Abstract
Enzymes that operate on nucleic acid substrates are faced with the unusual situation where the substrate is much larger than themselves. Despite the potential to promote catalysis by utilizing the significant binding energy available through their interaction with substrate, ATP hydrolysis is frequently a part of the mechanism of these enzymes. The reasons for this have become clearer in recent years, and a surprising range of ways that these enzymes utilize the free energy of hydrolysis of ATP has been revealed. This review describes these different mechanisms in the context of the biochemical reactions that they support.
Keywords: ATP hydrolysis/binding energy/catalysis/nucleic acid modifying enzymes
Introduction
Enzymes promote catalysis by lowering the activation energy of the reactions that they catalyse. Although many enzymes provide chemical means such as acid/base catalysis to speed up reactions, others are able to promote catalysis by binding energy alone (Haldane, 1930). A classic example of this mechanism is the formation of tyrosyl-adenylate by tyrosyl-tRNA synthetase. Work from the Fersht laboratory over several years (reviewed in Fersht et al., 1986) has revealed how the enzyme can reduce the activation energy of the reaction simply by binding tightly to the transition state. Many other enzymes have also been shown to be able to utilize binding energy in this way. Perhaps even more striking are studies that show how antibody molecules raised against transition state analogues are effective catalysts (Schultz and Lerner, 1995). These studies demonstrate that the principle is widely applicable and that stabilization of the transition state can be a very effective way to produce a catalyst. A natural corollary of this proposal is that enzymes with large substrates (and therefore a greater potential binding energy) should be able to exploit binding energy to catalyse reactions. There are few enzymes whose substrates are larger than that of DNA modifying enzymes, and one might conclude that these enzymes should be very effective at utilizing binding energy. In fact, there are surprisingly few examples of DNA modifying enzymes utilizing binding energy to promote catalysis. One elegant and well-studied exception is the EcoRV restriction endonuclease (Vipond and Halford, 1993). In this case, binding energy is used to discriminate between cognate and non-cognate sequences by catalysis rather than binding, thereby achieving the exquisite specificity that is a hallmark of these enzymes. Another recent example is uracil DNA glycosylase (Jiang et al., 2003), where binding energy is used not only to stabilize the transition state but also in the binding of substrate. However, these examples remain exceptions rather than the general rule.
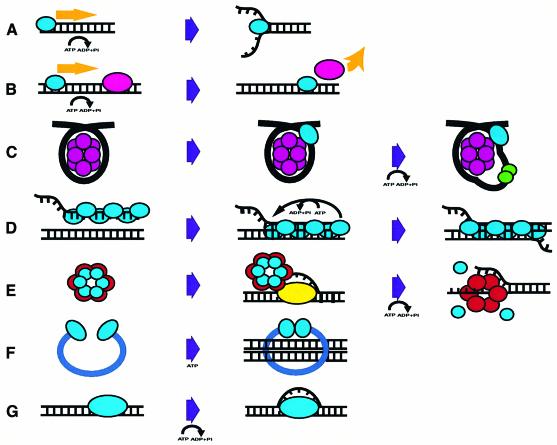
The observation that many DNA modifying enzymes hydrolyse ATP, despite catalysing reactions that are energetically favourable, leaves us with a conundrum. We are taught that evolution drives cells to utilize ATP by the most economical means, yet these enzymes appear to be wasting ATP. How can this apparent contradiction be explained? Of course, cells are not wasting ATP but are in fact using the free energy of ATP hydrolysis in a number of cunning ways to control reactions, thereby maintaining genomic integrity. Recent work has revealed how some of these systems operate, and this review sets out our present understanding of these processes (Figure 1).
Fig. 1. Examples of ATP-mediated reactions on nucleic acid substrates. (A) A helicase (blue) unwinds base-paired strands of DNA or RNA. (B) A motor protein (blue) dissociates another protein (pink) from the nucleic acid substrate. (C) A chromatin remodelling factor (blue) releases DNA from the nucleosome (purple) to allow the access of other factors (green). (D) RecA protein (blue) may be recycled on the same substrate with concomitant hydrolysis of ATP. (E) DnaB helicase (red) is bound to DnaC (blue). After recruitment to the origin-bound DnaA (yellow), ATP is hydrolysed to allow loading of the helicase onto single-stranded DNA. (F) Two SMC (Structural Maintenance of Chromosomes) head domains (blue) dimerize after binding ATP. (G) Origin unwinding is achieved by the origin recognition complex (ORC) complex (blue) hydrolysing ATP. More detailed descriptions of the mechanisms are given in the text.
