Abstract
The mitochondrial proteins Isu1p and Isu2p play an essential role in the maturation of cellular iron–sulfur (Fe/S) proteins in eukaryotes. By radiolabelling of yeast cells with 55Fe we demonstrate that Isu1p binds an oxygen-resistant non-chelatable Fe/S cluster providing in vivo evidence for a scaffolding function of Isu1p during Fe/S cluster assembly. Depletion of the cysteine desulfurase Nfs1p, the ferredoxin Yah1p or the yeast frataxin homologue Yfh1p by regulated gene expression causes a strong decrease in the de novo synthesis of Fe/S clusters on Isu1p. In contrast, depletion of the Hsp70 chaperone Ssq1p, its co-chaperone Jac1p or the glutaredoxin Grx5p markedly increased the amount of Fe/S clusters bound to Isu1p, even though these mitochondrial proteins are crucial for maturation of Fe/S proteins. Hence Ssq1p/Jac1p and Grx5p are required in a step after Fe/S cluster synthesis on Isu1p, for instance in dissociation of preassembled Fe/S clusters from Isu1p and/or their insertion into apoproteins. We propose a model that dissects Fe/S cluster biogenesis into two major steps and assigns its central components to one of these two steps.
Keywords: chaperones/ferredoxin/glutaredoxin/iron–sulfur proteins/mitochondria
Introduction
Proteins with iron–sulfur (Fe/S) cofactors are ubiquitous in bacteria, archaea and eukaryotes (Beinert et al., 1997). They play central roles in fundamental cellular processes including redox reactions, metabolic catalysis and the sensing of iron and ambient oxygen levels (Johnson, 1998; Beinert and Kiley, 1999). The process of Fe/S cluster biosynthesis is highly conserved in nature and is mediated by a complex assemblage of proteins (Lill and Kispal, 2000; Frazzon and Dean, 2003). In eukaryotes such as Saccharomyces cerevisiae, mitochondria play a central role in the biosynthesis of cellular Fe/S proteins. The iron–sulfur cluster (ISC) assembly machinery of mitochondria is required for the biosynthesis of mitochondrial Fe/S proteins that perform central roles in respiration (e.g. complexes I, II and III of the respiratory chain) and the citric acid cycle (e.g. aconitase). The yeast mitochondrial ISC components are also essential for the maturation of cytosolic Fe/S proteins such as isopropylmalate isomerase Leu1p (Kispal et al., 1999; Kaut et al., 2000; Lange et al., 2000; Li et al., 2001). In addition, components of the mitochondrial ‘ISC export machinery’ are needed for maturation of these cytosolic Fe/S proteins. Known members of this machinery are the ABC transporter of the mitochondrial inner membrane Atm1p, the intermembrane space protein Erv1p and glutathione (Kispal et al., 1999; Lange et al., 2001; Sipos et al., 2002).
So far, insight into the mechanism of Fe/S protein biosynthesis has mainly been gained from in vitro experiments. Fe/S cluster assembly is initiated by the abstraction of sulfur from cysteine in a reaction catalysed by cysteine desulfurases such as bacterial IscS, NifS, SufS or yeast Nfs1p (Zheng et al., 1993, 1994; Clausen et al., 2000). The highly conserved ISC assembly proteins IscU and NifU play a central role for Fe/S cluster formation in the bacterial ISC and NIF systems, respectively. In S.cerevisiae the homologous protein pair Isu1p/Isu2p is essential, and loss of function is associated with severe defects in Fe/S cluster maturation inside and outside mitochondria (Schilke et al., 1999; J.Gerber, unpublished data). IscU has been shown in vitro to bind iron in a labile fashion with micromolar affinity (Agar et al., 2000b; Nuth et al., 2002). A [2Fe–2S] cluster can be assembled on IscU and NifU by chemical or IscS/NifS-directed reconstitution in vitro, but at rather slow rates (reviewed by Frazzon and Dean, 2003). The preformed cluster can subsequently be transferred from IscU to an Fe/S apoprotein. Therefore it was proposed that IscU-like proteins serve as scaffolds for the de novo biosynthesis of cellular Fe/S clusters in general. However, so far no data have been provided to demonstrate that IscU-like proteins actually bind Fe/S clusters in vivo. IscU/Isu1p/Isu2p interacts with IscS/Nfs1p, the ATP-dependent Hsp70 chaperone HscA (yeast homologue Ssq1p) and its cognate J-type co-chaperone HscB (yeast homologue Jac1p) (Hoff et al., 2000; Silberg et al., 2001; Voisine et al., 2000, 2001; Kim et al., 2001; Dutkiewicz et al., 2003). Binding of IscU to HscA/HscB stimulates the ATPase activity of the Hsp70 chaperone, but the mechanistic meaning of the association remains unclear.
Besides these central components of biosynthesis, a number of other proteins have been identified with a crucial role in Fe/S protein assembly, yet their site and mode of action in biogenesis is enigmatic. For instance, the requirement of an electron transfer chain consisting of ferredoxin (yeast Yah1p) and ferredoxin reductase (yeast Arh1p) is well established, but the molecular reaction requiring electron flow is unknown (Nakamura et al., 1999; Lange et al., 2000; Li et al., 2001). Further, non-essential roles are contributed by bacterial IscA and its yeast homologues Isa1p and Isa2p (Jensen and Culotta, 2000; Kaut et al., 2000; Pelzer et al., 2000; Krebs et al., 2001; Ollagnier-de-Choudens et al., 2001; Wu et al., 2002a), Grx5p, a mitochondrial isoform of glutaredoxin (Rodriguez-Manzaneque et al., 2002), and Yfh1p, the yeast homologue of frataxin (Chen et al., 2002; Mühlenhoff et al., 2002b). In vitro reconstitution of mitochondrial Fe/S protein biogenesis revealed a requirement for cysteine, ATP, NADH and reduced iron as essential low-molecular-mass cofactors (Mühlenhoff et al., 2002a).
Although our knowledge about Fe/S cluster formation in biological systems has been increased considerably in recent years, fundamental problems have not yet been resolved. For instance, very little is known about the mechanistic function and the site of action of most ISC assembly proteins and the cooperation of the ISC proteins during Fe/S cluster assembly in vivo. In this work, we have developed a novel experimental system that establishes the de novo synthesis of an Fe/S cluster on S.cerevisiae Isu1p in vivo. By employing yeast mutants facilitating the rapid depletion of various ISC components, we were able to define the site of involvement of these proteins in either de novo Fe/S cluster formation on Isu1p or steps thereafter. The investigation provides for the first time a mechanistic insight into the participation of individual ISC components in the two major steps of Fe/S protein assembly in vivo.
Results
The mitochondrial matrix protein Isu1p binds an Fe/S cluster in vivo
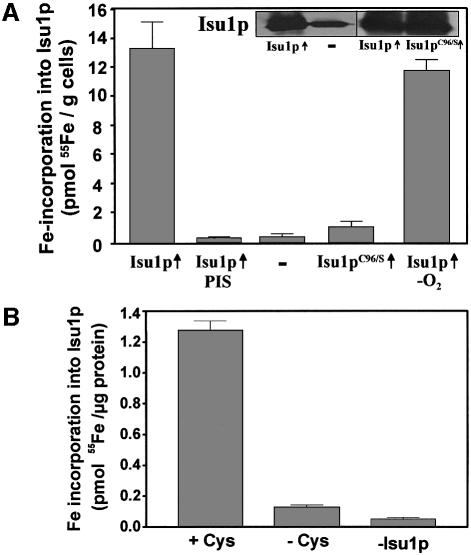
In order to analyse the potential association of iron (or an Fe/S cluster) with Isu1p in vivo, S.cerevisiae ISU1 was overexpressed in wild-type cells from a high copy vector under the control of the strong promoter of TDH3. As judged by immunostaining, an ∼10-fold overproduction of the protein could be obtained (Figure 1A, inset). Cells were radiolabelled with 55Fe in vivo, and after cell lysis Isu1p was isolated by immunoprecipitation with polyclonal antibodies raised against recombinant Isu1p. Immuno precipitation of Isu1p was almost quantitative (not shown). A significant amount of radioactive 55Fe co-immunoprecipitated with the immunobeads using cells overproducing Isu1p (Figure 1A). Iron association with Isu1p was highly specific, since only background levels of radioactivity (<0.3 pmol 55Fe/g cells) were detected when the immunoprecipitation was carried out with preimmune serum or when cells were used that did not overproduce Isu1p. As a further criterion for the specificity of iron binding, we took advantage of cells overproducing a site-directed mutant of Isu1p that carries a serine residue instead of a cysteine at position 96 (Isu1pC96S). Iron association with Isu1pC96S declined to almost background levels despite high levels of the mutant protein (Figure 1A). This result is consistent with findings on bacterial NifU in vitro, where cysteine 62 (corresponding to residue 96 of the precursor form of Isu1p) has been shown to be required for iron binding in vitro and hence was proposed to be one of the ligands for transient Fe/S cluster association (Agar et al., 2000b).
