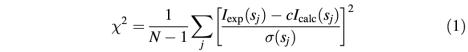Abstract
Brutons’s tyrosine kinase (Btk) is a non-receptor protein tyrosine kinase (nrPTK) essential for the development of B lymphocytes in humans and mice. Like Src and Abl PTKs, Btk contains a conserved cassette formed by SH3, SH2 and protein kinase domains, but differs from them by the presence of an N-terminal PH domain and the Tec homology region. The domain structure of Btk was analysed using X-ray synchrotron radiation scattering in solution. Low resolution shapes of the full-length protein and several deletion mutants determined ab initio from the scattering data indicated a linear arrangement of domains. This arrangement was further confirmed by rigid body modelling using known high resolution structures of individual domains. The final model of Btk displays an extended conformation with no, or little, inter-domain interactions. In agreement with these results, deletion of non-catalytic domains failed to enhance the activity of Btk. Taken together, our results indicate that, contrary to Src and Abl, Btk might not require an assembled conformation for the regulation of its activity.
Keywords: ab initio models/protein tyrosine kinase/rigid body refinement/Tec kinases/XLA
Introduction
Bruton’s protein tyrosine kinase (Btk) is a member of the Tec family of non-receptor protein tyrosine kinases (nrPTK) (Neet and Hunter, 1996; Robinson et al., 2000; Smith et al., 2001). In humans its activity is required for the normal maturation of B lymphocytes, which are the source for circulating antibodies. Naturally occurring mutations in the Btk gene result in X-linked agamma globulinaemia (XLA), a genetic disease characterized by the lack of mature B-cells, low levels of gammaglobulins and high susceptibility to bacterial infections (Smith et al., 1994b; Qiu and Kung, 2000; Cariappa and Pillai, 2002). Similar defects in mice produce a milder immunodeficiency (Xid) (Rawlings et al., 1993; Thomas et al., 1993). Tec kinases share with Src and Abl/Arg family PTKs a basic cassette composed of SH3, SH2 and kinase domains. The SH3 and SH2 domains have important modular functions targeting signalling molecules to specific locations and may also regulate intramolecular interactions (Pawson and Nash, 2003). However, while Src and Abl contain lipid modification signals at the N-terminus, Btk and other Tec family kinases display a Pleckstrin homology (PH) domain, which acts as a membrane targeting unit (Scharenberg et al., 1998; Baraldi et al., 1999). The region between the PH and the SH3 domains is unique to Tec kinases and has been called the Tec homology (TH) region (Smith et al., 1994b; Vihinen et al., 1994). It contains a zinc binding region called the Btk motif (BM), which docks against the PH domain and two proline-rich peptides. In some family members the proline-rich regions have been found to associate intramolecularly with the SH3 domain (Andreotti et al., 1997; Hansson et al., 2001).
Btk is expressed in all haematopoietic cell lineages except T cells (Smith et al., 1994a). A complete picture of Btk’s role in B-cell development does not yet exist, but like other nrPTKs it is involved in the transmission of signals from the B-cell receptor (BCR). BCR stimulation leads to activation of phosphatidyl-inositol-3-kinase resulting in the formation of cell membrane phosphoinositides, to which the PH domain of Btk binds (Salim et al., 1996; Rameh et al., 1997; Baraldi et al., 1999). Once Btk is recruited to the membrane it is phosphorylated at Tyr551 in the activation loop of the kinase domain by Src family kinases (Mahajan et al., 1995; Backesjo et al., 2002). Activation is completed by an autophosphorylation at Tyr223 in the SH3 domain (Park et al., 1996; Rawlings et al., 1996). Upon activation, Tec family members become enriched in membrane rafts and interact with proteins located in these structures (Vargas et al., 2002). Subsequently, Btk activates several downstream signalling components, including phospholipase Cγ (Takata and Kurosaki, 1996; Watanabe et al., 2001), Ca2+ (Takata and Kurosaki, 1996; Fluckiger et al., 1998), protein kinase Cβ (Kang et al., 2001) and NFkB (Bajpai et al., 2000; Petro et al., 2000).