Direct chemical coupling
Perhaps the simplest way that the free energy of hydrolysis of ATP can be utilized by enzymes is by direct chemical coupling. There are many examples of this mechanism in other areas of biochemistry (e.g. glycolysis), but surprisingly few in nucleic acid biochemistry. DNA (and RNA) ligases cleave ATP to AMP and PPi but trap the chemical energy directly via a covalent linkage of the AMP to a lysine side chain in the active site (Engler and Richardson, 1982). This ‘activated enzyme’ then transfers the AMP moiety to the free 5′-hydroxyl group on the nucleic acid substrate. Finally, the enzyme catalyses phosphodiester bond formation in the substrate with the concomitant release of free AMP. Thus, the free energy of hydrolysis of ATP is coupled directly via chemical means to the formation of a new phosphodiester bond. A similar mechanism is adopted by mRNA capping enzymes (Shuman and Hurwitz, 1981) but with GTP as co-factor to form the covalently ‘capped’ GMP-mRNA adduct.
Aminoacyl-tRNA synthetases also utilize ATP hydrolysis in a direct coupling mechanism to create an activated aminoacyl adenylate intermediate (also an AMP adduct), thus facilitating the formation of the final amino acid charged tRNA product. In this case, however, the transfer occurs via a concerted mechanism and does not involve any covalent enzyme adducts.
Motor proteins
Although direct chemical conversion is relatively rare in nucleic acid metabolizing enzymes, there are a great number of ways that ATP hydrolysis is used by these proteins to drive motor processes (Figure 1). Among the best-studied examples are helicases, enzymes that catalyse the energetically unfavourable separation of nucleic acid duplexes into their component strands. Whereas binding energy could be utilized by a stoichiometric helicase to promote unwinding, a catalytic helicase requires additional energy to drive the reaction. At first sight, this seems to be a simple enough idea but, as is so often the case, there is more to the story than first meets the eye. Recent structural and biochemical data for the PcrA helicase (Velankar et al., 1999; Soultanas et al., 2000) have revealed that the helicase activity should be considered as two separate components, a duplex destabilizing activity and a DNA translocase, that can be uncoupled from each other. ATP hydrolysis promotes both processes, not only destabilizing the duplex but also allowing the enzyme to translocate unidirectionally along one of the product single-stranded DNA strands to prevent reannealing of the duplex, itself an energetically unfavourable process. So far so good, but other work has revealed the greater complexity of the situation. The translocation process can be studied independently by a number of means. Experiments using 5′-biotinylated DNA have shown that helicases can produce enough force to displace streptavidin (Morris and Raney, 1999). Other studies have shown that PcrA hydrolyses one ATP for each base along which it translocates (Dillingham et al., 2000). This is surprising, given that estimates of the free energy of association of DNA base pairs suggest that hydrolysis of one ATP should be sufficient to separate at least four base pairs or even as many as 12 (reviewed in Wigley, 2000). The observation that helicases can displace proteins from nucleic acids (Morris and Raney, 1999; Jankowsky et al., 2001) provides a clue as to why such a large force can be generated yet combined with an apparent wastage of ATP. There are times when bound proteins need to be displaced from the nucleic acid to allow other more urgent reactions (e.g. DNA repair or ribosomal RNA folding) to occur. Helicases can, therefore, have this additional role as biochemical ‘snow ploughs’ and need to have sufficient ‘excess’ energy available to them to be able to drive proteins, such as tightly bound transcription factors, from their nucleic acid targets.