Fig. 1. Stable binding of an Fe/S cluster to Isu1p. (A) Yeast cells overproducing (↑) wild-type Isu1p or the site-directed mutant Isu1pC96S from the plasmid p426GPD were grown in ‘iron-poor’ minimal medium with glucose for 16 h. Control cells (–) contained the empty plasmid. Cells were radiolabelled with 55Fe for 2 h and cell lysates were prepared (Kispal et al., 1999). Isu1p was immunoprecipitated with anti-Isu1p antibodies under aerobic or anaerobic (–O2) conditions and the amount of radioactivity coprecipitated with the immuno-beads was quantitated by liquid scintillation counting. A control immuno precipitation was performed with preimmune serum (PIS). The inset shows an immunostaining of Isu1p in extracts from wild-type cells (–) or cells overproducing Isu1p or Isu1pC96S (↑). Error bars indicate the standard deviation of the measurements. (B) Isu1p and Nfs1p were purified after overproduction in Escherichia coli. Isu1p (10 µg) and Nfs1p (5 µg) were incubated with 55Fe in the presence or absence of cysteine (Cys) for 150 min under anaerobic conditions developed for in vitro reconstitution of mitochondrial Fe/S protein biogenesis (Mühlenhoff et al., 2002a). A labelling reaction lacking Isu1p (–Isu1p) served as control. Reactions were terminated with EDTA, Isu1p was immunoprecipitated and the amounts of co-immunoprecipitated 55Fe were determined by scintillation counting.
Iron binding to Isu1p was remarkably stable, as bound iron was not removed by treatment with EDTA and Triton X-100 which were routinely present in our immunoprecipitation buffer. No significant increase of iron binding was observed when these two compounds were omitted (not shown). Moreover, iron association with Isu1p was unchanged when the entire purification proce dure (taking 1.5 h) was carried out under anaerobic conditions (Figure 1A). This indicates that the iron bound to Isu1p was not removed by chelators and remained stably associated even under aerobic conditions.
To investigate whether iron is bound to Isu1p as mononuclear iron or in the form of an Fe/S cluster, we purified both Isu1p and the cysteine desulfurase Nfs1p and used these proteins for in vitro reconstitution of iron binding to Isu1p. Both proteins were incubated in the presence of radioactive 55Fe under anaerobic conditions followed by immunoprecipitation of Isu1p and scintillation counting. Efficient 55Fe binding to the immunobeads was strictly dependent on the presence of Isu1p and cysteine (Figure 1B). Apparently, iron binding to Isu1p depended on the production of sulfide from cysteine by Nfs1p. These results and the non-chelatable character of Isu1p-bound iron suggest that bound iron was part of an Fe/S cluster (see also below).
Fe/S cluster binding to Isu1p in vivo requires Nfs1p
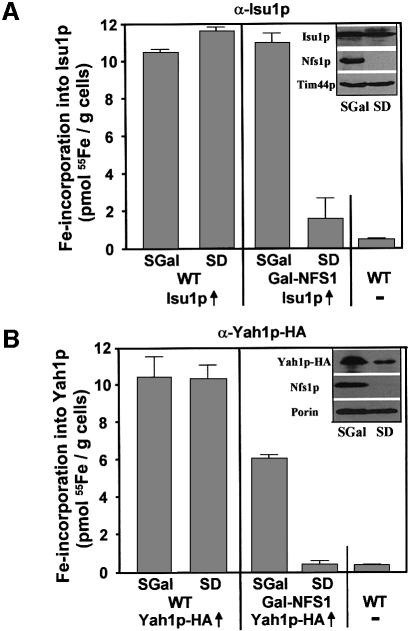
In order to identify which components of the ISC assembly machinery are needed for stable association of iron with Isu1p in vivo, the ISU1 reporter plasmid was used to transform various yeast mutant cells in which the ISC proteins can be depleted rapidly by regulated gene expression. In parallel, the mutant cells were transformed with plasmids that supported the overproduction of a haemagglutinin (HA) tagged version of the [2Fe-2S] ferredoxin Yah1p (termed Yah1p-HA) serving as a model Fe/S protein. At first, we investigated the effect of depletion of the cysteine desulfurase Nfs1p on iron binding to Isu1p. To this end, we used the strain Gal-NFS1 that carries the essential NFS1 gene under the control of the regulatable GAL1-10 promoter which is induced in the presence of galactose (SGal medium) and repressed in the presence of glucose (SD medium). The latter condition resulted in a strong depletion of Nfs1p relative to other mitochondrial proteins (Figure 2, inset; Kispal et al., 1999) and an eventual growth arrest of these cells. After cultivation in these media, cells were radiolabelled with 55Fe, and cell extracts were analysed for radioactivity associated with immunoprecipitated Isu1p and Yah1p-HA. Similar amounts of radioactive 55Fe were co-immunoprecipitated with either of these proteins in wild-type cells grown in glucose- or galactose-containing media or with Gal-NFS1 cells cultivated in the presence of galactose (Figure 2). Iron association was highly specific, as only background signals (<0.5 pmol 55Fe/g cells) were obtained with cells that did not overproduce Isu1p or Yah1p-HA.
Fig. 2. Assembly of an Fe/S cluster on Isu1p in vivo requires the cysteine desulfurase Nfs1p. Wild-type (WT) and Gal-NFS1 cells overproducing (↑) (A) Isu1p or (B) an HA-tagged version of Yah1p (Yah1p-HA) were incubated in iron-poor medium supplemented with galactose (SGal) or glucose (SD) in order to downregulate the expression of NFS1 in Gal-NFS1 cells. Cells were radiolabelled with 55Fe, Isu1p and Yah1p-HA were immunoprecipitated from the cell lysates and the amount of co-immunoprecipitated 55Fe was quantified by scintillation counting. As a control, wild-type cells that did not overproduce the two proteins were analysed. The insets show the immunostaining of the indicated mitochondrial proteins in extracts derived from Gal-NFS1 cells grown in the different minimal media.
On the contrary, upon growth in the presence of glucose the amount of iron associated with both proteins strongly declined to almost background levels, although they were still overproduced under these conditions (Figure 2, inset). These data indicate that Nfs1p, in addition to its essential role in the maturation of standard Fe/S proteins, is required for stable binding of iron to Isu1p in vivo. Therefore we conclude from the in vitro and in vivo dependence of iron binding upon the sulfur donor Nfs1p that the iron associated with Isu1p is bound in form of an Fe/S cluster. The results for the first time provide in vivo evidence that an Fe/S cluster can be assembled on IscU-like proteins. Hence, they extend the pioneering in vitro studies on the transient binding of an Fe/S cluster on bacterial NifU and IscU (Agar et al., 2000a; Yuvaniyama et al., 2000). In these studies, only an Fe/S cluster, and not mononuclear iron, remained stably associated with the rubredoxin-like binding site of the scaffold protein (Agar et al., 2000b; Nuth et al., 2002). These results are similar to our findings, which did not detect stably bound iron without the activity of the sulfur donor Nfs1p. We further conclude that the extreme oxygen sensitivity of Fe/S cluster synthesis observed in vitro (Figure 1B; Mühlenhoff et al., 2002a) is not due to damage of the transient Fe/S cluster on Isu1p, but rather is explained by synthesis of the cluster.