Structural studies on members of the Src and Abl families of nrPTKs have revealed that the SH3 and SH2 modules are not only involved in target recognition, but also participate in the regulation of the catalytic activity through interaction with the kinase domain. In the off state, the SH3 and SH2 domains dock on the back of the kinase domain in what has been described as an assembled conformation (Sicheri et al., 1997; Xu et al., 1997; Hantschel et al., 2003; Nagar et al., 2003). Although these interactions do not directly block access to the catalytic site they seem to prevent the assembly of key catalytic residues through subtle mechanisms. A crucial component of this regulatory machinery is the linker region between the SH2 and kinase domain, which adopts a polyproline type II helix conformation interacting both with the SH3 and kinase domains. Disruption of these interactions either by point mutation or by competition with high affinity ligands of the SH3 domain leads to kinase activation (Gonfloni et al., 1997). Given the domain conservation it has been proposed that a similar regulatory mechanism could operate in Tec family kinases as well (Harrison, 2003).
In the present work we describe the functional and structural characterization of full-length Btk, as well as several mutant and deletion variants. Our results indicate that in contrast to Src and Abl, the regulatory domains of Btk have less capacity to influence the activity of the catalytic domain. The three-dimensional arrangement of domains in Btk in solution was investigated at low resolution using synchrotron radiation small-angle X-ray scattering (SAXS). A model of the domain structure of the human Btk kinase—the first full-length structural model from a member of the Tec family—is built based on the scattering from full-length protein and three deletion mutants. The protein displays an extended rather than an assembled conformation suggesting that Btk has different regulatory mechanisms compared with Src or Abl.
Results
Expression and purification of full-length Btk
We expressed and purified full-length recombinant human Btk (wt-Btk) as well as two full-length mutant variants: Btk-R28C, which displays a mutation inactivating the ligand binding surface of the PH domain (Baraldi et al., 1999) and Btk-K430R, a kinase-dead mutant where a critical Lys at the catalytic domain has been replaced by Arg. Both mutations are known to disrupt the activity without affecting the overall fold of the individual domains. The three protein variants were expressed in insect cells with the use of a baculovirus protein expression system (see Materials and methods).
Mass spectrometry analysis of tryptic digests from insect cell-expressed wt-Btk and Btk-R28C revealed the presence of a number of phosphorylated peptides. Many of the phosphopeptides were absent in the kinase-dead mutant Btk-K430R, indicating that these were a consequence of autocatalytic activity. However two phosphopeptides, one containing phospho-Thr191 and one spanning amino acids (aa) 407–420 were still present in this mutant, indicating that cellular protein kinases present in insect cells are also capable of phosphorylating Btk. In order to ensure homogeneity all the samples for structural analysis were enzymatically dephosphorylated (see Materials and methods). However, the phosphorylation at Thr191, which lies in the TH domain, proved to be particularly resistant being the only phosphorylated residue that could be detected after the treatments. The samples were free from phosphorylation at Tyr551 of the activation loop as well as at Tyr223.