A subset of DNA ‘helicases’ has been found to have a role in chromatin remodelling. Although initially identified as helicases by sequence analysis, purified enzymes have failed to show any helicase activity. More recent work (reviewed in Becker, 2002) has revealed that these ‘helicases’ are in fact DNA translocases. By fixing the translocase with respect to a nucleosome, translocation of the DNA against this produces a twist that, in turn, creates a transient loop that protrudes from the surface. This loop can move around the nucleosome, resulting in sliding of the nucleosome and hence remodelling of the bound DNA. DNA translocation is only a part of the activity required for a bona fide DNA helicase, which also requires a duplex destabilization activity (Soultanas et al., 2000). Another DNA translocase is the RuvAB complex that utilizes ATP hydrolysis to drive the migration of four-way (Holliday) junctions during recombination (West, 1996), a reaction for which there is no net energy change other than the entropic aspect of unidirectional translocation. Other enzymes such as type I restriction endonucleases are also DNA translocases (Firman and Szczelkun, 2000).
DNA supercoiling
Topoisomerases are enzymes that control the level of supercoiling in cells (Wang, 1996). The type II enzyme subclass (e.g. bacterial DNA gyrase and eukaryotic topoisomerase II) utilizes ATP to drive alteration in supercoiling by steps of two. The reaction mechanism requires a double-stranded break to be introduced into the duplex DNA substrate that is then opened up by >20Å by the enzyme. A further substantial conformational change in the protein then results in the capture and passing of another double-stranded DNA segment through this break before it is re-sealed by the enzyme. ATP hydrolysis drives the remarkable series of conformational changes that take place during the reaction.
However, despite this extraordinary reaction mechanism, it seems that ATP utilization by topoisomerases is even smarter than first thought (Rybenkov et al., 1997). It turns out that when the global thermodynamics of DNA supercoiling in cells are investigated, the reactions are not at equilibrium. Neither the distribution of topological isomers nor the steady-state fraction of knotted and catenated DNA molecules is as expected. The distribution of isomers is much tighter than at thermodynamic equilibrium, whereas the degree of knotting and catenation is as much as 80 times lower than expected. Topo isomerases do not, therefore, merely utilize ATP hydrolysis to drive the supercoiling reaction but, remarkably, also use it to control the global topology of the DNA by removing topological links that are barriers to DNA segregation after replication.
Proofreading
Mismatched base pairs in DNA result from misincorporation by polymerases. The recognition and repair of mismatched base pairs in DNA is essential, and failure of this process is a major cause of genome instability and, hence, cancer (Modrich and Lahue, 1996). Homologues of the MutHLS protein family are found in nearly all organisms and are required for mismatch repair. The MutS protein recognizes the mismatch and controls the activity of the other proteins. MutS is an ATPase and recent evidence suggests that ATP hydrolysis is used by the protein both to verify mismatches and activate initiation of the repair process by the MutH and MutL proteins (Junop et al., 2001). The mechanism is proposed to be a form of proofreading of the mismatch such that activation only occurs if ATP and a mismatched base pair are bound simultaneously. Furthermore, it has been shown that the two ATPase sites in MutS are asymmetric despite being chemically identical (Lamers et al., 2003) and that alternation of ATP hydrolysis between these sites controls the timing of the different steps in mismatch repair.
Interestingly, the MutL protein is also an ATPase, but in this case ATP hydrolysis serves as a switch to control the activation of MutH endonuclease activity (Hall and Matson, 1999).
Protein recycling
Homologous recombination is initiated by RecA in bacteria and by Rad51 in eukaryotes. The reactions catalysed by both enzymes involve invasion of a free 3′-end of DNA into a homologous duplex region, followed by strand exchange and migration to eventually establish a four-way junction in the DNA. RecA can promote strand exchange over a distance of around 3 kbp even in the presence of non-hydrolysable ATP analogues such as ATP-γ-S (Menetski et al., 1990). The obvious conclusion from this observation is that ATP hydrolysis is not required for the strand exchange process per se, but for some other function. This led the authors to conclude that this alternative function was to allow ordered disassembly of the RecA filament after the reaction or, more importantly, at the tail end of the filament (Figure 1). This would provide directionality to the reaction, thereby driving an energetically unfavourable process. This process has some analogies with the helicase reaction, in that energetically unfavourable unidirectional translocation is a part of the reaction. However, whereas helicases achieve this via a catalytic process with a single protein molecule translocating many base pairs at a time, RecA can be considered to be a stoichiometric helicase because each molecule binds once and then releases the DNA with the next step being taken by another protein molecule that contacts the first. Rad51 is a very much poorer ATPase than RecA, and it is probably no coincidence that Rad51-catalysed strand exchange activity is comparable with that of RecA with non-hydrolysable ATP analogues.