Mutual requirement of Isu1p and Yah1p for their assembly with Fe/S clusters
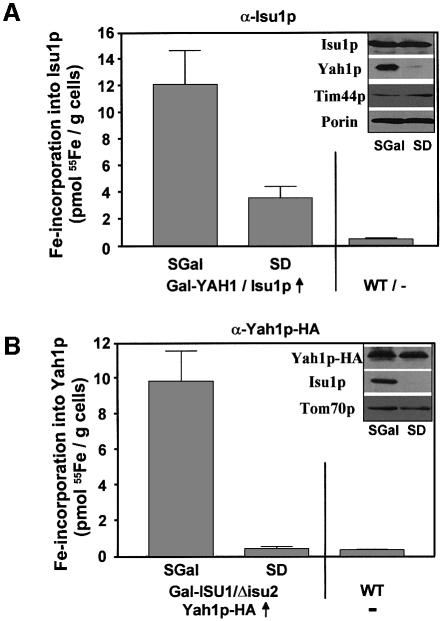
We asked whether the Fe/S cluster formation on Isu1p was dependent on the electron transfer chain comprised of the ferredoxin Yah1p and its reductase Arh1p. To this end, we analysed the Fe/S cluster formation on Isu1p in the conditional yeast strain Gal-YAH1 (Lange et al., 2000). The amount of 55Fe bound to overproduced Isu1p drastically declined upon downregulation of Yah1p in this strain, although the protein level of Isu1p and other mitochondrial proteins remained unchanged under these conditions (Figure 3A, inset). The decline in Fe/S cluster formation on Isu1p upon depletion of Yah1p is comparable to the defects in the maturation of mitochondrial Fe/S-cluster-containing proteins such as aconitase in the absence of this protein (Lange et al., 2000). These data indicate that Yah1p is directly involved in the de novo synthesis of Fe/S clusters on Isu1p. Apparently, a reduction step involving Yah1p, and presumably Arh1p, is required for the formation of the transient Fe/S cluster on Isu1p.
Fig. 3. Fe/S cluster assembly on Isu1p depends on the ferredoxin Yah1p. (A) Gal-YAH1 cells overproducing (↑) Isu1p and (B) Gal-ISU1/Δisu2 cells overproducing HA-tagged Yah1p were cultivated in iron-poor SGal or SD minimal media in order to allow or repress, respectively, synthesis of Yah1p and Isu1p. In parallel, wild-type (WT) cells were analysed. After radiolabelling with 55Fe, Isu1p or Yah1p-HA were immunoprecipitated from cell lysates and the amounts of co-immunoprecipitated 55Fe were determined. The insets show immunostaining of various mitochondrial proteins in extracts derived from the two mutant cells after growth in the indicated media.
For the analysis of the consequences of Isu1p depletion on Fe/S cluster formation on Yah1p we made use of a conditional mutant strain (Gal-ISU1/Δisu2) that carries the ISU1 gene under control of the GAL1-10 promoter and has the ISU2 gene deleted (Gerber et al., 2003). When this strain carrying an HA-tagged YAH1-overexpressing plasmid was grown under non-permissive conditions in order to downregulate ISU1 expression, the amount of radioactive iron co-immunoprecipitated with Yah1p-HA declined to background levels, although the level of the Yah1p-HA apoprotein was not affected (Figure 3B). This finding indicates that the Isu proteins are essential for the assembly of this [2Fe-2S] protein. Therefore we conclude that the [2Fe-2S] cluster on Yah1p is formed by the same general Isu-protein-dependent pathway as the [4Fe-4S] cluster of other cellular Fe/S proteins, even though Yah1p is a peculiar Fe/S protein in that it is required for its own biosynthesis.
Fe/S cluster assembly on Isu1p and Yah1p requires frataxin
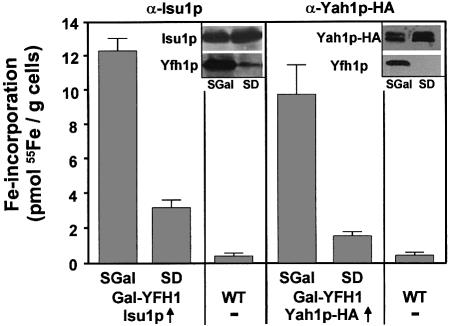
Further, we analysed the involvement of the yeast frataxin homologue Yfh1p in Fe/S cluster formation on Isu1p and Yah1p. Recently, Yfh1p has been shown to be necessary for Fe/S protein maturation in mitochondria (Mühlenhoff et al., 2002b). However, the precise role of Yfh1p in Fe/S protein formation remains unknown. In order to investigate a possible function of Yfh1p in early steps of Fe/S cluster biogenesis, Isu1p and Yah1p-HA were overproduced in the promoter-exchange mutant Gal-YFH1 (Mühlenhoff et al., 2002b) and cells were radiolabelled with 55Fe. The amount of 55Fe that was co-immunoprecipitated with Isu1p and Yah1p-HA strongly declined in this strain upon downregulation of Yfh1p (Figure 4). Since the expression of the respective proteins did not change upon depletion of Yfh1p (Figure 4, insets), we conclude that Yfh1p, similar to Nfs1p and Yah1p, participates in Fe/S cluster formation on the Isu1p scaffold.
Fig. 4. Depletion of frataxin (Yfh1p) results in decreased Fe/S cluster formation on Isu1p. Gal-YFH1 cells overproducing (↑) Isu1p and Yah1p-HA or wild-type (WT) cells were grown under conditions permissive (SGal) and repressive (SD) for synthesis of Yfh1p. After radiolabelling with 55Fe and cell lysis, immunoprecipitations were carried out using specific antisera and the amounts of co-immuno precipitated radioactivity were quantified. The insets show immuno staining of various proteins in extracts derived from Gal-YFH1 cells.
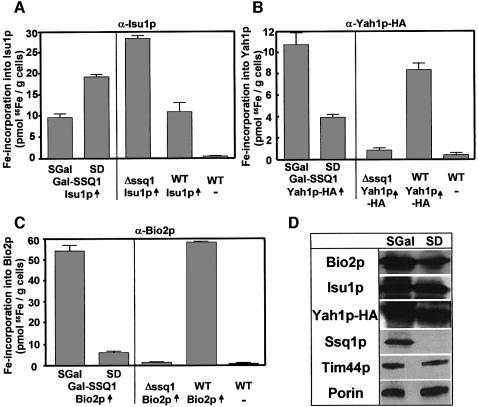
Fe/S clusters accumulate on Isu1p upon depletion of the chaperones Ssq1p and Jac1p
The site of action of the mitochondrial Hsp70 chaperone Ssq1p and its J-type co-chaperone Jac1p in Fe/S protein biogenesis is not known. Therefore we examined their potential roles in Fe/S cluster synthesis on Isu1p and compared it with the de novo formation of Yah1p-HA and Bio2p holoproteins. We used an SSQ1 deletion strain (Δssq1) and the promoter exchange mutant Gal-SSQ1 that had both been transformed with the ISU1 overexpression plasmid (see above). After 55Fe radiolabelling and cell lysis as described above, approximately wild-type levels of 55Fe could be immunoprecipitated by Isu1p-specific antibodies in Gal-SSQ1 cells under growth conditions enabling synthesis of Ssq1p (Figure 5A). Strikingly, the amount of 55Fe labelling of Isu1p doubled when the Gal-SSQ1 strain was grown in glucose-containing SD media in order to deplete Ssq1p levels. In Δssq1 cells, the amount of Isu1p-bound 55Fe/S cluster increased even threefold compared with the wild-type situation. Since the levels of the Isu1p polypeptide chain remained unaffected upon depletion of Ssq1p (Figure 5D), these data clearly show that the synthesis of Fe/S clusters on Isu1p does not involve functional Ssq1p. Rather, the observed ‘trapping’ of Fe/S clusters on Isu1p indicates a critical role of Ssq1p in a step subsequent to Fe/S cluster assembly on Isu1p.