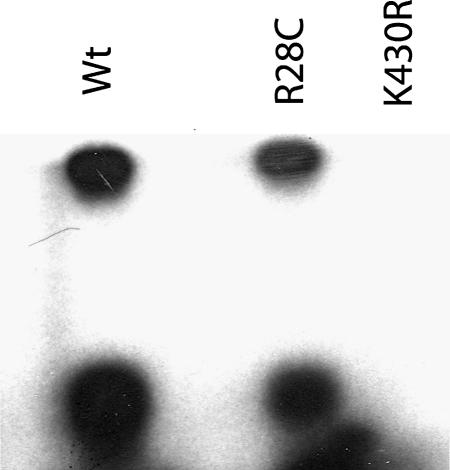
As shown in Figure 1, purified wt-Btk was able in vitro to autophosphorylate and to phosphorylate a target peptide corresponding to aa 217–229 of human Btk, which contains the natural phosphorylation target Tyr223. Btk-R28C showed an activity level similar to that of wt-Btk. Indeed enzyme kinetic studies gave values of the apparent Km for ATP of 56 (± 11) µM and turnover rates of 0.44 min–1 for both proteins. These values are in agreement with those obtained for other nrPTKs, for example, Amrein et al. (1995) reported Km values of 4 and 46 µM and turnover rates of 1.5 and 2 min–1 for c-Src and Csk, respectively. These values indicate that mutation of the phosphoinositide binding surface on the PH domain does not affect the intrinsic catalytic activity of the protein. R28C is one of the mutations producing XLA in humans and our results confirm that the R28C phenotype is likely to be due to the inability of the protein to localize to the plasma membrane but not due to the lack of kinase activity. In agreement with these results, the catalytic activity of purified wt-Btk was not altered by addition of inositol-(1-3-4-5) tetraphosphate in micromolar amounts, a soluble ligand for the Btk PH domain (data not shown).
Fig. 1. In vitro kinase activity assays were performed with wt-Btk, Btk-R28C and Btk-K430R using a substrate peptide containing the natural phosphorylation target Btk-Tyr223. The products of the reaction were separated by SDS–PAGE and autoradiographed. The fast migrating bands correspond to the substrate peptide, while the upper bands correspond to full-length Btk and indicate autophosphorylation activity.
Extended conformation of Btk from hydrodynamics
In dynamic light scattering (DLS) experiments (data not shown), wt-Btk samples displayed larger hydrodynamic radii than those expected for globular proteins with this molecular mass (Mr = 76 kDa). This suggested that Btk adopted an extended rather than globular conformation. In analytical ultracentrifugation (AUC) experiments, both wt-Btk and Btk-R28C yielded a single sedimentation boundary indicating sample homogeneity, and the sedimentation coefficient (S20,w) equal to 3.3S. The expected coefficient for globular proteins with Mr = 76 kDa would be significantly higher (4.15S). In particular, regulated Src (Mr = 49 kDa), in the assembled conformation, yielded S20,w = 3.9 under similar experimental conditions (Weijland et al., 1997). The whole-body modelling approach indicated that the observed S20,w of the wt-Btk is compatible with a prolate ellipsoid with the axial ratio ∼1:4. These results confirm the high anisometry of wt-Btk and Btk-R28C observed by DLS.
Overall parameters from SAXS
Wt-Btk and Btk-R28C displayed identical sedimentation velocity and catalytic activity indicating that protein conformation and regulatory state were not altered by this mutation. As Btk-R28C was more amenable to expression and purification, this mutant was selected for the SAXS experiments. To obtain additional information about the domain structure of Btk, SAXS data were also collected from three deletion mutants: Btk.K (aa 386–659) representing the kinase domain, SH3–SH2 (aa 212–382) and PH–SH2 (aa 1–394) comprising the PH, TH, SH3 and SH2 domains. The Btk.K construct displayed catalytic activity and, according to mass spectrometry, did not contain phosphorylated residues or other post-translational modifications. The SH3–SH2 construct contained the mutation C337M and two additional lysines at the C-terminus, which was required to increase its solubility (Nera et al., 2000).
Figure 2A displays the scattering data as a function of momentum transfer (s = 4π sin(θ)/λ, where 2θ is the scattering angle and λ is the X-ray wavelength), and Table I presents the structural parameters computed from the data. The estimated Mr values of all the samples agree within the errors with the values predicted from their sequences indicating that they are monomeric in solution. This was also supported by the volumes of the hydrated particles computed from the scattering curves (not shown). The values of the radius of gyration Rg and maximum diameter Dmax increase significantly from the construct SH3–SH2 (two domains) to PH–SH2 (three domains) and further to the full-length protein, which points to a linear arrangement of domains rather than to an assembled configuration. This suggestion is further supported by the shape of the distance distribution functions p(r) computed from the experimental data (Figure 2B). The p(r) function reveals the distribution histogram of interatomic distances within the molecule yielding visual information about its shape. The p(r) of Btk.K and SH3–SH2 are bell-shaped—typical for globular particles—but SH3–SH2 displays a hump at about r = 4 nm indicating a two-domain structure. PH–SH2 and especially the full-length protein display p(r) with an extended slope at higher distances characteristic for elongated particles. This indicates that longer constructs are formed by sequentially attaching domains to extremities of shorter constructs like in a string of beads.