Clamp loaders
Replicative DNA polymerases are responsible for synthesis of the leading strand during DNA replication. These enzymes catalyse the synthesis of many thousands of base pairs of DNA in a single processive reaction. In order to achieve this, replicative polymerases from both prokaryotes and eukaryotes have a component that endows processivity to the enzyme complex. In both cases, this is achieved by loading a toroidal protein ring onto the DNA, thus preventing disassociation of the enzyme from the DNA template by a simple topological constraint (Jeruzalmi et al., 2002). However, there is a problem with this approach, in that the rings need to be opened so that the DNA can be threaded through them. The proteins that catalyse this reaction are similar in both groups of enzymes and utilize ATP to promote this process. In principle, the opening and closing of a protein ring need not require ATP, since the protein itself is not altered by the reaction, but it appears that the process is controlled by ATP hydrolysis. In this case, the nucleotide appears to be utilized to drive conformational changes in the protein complex to control the loading of protein rings at template primer sites. Recent work on the E.coli clamp loader (Ason et al., 2003) suggests that the control is at the level of DNA binding. The ATP-bound complex has a high affinity for DNA, but only elongation-proficient DNA substrates promote rapid ATP hydrolysis and conversion to the ADP-bound state that has a lower affinity for DNA. Thus, ATP hydrolysis apparently serves as a control mechanism in this case, although an alternative role in protein complex recycling cannot be ruled out.
DnaB/DnaC and helicase loading
Separation of the DNA strands is one of the essential steps during DNA replication. Formation of the replication fork requires a processive helicase that can unwind many thousands of base pairs in a continuous process. In a similar manner to that described above for the polymerases, the processivity of replicative helicases is achieved by the formation of a toroidal protein that encircles the DNA. Consequently, there is a similar topological problem in loading the rings. Loading of the replicative helicase (DnaB) in prokaryotes is controlled by an ATPase, DnaC. Surprisingly, the loading process itself does not require the hydrolysis of ATP (Davey et al., 2002). Instead, it appears that the ATPase activity controls a sequence of events after loading of the helicase (Figure 1). The DnaC–ATP complex actually inhibits the helicase activity of DnaB and promotes opening of the replication bubble. However, the ATPase activity of DnaC is stimulated by both DnaB and single-stranded DNA, thus ensuring that helicase release is averted unless a fully competent initiation complex is correctly formed at the origin.
SMC family proteins
The SMC (Structural Maintenance of Chromosomes) family of proteins is characterized by an ATPase domain that is interrupted by a long flexible, coiled-coil region (Strunnikov and Jessberger, 1999). Several members of this family, including Rad50, SbcCD and cohesins (see below), interact with nucleic acids. Crystal structures of the ATPase domains of Rad50 alone and in complex with ATP (Hopfner et al., 2000) suggest that ATP binding stabilizes the dimerization of these domains. ATP hydrolysis only occurs after dimerization, which then allows disassociation of the dimer. Thus, ATP binding and hydrolysis controls the association and disassociation of the ‘heads’ of these SMC family molecules. This mechanism is thought to be important for the function of cohesin (Nasmyth, 2002), a multi-subunit complex that holds sister chromatids together during cell division. The cohesin complex comprises several proteins, including an SMC heterodimer, and it is suggested that the complex works by encircling the sister chromatids and holding them together until ATP hydrolysis allows disassociation of the SMC proteins so that DNA can be released from the cohesin ring (Figure 1).
Other systems, other mechanisms?
Although we now understand a great deal about the reactions catalysed by many ATP-dependent nucleic acid modifying enzymes, there are others about which we understand much less. One example is the initiation of DNA replication in eukaryotes, a process controlled by the origin recognition complex (ORC) proteins (Figure 1). An important requisite of DNA replication is that it occurs once, and only once, during each cell cycle. This process is, therefore, tightly controlled, and once again a role for ATP hydrolysis has been revealed in this process. Mutations in the ORC proteins that disable ATPase activity are not functional in vivo (Klemm and Bell, 2001). However, although proven to be essential, the mechanism for ATP utilization remains unclear.
Another group of DNA-dependent ATPases are the Rad51 paralogues, which appear to play an important role in DNA repair (Thacker, 1999). These proteins have a very low intrinsic ATPase activity whose function(s) also remain unclear at the present time.
