Abstract
Mnt is a Max-interacting transcriptional repressor that has been hypothesized to function as a Myc antagonist. To investigate Mnt function we deleted the Mnt gene in mice. Since mice lacking Mnt were born severely runted and typically died within several days of birth, mouse embryo fibroblasts (MEFs) derived from these mice and conditional Mnt knockout mice were used in this study. In the absence of Mnt, MEFs prematurely entered the S phase of the cell cycle and proliferated more rapidly than Mnt+/+ MEFs. Defective cell cycle control in the absence of Mnt is linked to upregulation of Cdk4 and cyclin E and the Cdk4 gene appears to be a direct target of Mnt–Myc antagonism. Like MEFs that overexpress Myc, Mnt–/– MEFs were prone to apoptosis, efficiently escaped senescence and could be transformed with oncogenic Ras alone. Consistent with Mnt functioning as a tumor suppressor, conditional inactivation of Mnt in breast epithelium led to adenocarinomas. These results demonstrate a unique negative regulatory role for Mnt in governing key Myc functions associated with cell proliferation and tumorigenesis.
Keywords: breast cancer/Cdk4/cyclin E/Mnt/Myc
Introduction
The Myc family of proteins, particularly c-Myc and N-Myc, play fundamental roles in promoting cell proliferation and can contribute to tumorigenesis when their expression is deregulated and/or abnormally elevated (for review, see Grandori et al., 2000; Pelengaris et al., 2002). Although ectopic Myc expression can elicit a strong proliferative response, it also renders cells highly sensitive to apoptosis (Askew et al., 1991; Evan et al., 1992). Indeed, the ability of Myc overexpression to promote tumorigenesis is greatly enhanced in most cell types tested by coexpression of proteins that inhibit apoptosis such as Bcl2 or BclXL, or in genetic backgrounds in which proteins that positively regulate apoptosis, such as p53 and Bax, are absent (Pelengaris et al., 2002). From these experimental systems has emerged the paradigm of a simple two-step process (Myc overexpression and suppression of apoptosis) as sufficient for malignant conversion of at least some, and perhaps most, cell types (Pelengaris et al., 2002). Additional mechanistic support for this type of two-step process is provided by experiments showing that apoptosis induced by excessive Myc levels imposes strong selective pressure on primary mouse fibroblasts for the acquisition of disabling mutations in p53 or the p53-inducing protein p19ARF (Zindy et al., 1998). These and other studies demonstrate that mutations that disrupt p53 pathway function alleviate, at least partially, the apoptotic response associated with sustained high levels of Myc and thus allow its strong proliferative signal to predominate (Pelengaris et al., 2002). Since the p53 pathway also plays an important function in mediating cellular senescence (Sherr, 2001), events that promote selection of cells lacking p53 function are expected to predispose cells to bypass senescence and acquire an immortal phenotype, an important characteristic of malignant tumor cells. Indeed, the long recognized ability of c-Myc to promote senescence escape (Land et al., 1986 and references therein) is consistent with this idea.
The biological activity of Myc proteins is largely, if not wholly, dependent on heterodimerization with Max through their related bHLHZip domains (Eisenman, 2001). The Myc–Max heterodimer binds to E-box DNA sequences and can activate transcription through a tethered complex of proteins that contains histone acetyltransferase and other activities that remodel chromatin (Eisenman et al., 2001). Max also heterodimerizes with several other proteins that contain Myc-like bHLHZip domains, including the Mad family of proteins, the Mad-related protein Mnt, and Mga (Grandori et al., 2000; Zhou and Hurlin, 2001). The findings that Mad–Max, Mnt–Max and Mga–Max complexes appear to function as dedicated transcriptional repressors and can suppress Myc-dependent cell transformation in cell culture experiments (Grandori et al., 2000) raised the possibility that they might antagonize Myc function in vivo. This has been only partially borne out in studies of mice and cells that contain targeted deletion of Mad1, Mxi1 and Mad3 (Grandori et al., 2000; Zhou and Hurlin, 2001). Surprisingly, however, each of these mice is viable, fertile and fail to spontaneously give rise to tumors, an outcome predicted for a Myc antagonist.
Unlike Mad proteins, Mnt is ubiquitously expressed and readily detected as Mnt–Max heterodimers along with Myc–Max in proliferating cells (Zhou and Hurlin, 2001). Thus, the widespread coexpression of Myc and Mnt in cells suggests that Mnt might play a more general role than individual Mad proteins as a negative modulator of Myc biological activity. To explore this possibility, we mutated the Mnt gene in mice. Mice lacking Mnt differ dramatically from mice lacking Mad family members in that they are born runted and typically die within a few days of birth (K.Toyo-oka, S.Hirotsune, Z.-Q.Zhou, M.Gambello, R.N.Eisenman, P.J.Hurlin and A.Wynshaw-Boris, manuscript in preparation). The small size of embryos lacking Mnt is apparent as early as embryonic day 13.5 and demonstrate an important role for Mnt in embryonic development. Because of the high incidence of embryonic lethality, in this study we used mouse embryo fibroblasts (MEFs) derived from Mnt–/– mice and conditional Mnt knockout mice to investigate Mnt function. In contrast to loss of Mad proteins, Mnt deficiency was found to cause a phenotype remarkably similar to that caused by Myc overexpression and to predispose cells to tumorigenesis in vivo.
Results
MEFs lacking Mnt exhibit a hyperproliferative phenotype
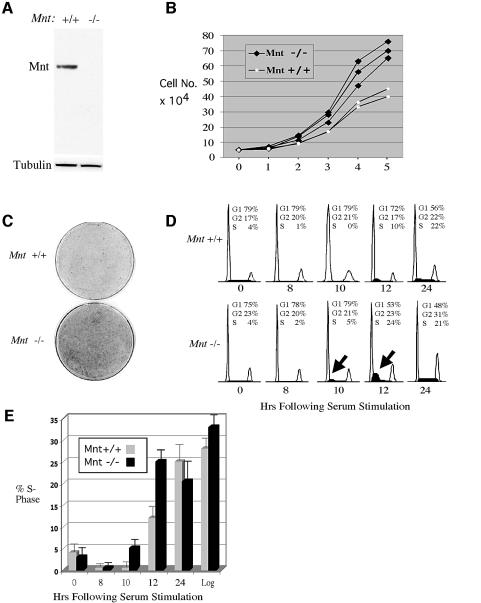
Because cellular Myc levels have profound consequences on cell proliferation, we first investigated whether cell proliferation was affected by loss of Mnt in MEFs. For these and subsequent experiments, MEFs were obtained from Mnt+/+ and Mnt–/– embryos at embryonic day 13.5. Mnt is easily detectable in MEFs derived from mutant mice, but undetectable in mutant mice (Figure 1A). In an apparent contradiction to the small size and lethality caused by loss of Mnt, early passage Mnt–/– MEFs proliferated at a higher rate and grew to higher density than age-matched Mnt+/+ MEFs (Figure 1B and C). The important role for Myc in S-phase progression (Amati et al., 1998; Grandori et al., 2000) prompted a comparison of the kinetics of S-phase entry upon serum stimulation of quiescent Mnt–/– MEFs and Mnt+/+ MEFs. Using fluorescence activated cell sorting (FACS) and bromodeoxyuridine (BrdU) incorporation, Mnt–/– MEFs were con sistently found to enter into S phase in advance of Mnt+/+ MEFs (Figure 1D and E).