Fig. 5. The Hsp70 chaperone Ssq1p is involved in the de novo maturation of mitochondrial Fe/S proteins, but not required for Fe/S cluster assembly on Isu1p in vivo. Iron-starved wild-type (WT), Δssq1 and Gal-SSQ1 cells overproducing (↑) (A) Isu1p, (B) Yah1p-HA and (C) Bio2p (grown as in Figure 2) were radiolabelled with 55Fe. Immunoprecipitation was carried out using specific antisera against these proteins and the amounts of co- immunoprecipitated radioactivity were determined by liquid scintillation counting. (D) Immunostaining of various mitochondrial proteins in extracts from Gal-SSQ1 cells grown in galactose-containing (SGal) or glucose-containing (SD) minimal media.
So far, a role of Ssq1p in the de novo assembly of an Fe/S cluster has not been demonstrated. In order to verify such a function of Ssq1p in our assay system, both Yah1p-HA and Bio2p were overproduced in Gal-SSQ1, Δssq1 and wild-type cells. Upon growth of wild-type or Gal-SSQ1 cells in the presence of galactose, large amounts of 55Fe could be immunoprecipitated using HA-tag- or Bio2p-specific antibodies (Figure 5B and C), whereas only background signals (<1 pmol 55Fe/g cells) were obtained with cells that did not overproduce these proteins. On the contrary, a strong reduction in the amounts of 55Fe associated with both Yah1p-HA and Bio2p was observed upon depletion of Ssq1p by growth in the presence of glucose. A similar effect was observed in the Δssq1 strain. Since the synthesis of the Bio2p and Yah1p-HA apoproteins remained similar in the Gal-SSQ1 and Δssq1 strains under both growth conditions (Figure 5D; data not shown), these data establish a crucial function of Ssq1p in the de novo maturation of mitochondrial Fe/S proteins. Thus the findings extend earlier reports that demonstrated defects of mitochondrial Fe/S enzyme activities in strains defective in SSQ1 (Strain et al., 1998; Schilke et al., 1999). Apparently, these defects were direct consequences of impaired Fe/S cluster maturation rather than of decreased stability of the Fe/S cluster-containing proteins.
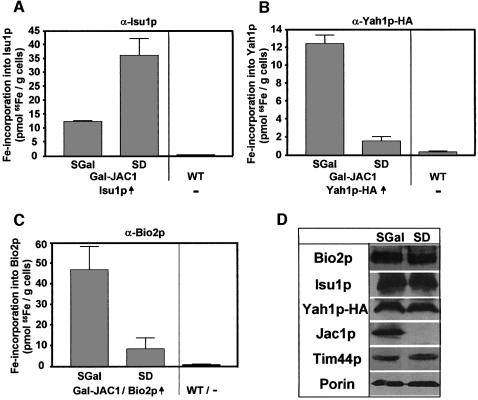
In addition, we analysed the de novo Fe/S cluster formation upon depletion of Jac1p in the promoter exchange mutant Gal-JAC1. Since Ssq1p and Jac1p functionally cooperate (Dutkiewicz et al., 2003), a phenotype of this mutant similar to that of the Gal-SSQ1 strain seemed likely. Isu1p, Yah1p-HA and Bio2p were overproduced in Gal-JAC1 cells carrying the GAL1-10 promoter upstream of the JAC1 gene. The association of 55Fe with the overproduced proteins was tested after radiolabelling, cell lysis and immunoprecipitation. The amount of 55Fe co-immunoprecipitated with Isu1p increased 3-fold in the Gal-JAC1 strain upon depletion of Jac1p (Figure 6A). In contrast, a strong decline of 55Fe associated with both Yah1p-HA and Bio2p almost to the levels detected with cells that did not overproduce these proteins was found under these conditions (Figure 6B and C), although the levels of the various overproduced Fe/S proteins did not change significantly upon downregulation of Jac1p (Figure 6D). Hence the consequences of depleting the two chaperones Ssq1p and Jac1p on the association of Fe/S clusters with Isu1p, Yah1p and Bio2p are strikingly similar. Taken together, our data clearly show that the Ssq1p and Jac1p chaperones are crucial for the general process of Fe/S protein maturation. However, they are not required for the de novo synthesis of an Fe/S cluster on Isu1p as their depletion leads to an accumulation of Fe/S clusters on the Isu1p scaffold. Therefore the chaperones seem to be involved in a later step of Fe/S protein biogenesis.
Fig. 6. Increased accumulation of an Fe/S cluster on Isu1p upon depletion of Jac1p in vivo. Gal-JAC1 cells overproducing (↑) (A) Isu1p, (B) Yah1p-HA and (C) Bio2p (grown as in Figure 2) were labelled with radioactive 55Fe in vivo, immunoprecipitations were carried out using specific antibodies against the three overproduced proteins and the amounts of co-immunoprecipitated radioactivity were quantified. As a control, wild-type (WT) cells that did not overproduce these proteins were analysed. (D) Immunostaining of various mitochondrial proteins in extracts from Gal-JAC1 cells grown in the presence of galactose (SGal) or glucose (SD).
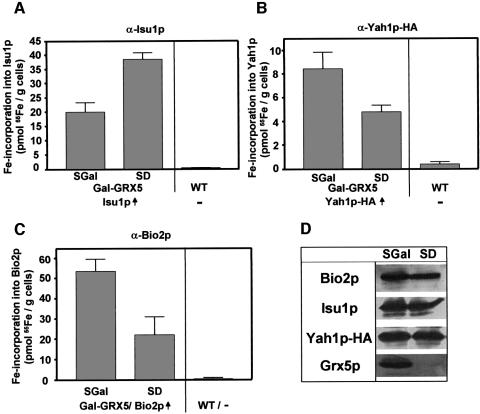
Depletion of mitochondrial glutaredoxin Grx5p results in accumulation of Fe/S clusters on Isu1p
Finally, we investigated the role of the mitochondrial glutaredoxin Grx5p in the de novo Fe/S cluster synthesis. Grx5p has recently been shown to be required for the maintenance of Fe/S protein activity in mitochondria (Rodriguez-Manzaneque et al., 2002), but its function has remained unclear. A GAL1-10 promoter exchange mutant (termed Gal-GRX5), allowing the depletion of Grx5p, was constructed and transformed with overexpression plasmids carrying the ISU1, HA-tagged YAH1 and BIO2 genes. Upon growth in glucose-containing minimal media, Grx5p was depleted to levels undetectable by immunostaining while the amounts of Isu1p, Yah1p-HA and Bio2p were comparable to those found after cultivation in the presence of galactose allowing synthesis of Grx5p (Figure 7D). When these cells were radiolabelled with 55Fe, a 2-fold increase of Fe/S clusters associated with Isu1p was found upon depletion of Grx5p (Figure 7A). Concomitantly, the de novo biosynthesis of the Fe/S proteins Yah1p-HA and Bio2p was decreased 2- to 3-fold (Figure 7B and C). Even though this defect is rather weak compared with the ISC mutants examined above, the data support the suggestion that Grx5p performs a direct function in Fe/S protein biogenesis (Rodriguez-Manzaneque et al., 2002). Clearly, such a role is confined to a step after the assembly of an Fe/S cluster on Isu1p.
Fig. 7. Depletion of the mitochondrial glutaredoxin Grx5p leads to accumulation of Fe/S clusters on Isu1p. Gal-GRX5 cells were transformed with plasmids for overproduction (↑) of (A) Isu1p, (B) Yah1p-HA and (C) Bio2p. Analyses for 55Fe radiolabelling of these proteins by immunoprecipitation and the evaluation of the data was carried out as described in Figure 5. (D) Immunostaining of the indicated proteins after growth in the media used.