Fig. 2. (A) X-ray scattering patterns and the scattering computed from the models of the Btk constructs. (1) Kinase domain, (2) and (3) SH3–SH2 and PH–SH2 constructs, (4) full-length protein. Dots with error bars: experimental data; red solid lines: scattering from ab initio models; blue dashed lines: scattering from the models obtained by rigid body refinement (for the kinase domain from its atomic structure); green triangles: models with added loops (for PH–SH2 and full-length protein). The scattering patterns are appropriately displaced in the logarithmic scale for better visualization. (B) Distance distribution functions of different Btk constructs computed from the X-ray scattering patterns. The sequence of samples is as in (A), the p(r) functions are normalized to unity at their maximum.
Table I. Summary of structural parameters of Btk constructs obtained by SAXS.
| Sample | Mr sas (kDa)a | Mr seq (kDa)a | Rg (nm)b | RgGN (nm)b | RgLP (nm)b | Dmax, (nm)c | DmaxLP, (nm)c | χDRd | χRBd | χLPd |
|---|---|---|---|---|---|---|---|---|---|---|
| Kinase domain | 34 ± 5 | 30 | 2.5 ± 0.1 | 2.4 | 2.0 | 8 ± 0.5 | 6 | 0.84 | 1.80 | 1.80 |
| SH3–SH2 | 24 ± 4 | 19 | 2.5 ± 0.1 | 2.6 | 2.2 | 9 ± 1 | 7 | 0.52 | 1.17 | 1.17 |
| PH–SH2 | 45 ± 7 | 43 | 3.4 ± 0.3 | 3.8 | 3.7 | 14 ± 1 | 13 | 0.47 | 0.81 | 0.55 |
| Full construct | 76 ± 9 | 76 | 5.0 ± 0.4 | 5.5 | 5.2 | 20 ± 2 | 16 | 0.83 | 1.32 | 1.19 |
aMr sas and Mr seq are molecular masses calculated from the scattering data and predicted from the primary sequence.
bRg, RgGN and RgLP are the experimental radii of gyration using Guinier approximation and program GNOM, and the value calculated from the rigid body models with added loops, respectively.
cDmax and DmaxLP are experimental maximum diameters and those calculated from the models with added loops.
dχDR, χRB and χLP are the discrepancies between the experimental data and the fits yielded by ab initio models computed by GASBOR, models obtained by rigid body refinement (atomic model for kinase domain) and models with added loops, respectively.
Molecular modelling
The domain structure of Btk was modelled using a two-step procedure. Firstly, low-resolution models of the full-length protein and deletion mutants were independently reconstructed ab initio from the scattering data. Secondly, atomic models of individual domains were docked into the low-resolution shapes and their position and orientation was further refined using the program MASSHA (Konarev et al., 2001) working with two domains at a time to fit the appropriate SAXS pattern. The high resolution models were retrieved from the Brookhaven Protein Data Bank (PDB): PH–BM domain 1BTK chain A (Hyvonen and Saraste, 1997), SH3 domain 1AWX (Hansson et al., 1998) and kinase domain 1K2P chain A (Mao et al., 2001). For the SH2 domain of Btk a homology model was generated with Swiss modeller (Guex and Peitsch, 1997) using the SH2 domain of c-Src and Hck as templates (Sicheri et al., 1997; Xu et al., 1997).