Fig. 1. Primary mouse embryonic fibroblasts (MEFs) lacking Mnt proliferate at an accelerated rate, grow to high density and prematurely enter S phase. (A) Western blot analysis of Mnt in MEFs obtained from Mnt+/+ (+/+) and Mnt–/– (–/–) embryos. (B) Passage 3 Mnt–/– and Mnt+/+ MEFs were plated at 50 000 cells per 60 mm dish (day 0) and cell numbers per plate determined on six consecutive days. (C) Dishes stained 8 days after plating 50 000 cells showing high-density growth by Mnt–/– MEFs. (D) Fluorescence activated cell sorting (FACS) profiles of propidium iodide stained cells collected at the indicated times following serum stimulation of Mnt+/+ and Mnt–/– MEFs (passage 3), made quiescent by confluence inhibition and serum withdrawal. Deduced percentages of cells in G1, G2 and S phase are shown. The arrows indicate the elevated S-phase fraction of Mnt–/– MEFs at 10 and 12 h. (E) The percentage of cells in S phase determined by bromodeoxyuridine (BrdU) incorporation for three different sets of Mnt+/+ and Mnt–/– MEFs. (F) Western blot analysis of the indicated proteins following serum stimulation of quiescent passage 3 Mnt+/+ and cells. (G) Cyclin E1 and Cdk2 kinase assays. Cyclin E1 and Cdk2 were immunoprecipitated from equal amounts of protein from lysates obtained at the indicated times following serum stimulation. Kinase assays were performed using histone H1 as substrate. Total Cdk2 levels were determined from the same lysates by western blot.
To gain insight into the mechanism by which loss of Mnt promotes S-phase entry, expression patterns of Mnt, c-Myc and a number of proteins that play important roles in S-phase progression were determined in Mnt+/+ and Mnt–/– MEFs during cell cycle entry. Whereas Mnt levels fluctuated little during the 24 h period following serum stimulation of quiescent Mnt+/+ MEFs, c-Myc exhibited its highly characteristic early induction pattern (Figure 1F). The c-Myc expression pattern was normal in Mnt–/– MEFs (Figure 1F). However, the normal expression pattern of cyclin E1 was lost in Mnt–/– MEFs (Figure 1F). In Mnt–/– MEFs, cyclin E1 was constitutively expressed at high levels at all time points examined (Figure 1F). In contrast, Cdk2, E2f2, cyclin D1, cyclin D2 and Cdk4 regulation was not strongly affected by loss of Mnt, although Cdk4 levels appeared higher (Figure 1F).
To determine whether deregulated expression of cyclin E1 resulted in increased cyclin E1–Cdk2 kinase activity, cyclin E1 and Cdk2 were immunoprecipitated from lysates collected following serum stimulation of quiescent Mnt–/– and Mnt+/+ MEFs, and kinase assays were performed using histone H1 as substrate (Figure 1G). cyclin E1-associated kinase activity was elevated in Mnt–/– MEFS at all time points analyzed, but particularly at 10 and 24 h after serum stimulation (Figure 1G), time points when p27KIP1 levels had dropped to their lowest point (Figure 1F). Consistent with the cyclin E kinase activity, Cdk2 kinase activity was significantly higher in Mnt–/– MEFs at the 12 and 24 h time points (Figure 1G). Cyclin E and Cdk2 kinase activity in Mnt–/– MEFs was not due to differences in Cdk2 protein levels (Figure 1G). These results point to deregulation of cyclin E1 and elevated cyclin E1–Cdk2 activity as a key mechanism underlying the robust S-phase entry and accelerated proliferation rate of early passage Mnt–/– MEFs. Moreover, upregulation of cyclin E and premature S-phase entry are defects similar to those caused by ectopic Myc expression (Amati et al., 1998).
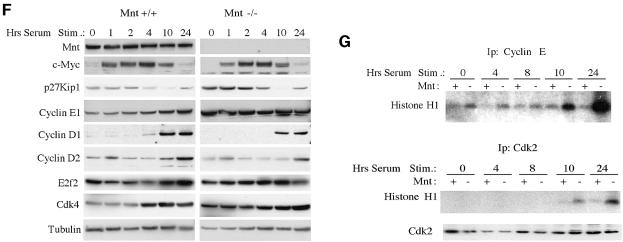
While ectopic c-Myc expression promotes proliferation, it also sensitizes cells to apoptosis, particularly primary cells and under conditions where serum-provided survival factors are limiting (Askew et al., 1991; Evan et al., 1992). Therefore, we tested whether logarithmically growing Mnt–/– MEFs at passage 3 were sensitized to apoptosis following reduction of serum levels in growth medium from 10 to 0.1%. Whereas apoptosis levels [as measured by terminal deoxynucleotidyl transferase (TUNEL) assay and caspase 3 cleavage] were low in 10% serum medium for both Mnt+/+ and Mnt–/– MEFs, Mnt–/– MEFs showed a several fold higher level of apoptosis 24 h after serum withdrawal (Figure 2A and B). Apoptosis levels in Mnt–/– MEFs declined on subsequent days (Figure 2B) and correlated with a decline in the population of viable cells (data not shown). The increased sensitivity to apoptosis of Mnt–/– MEFs is consistent with a hyper-Myc phenotype.
Fig. 2. MEFs lacking Mnt are sensitized to apoptosis. (A) DAPI and TUNEL staining of passage 3 Mnt+/+ and Mnt–/– MEFs 24 h following the replacement of growth medium containing 10% fetal bovine serum (FBS) with medium containing 0.1% FBS. (B) Western blot showing caspase cleavage (activation) is more robust in Mnt–/– MEFs when treated as in (A). The corresponding percentages of TUNEL positive cells in cultures of Mnt+/+ and Mnt–/– MEFs are shown. Percentages were determined from two independent experiments performed in triplicate. Results are presented as mean ± SEM.
Mnt deficiency promotes escape from senescence
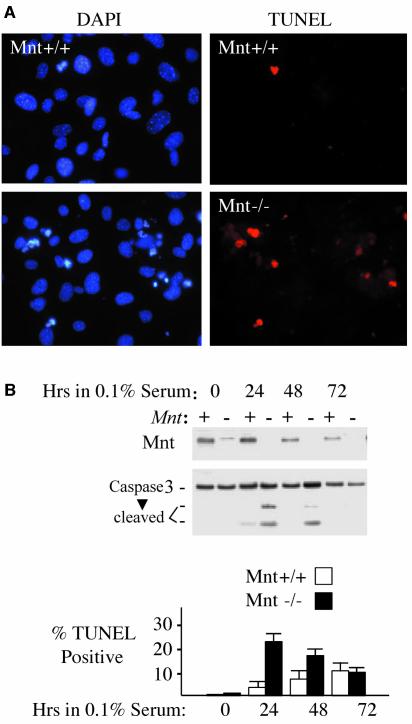
When primary MEFs are serially passaged in culture, they undergo a limited number of population doublings (typically 15–25) before entering senescence, a prolonged slow or no growth phase (Todaro and Green, 1963). Rare MEF variants eventually escape senescence, a process that is typically linked to the acquisition of inactivating p53 mutations or biallelic deletion of the INK4A locus (Sherr et al., 2001). When Mnt–/– MEFs were serially cultured according to a 3T9 format, faster growing and apparently immortal populations emerged 5–10 passages before similar Mnt+/+ populations emerged (Figure 3A). Of four Mnt–/– MEF strains analyzed at passage 25, all showed either loss of p19ARF expression or very high levels of p53 and p19ARF (Figure 3B), a pattern highly predictive of inactivating p53 mutations (Quelle et al., 1995; Sherr, 2001). Further, these cells no longer exhibited the acute sensitivity to apoptosis displayed by Mnt–/– MEFs at passage 3 or 4 (data not shown), consistent with abrogated p53 signaling (Zindy et al., 1998). Similar to immortal Mnt–/– MEFs, Mnt+/+ MEF strains analyzed at passage 25 showed either high p53 and p19ARF levels, diagnostic of p53 mutation, or undetectable p19ARF (Figure 3B). Thus, the mechanism of immortalization appears similar in Mnt–/– and Mnt+/+ cells, but the process is accelerated in Mnt–/– MEFs.