Discussion
In this communication, we have investigated the role of Isu1p in Fe/S protein biogenesis in mitochondria. Together with its homologue Isu2p the protein plays an essential role in this recently discovered biosynthetic process in that all Fe/S proteins of S.cerevisiae investigated so far require this protein pair for their maturation (Garland et al., 1999; Schilke et al., 1999; J.Gerber, unpublished data). Therefore insights into the function of Isu1/2p are crucial for a better molecular understanding of Fe/S protein assembly in the living cell. By using radiolabelling of yeast cells, we were able to detect iron associated with Isu1p in vivo. Iron binding to Isu1p strictly depended on the function of the sulfur donor Nfs1p. Therefore we conclude that iron is bound to Isu1p in the form of an Fe/S cluster. This is further confirmed by in vitro data showing that stable iron binding to purified Isu1p required the release of sulfur from cysteine by Nfs1p. It was not possible to directly detect Nfs1p-generated sulfur in association with Isu1p. Cysteine (or sulfur) cannot be depleted in vivo, since it is needed for protein synthesis. Therefore the specific radioactivity for cysteine/sulfur was too low to detect a 35S-labelled Fe/S cluster on Isu1p or other Fe/S proteins.
Association of an Fe/S cluster rather than mononuclear iron with Isu1p is further supported by the stable and non-chelatable binding characteristics which fit well to the properties of purified NifU and IscU, the bacterial homologues of Isu1p (Agar et al., 2000a,b,c; Yuvaniyama et al., 2000; Nuth et al., 2002). Binding of mononuclear iron to these proteins is detectable only by spectroscopic means at micromolar affinity, yet bound iron is readily displaced from the proteins upon purification. In contrast, after in vitro reconstitution of an Fe/S cluster, NifU/IscU holoproteins were stable and could be isolated. Furthermore, the Fe/S cluster binding properties of the two bacterial proteins are rather similar to those of in vivo labelled Isu1p in that the clusters are remarkably stable in the presence of the metal chelator EDTA or the detergent Triton X-100. Taken together, our findings provide the first physiological evidence for the pioneering in vitro studies demonstrating that IscU-like proteins may provide a scaffold for the de novo synthesis of an Fe/S cluster (Frazzon and Dean, 2003).
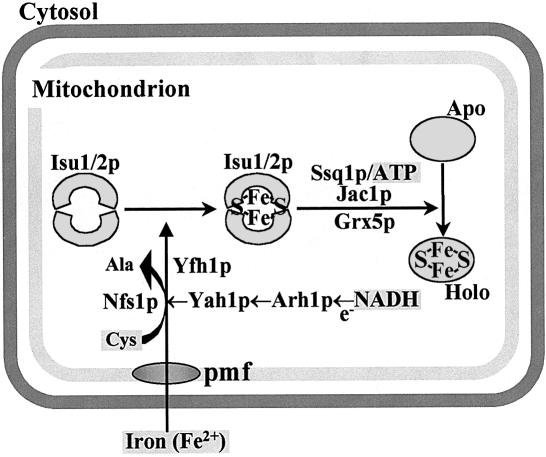
Mitochondrial Fe/S protein biogenesis is a remarkably complex process involving ∼10 ISC assembly proteins (Mühlenhoff and Lill, 2000; Gerber and Lill, 2002). The exact site of requirement of the various proteins in different steps of biogenesis was unknown hitherto. Using various conditional yeast mutants allowing the depletion of the components of the mitochondrial ISC assembly machinery, we were able to dissect the pathway into two major parts and to assign a function of the individual ISC proteins to either the de novo assembly of the Fe/S cluster on Isu1p or to cluster dislocation from Isu1p. Our findings are summarized in a model of Fe/S protein biogenesis which reserves a central role for Isu1p, and presumably Isu2p (Figure 8). After the membrane-potential-dependent import of reduced iron into the mitochondrial matrix (Lange et al., 1999) the metal ions associate with Isu1p. For stable binding to the scaffold protein, sulfur has to be provided by the cysteine desulfurase Nfs1p and is used for synthesis of an Fe/S cluster. The relative order of association of iron and sulfur with Isu1p during cluster synthesis could not be resolved by our in vivo studies, and likewise has not yet become clear from recent in vitro examinations (Smith et al., 2001; Nuth et al., 2002). Fe/S cluster synthesis on Isu1p further requires the function of the ferredoxin Yah1p and presumably the ferredoxin reductase Arh1p which receive electrons from NADH (Lange et al., 2000; Li et al., 2001; Mühlenhoff et al., 2002a). As an additional component, Yfh1p is crucial for the assembly of the Fe/S cluster on Isu1p. The order of action of the proteins required early in biogenesis is beyond the resolution of our in vivo assay and will need further detailed molecular studies.
Fig. 8. Working model for the biogenesis of Fe/S proteins in mitochondria. The essential protein pair Isu1p and Isu2p plays a crucial role in biogenesis by serving as a scaffold for de novo assembly of an Fe/S cluster. As defined here, this reaction crucially involves the cysteine desulfurase Nfs1p, the electron transport chain Yah1p/Arh1p and Yfh1p (frataxin) providing an unknown function. The chaperones Ssq1p/Jac1p and the glutaredoxin Grx5p are not needed for Fe/S cluster synthesis on Isu1/2p; rather, they may function in the dislocation of the Fe/S cluster from Isu1/2p and/or its insertion into the apoproteins. The requirement of reduced iron, NADH, ATP and cysteine has been documented by in vitro reconstitution of the Fe/S protein assembly reaction (Mühlenhoff et al., 2002a). Iron import into mitochondria by an unknown transporter is driven by a proton motive force (pmf) (Lange et al., 1999).
Proteins involved in steps later in biogenesis are the ATP-dependent Ssq1p, Jac1p and the monothiol glutaredoxin Grx5p. While depletion of Nfs1p, Yah1 and Yfh1p strongly interferes with the formation of the Fe/S cluster on Isu1p, a 2- to 5-fold increase of bound iron is detected upon depletion of those proteins. This finding demonstrates that their function in Fe/S protein biogenesis is dispensable for the generation of the Fe/S cluster-containing Isu1p holoprotein. Rather, the accumulation of Fe/S clusters on Isu1p suggests that the Fe/S cluster remains trapped after its assembly on the scaffold protein, most likely due to a block in subsequent steps of Fe/S protein biosynthesis. This strongly argues for a role of the Ssq1p/Jac1p chaperones and Grx5p in either the dissociation of the Isu1p–Fe/S cluster complex and/or the transfer of the Fe/S cluster to the apoproteins. In summary, the model depicted in Figure 8 provides for the first time distinct sites of action for the known major components of Fe/S protein biogenesis. It further allows us to deduce where the low-molecular-mass cofactors identified through in vitro reconstitution of Fe/S protein biogenesis (cysteine, ATP and NADH) are utilized (Mühlenhoff et al., 2002a).
Our data strongly support a role for Yah1p early in biogenesis during the assembly of the transiently bound Fe/S cluster on Isu1p. Yah1p, in conjunction with its reductase Arh1p, forms a one-electron chain in mitochondria. Most probably, electrons are required for reducing the sulfan sulfur (S0) released by Nfs1p to a sulfide (S2–) present in the Fe/S cluster. Other less likely possibilities are the reduction of iron or potential disulfide bonds in Isu1p and other ISC components. Iron is known to be imported in the reduced form (Lange et al., 1999) (Figure 8), and the mitochondrial matrix is a highly reducing environment that should not give rise to oxidation of ferrous iron before its utilization in biogenesis. Our results do not exclude an additional role for the electron transfer chain in later steps of biogenesis. Evidence for the importance of reducing conditions for Fe/S cluster dissociation from scaffold proteins has been collected from in vitro studies on bacterial NifU where the displacement of the transient Fe/S cluster from the N-terminal domain of NifU is stimulated by reducing conditions (Yuvaniyama et al., 2000). Since NifU carries a permanent [2Fe-2S] cluster on its central ferredoxin-like domain, it was proposed that this domain may catalyse a reductive dissociation of the transient Fe/S cluster and thus initiate its insertion into an Fe/S apoprotein. On the other hand, ferredoxin and ferredoxin reductase did not induce the dissociation of the Fe/S cluster from purified human IscU in vitro (Wu et al., 2002b).