Shape analysis
Ab initio low-resolution models were obtained by simulated annealing programs DAMMIN (Svergun, 1999) and GASBOR (Svergun et al., 2001). Repetitive ab initio runs for each construct yielded superimposable models neatly fitting the experimental data (Figure 2A; Table I). These models were analysed to select the most typical model and to construct the average model.
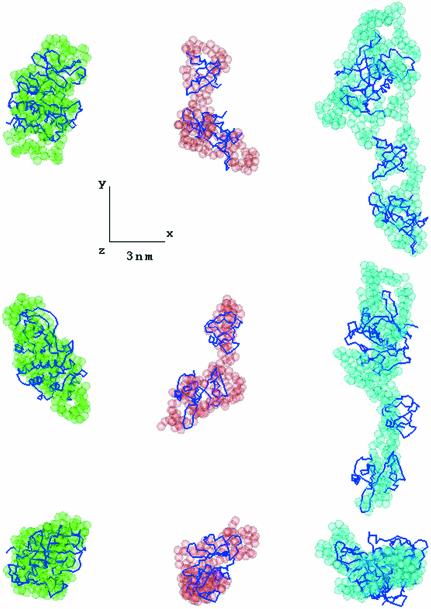
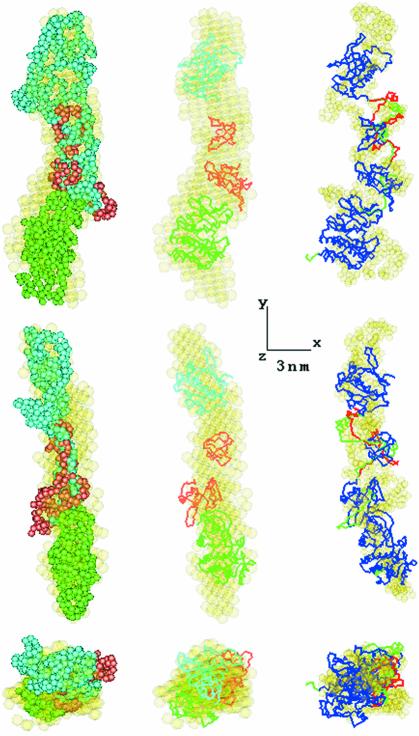
The most typical ab initio models of the partial constructs are displayed in Figure 3, and the most typical and averaged models of the full-length Btk are presented in Figure 4 (semi-transparent beads). Btk.K, the only single domain construct, shows the most globular shape. All other constructs display elongated conformations with a direct correlation between the number of domains and the length of the models. This further confirms the linear arrangement of the individual domains established in the analysis of the overall parameters and p(r) functions. The models of the three partial constructs could be combined and superimposed with the model of the full-length protein (Figure 4, left panel), suggesting a similar domain arrangement in the full-length Btk and in the deletion mutants.
Fig. 3. Typical low resolution models of the Btk constructs obtained by GASBOR (semi-transparent spheres) and the models constructed by rigid body refinement (Cα traces). Left column, kinase domain; middle column, SH3–SH2 domain; right column, PH–SH2 domain. For the kinase domain its high resolution model is displayed. In all panels, the middle and bottom rows are rotated counter clockwise by 90° around the y and x-axis, respectively. All three-dimensional models were displayed using the program ASSA (Kozin et al., 1997).
Fig. 4. Superposition of the averaged low resolution model of the full-length Btk construct (in yellow) with typical low resolution models of the partial constructs (left panel) and with Btk model obtained by rigid body refinement (middle). Low resolution models are colored as in Figure 3, atomic structures of PH and kinase domains are given in cyan and green, respectively, SH3–SH2 partial model in tomato. Typical conformations of inter-domain linkers constructed by program CREDO for PH–SH2 construct and full-length BTK are displayed on the right panel in red and green, respectively. The Btk model without linkers is shown in blue, its typical low resolution model in yellow.