Fig. 3. MEFs lacking Mnt efficiently escape senescence. (A) Proliferation curves of two unique Mnt+/+ MEF and three unique Mnt–/– populations passaged continuously using a 3T9 protocol. Cell numbers were determined at the passages indicated and converted into population doubling level (the number of times the original starting number of cells has doubled). (B) Expression of p53 and p19ARF in two passage 25 Mnt+/+ and three passage 25 Mnt–/– MEFs. The highly elevated p53 in one Mnt+/+ and one Mnt–/– cell line is indicative of inactivating mutation in p53. The other Mnt+/+ cell line and other two Mnt –/– samples fail to express detectable p19ARF. NIH 3T3 fibroblasts, which lack p19ARF and p53–/– MEFs were used as controls. The asterisk indicates a non-specific protein recognized by p19ARF antiserum. (C) Western blot showing expression levels of the indicated proteins in Mnt+/+ and Mnt–/– MEFs as a function of passage level. (D) Comparison of p53 and p19ARF levels in multiple independent Mnt+/+ and Mnt–/– MEFs at passage 4. (E) Senescence-associated β-galactosidase assays showing intense activity in Mnt–/– MEFs at passage 4 and a reduced activity by passage 12, when activity in Mnt+/+ MEFs was present. Similar results were observed in several different Mnt+/+ and Mnt–/– strains. (F) Morphology of Mnt+/+ and Mnt–/– MEFs at the indicated passage levels.
To examine the immortalization process in more detail, the expression of p19ARF and p53 was followed during serial passage of Mnt–/– MEFs. Compared with Mnt+/+ MEFs, Mnt–/– MEFs showed a premature elevation in both p19ARF and p53 (Figure 3C and D), proteins closely associated with culture-induced senescence (Kamijo et al., 1997) that are upregulated by Myc overexpression (Lowe and Ruley, 1993; Zindy et al., 1998). The elevated p19ARF and p53 levels generally coincided with decreased proliferation of Mnt–/– MEFs (Figure 3A) and with strong senescence-associated β-galactosidase activity in many of these cells (Figure 3E). Despite entering a phase of slowed proliferation, many or most Mnt–/– MEFs failed to adopt the flattened, stress-fiber-filled morphology characteristic of their senescent Mnt+/+ MEFs counterparts (Figure 3F). The emergence of faster growing Mnt–/– MEFs coincided with a decline in p19ARF and p53 levels, and by passage 10–12, p19ARF was difficult to detect (Figure 3C). Likewise, levels of p16INK4A, an inhibitor of CDK4/6 kinase activity that is encoded at the same genetic locus as p19Arf (INK4A locus; Quelle et al., 1995), could not be detected in immortal (p31) populations of the same Mnt–/– MEF strain (Figure 3C). The presence of p53, but lack of detectable p19ARF and p16INK4A in this particular Mnt–/– strain, is consistent with deletion, or epigenetic silencing of the INK4A locus, an event sufficient for MEFs to bypass senescence (Kamijo et al., 1997). In contrast, levels of p19ARF and p16INK4A incrementally increased in Mnt+/+ MEFs analyzed in parallel, and in our experiments reached highest levels after passage 9 (Figure 3C), when these cells entered into a protracted senescence state (Figure 3A). However, when examined at passage 31, neither p19ARF, nor p16INK4A could be detected in this strain (Figure 3C), linking disruption of the INK4A locus with their immortalization.
These results suggest that, like Myc overexpression (Zindy et al., 1998), loss of Mnt predisposes MEFs to senescence escape through a mechanism involving accelerated selection for cells with compromised p53 function.
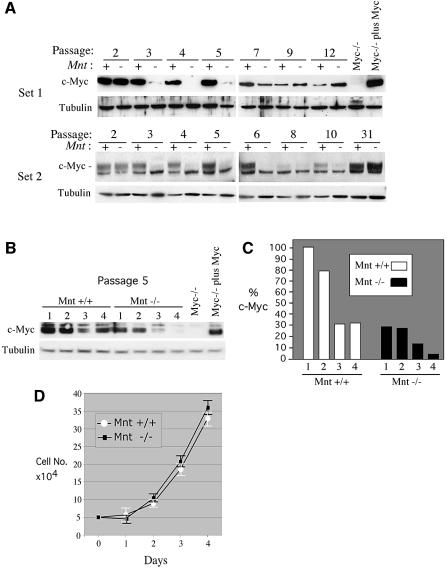
Downregulation of c-Myc in Mnt-deficient MEFs
Although c-Myc was properly induced in Mnt–/– MEFs during cell cycle entry (Figure 1F), when these cells were serially cultured, a striking decline in c-Myc protein was observed following two passages in culture (Figure 4A). c-Myc levels in Mnt+/+ cells declined much later (Figure 4A) and correlated with senescence. The decline in c-Myc levels at passage 3 was highly reproducible, occurring in all five unique cultures of Mnt–/– MEFs examined. Since primary MEF proliferation is profoundly diminished when c-Myc levels are decreased or eliminated (de Alboran et al., 2001; Trumpp et al., 2001), we examined the relationship between c-Myc levels and proliferation in Mnt–/– MEFs. For these experiments, growth curves were carried out with passage 5 cells while c-Myc levels were monitored. Remarkably, c-Myc levels were found to be between 4- and 10-fold lower in Mnt–/– MEFs than in Mnt+/+ MEFs on four consecutive days tested (Figure 4B). Despite the low c-Myc levels, Mnt–/– MEFs proliferated at a rate comparable with age-matched Mnt+/+ MEFs (Figure 4B). Since neither N-myc nor L-Myc mRNA could be detected by RT–PCR in Mnt–/– MEFs (data not shown), these results raise the possibility that MEFs lacking Mnt have reduced c-Myc requirements for proliferation.
Fig. 4. Downregulation of c-Myc protein levels in Mnt–/– MEFs. (A) Analysis of c-Myc levels in Mnt+/+ (+) and Mnt–/– (–) MEFs during serial 3T9 passaging. Lysates were collected on every third day following re-dilution and plating of 9 × 105 cells. Western blots performed using two different c-Myc specific antibodies [Set 1, anti-c-Myc, N-262 (Santa Cruz); Set 2, anti c-Myc, 06-213 (Upstate)]. Set 1 and Set 2 lysates were collected from unique sets of Mnt+/+ and Mnt–/– MEFs. (B) c-Myc protein levels in passage 5 MEFs determined on consecutive days following plating of 5 × 104 cells in 60 mm dishes. (C) c-Myc levels, determined by densitometry, as percentage of c-Myc in Mnt+/+ cells on day 1. (D) Growth curves for MEFs used in (B). Cell counts were performed from triplicate plates.
Although c-Myc remained at very low levels in Mnt–/– MEFs for several passages, it returned to easily detectable levels by passage 9–12 and in immortal populations (Figure 4A). Thus, the re-emergence of c-Myc expression in Mnt–/– MEFs appears to be linked to escape from senescence (Figure 3).
Cdk4 is a Mnt target gene
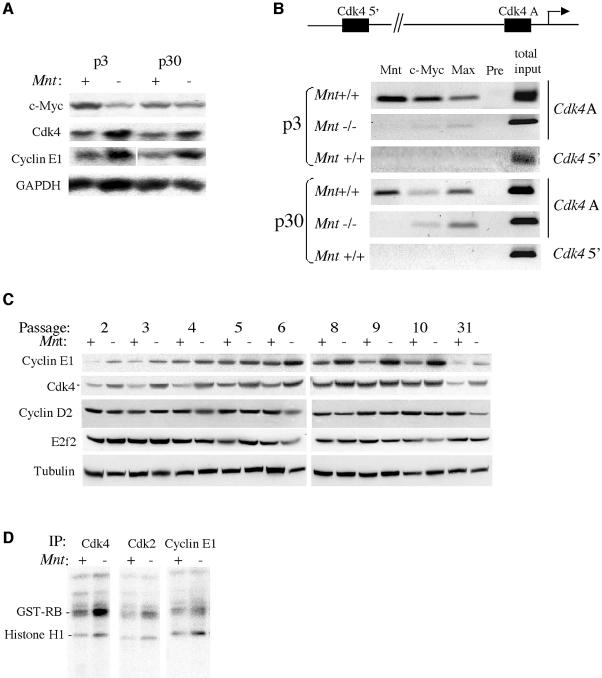
The ability of Mnt–/– MEFs to proliferate in the presence of low Myc levels suggested that loss of Mnt might cause upregulation of key Myc target genes capable of compensating for low c-Myc. Northern analysis was initially used to screen candidate Myc target genes. A number of putative Myc target genes, including ODC, MrDb, Cad, E2f2, E2f3 and nucleolin, were not significantly affected by loss of Mnt (Z.-Q.Zhou, W.Walker, A.Wynshaw-Boris and P.J.Hurlin, manuscript in preparation). A notable exception was the Myc target gene Cdk4 (Hermeking et al., 2000; Menssen and Hermeking, 2002), which was upregulated in Mnt–/– MEFs at both passage 3, when Myc levels were low, and at passage 30, when c-Myc levels were near normal (Figure 5A). In addition to Cdk4, the cyclin E gene, which is upregulated by ectopic c-Myc expression (Amati et al., 1998), was elevated in both passage 3 and 30 Mnt–/– MEFs. The elevated cyclin E mRNA levels are in general agreement with the deregulation of cyclin E protein levels observed in cell cycle entry experiments (Figure 1E).