In addition to Nfs1p and Yah1p, the frataxin homologue Yfh1p is necessary for Fe/S cluster assembly on Isu1p. This role strengthens the idea of a primary function of frataxin in Fe/S protein biogenesis (Mühlenhoff et al., 2002b). Such a function would also explain why frataxin-deficient mitochondria accumulate iron, as this behaviour appears to be a general consequence of an impairment of Fe/S protein biogenesis (Mühlenhoff and Lill, 2000). What might be the function of frataxin? We have recently identified a physical interaction of Yfh1p with the core Fe/S cluster assembly complex Isu1p/Nfs1p (Gerber et al., 2003). Therefore it is tempting to speculate that frataxin might serve a role in iron loading of Isu1p.
A striking finding of our investigation is the mutual requirement of Isu1p and Yah1p for their conversion into Fe/S holoproteins. Fe/S cluster formation on Isu1p depends on Yah1p as a source of electrons, while Yah1p requires Isu1p as a provider of its native [2Fe-2S] cluster. Despite this ‘chicken and egg’ situation, our study revealed no conspicuous differences between the formation of the [2Fe-2S] cofactor on Yah1p and the synthesis of [4Fe-4S] clusters on other mitochondrial Fe/S proteins such as aconitase or Bio2p (Lange et al., 2000). Hence, although Yah1p is special in that it is required for its own biosynthesis, there is no separate route for biosynthesis of its own [2Fe-2S] cluster. This contention fits well with recent in vitro studies which have shown that IscU can serve as a scaffold for the assembly of both types of clusters (Agar et al., 2000a; Wu et al., 2002b). Even though we used overproduced Isu1p for our analyses, we believe that the results faithfully reflect the physiological situation. The ISC proteins are of low abundance and overexpression in yeast is moderate. Therefore an increase in Isu1p may only lead to a somewhat slower dissociation of Fe/S clusters from Isu1p because factors catalysing this reaction may be limiting. In fact, this may facilitate detection of the transient Fe/S cluster by our procedure.
The mitochondrial Hsp70 chaperone Ssq1p and its J-type co-chaperone Jac1p form a specialized chaperone system that is dedicated to the formation of Fe/S proteins (Strain et al., 1998; Voisine et al., 2000, 2001; Kim et al., 2001; Lutz et al., 2001; Dutkiewicz et al., 2003). Inactivation of either protein results in loss of function of mitochondrial Fe/S proteins and the accumulation of iron in mitochondria, both criteria representing discrete indicators for defects in cellular Fe/S protein maturation (Lill and Kispal, 2000; Craig and Marszalek, 2002). Our data show that the depletion of both Jac1p and Ssq1p further results in a combination of a strong deficiency in the de novo Fe/S protein maturation on mitochondrial Fe/S proteins and, most strikingly, the accumulation of Fe/S clusters on Isu1p. Apparently, in the absence of the chaperones, Isu1p can still be loaded with Fe/S clusters, and its holoform will accumulate as a reaction intermediate. These results substantiate that Ssq1p and Jac1p form a functional unit in Fe/S protein biogenesis, yet rule out an involvement of this chaperone system in de novo Fe/S cluster assembly on Isu1p. Three alternative scenarios may explain the apparent increase of Fe/S cluster formation on Isu1p. First, the Ssq1p/Jac1p chaperone system may perform a regulatory function in that it controls the rate of Fe/S cluster assembly on Isu1p. Thus its impairment may result in an uncontrolled overloading of Isu1p with Fe/S clusters. Second, the Fe/S cluster on Isu1p may be synthesized in an incorrect transport-incompetent form in the absence of a chaperone. Third, and we believe most likely, the chaperone system is required for the dislocation of a preassembled Fe/S cluster from Isu1p (Figure 8). In support of this idea, a specific interaction between Isu1p/IscU and the two chaperones has been amply documented, for example by stimulation of the ATPase activity of Hsc66/Ssq1p by IscU/Isu1p (Hoff et al., 2000; Silberg et al., 2001; Dutkiewicz et al., 2003). How the chaperone proteins may facilitate dislocation of the Fe/S clusters from Isu1p remains enigmatic, the more so as they appear to bind to both holo- and apoforms of IscU (Hoff et al., 2000). Further studies designed to follow the displacement of preassembled Fe/S clusters from Isu1p should help to unravel the molecular function of the chaperones. Notably, a dedicated chaperone system has not yet been identified for the formation of nitrogenase (Frazzon and Dean, 2002). Hence, Fe/S cluster transfer from NifU to the nitrogenase must be either assisted by unspecialized generic chaperones or occur spontaneously.
Little is known to date about how the glutaredoxin Grx5p may contribute to Fe/S protein biogenesis. Moreover, it is not known whether this atypical isoform of glutaredoxin interacts with glutathione. Our data provide the first clue that the protein performs its crucial task late in biogenesis (Figure 8), e.g. during Fe/S cluster dissociation from Isu1p and/or insertion into the apo proteins. This knowledge will be invaluable for future in vitro studies on Fe/S cluster dislocation from Isu1p. Interestingly, overexpression of SSQ1, and to a lesser extent ISA2, suppresses the defects arising from deletion of GRX5 (Rodriguez-Manzaneque et al., 2002). This result fits nicely with our observation that Ssq1p and Grx5p participate in the same step of Fe/S protein biogenesis.
The model depicted in Figure 8 defines discrete sites of action for the known crucial components of Fe/S protein biogenesis. Two additional ISC components termed Isa1p and Isa2p have been omitted in this scheme, even though they fulfil a discrete function in Fe/S cluster assembly in yeast (Jensen and Culotta, 2000; Kaut et al., 2000; Pelzer et al., 2000). Deletion of ISA1 and ISA2 genes is associated with comparably mild phenotypes, indicating that these proteins are dispensable for Fe/S protein biogenesis, unlike, for example, Nfs1p, the Isu proteins, Yah1p or Arh1p. A potential role of these proteins seems likely from recent in vitro studies on various bacterial Isa homologues (e.g. Krebs et al., 2001; Ollagnier-de-Choudens et al., 2001; Wu et al., 2002a). An Fe/S cluster can be assembled by in vitro reconstitution methods on these Isa protein homologues, suggesting that the Isa protein family may also serve a scaffolding function and thus provide an alternative to the Isu proteins. Experiments to test this potential role in vivo are currently under way and will be reported separately (U.Mühlenhoff, unpublished data).
Our in vivo study assigns the known major components of the mitochondrial ISC assembly machinery to different partial reactions of Fe/S protein biogenesis. The high conservation between eukaryotic and prokaryotic ISC proteins predicts a similar pathway in both systems. Hence our investigation provides the basis for further studies both in intact cells and in vitro to dissect further the various steps of biogenesis and to unravel the precise molecular role of the ISC components. It is evident from our study that the chemistry of Fe/S cluster synthesis on Isu1p will represent one of the main challenges for future mechanistic analyses.
Materials and methods
Yeast strains and cell growth
The following strains of Saccharomyces cerevisiae were used: W303–1A (MATa, ura3-1, ade2-1, trp1-1, his3-11,15, leu2-3112), which served as the wild type from which all mutant strains (except Δssq1) were derived, Δssq1 (BY4741, SSQ1:kan) (Euroscarf), Gal-YAH1 (Lange et al., 2000), Gal-ISU1/Δisu2 (Gerber et al., 2003), Gal-YFH1 (Mühlenhoff et al., 2002b), and Gal-NFS1, Gal-SSQ1, Gal-JAC1 and Gal-GRX5 (this work). In the last four strains, the following upstream regions (promoter) were exchanged for the galactose-inducible GAL1-10 promoter (Mühlenhoff et al., 2002b): Gal-NFS1, nucleotides –250 to –24; Gal-JAC1, nucleotides –150 to –5; Gal-SSQ1, nucleotides –158 to –3; Gal-GRX5, nucleotides –291 to –2. Cells were grown in rich (YP) and minimal (SC) media, or minimal (‘iron-poor’) medium lacking added iron chloride, containing the required carbon sources (Sherman, 1991).