Conclusions
Classically, coupling ATP hydrolysis to a reaction has been seen as a mechanism to drive energetically unfavourable processes so that the overall energy change of the system remains favourable. Although this is important for many enzyme-catalysed reactions, such as those in the glycolytic pathway, recent research has revealed many other ways that the free energy of ATP hydrolysis can be utilized by enzymes. There are examples of ATP hydrolysis being used for chemical coupling to produce ‘high energy’ intermediates and to drive conformational changes in a variety of protein machines, from motors to clamp loaders. Other processes, such as protein recycling, proofreading of substrates and the alteration of global DNA topology, can also be linked to ATP hydrolysis. This apparent extravagance becomes less of an issue when one considers the overall ATP turnover in a cell and the relatively small amount of ATP that is hydrolysed by these enzymes. This is a cost that cells are evidently willing to pay in order to maintain integrity of their genome to be passed on to the next generation.
Although this review focuses on nucleic acid modifying enzymes, the principles are by no means unique to these proteins. Many other systems (such as F1-ATPase or actin) utilize ATP in complex ways to attain biological goals. It is unlikely that we have uncovered all of these mechanisms, and the next few years will undoubtably reveal further insights into the complexities of ATPases.
Acknowledgments
Acknowledgement
We thank M.Dillingham for critical reading of the manuscript.
References
- Ason B., Handayani,R., Williams,C.R., Bertram,J.G., Hingorani,M.M., O’Donnell,M., Goodman,M.F. and Bloom,L.B. (2003) Mechanism of loading the Escherichia coli DNA polymerase IIIβ sliding clamp: bona fide primer/templates preferentially trigger the γ complex to hydrolyze ATP and load the clamp. J. Biol. Chem., 278, 10033–10040. [DOI] [PubMed] [Google Scholar]
- Becker P.B. (2002) Nucleosome sliding: facts and fiction. EMBO J., 21, 4749–4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey M.J., Fang,L., McInerney,P., Georgescu,R.E. and O’Donnell,M. (2002) The DnaC helicase loader is a dual ATP/ADP switch protein. EMBO J., 21, 3148–3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillingham M.S., Wigley,D.B. and Webb,M.R. (2000) Demonstration of unidirectional single stranded DNA translocation by PcrA helicase: measurement of step size and translocation speed. Biochemistry, 39, 205–212. [DOI] [PubMed] [Google Scholar]
- Engler M.J., and Richardson,C.C. (1982) DNA ligases. In Boyer,P.D. (ed.), The Enzymes, Vol. 15. Academic Press, New York, NY, pp. 3–29. [Google Scholar]
- Fersht A.R., Leatherbarrow,R.J. and Wells,T.N.C. (1986) Binding energy and catalysis: a lesson from protein engineering of the tyrosyl-tRNA synthetase. Trends Biochem. Sci., 11, 321–325. [Google Scholar]
- Firman K. and Szczelkun,M.D. (2000) Measuring motion on DNA by the type I restriction endonuclease EcoR124I using triplex displacement. EMBO J., 19, 2094–2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldane J.B.S. (1930) Enzymes. Longman Green, London, UK, p. 182. [Google Scholar]
- Hall M.C. and Matson,S.W. (1999) The Escherichia coli MutL protein physically interacts with MutH and stimulates the MutH-associated endonuclease activity. J. Biol. Chem., 274, 1306–1312. [DOI] [PubMed] [Google Scholar]
- Hopfner K.P., Karcher,A., Shin,D.S., Craig,L., Arthur,L.M., Carney,J.P. and Tainer J.A. (2000) Structural biology of Rad50 ATPase: ATP-driven conformational control in Dna double-strand break repair and in the ABC-ATPase superfamily. Cell, 101, 789–800. [DOI] [PubMed] [Google Scholar]
- Jankowsky E., Gross,C.H., Shuman,S. and Pyle,A.M. (2001) Active disruption of an RNA–protein interaction by a DexH/D RNA helicase. Science, 291, 121–125. [DOI] [PubMed] [Google Scholar]