Fig. 5. The Cdk4 gene is upregulated in the absence of Mnt. (A) Northern blot showing c-Myc, Cdk4 and cyclin E mRNA levels in Mnt+/+ (+) and Mnt–/– (–) MEFs at passage 3, and passage 30 cells. Whereas Cdk4 and cyclin E1 levels are elevated in Mnt–/– MEFs at both passage 3 and 30, c-Myc mRNA levels are reduced in Mnt–/– MEFs at passage 3, but expressed at comparable levels to Mnt+/+ cells at passage 30. GAPDH was used as a loading control. (B) ChIP assay for Cdk4 promoter occupancy by Mnt, Myc and Max at passage 3 and passage 30. Immunoprecipitations were performed using near confluent cells collected 3 days after plating cells in medium containing 10% serum. The cell culture conditions used in these assays were designed to approximate conditions in which c-Myc levels are strongly reduced in Mnt–/– MEFs at passage 3–6 (Figure 4). Antibodies against the indicated proteins were used and Mnt preimmune (pre) serum served as negative control. For PCRs, primers that amplify a 140 bp, E-box-containing region (designated Cdk4A) were used. This E-box lies 119 bp from the Cdk4 transcriptional start site. PCR primers that amplify an E-box-containing region ∼4.3 kb upstream from the Cdk4 transcriptional start site were used as a control. (C) Western blot analysis of the indicated proteins during 3T9 passaging of representative sets of Mnt+/+ (+) and Mnt–/– (–) MEFs. Tubulin was used as a loading control. (D) Kinase assays performed with immunoprecipitated Cdk4, Cdk2 and cyclin E1 prepared from lysates of logarithmically growing Mnt+/+ (+) and Mnt–/– (–) MEFs at passage 4. Equal protein amounts were used for immunoprecipitations. GST–Rb (Rb amino acids 769–921) and histone H1 were used as substrates.
The Cdk4 gene was of particular interest as a direct Mnt target gene, since it was previously shown to be capable of partially rescuing the slow proliferation phenotype of Rat1A fibroblasts lacking c-Myc (Hermeking et al., 2000). Chromatin immunoprecipitation (ChIP) experiments carried out using Mnt–/– and Mnt+/+ MEFs at passage 3 and 30 showed specific Mnt binding to an E-box-containing, Cdk4 promoter region (Cdk4 proA) in Mnt+/+ MEFs but not Mnt+/+ (Figure 5B). c-Myc and Max antibodies were also effective in immunoprecipitating the same region of the Cdk4 promoter (Cdk4 proA) bound by Mnt in Mnt+/+ cells (Figure 5B), suggesting that Mnt–Max and Myc–Max complexes compete for binding to this site. Binding of Mnt to Cdk4 proA was specific, since Mnt antibody failed to immunoprecipitate Cdk4 proA in Mnt–/– MEFs and Mnt preimmune serum, which served as a negative control in these experiments, failed to immunoprecipitate Cdk4 proA in Mnt+/+ MEFs (Figure 5B). Further, antibodies to Mnt, c-Myc and Max failed to immunopreciptate an E-box-containing region 4.3 kb upstream of the Cdk4 transcriptional start site (Figure 5B).
Consistent with the low level of c-Myc in Mnt–/– MEFs at passage 4 (Figure 4), it was difficult to detect c-Myc and Max bound to Cdk4 proA in these cells (Figure 5B). However, both c-Myc and Max were bound to Cdk4 proA in immortal (passage 35) Mnt–/– MEFs (Figure 5B). Together with the increased Cdk4 mRNA levels found in passage 4 and immortal Mnt–/– MEFs, these results argue that the Cdk4 gene is a direct target of Mnt and that it is derepressed in the absence of Mnt. In addition to increased mRNA levels and the apparent loss of Mnt repression at the Cdk4 promoter, Cdk4 protein levels were elevated at all passage levels of Mnt–/– MEFs examined (Figure 5C). Further, Cdk4-associated kinase activity was high in logarithmically growing passage 4 Mnt–/– MEFs (Figure 5D). In addition to Cdk4, cyclin E1 was elevated in Mnt–/– MEFs at all passage levels (Figure 5C) and cyclin E1–Cdk4 activity was high in passage 4 MEFs (Figure 5D). In contrast to Cdk4 and cyclin E1, cyclin D2 and E2f2 levels did not appear to be affected by loss of Mnt (Figure 5C).
Mnt–/– MEFs are sensitive to transformation by oncogenic Ras alone
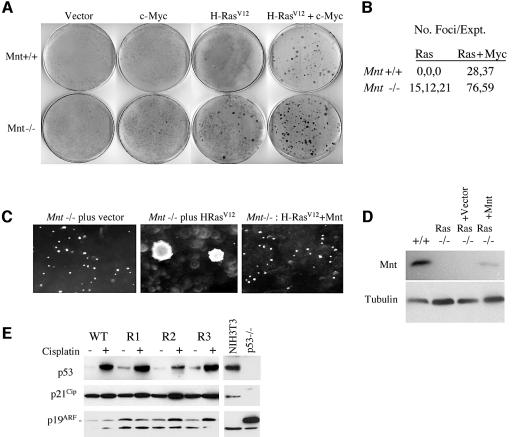
Whereas constitutive overexpression of Myc sensitizes primary fibroblasts to apoptosis and fails to transform these cells, coexpression of c-Myc with oncogenic Ras (e.g. H-RasV12) transforms them into cells that form foci and grow in an anchorage-independent manner (Land et al., 1986). To determine whether Mnt deficiency rendered MEFs more susceptible to transformation, MEFs were infected with viruses expressing c-Myc, H-RasV12 or c-Myc plus H-RasV12 and focus formation measured 2 weeks later. Whereas infection of H-RasV12 alone failed to produce foci in Mnt+/+ MEFs, a significant number of foci were produced in Mnt–/– MEFs (Figure 6A and B). Further, when sequentially infected with c-Myc and H-RasV12 viruses, Mnt–/– MEFs produced more transformed foci than Mnt+/+ MEFs (Figure 6A and B). Thus, loss of Mnt sensitized MEFs to transformation by H-RasV12 alone and made them super-transformable by c-Myc plus H-RasV12.
Fig. 6. Mnt–/– MEFs can be transformed by H-RasV12 alone. (A) Equal numbers of Mnt+/+ and Mnt–/– MEFs at passage 2 were infected either alone with pBabe, pBabe-H-RasV12 or pBabe-c-Myc virus or sequentially with c-Myc and H-RasV12 virus as indicated. Following infection, cells were fed growth medium every other day for 2 weeks and then stained with methylene blue. (B) The average number of foci produced on duplicate plates is shown for three (H-RasV12 alone) and two (c-Myc + H-RasV12) experiments. (C) Soft agar growth assay showing robust colony formation by H-RasV12-transformed Mnt–/– MEFs, but not by Mnt–/– without H-RasV12, and failure of Mnt–/– MEFs to form colonies following retroviral introduction of Mnt. (D) Western blot showing Mnt expression following virus infection of Mnt–/– MEFs. (E) Expression of p53 and p19ARF in three clonal populations of H-RasV12-transformed Mnt–/– MEFs (R1–3). Levels of p53, p21Cip1 and p19ARF were determined by western blot with the indicated antibodies before and 16 h after treatment with 5 0 µM cisplatin. NIH 3T3 fibroblasts, which lack p19ARF and p53–/– MEFs, were used as controls.