Validation of the kinase domain model
The scattering curve from the atomic model of the kinase domain Btk.K computed by the program CRYSOL (Svergun et al., 1995) yields the fit to the experimental data in Figure 2A (curve 1). By superposition of the atomic model of Btk.K with the most typical dummy residue (DR) model, the latter seems somewhat less compact, but the overall agreement is reasonably good (Figure 3, left column). The observed difference may arise from minor alterations in the tertiary structure between the crystal and solution but also from minor aggregation of Btk.K in solution. As these small differences were irrelevant for the analysis of the domain structure of Btk, its crystallographic model was taken as the best approximation to the structure of the kinase domain in solution for subsequent modelling.
Rigid body modelling
First, the relative position of the SH3 and SH2 domains was established to fit the SAXS data from this construct (Figure 2A, curve 2). The two domains were docked into the average ab initio model of SH3–SH2 with N-terminal of SH2 proximal to the C-terminal of SH3. Their rigid body modelling was done by the automated refinement option of MASSHA, but, given the short (7-residues) missing linker between the domains, the search for the positional parameters was restrained so that the distance between the two termini did not exceed 2.5 nm. The resulting model displayed as Cα trace in Figure 3 (middle column) yields a good fit to the experimental data (Figure 2A, curve 2; Table I). Moreover, its V-like shape agrees well with the most typical ab initio model of SH3–SH2 (Figure 3, superposition in middle column).
The SH3–SH2 model and the atomic structure of the PH–BM domain were treated as two rigid bodies to model the PH–SH2 construct. The PH–BM domain was placed along the line connecting the centres of SH3 and SH2 at a distance of 4 nm from the centre of masses of SH3 and rotated so that its C-terminal pointed to the SH3 domain. The modelling was restrained by imposing the upper limit of 6 nm on the distance between the centres of PH–BM and SH3. The refined model of PH–SH2 (Figure 3, Cα trace in right column) neatly fits the data (Figure 2A, curve 3; Table I) and has an elongated shape, similar to the most typical ab initio model (Figure 3, semi-transparent beads in right column).
Finally, the kinase domain was added to the PH–SH2 model (treated as a rigid body) to fit the entire assembly into the averaged model of the full-length protein in Figure 4. The refinement of the Btk.K position restrained by proximity of the C-terminal end of SH2 and N-terminal end of the kinase domain yields a good fit to the experimental data from the full-length protein (Figure 2A, curve 4; Table I) and preserves the overall elongated shape obtained ab initio (superposition in Figure 4, middle column).
Addition of inter-domain linkers
The high resolution models of the constructs positioned by rigid body refinement superimpose well with the ab initio models as displayed in Figures 3 and 4. However, discontinuities are observed in the regions between adjacent domains for PH–SH2 and the full-length protein, which are likely to be occupied by linker peptides. Indeed, 50-, 7- and 21-residues linkers between the PH–BM and SH3, the SH3 and SH2 and the SH2 and kinase domains, respectively, as well as a 5-residues C-terminal extension of Btk.K were not present in the atomic models. To obtain information about the linker regions the refined positions of the domains were fixed and missing loops were generated to fit the appropriate experimental data with the program suite CREDO (Petoukhov et al., 2002). The addition of linkers to complement the models significantly improved the fits of the PH–SH2 and full-length Btk models to the experimental data (Table I; Figure 2A, curves 3 and 4). Addition of the short linker to the SH3–SH2 domain did not change the fit. Although the restored fragments displayed somewhat different conformations in independent CREDO runs (starting from different random loops), they were confined in space and permitted to draw tentative volumes occupied by the missing portions (Figure 4, right column). Independent modelling of the BM–SH3 and SH3–SH2 linkers by fitting the data from PH–SH2 and those from the full-length protein yielded similar configurations. Moreover, the final model of full-length Btk with linkers and C-terminal portion super imposes well with the most probable ab initio model of the protein (Figure 4, right column). The structural parameters computed from the models with added linkers agree well with the experimental data (Table I).
