature (London) 1994;372:695–697. doi: 10.1038/372695a0. [DOI] [PubMed] [Google Scholar]
- 35.Yang F D, Spanevello R A, Celiker I, Hirschmann R, Rubin H, Cooperman B S. FEBS Lett. 1990;272:61–64. doi: 10.1016/0014-5793(90)80449-s. [DOI] [PubMed] [Google Scholar]
- 36.Consentino G, Lavallée P, Rakhit S, Plante R, Gaudette Y, Lawets C, Whitehead P W, Duceppe J S, Lépine- Frenette C, Dansereau N, Guilbalt C, Langelier Y, Gaudreau P, Thelander L, Guindon Y. Biochem Cell Biol. 1991;69:79–83. doi: 10.1139/o91-011. [DOI] [PubMed] [Google Scholar]
- 37.Climent I, Sjöberg B-M. Biochemistry. 1991;30:5164–5171. doi: 10.1021/bi00235a008. [DOI] [PubMed] [Google Scholar]
- 38.Studier F W, Rosenberg A H, Dunn J J, Dubendorff J W. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 39.Mann G J, Gräslund A, Ochiai E-I, Ingemarson R, Thelander L. Biochemistry. 1991;30:1939–1947. doi: 10.1021/bi00221a030. [DOI] [PubMed] [Google Scholar]
- 40.Davis R, Thelander M, Mann G J, Behravan G, Soucy F, Beaulieu P, Lavallée P, Gräslund A, Thelander L. J Biol Chem. 1994;269:23171–23176. [PubMed] [Google Scholar]
- 41.Hales B J. Methods Enzymol. 1993;227:384–395. [Google Scholar]
- 42.Ormö M, Sjöberg B-M. Anal Biochem. 1990;189:138–141. doi: 10.1016/0003-2697(90)90059-i. [DOI] [PubMed] [Google Scholar]
- 43.Engström Y, Eriksson S, Thelander L, Åkerman M. Biochemistry. 1979;18:2941–2948. doi: 10.1021/bi00581a004. [DOI] [PubMed] [Google Scholar]
- 44.Thelander L, Berg P. Mol Cell Biol. 1986;6:3433–3442. doi: 10.1128/mcb.6.10.3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carlson J, Fuchs J A, Messing J. Proc Natl Acad Sci USA. 1984;81:4294–4297. doi: 10.1073/pnas.81.14.4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nordlund P, Eklund H. J Mol Biol. 1993;232:123–164. doi: 10.1006/jmbi.1993.1374. [DOI] [PubMed] [Google Scholar]
- 47.Kauppi B, Nielsen B A, Ramaswamy S, Larssen I K, Thelander M, Thelander L, Eklund H. J Mol Biol. 1996;262:706–720. doi: 10.1006/jmbi.1996.0546. [DOI] [PubMed] [Google Scholar]
- 48.Caras I W, Levinson B B, Fabry M, Williams S R, Martin D W., Jr J Biol Chem. 1985;260:7015–7022. [PubMed] [Google Scholar]
- 49.Mao S S, Holler T P, Yu G X, Bollinger J-M, Jr, Booker S, Johnston M I, Stubbe J. Biochemistry. 1992;31:9733–9743. doi: 10.1021/bi00155a029. [DOI] [PubMed] [Google Scholar]
- 50.Åberg A, Hahne S, Karlsson M, Larsson Å, Ormö M, Åhgren A, Sjöberg B-M. J Biol Chem. 1989;264:12249–12252. [PubMed] [Google Scholar]
- 51.Atkin C L, Thelander L, Reichard P, Lang G. J Biol Chem. 1973;248:7464–7472. [PubMed] [Google Scholar]
- 52.Galli C, Atta M, Andersson K K, Gräslund A, Brudvig G W. J Am Chem Soc. 1995;117:740–746. [Google Scholar]
- 53.Sahlin M, Gräslund A, Ehrenberg A. J Magn Reson. 1986;67:135–137. [Google Scholar]
- 54.Pentreath V W, Baugh P J, Lavin D R. Onderstepoort J Vet Res. 1994;61:369–377. [PubMed] [Google Scholar]
- 55.Eliasson R, Pontis E, Jordan A, Reichard P. J Biol Chem. 1996;271:26582–26587. doi: 10.1074/jbc.271.43.26582. [DOI] [PubMed] [Google Scholar]
- 56.Beck W S. J Biol Chem. 1967;242:3148–3158. [PubMed] [Google Scholar]
- 57.Chen A K, Bhan A, Hopper S, Abrams R, Franzen J S. Biochemistry. 1974;13:654–661. doi: 10.1021/bi00701a004. [DOI] [PubMed] [Google Scholar]
- 58.Berglund O, Eckstein F. Eur J Biochem. 1972;28:492–496. doi: 10.1111/j.1432-1033.1972.tb01936.x. [DOI] [PubMed] [Google Scholar]
- 59.Eliasson R, Pontis E, Sun X, Reichard P. J Biol Chem. 1994;269:26052–26057. [PubMed] [Google Scholar]
- 60.Eriksson S, Sjöberg B-M, Jörnvall H, Carlquist M. J Biol Chem. 1986;261:1878–1882. [PubMed] [Google Scholar]
- 61.Parker N J, Begley C G, Fox R M. Nucleic Acids Res. 1991;19:3741. doi: 10.1093/nar/19.13.3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fernandez-Sarabia M J, McInerny C, Harris P, Gordon C, Fantes P. Mol Gen Genet. 1993;238:241–251. doi: 10.1007/BF00279553. [DOI] [PubMed] [Google Scholar]
- 63.Rubin H, Salem J S, Li L-S, Yang F-D, Mama S, Wang Z-M, Fisher A, Hamann C S, Cooperman B S. Proc Natl Acad Sci USA. 1993;90:9280–9284. doi: 10.1073/pnas.90.20.9280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thelander L, Larsson B. J Biol Chem. 1976;251:1398–1405. [PubMed] [Google Scholar]