Clones of H-RasV12-transformed Mnt–/– MEFs were also capable of anchorage-independent growth (Figure 6C), a phenotype that closely corresponds to malignant transformation. To determine whether loss of Mnt was sufficient for transformation by H-RasV12, H-RasV12-transformed Mnt–/– MEFs were infected with Mnt-expressing retrovirus, selected for virus-infected cells and tested growth in soft agar. Introduction of Mnt inhibited soft agar colony formation (Figure 6C and D). Further, introduction of Mnt into H-RasV12-transformed Mnt–/– MEFs caused these cells to proliferate more slowly and exhibit characteristics typical of senescent MEFs, such as enlarged cytoplasm containing extensive stress fibers and senescence-associated β-galactosidase activity (data not shown). Thus, introduction of Mnt into Ras-transformed Mnt–/– MEFs restores the normal, senescence-like response caused by oncogenic Ras (Serrano et al., 1997) and suggest that events secondary to loss of Mnt are not responsible for susceptibility to H-RasV12 transformation.
Since Mnt–/– MEFs with disrupted p19ARF–p53 pathway function rapidly emerge during serial passaging (Figure 3B and C), it remained possible that susceptibility to Ras transformation may have been due to defects in this pathway (Sherr et al., 2001). However, when treated with the DNA damage agent cisplatin, H-RasV12-transformed Mnt–/– MEFs, like Mnt+/+ MEFs, induced p53, and p53 function appeared to be intact, since levels of its target gene-encoded protein, p21Cip1, were also induced in these cells (Figure 6D). Consistent with the normal response of MEFs to oncogenic Ras (Sherr, 2001), basal levels of p53 and p19ARF levels were elevated in Ras-transformed Mnt–/– MEFs. We conclude that susceptibility of Mnt–/– MEFs to transformation by H-RasV12 is not due to compromised p53 pathway function. Instead, we propose that Mnt–/– MEFs behave similarly to primary MEFs that overexpress Myc, which confers susceptibility to cooperative transformation by H-RasV12 (Land et al., 1986).
Mammary adenocarcinoma formation in the absence of Mnt
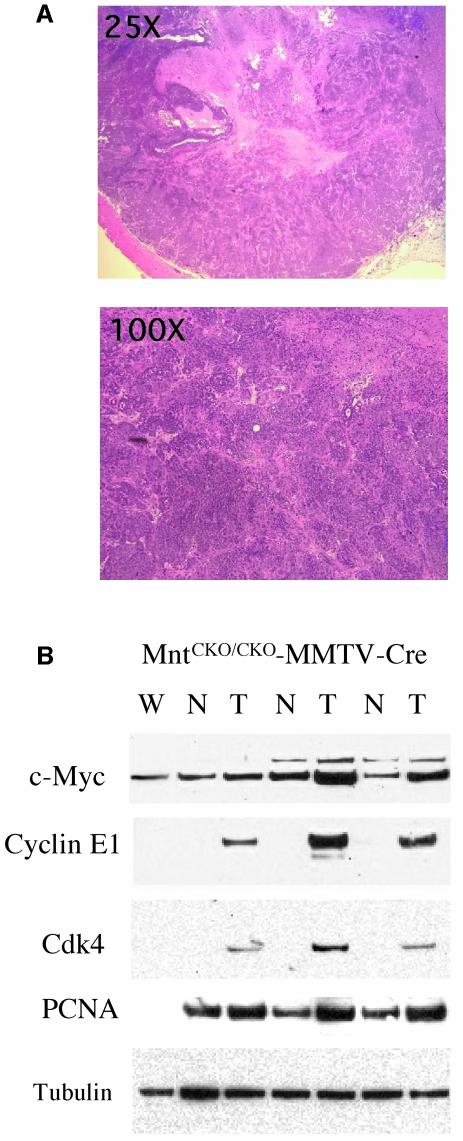
The chromosomal location of the human MNT gene at 17p13.3 (Hurlin et al., 1997; Meroni et al., 1997) is a hot spot for loss of heterozygosity (LOH) in breast and other cancers, and it has been estimated that 20–30% of sporadic breast cancers have LOH at 17p13.3 (Cornelis et al., 1994). Since homozygous Mnt deletion is usually lethal (K.Toyo-oka, M.Gambello, Z.Q.Zhou, R.N.Eisenman, P.J.Hurlin, S.Hirotsune and A.Wynshaw-Boris, manuscript in preparation), the potential for loss of Mnt in mammary epithelium to cause breast cancer was tested by crossing MMTV-Cre transgenic mice with MntCKO mice to generate MntCKO/CKO-MMTV-Cre mice (see Materials and methods). To promote Cre transcription from the pregnancy hormone-responsive MMTV promoter, 10 female control MntCKO/CKO mice and 10 MntCKO/CKO-MMTV-Cre mice were continuously mated and monitored for tumor formation. Whereas no tumors developed in MntCKO/CKO mice, 6 of the 10 MntCKO/CKO-MMTV-Cre mice developed adenocarcinomas (Figure 7A). Tumor latency ranged from 6 months to 20 months, a range similar to that caused by ectopic expression of c-Myc in mammary epithelium (Hutchinson and Muller, 2000).
Fig. 7. Formation of adenocarcinomas in mammary epithelium targeted for disruption of Mnt. (A) Hematoxylin and eosin sections of adeno carcinomas formed in MMTV-Cre;MntCKO/CKO mice. (B) Western blot analysis of Mnt+/+ (W) breast, phenotypically normal (N) breast in MMTV-Cre;MntCKO/CKO mice and tumor (T) tissue for the indicated proteins.
Similar to Mnt–/– MEFs, both cyclin E1 and Cdk4 levels were dramatically higher in three tumors examined compared with surrounding normal tissue in MMTV-MntCKO/CKO mice and to normal tissue in Mnt+/+ mice (Figure 7B). Very long exposures of the Western blot shown in Figure 7B, showed that phenotypically normal MMTV-MntCKO breast tissue expressed higher levels of cyclin E1 and Cdk4 than in Mnt+/+ breast tissue (data not shown). Interestingly, levels of proliferating cell nuclear antigen (PCNA), a subunit of DNA polymerase δ and E2f target gene, was elevated in phenotypically normal breast tissue samples of MMTV-MntCKO mice compared with Mnt+/+ breast tissue (Figure 7B). These expression data are consistent with the presence of a cryptic subset of cells within phenotypically normal breast tissue of MMTV-MntCKO/CKO-Cre mice that are predisposed to tumorigenesis.
Discussion
Deregulation and/or overexpression of Myc proteins in cells cause a complex phenotype characterized by enhanced proliferation and tumorigenesis, but also by apoptosis and/or cell cycle arrest (Grandori et al., 2000; Pelengaris et al., 2002). In this study, we show that deletion of Mnt mimics many of the hallmark characteristics of Myc overexpression, including tumorigenesis. Although our results demonstrate a unique role for Mnt as a Myc antagonist, they also reveal a potential function for Mnt in mediating feedback regulation of c-Myc transcription. Thus, Mnt appears to govern Myc activities on multiple levels.