- Jeruzalmi D., O’Donnell,M. and Kuriyan,J. (2002) Clamp loaders and sliding clamps. Curr. Opin. Struct. Biol., 12, 217–224. [DOI] [PubMed] [Google Scholar]
- Jiang Y.L., Ichikawa,Y., Song,F. and Stivers,J.T. (2003) Powering DNA repair through substrate electrostatic interactions. Biochemistry, 42, 1922–1929. [DOI] [PubMed] [Google Scholar]
- Junop M.S., Obmolova, Rausch,K., Hseih,P. and Yang,W. (2001) Composite active site of an ATPase: MutS uses ATP to verify mismatch recognition and authorise DNA repair. Mol. Cell, 7, 1–12. [DOI] [PubMed] [Google Scholar]
- Klemm R.D. and Bell,S.P. (2001) ATP bound to the origin recognition complex is important for preRC formation. Proc. Natl Acad. Sci. USA, 98, 8361–8367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamers M.H., Winterwerp,H.H.K. and Sixma,T. (2003) The alternating ATPase domains of MutS control DNA mismatch repair. EMBO J., 22, 746–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menetski J.P., Bear,D.G. and Kowalczykowski,S.C. (1990) Stable DNA heteroduplex formation catalyzed by the Escherichia coli RecA protein in the absence of ATP hydrolysis. Proc. Natl Acad. Sci. USA, 87, 21–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrich P. and Lahue,R. (1996) Mismatch repair in replication fidelity, genetic recombination and cancer biology. Annu. Rev. Biochem., 65, 101–133. [DOI] [PubMed] [Google Scholar]
- Morris P.D. and Raney,K.D. (1999) DNA helicases displace streptavidin from biotin-labeled oligonucleotides. Biochemistry, 38, 5164–5171. [DOI] [PubMed] [Google Scholar]
- Nasmyth K. (2002) Segregating sister genomes: the molecular biology of chromosome separation. Science, 297, 559–565. [DOI] [PubMed] [Google Scholar]
- Rybenkov V.V., Ullsperger,C., Vologodskii,A.V. and Cozzarelli,N.R. (1997) Simplification of DNA topology below equilibrium values by type II topoisomerases. Science, 277, 690–693. [DOI] [PubMed] [Google Scholar]
- Schultz P.G. and Lerner,R.A. (1995) From molecular diversity to catalysis: lessons from the immune system. Science, 269, 1835–1842. [DOI] [PubMed] [Google Scholar]
- Shuman S., and Hurwitz,J. (1981) Mechanism of mRNA capping by vaccinia virus guanylyltransferase: characterization of an enzyme-guanylate intermediate. Proc. Natl Acad. Sci. USA, 78, 187–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soultanas P., Dillingham,M.S., Wiley,P., Webb,M.R. and Wigley,D.B. (2000) Uncoupling DNA translocation and helicase activity in PcrA: direct evidence for an active mechanism. EMBO J., 19, 3799–3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strunnikov A.V. and Jessberger,R. (1999) Structural maintenance of chromosomes (SMC) proteins: conserved molecular properties for multiple biological functions. Eur. J. Biochem., 263, 6–13. [DOI] [PubMed] [Google Scholar]
- Thacker J. (1999) A surfeit of Rad51-like genes? Trends Genet., 15, 166–168. [DOI] [PubMed] [Google Scholar]
- Velankar S.S., Soultanas,P., Dillingham,M.S., Subramanya,H.S. and Wigley,D.B. (1999) Crystal structures of complexes of PcrA helicase with a DNA substrate indicate an inchworm mechanism. Cell, 97, 75–84. [DOI] [PubMed] [Google Scholar]
- Vipond I.B. and Halford S.E. (1993) Structure-function correlation for the EcoRV restriction enzyme: from non-specific binding to specific DNA cleavage. Mol. Microbiol., 9, 225–231. [DOI] [PubMed] [Google Scholar]
- Wang J.C. (1996) DNA topoisomerases. Ann. Rev. Biochem., 65, 635–692. [DOI] [PubMed] [Google Scholar]
- West S.C. (1996) The RuvABC proteins and Holliday junction processing in E.coli. J. Bacteriol., 178, 1237–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigley D.B. (2000) DNA helicases: one small step for PcrA, one giant leap for RecBC? Curr. Biol., 10, R444–R445. [DOI] [PubMed] [Google Scholar]