55Fe incorporation into Fe/S cluster apoproteins in vivo
For analysing iron binding of Isu1p in vivo, ISU1 was amplified by PCR and inserted into the BamHI and XhoI restriction sites of the 2µ plasmid p426GPD, which are located immediately downstream of the strong constitutive TDH3 promoter (Mumberg et al., 1995). The following Fe/S cluster-containing reporter proteins were used: mitochondrial biotin synthase (Bio2p) and an HA-tagged version of Yah1p (Lange et al., 2000), both overexpressed from vector p426GPD under the control of the TDH3 promoter. For 55Fe labelling, yeast strains were grown overnight at 30°C in glucose-containing (SD) or galactose-containing (SGal) minimal media lacking added iron chloride (Sherman, 1991). Subsequently, cells were radiolabelled with 10 µCi of 55FeCl3 (ICN) and 1 mM ascorbate for 2 h at 30°C. Cells were collected, resuspended in TNETG buffer (20 mM Tris pH 7.4, 150 mM NaCl, 2.5 mM EDTA, 0.5% Triton X-100, 10% glycerol) and lysed mechanically by vortexing with glass beads. The Fe/S reporter proteins were immunoprecipitated from the clarified lysate using specific antibodies under both aerobic and anaerobic conditions (Harlow and Lane, 1998; Mühlenhoff et al., 2002a). Radioactivity associated with immunoprecipitated material was quantified by liquid scintillation counting as described previously (Kispal et al., 1999). Samples used for immunoprecipitation contained equal amounts of protein. Individual experiments were performed at least three times.
Miscellaneous methods
Site-directed mutagenesis was performed by unique site elimination (Deng and Nickoloff, 1992). All other methods were as described earlier (Mühlenhoff et al., 2002b).
Acknowledgments
Acknowledgements
Our work is dedicated to our friend and colleague Dr Gyula Kispal who died in a tragic car accident on March 20, 2003. His scientific contributions were ground-breaking for our understanding of Fe/S protein biogenesis. Our study was supported by grants from the Sonder forschungsbereiche 286 and 593, Deutsche Forschungsgemeins chaft, Fonds der Chemischen Industrie, Deutsches Humangenomprojekt and the Fritz-Thyssen-Stiftung.
References
- Agar J.N., Krebs,C., Frazzon,J., Huynh,B.H., Dean,D.R. and Johnson,M.K. (2000a) IscU as a scaffold for iron-sulfur cluster biosynthesis: sequential assembly of [2Fe-2S] and [4Fe-4S] clusters in IscU. Biochemistry, 39, 7856–7862. [DOI] [PubMed] [Google Scholar]
- Agar J.N., Yuvaniyama,P., Jack,R.F., Cash,V.L., Smith,A.D., Dean,D.R. and Johnson,M.K. (2000b) Modular organization and identification of a mononuclear iron-binding site within the NifU protein. J. Biol. Inorg. Chem., 5, 167–177. [DOI] [PubMed] [Google Scholar]
- Agar J.N., Zheng,L., Cash,V.L., Dean,D.R. and Johnson,M.K. (2000c) Role of the IscU protein in iron–sulfur cluster biosynthesis: IscS-mediated assembly of a [Fe2S2] cluster in IscU. J. Am. Chem. Soc., 122, 2136–2137. [Google Scholar]
- Beinert H. and Kiley,P.J. (1999) Fe–S proteins in sensing and regulatory functions. Curr. Opin. Chem. Biol., 3, 152–157. [DOI] [PubMed] [Google Scholar]
- Beinert H., Holm,R.H. and Münck,E. (1997) Iron–sulfur clusters: Nature’s modular, multipurpose structures. Science, 277, 653–659. [DOI] [PubMed] [Google Scholar]
- Chen O.S., Hemenway,S. and Kaplan,J. (2002) Inhibition of Fe-S cluster biosynthesis decreases mitochondrial iron export: evidence that Yfh1p affects Fe–S cluster synthesis. Proc. Natl Acad. Sci. USA, 99, 12321–12326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen T., Kaiser,J.T., Steegborn,C., Huber,R. and Kessler,D. (2000) Crystal structure of the cystine C-S lyase from Synechocystis: stabilization of cysteine persulfide for FeS cluster assembly. Proc. Natl Acad. Sci. USA, 97, 3856–3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig E.A. and Marszalek,J. (2002) A specialized mitochondrial molecular chaperone system: a role in formation of Fe/S centers. Cell. Mol. Life Sci., 59, 1658–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W.P. and Nickoloff,J.A. (1992) Site-directed mutagenesis of virtually any plasmid by eliminating a unique site. Anal. Biochem., 200, 81–88. [DOI] [PubMed] [Google Scholar]
- Dutkiewicz R., Schilke,B., Kneiszner,H., Walter,W., Craig,E.A. and Marszalek,J. (2003) Ssq1, a mitochondrial Hsp70 involved in iron–sulfur (Fe/S) center biogenesis: similarities to and differences from its bacterial counterpart. J. Biol. Chem., in press. [DOI] [PubMed] [Google Scholar]
- Frazzon J. and Dean,D.R. (2002) Biosynthesis of the nitrogenase iron-molybdenum-cofactor from Azotobacter vinelandii. Met. Ions Biol. Syst., 39, 163–186. [PubMed] [Google Scholar]
- Frazzon J. and Dean,D.R. (2003) Formation of iron–sulfur clusters in bacteria—an emerging field in bioinorganic chemistry. Curr. Opin. Chem. Biol., 7, 166–173. [DOI] [PubMed] [Google Scholar]
- Garland S.A., Hoff,K., Vickery,L.E. and Culotta,V.C. (1999) Saccharomyces cerevisiae ISU1 and ISU2: members of a well-conserved gene family for iron–sulfur cluster assembly. J. Mol. Biol., 294, 897–907. [DOI] [PubMed] [Google Scholar]
- Gerber J. and Lill,R. (2002) Biogenesis of iron–sulfur proteins in eukaryotes: components, mechanism and pathology. Mitochondrion, 2, 71–86. [DOI] [PubMed] [Google Scholar]
- Gerber J., Mühlenhoff,U. and Lill,R. (2003) An interaction between frataxin and Isu1/Nfs1 that is crucial for Fe/S cluster synthesis on Isu1. EMBO Rep., in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow E. and Lane,D. (1998) Using Antibodies: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Hoff K.G., Silberg,J.J. and Vickery,L.E. (2000) Interaction of the iron–sulfur cluster assembly protein IscU with the Hsc66/Hsc20 molecular chaperone system of Escherichia coli. Proc. Natl Acad. Sci. USA, 97, 7790–7795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen L.T. and Culotta,V.C. (2000) Role of Saccharomyces cerevisiae ISA1 and ISA2 in iron homeostasis. Mol. Cell. Biol., 20, 3918–3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson M.K. (1998) Iron–sulfur proteins: new roles for old clusters. Curr. Opin. Chem. Biol., 2, 173–181. [DOI] [PubMed] [Google Scholar]
- Kaut A., Lange,H., Diekert,K., Kispal,G. and Lill,R. (2000) Isa1p is a component of the mitochondrial machinery for maturation of cellular iron–sulfur proteins and requires conserved cysteine residues for function. J. Biol. Chem., 275, 15955–15961. [DOI] [PubMed] [Google Scholar]
- Kim R., Saxena,S., Gordon,D.M., Pain,D. and Dancis,A. (2001) J-domain protein, Jac1p, of yeast mitochondria required for iron homeostasis and activity of Fe–S cluster proteins. J. Biol. Chem., 276, 17524–17532. [DOI] [PubMed] [Google Scholar]