Our previous observation that Mnt can inhibit Myc-dependent transformation, and that a weakly activating mutant version of Mnt (ΔSIDMnt) can, like Myc, cooperate with H-RasV12 in cell transformation, provided evidence to indicate that Mnt can function as a Myc antagonist (Hurlin et al., 1997). The ability of Mnt to antagonize Myc is thought to arise due to (i) competition for binding to their shared and obligate DNA-binding cofactor Max, (ii) competition between Mnt–Max and Myc–Max complexes for binding to E-box sites in shared target genes and (iii) transcriptional repression of target genes that Myc–Max activates (Hurlin et al., 1997). According to this model, the relative levels of Mnt–Max and Myc–Max in cells at any given time will govern expression of their shared target genes (Orian et al., 2003). The same is thought to be true for the Mad family of proteins and Mga, which also interact with Max and are transcriptional repressors (Grandori et al., 2000; Zhou and Hurlin, 2001). However, whereas Mnt is ubiquitously expressed (Hurlin et al., 1997), the expression patterns of Mad and Mga genes appear to be more restricted, both temporally and spatially, in developing and adult tissues (Hurlin et al., 1995, 1999; Queva et al., 1998). Perhaps most important to its apparent function as a Myc antagonist, Mnt is expressed in both quiescent cells and throughout the cell cycle (Figure 1F). Although little is known about Mad and Mga protein levels as a function of the cell cycle, the data available suggest that they are expressed at very low level in most proliferating cells (Zhou and Hurlin, 2001). Importanly, Mnt–Max and Myc–Max complexes appear to be the predominant Max complexes in proliferating cells (Zhou and Hurlin, 2001). Thus, expression of Mnt throughout the cell cycle is predicted to result uniquely in Mnt–Max antagonism of critical Myc–Max functions involved in cell cycle entry and the normal proliferative cell cycle (Amati et al., 1998). The observation that early passage Mnt–/– MEFs have an accelerated rate of proliferation (Figure 1B), fail to efficiently arrest by contact inhibition (Figure 1C) and prematurely enter S phase (Figure 1D and E) is consistent with this prediction. Further, the elevated levels of cyclin E1 and Cdk4 and associated kinase activity in Mnt–/– MEFs (Figures 1F and G and 5A, C and D) closely resembles the consequences of deregulated expression of Myc (Amati et al., 1998).
Mnt as a component in c-Myc feedback regulation
A striking difference between Mnt+/+ and Mnt–/– MEFs is the severe drop in c-Myc levels that occurs in the latter cells soon after being established in culture (between passage 2 and 3, Figure 4A). The drop in c-Myc protein levels correspond to a drop in c-Myc RNA levels (Figure 5A) and precedes the decline in the proliferation rate of Mnt–/– MEFs that typically occurs after passage 5 (Figure 3A). Indeed, we find that Mnt–/– MEFs at passage 5 continue to proliferate in the presence of extremely low levels of c-Myc (Figure 4A and B). Since deletion of c-Myc levels in MEFs results in the cessation of proliferation (de Alboran et al., 2001; Trumpp et al., 2001), our results raise the possibility that cells lacking Mnt may be less dependent on Myc for proliferation. However, it is important to emphasize that c-Myc levels are induced in quiescent Mnt–/– MEFs following mitogenic stimulation (Figure 1F) and that c-Myc is transiently induced in passage 5 Mnt–/– MEFs immediately following their replating in fresh medium (Figure 4B). Thus, growth factor-mediated induction of c-Myc appears not to be affected in Mnt–/– MEFs. One possiblility is that c-Myc is still required for quiescent Mnt–/– MEFs to enter the cell cycle, but that it is no longer required, and perhaps detrimental, to sustained proliferation (see below). Interestingly, preliminary results indicate that c-Myc levels are also very low in embryonic day 13.5 Mnt–/– embryos (Z.-Q.Zhou, A.Wynshaw-Boris and P.JHurlin, data not shown). Thus, although it remains to be formally proven, reduced c-Myc levels may be related to the runting and early postnatal lethality caused by loss of Mnt (K.Toyo-oka, S.Hirotsune, Z.-Q.Zhou, R.N.Eisenman, P.J.Hurlin and A.Wynshaw-Boris, manuscript in preparation).
Ectopic overexpression of c-Myc in a variety of cell types leads to downregulation of endogenous c-Myc, and it is postulated that Myc negative autoregulation functions to counter the potentially deleterious consequences of Myc-dependent hyperproliferation (Penn et al., 1990). Thus, one possibility is that c-Myc downregulation in early passage Mnt–/– MEFs is a consequence of sustained hyperactive Myc signaling, caused by loss of Myc antagonism and activation of the Myc autosuppression pathway. Perhaps similar to Cdk4 regulation, loss of Mnt may lead to derepression of an unknown Myc target gene(s), whose encoded protein mediates Myc autosuppression. Although the mechanism responsible for c-Myc downregulation in Mnt–/– MEFs is unknown, these cells will provide a useful model system to study Myc autosuppression.
Mnt/Myc regulation of Cdk4 and cyclin E
The ability of Mnt–/– MEFs to proliferate in the presence of abnormally low Myc levels may be at least partially explained by upregulation of Cdk4 and cyclin E. Consistent with this idea, Cdk4 overexpression has been demonstrated to partially rescue the slow growth of immortal Rat1 fibroblasts that lack c-Myc (Hermeking et al., 2000). Cdk4 and cyclin E were consistently found elevated in Mnt–/– MEFs independent of c-Myc levels, and also appeared elevated in breast tumors lacking Mnt (Figures 1F, 5A and C and 7B). The Cdk4 gene appears to be directly regulated by Mnt and Myc, since they both bind to at least one site in the Cdk4 promoter region (Figure 5B; see also Menssen and Hermeking, 2002). The inability to detect c-Myc binding to the Cdk4 promoter in passage 4 Mnt–/– MEFs correlates with the very low levels of c-Myc in these cells (Figure 4A). Since Cdk4 mRNA levels are elevated in the absence of c-Myc binding (Figure 5A), these results suggest that the Cdk4 promoter is derepressed in the absence of Mnt. Moreover, these data are consistent with a model in which c-Myc normally upregulates Cdk4 transcription (Hermeking et al., 2000), at least in part, by displacing Mnt–Max complexes.
It is possible that upregulation of Cdk4 is responsible for deregulation of cyclin E transcription (Figure 5A), since Cdk4 phosphorylation and inactivation of Rb and other pocket proteins relieves E2f-dependent repression of cyclin E transcription (Dyson, 1998). Alternatively, the cyclin E gene may be a direct target of Mnt repression. In either case, the increased cyclin E- and Cdk4-associated kinase activity in Mnt–/– cells and presumed disruption of Rb pathway activity may function to sensitize these cells to transformation by oncogenic Ras. In wild-type cells, oncogenic Ras induces a senescence-like growth arrest, and the Rb pathway, functioning downstream of p53, is required for this activity (Sherr, 2001). Like overexpression of Myc, our results suggest that loss of Mnt subverts this pathway. Further, our results showing that p19ARF and p53 are present, and that p53 is functional, in Ras-transformed Mnt–/– MEFs is consistent with the notion that the Rb pathway is crippled in these cells. However, if Rb function was completely abrogated by loss of Mnt, the expectation would be that these cells would bypass senescence without the need to acquire mutations that disrupt p53 pathway function (Chau and Wang, 2003). On the contrary, immortalization caused by overexpression of Myc (Zindy et al., 1998) and by loss of Mnt is closely linked to disrupted p53 pathway function. One possible explanation is that transformation by Myc plus oncogenic Ras is an acute readout and that these cells are subject to selection for p53/ARF mutation for long-term survival. This interpretation is consistent with our observation that the percentage of Mnt–/– MEFs sensitive to transformation by oncogenic Ras increases as a function of passage level and that immortal Mnt–/– MEFs are more susceptible to transformation by oncogenic Ras than primary Mnt–/– MEFs (data not shown). An analysis of p53 pathway status in long-term cultures of Mnt–/– cells transformed with oncogenic Ras and long-term cultures of cells transformed by Myc and oncogenic Ras may help resolve this issue.
Tumor progression in the absence of Mnt
The finding that conditional disruption of Mnt in mouse mammary epithelium leads to malignant adenocarcinomas (Figure 7A) underscores the relevance of characteristics defined in Mnt–/– MEFs to tumorigenesis and further supports the idea that Mnt functions as a Myc antagonist. These results also raise the possibility that Mnt may function as a tumor suppressor in human breast cancer. Indeed the MNT gene resides at17p13.3, a hotspot for LOH in breast and other cancers (Cornelis et al., 1994; Hoff et al., 2000). However, a previous study of nine sporadic breast cancers showing LOH specifically at the MNT locus failed to identify mutations in the remaining allele (Nigro et al., 1998). Nonetheless, the large body of evidence indicating that a breast cancer tumor suppressor gene resides at 17p13.3, together with results shown here, should provide impetus for a more extensive study to determine whether Mnt is inactivated in some breast cancers.
The latency of tumor formation in MntCKO/CKO-MMTV-Cre mice indicates that secondary events must cooperate with loss of Mnt in tumorigenesis. Our analysis of Mnt–/– MEFs during the process of senescence escape (Figure 3) predicts that loss of p19ARF or mutation of p53 might function as cooperating oncogenic events, as they do for Myc-driven tumorigenesis (Pelengaris et al., 2002). This prediction can be addressed using mouse genetic strategies that have been used to successfully define the relationship between p53 and Myc, and more generally between Myc and apoptosis, in tumor progression (Pellengaris et al., 2002). Furthermore, although our results are consistent with loss of Mnt and overexpression of Myc having similar or overlapping consequences in MEFs and in breast epithelium, it will be important to extend these findings to other tissues and cell types in vivo to determine whether loss of Mnt, like overexpression of Myc, can widely contribute to tumorigenesis.
Materials and methods
Mnt knockout mice and conditional knockout mice
A conditional knockout construct for Mnt was designed to flank exons 4 and 6 with loxP sites, and a Hygro selection cassette was placed inside of the loxP site in intron 3 (K.Toyo-oka, S.Hirotsune, Z.-Q.Zhou, R.N.Eisenman, P.J.Hurlin and A.Wynshaw-Boris, manuscript in preparation). This construct was transfected into embryonic stem cells, and targeted clones were identified and used to produce Mnt+/cko mice. Using the female germline deleter EIIa-Cre transgenic mice (Lakso et al., 1996), Cre-mediated deletion of Mnt-loxP allele was performed in vivo and resulted in the deletion of the C-terminal amino acid residues from 235 to the end of Mnt at 591 amino acids, eliminating the Hygro gene as well. No Mnt-specific transcripts could be detected by RT–PCR in RNA obtained from embryos homozygous for the disrupted Mnt gene, indicating that this disruption produced a –/– allele. To delete Mnt only in the breast, MMTV-Cre mice (Wagner et al., 1997) were mated with Mnt+/cko mice to ultimately produce MMTV-Cre;Mntcko/cko mice.
Cell culture
MEFs were prepared essentially as described previously (Zindy et al., 1998). For cell cycle entry experiments, cells were plated at 2 × 106 cells per 100 mm dish and grown for 5 days, with fresh medium being added every other day. Confluence-arrested cells were replated at 2 × 106 cells per 100 mm dish in medium containing 0.1% serum for 24 h prior to adding medium containing 20% serum. To monitor entry into the cell cycle, cells were harvested at specific time points following serum stimulation, fixed in cold 70% ethanol, washed three times with PBS containing 0.02 mg/ml propidium iodide, 0.1% Triton X-100 and 0.2 mg/ml DNase-free RNase and analyzed by FACS using a Becton Dickinson FACSAN apparatus and Multicycle and Cellquest software. For BrdU incorporation, cells growing on cover slips were incubated for 30 min with 20 µM BrdU prior to washing in PBS and fixing in ethanol. Cells were then incubated with mouse anti-BrdU antibody (Roche), and BrdU antibody was visualized using a secondary fluorescein-conjugated anti-mouse Ig antibody (Molecular Probes).
For apoptosis assays, subconfluent, logarithmically growing MEFs were subjected to a drop in fetal bovine serum (FBS) levels from 10 to 0.1%. Apoptotic cells were identified by TUNEL assays using a kit (Roche). Caspase 3 activation was determined by western blot using an anti-caspase 3 antibody (Cell Signaling) recognizing both full-length and cleaved (activated) forms.
Retroviral infection and transformation assays
Recombinant retroviruses were generated by either stable or transient transfection of HEK 293Eco-packaging cells (Stratagene) according to the manufacturer’s protocols. Retroviral constructs included pBabe-puro, pBabe-puro-Myc, pBabe-puro-Hrasv12 and pFb-Mnt. The latter virus contains the mouse cDNA in the vector pFb (Stratagene). For infection, subconfluent MEFs were incubated at 37°C with viral supernatants in the presence of 4 µg/ml polybrene (Sigma). Viral infection efficiencies were determined by performing parallel infections with virus expressing green fluorescent protein (pBabe-puro-EGFP) and visual inspection. For Ras transformation experiments, MEFs were infected with pBabe-HRasV12 virus for 16–24 h. For c-Myc plus Ras transformation experiments, cells were first infected with Myc virus and, 48 h later, infected with H-Ras virus. Transformation experiments were stained with methylene blue 14 days after infections. For soft agar assays, cells were suspended in 0.3% agar in DMEM containing 10% FBS and plated on top of solidified agar (0.5%).
Western blots, kinase assays and antibodies
Western blots and kinase assays were performed as described previously (Mateyak et al., 1999). Histone H1 (Sigma) and GST–RB (RB amino acids 769–921) were used as substrate. Antibodies used in western blots and kinase assays included affinity purified anti-Mnt (Hurlin et al., 1997) and antibodies against p19ARF (ab80; Abcam), p27KIP1 (57; BD Pharmingen), p53 (FL-393; Santa Cruz), p21Cip1 (F-5; Santa Cruz) Tubulin (AC-40; Sigma), caspase 3 (9662; Cell Signaling), cleaved caspase 3 (9661; Cell Signaling), cyclin D1 (AB-3; Oncogene Sciences), cyclin D2 (H-289; Santa Cruz), cyclin E1 (M-20; Santa Cruz), Cdk2 (M2; Santa Cruz), Cdk4 (H-22; Santa Cruz), c-Myc N-262 (Santa Cruz), c-Myc 06-213 (Upstate), PCNA (PC10; Santa Cruz) and E2f2 (C-20; Santa Cruz).
Northern blot and ChIP analysis
Northern blots were performed using Hybond+ nylon membranes (Amersham) according to the manufacturer’s directions. cDNA probes were generated using RT–PCR and labeled with [32P]CTP by standard random primer method. ChIP assays were performed as described previously (Shang et al., 2000), using 2 × 107 cells per immunoprecipitation. The PCR primer pair for Cdk4 promoter region A (Cdk4 pro A) was CACCACACCCCTCCCATCAAA and CCCACCCTCACCATGTGA CCA. These primers amplify a 140 bp region containing an E-box (CACGTG) 119 bp from the transcriptional start site. The PCR primer pair for a 167 bp control region containing a CACGTG E-box 4292 bp upstream of the transcriptional start site was GGTTCGGAGCCC ACTGTCCAG and ACGGCGACGAGGAGGAGTACTA.
Acknowledgments
Acknowledgements
This work was supported by grants from the NIH to A.W.-B., and from the NIH and Shriners Hospitals for Children to P.J.H.