- Kispal G., Csere,P., Prohl,C. and Lill,R. (1999) The mitochondrial proteins Atm1p and Nfs1p are required for biogenesis of cytosolic Fe/S proteins. EMBO J., 18, 3981–3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs C., Agar,J.N., Smith,A.D., Frazzon,J., Dean,D.R., Huynh,B.H. and Johnson,M.K. (2001) IscA, an alternate scaffold for Fe–S cluster biosynthesis. Biochemistry, 40, 14069–14080. [DOI] [PubMed] [Google Scholar]
- Lange H., Kispal,G. and Lill,R. (1999) Mechanism of iron transport to the site of heme synthesis inside yeast mitochondria. J. Biol. Chem., 274, 18989–18996. [DOI] [PubMed] [Google Scholar]
- Lange H., Kispal,G., Kaut,A. and Lill,R. (2000) A mitochondrial ferredoxin is essential for biogenesis of intra- and extra-mitochondrial Fe/S proteins. Proc. Natl Acad. Sci. USA, 97, 1050–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange H., Lisowsky,T., Gerber,J., Mühlenhoff,U., Kispal,G. and Lill,R. (2001) An essential function of the mitochondrial sulfhydryl oxidase Erv1p/ALR in the maturation of cytosolic Fe/S proteins. EMBO Rep., 2, 715–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Saxena,S., Pain,D. and Dancis,A. (2001) Adrenodoxin reductase homolog (Arh1p) of yeast mitochondria required for iron homeostasis. J. Biol. Chem., 276, 1503–1509. [DOI] [PubMed] [Google Scholar]
- Lill R. and Kispal,G. (2000) Maturation of cellular Fe/S proteins: the essential function of mitochondria. Trends Biochem. Sci., 25, 352–356. [DOI] [PubMed] [Google Scholar]
- Lutz T., Westermann,B., Neupert,W. and Herrmann,J.M. (2001) The mitochondrial proteins Ssq1 and Jac1 are required for the assembly of iron sulfur clusters in mitochondria. J. Mol. Biol., 307, 815–825. [DOI] [PubMed] [Google Scholar]
- Mühlenhoff U. and Lill,R. (2000) Biogenesis of iron–sulfur proteins in eukaryotes: a novel task of mitochondria that is inherited from bacteria. Biochim. Biophys. Acta, 1459, 370–382. [DOI] [PubMed] [Google Scholar]
- Mühlenhoff U., Richhardt,N., Gerber,J. and Lill,R. (2002a) Characterization of iron–sulfur protein assembly in isolated mitochondria: A requirement for ATP, NADH and reduced iron. J. Biol. Chem., 277, 29810–29816. [DOI] [PubMed] [Google Scholar]
- Mühlenhoff U., Richhardt,N., Ristow,M., Kispal,G. and Lill,R. (2002b) The yeast frataxin homologue Yfh1p plays a specific role in the maturation of cellular Fe/S proteins. Hum. Mol. Genet., 11, 2025–2036. [DOI] [PubMed] [Google Scholar]
- Mumberg D., Müller,R. and Funk,M. (1995) Yeast vectors for controlled expression of heterologous proteins in different genetic backgrounds. Gene, 156, 119–122. [DOI] [PubMed] [Google Scholar]
- Nakamura M., Saeki,K. and Takahashi,Y. (1999) Hyperproduction of recombinant ferredoxins in Escherichia coli by coexpression of the ORF1–ORF2–iscS–iscU–iscA–hscB–hscA–fdx–ORF3 gene cluster. J. Biochem., 126, 10–18. [DOI] [PubMed] [Google Scholar]
- Nuth M., Yoon,T. and Cowan,J.A. (2002) Iron–sulfur cluster biosynthesis: characterization of iron nucleation sites for assembly of the [2Fe–2S]2+ cluster core in IscU proteins. J. Am. Chem. Soc., 124, 8774–8775. [DOI] [PubMed] [Google Scholar]
- Ollagnier-de-Choudens S., Mattioli,T., Takahashi,Y. and Fontecave,M. (2001) Iron–sulfur cluster assembly: characterization of IscA and evidence for a specific and functional complex with ferredoxin. J. Biol. Chem., 276, 22604–22607. [DOI] [PubMed] [Google Scholar]
- Pelzer W., Mühlenhoff,U., Diekert,K., Siegmund,K., Kispal,G. and Lill,R. (2000) Mitochondrial Isa2p plays a crucial role in the maturation of cellular iron–sulfur proteins. FEBS Lett., 476, 134–139. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Manzaneque M.T., Tamarit,J., Belli,G., Ros,J. and Herrero,E. (2002) Grx5 is a mitochondrial glutaredoxin required for the activity of iron/sulfur enzymes. Mol. Biol. Cell, 13, 1109–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilke B., Voisine,C., Beinert,H. and Craig,E. (1999) Evidence for a conserved system for iron metabolism in the mitochondria of Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 96, 10206–10211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F. (1991) Getting started with yeast. Methods Enzymol., 194, 3–21. [DOI] [PubMed] [Google Scholar]
- Silberg J.J., Hoff,K.G., Tapley,T.L. and Vickery,L.E. (2001) The Fe/S assembly protein IscU behaves as a substrate for the molecular chaperone Hsc66 from Escherichia coli. J. Biol. Chem., 276, 1696–1700. [DOI] [PubMed] [Google Scholar]
- Sipos K., Lange,H., Fekete,Z., Ullmann,P., Lill,R. and Kispal,G. (2002) Maturation of cytosolic iron–sulfur proteins requires glutathione. J. Biol. Chem., 277, 26944–26949. [DOI] [PubMed] [Google Scholar]
- Smith A.D., Agar,J.N., Johnson,K.A., Frazzon,J., Amster,I.J., Dean,D.R. and Johnson,M.K. (2001) Sulfur transfer from IscS to IscU: the first step in iron–sulfur cluster biosynthesis. J. Am. Chem. Soc., 123, 11103–11104. [DOI] [PubMed] [Google Scholar]
- Strain J., Lorenz,C.R., Bode,J., Garland,S., Smolen,G.A., Ta,D.T., Vickery,L.E. and Culotta,V.C. (1998) Suppressors of superoxide dismutase (SOD1) deficiency in Saccharomyces cerevisiae. Identification of proteins predicted to mediate iron–sulfur cluster assembly. J. Biol. Chem., 273, 31138–31144. [DOI] [PubMed] [Google Scholar]
- Voisine C., Schilke,B., Ohlson,M., Beinert,H., Marszalek,J. and Craig,E.A. (2000) Role of the mitochondrial Hsp70s, Ssc1 and Ssq1, in the maturation of Yfh1. Mol. Cell. Biol, 20, 3677–3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voisine C., Cheng,Y.C., Ohlson,M., Schilke,B., Hoff,K., Beinert,H., Marszalek,J. and Craig,E.A. (2001) Jac1, a mitochondrial J-type chaperone, is involved in the biogenesis of Fe/S clusters in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 98, 1483–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Mansy,S.S., Hemann,C., Hille,R., Surerus,K.K. and Cowan,J.A. (2002a) Iron–sulfur cluster biosynthesis: characterization of Schizosaccharomyces pombe Isa1. J. Biol. Inorg. Chem., 7, 526–532. [DOI] [PubMed] [Google Scholar]
- Wu S.P., Wu,G., Surerus,K.K. and Cowan,J.A. (2002b) Iron–sulfur cluster biosynthesis. Kinetic analysis of [2Fe-2S] cluster transfer from holo ISU to apo Fd: role of redox chemistry and a conserved aspartate. Biochemistry, 41, 8876–8885. [DOI] [PubMed] [Google Scholar]
- Yuvaniyama P., Agar,J.N., Cash,V.L., Johnson,M.K. and Dean,D.R. (2000) NifS-directed assembly of a transient [2Fe-2S] cluster within the NifU protein. Proc. Natl Acad. Sci. USA, 97, 599–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L., White,R.H., Cash,V.L., Jack,R.F. and Dean,D.R. (1993) Cysteine desulfurase acitivity indicates a role for NifS in metallocluster biosynthesis. Proc. Natl Acad. Sci. USA, 90, 2754–2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L., White,R.H., Cash,V.L. and Dean,D.R. (1994) Mechanism for the desulfurization of l-cysteine catalyzed by the nifS gene product. Biochemistry, 33, 4714–4720. [DOI] [PubMed] [Google Scholar]