References
- Amati B., Alevizopoulos,K. and Vlach,J. (1998) Myc and the cell cycle. Front. Biosci., 3, D250–D268. [DOI] [PubMed] [Google Scholar]
- Askew D.S.,R.A Ashmun,B.C., Simmons, and Cleveland,J.L. (1991) Constitutive c-myc expression in an IL3-dependent myeloid cell line suppresses cell cycle arrest and accelerates apoptosis. Oncogene, 6, 1915–1922. [PubMed] [Google Scholar]
- Chau B.N. and Wang,J.Y. (2003) Coordinated regulation of life and death by RB. Nat. Rev. Cancer, 3, 130–138. [DOI] [PubMed] [Google Scholar]
- Cornelis R.S. et al. (1994) Evidence for a gene on 17p13.3, distal to TP53, as a target for allele loss in breast tumors without p53 mutations. Cancer Res., 54, 4200–4206. [PubMed] [Google Scholar]
- de Alboran I.M., O’Hagan,R.C., Gartner,F., Malynn,B., Davidson,L., Rickert,R., Rajewsky,K., DePinho,R.A. and Alt,F.W. (2001) Analysis of C-MYC function in normal cells via conditional gene-targeted mutation. Immunity, 14, 45–55. [DOI] [PubMed] [Google Scholar]
- Dyson N. (1998) The regulation of E2F by pRB-family proteins. Genes Dev., 12, 2245–2262. [DOI] [PubMed] [Google Scholar]
- Eberhardy S.R., D’Cunha,C.A. and Farnham,P.J. (2000) Direct examination of histone acetylation on Myc target genes using chromatin immunoprecipitation. J. Biol. Chem., 275, 33798–33805. [DOI] [PubMed] [Google Scholar]
- Eisenman RN. (2001) Deconstructing myc. Genes Dev., 15, 2023–2030. [DOI] [PubMed] [Google Scholar]
- Evan G.I., Wyllie,A.H., Gilbert,C.S., Littlewood,T.D., Land,H., Brooks,M., Water,C.M., Penn,L.Z., and Hancock,D.C. (1992) Induction of apoptosis in fibroblasts by c-myc protein. Cell, 69, 119–128. [DOI] [PubMed] [Google Scholar]
- Grandori C., Cowley,S.M., James,L.P., and Eisenman,R.N. (2000) The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu. Rev. Cell Dev. Biol., 16, 653–699. [DOI] [PubMed] [Google Scholar]
- Hermeking H. et al. (2000) Identification of CDK4 as a target of c-MYC. Proc. Natl Acad. Sci. USA, 97, 2229–2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoff C., Seranski,P., Mollenhauer,J., Korn,B., Detzel,T., Reinhardt,R., Ramser,J. and Poustka,A. (2000) Physical and transcriptional mapping of the 17p13.3 region that is frequently deleted in human cancer. Genomics, 70, 26–33. [DOI] [PubMed] [Google Scholar]
- Hurlin P.J., Queva,C., Koskinen,P.J., Steingrimsson,E., Ayer,D.E., Copeland,N.G., Jenkins,N.A. and Eisenman,R.N. (1995) Mad3 and Mad4: novel Max-interacting transcriptional repressors that suppress c-Myc dependent transformation and are expressed during neural and epidermal differentiation. EMBO J., 15, 2030–2041. [PMC free article] [PubMed] [Google Scholar]
- Hurlin P.J., Queva,C., and Eisenman,R.N. (1997) Mnt, a novel Max-interacting protein is coexpressed with Myc in proliferating cells and mediates repression at Myc binding sites. Genes Dev., 11, 44–58. [DOI] [PubMed] [Google Scholar]
- Hurlin P.J., Steingrimsson,E., Copeland,N.G., Jenkins,N.A. and Eisenman,R.N. (1999) Mga, a dual-specificity transcription factor that interacts with Max and contains a T-domain DNA-binding motif. EMBO J., 18, 7019–7028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson J.N. and Muller,W.J. (2000) Transgenic mouse models of human breast cancer. Oncogene, 19, 6130–6137. [DOI] [PubMed] [Google Scholar]
- Kamijo T., Zindy,F., Roussel,M.F., Quelle,D.E., Downing,J.R., Ashmun,R.A., Grosveld,G. and Sherr,C.J. (1997) Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell, 91, 649–659. [DOI] [PubMed] [Google Scholar]
- Karsunky H., Geisen,C., Schmidt,T., Haas,K., Zevnik,B., Gau,E. and Moroy,T. (1999) Oncogenic potential of cyclin E in T-cell lymphomagenesis in transgenic mice: evidence for cooperation between cyclin E and Ras but not Myc. Oncogene, 18, 7816–7824. [DOI] [PubMed] [Google Scholar]
- Lakso M., Pichel,J.G., Gorman,J.R., Sauer,B, Okamoto,Y., Lee,E., Alt,F.W. and Westphal,H. (1996) Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc. Natl Acad. Sci. USA, 93, 5860–5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Land H, Chen,A.C., Morgenstern,J.P., Parada,L.F., and Weinberg,R.A. (1986) Behavior of myc and ras oncogenes in transformation of rat embryo fibroblasts. Mol. Cell. Biol., 6, 1917–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe S.W. and Ruley,H.E. (1993) Stabilization of the p53 tumor suppressor is induced by adenovirus 5 E1A and accompanies apoptosis. Genes Dev., 7, 535–545. [DOI] [PubMed] [Google Scholar]
- Mateyak M.K., Obaya,A.J., and Sedivy,J.M. (1999) c-Myc regulates cyclin D-Cdk4 and -Cdk6 activity but affects cell cycle progression at multiple independent points. Mol. Cell. Biol., 19, 4672–4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menssen A. and Hermeking,H. (2002) Characterization of the c-MYC-regulated transcriptome by SAGE: identification and analysis of c-MYC target genes. Proc. Natl Acad. Sci. USA, 99, 6274–6279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meroni G. et al. (1997) Rox, a novel bHLHZip protein expressed in quiescent cells that heterodimerizes with Max, binds a non-canonical E box and acts as a transcriptional repressor. EMBO J., 16, 2892–2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigro C.L. et al. (1998) The human ROX gene: genomic structure and mutation analysis in human breast tumors. Genomics, 49, 275–282. [DOI] [PubMed] [Google Scholar]
- Orian A. et al. (2003) Genomic binding by the Drosophila Myc, Max, Mad/Mnt transcription factor network. Genes Dev., 17, 1101–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelengaris S., Khan,M. and Evan,G. (2002) c-MYC: more than just a matter of life and death. Nat. Rev. Cancer, 2, 764–776. [DOI] [PubMed] [Google Scholar]
- Penn L.J., Laufer,E.M. and Land,H. (1990) C-MYC: evidence for multiple regulatory functions. Semin. Cancer Biol., 1, 69–80. [PubMed] [Google Scholar]
- Quelle D.E., Zindy,F., Ashmun,R.A. and Sherr,C.J. (1995) Alternative reading frames of the INK4a tumor suppressor gene encode two unrelated proteins capable of inducing cell cycle arrest. Cell, 83, 993–1000. [DOI] [PubMed] [Google Scholar]
- Queva C., Hurlin,P.J., Foley,K.P. and Eisenman,R.N. (1998) Sequential expression of the MAD family of transcriptional repressors during differentiation and development. Oncogene, 16, 967–977. [DOI] [PubMed] [Google Scholar]
- Serrano M., Lin,A.W., McCurrach,M.E., Beach,D. and Lowe,S.W. (1997) Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell, 88, 593–602. [DOI] [PubMed] [Google Scholar]
- Shang Y., Hu,X., DiRenzo,J., Lazar,M.A. and Brown,M. (2000) Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell, 103, 843–852. [DOI] [PubMed] [Google Scholar]
- Sherr C.J. (2001) The INK4a/ARF network in tumour suppression. Nat. Rev. Mol. Cell Biol., 2, 731–737. [DOI] [PubMed] [Google Scholar]
- Todaro G.J. and Green,H. (1963) Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J. Cell Biol., 17, 299–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trumpp A., Refaeli,Y., Oskarsson,T., Gasser,S., Murphym,M, Martin,G.R. and Bishop,J.M. (2001) c-Myc regulates mammalian body size by controlling cell number but not cell size. Nature, 414, 768–773. [DOI] [PubMed] [Google Scholar]
- Wagner K.U., Wall,R.J., St’Onge,L., Gruss,P., Garrett,L., Wynshaw-Boris,A., Li,M., Furth,P.A. and Hennighausen,L. (1997) Cre-mediated gene deletion in the mammary gland. Nucleic Acids Res., 25, 4323–4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z.Q. and Hurlin,P.J. (2001) The interplay between Mad and Myc in proliferation and differentiation. Trends Cell Biol., 11, S10–S14. [DOI] [PubMed] [Google Scholar]
- Zindy F., Eischen,C.M., Randle,D.H., Kamijo,T., Cleveland,J.L., Sherr,C.J. and Roussel,M.F. (1998) Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev., 12, 2424–2433. [DOI] [PMC free article] [PubMed] [Google Scholar]