Assembled Btk models are not compatible with the experimental evidence
As a further test, we retrieved the coordinates of assembled c-Src (Xu et al., 1997; PDB entry 1fmk), added the PH–BM domain of Btk and performed rigid body refinement to fit the experimental data from the full-length Btk. All the models constructed in this way yielded significant systematic deviations from the experiment, and even the best (completely disconnected) model built by an exhaustive global search procedure had a rather high discrepancy χ =1.49. Moreover, the computed S20,w values of all these models were in the range 3.8–4.4S, incompatible with the experimental value 3.3S from AUC. In contrast, elongated models of the full-length Btk in Figure 4 yield S20,w ≈ 3.4–3.5S, in excellent agreement with the hydrodynamic data.
The catalytic activity of Btk is not affected by deletion of the regulatory regions
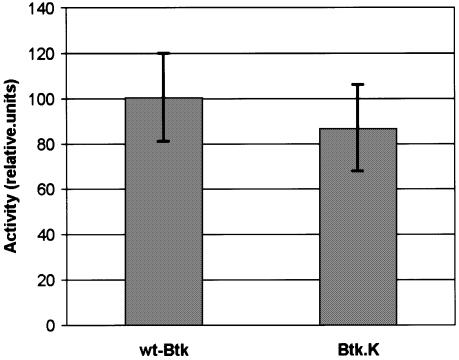
As mentioned above, in their inactive states c-Src and Abl adopt assembled conformations where the SH3, SH2 and SH2–kinase linker regions interact with the kinase domain. Competition of these regulatory interactions with ligands of the SH3 and SH2 domains or mutations in the SH2–kinase linker lead to activation (Gonfloni et al., 1997, 1999). However, the extended conformation of Btk suggested that the regulatory domains would have limited capacity to influence the activity of the kinase domain. In agreement with our results, we found that full-length phosphatase-treated wt-Btk and the Btk.K construct, where the regulatory regions were deleted have very similar activities (Figure 5).
Fig. 5. In vitro kinase activity assays were performed with full-length phosphatase-treated Btk (wt-Btk) and the Btk kinase domain (Btk.K) using a substrate peptide containing the natural phosphorylation target Btk-Tyr223. The incorporated radioactivity was separated with a [14C]resin and quantified in a liquid scintillation counter (see Materials and methods). The graph shows the average of three independent measures in arbitrary units with standard deviations.
Discussion
We have developed methods for the expression and purification of Btk and other Tec protein kinases. Biochemical characterization indicated that our preparations were pure and contained a single species. However, extensive crystallization screenings performed on several protein variants and family members failed to produce crystals. In the present work we have performed SAXS measurements on solutions of full-length Btk and three deletion constructs. By combining ab initio methods yielding overall shapes of these particles with the rigid body refinement to position atomic models of individual domains, a consistent model of the domain organization of Btk was obtained. This model displays a linear arrangement of domains and reconciles four independent scattering data sets, being also in good agreement with the results of hydrodynamic experiments.
The rigid body refinement results, although represented by Cα traces in Figures 3 and 4, should still be considered as low-resolution models, since they were constructed against low-resolution scattering data (∼1.5 nm). This resolution is, however, sufficient to reliably distinguish between extended and compact arrangements of domains in human Btk. The relative position of SH3 and SH2 domains is similar to that in the c-Src crystal structure (PDB entry 1fmk), although no information about the latter structure was used during the rigid body refinement of the SH3–SH2 construct. This suggests that the arrangement of SH3–SH2 domains in human Btk is similar to that in c-Src. In contrast, the assembled conformation of SH3–SH2 and kinase domains observed in the crystal structure of c-Src is incompatible with the available experimental evidence on human Btk.
Scattering from dissolved particles yields an average over the ensemble in the irradiated volume, and if the particles are flexible, modelling of SAXS data provides an average conformation in solution. As one cannot exclude that the extended human Btk molecule is flexible, the rigid body model in Figure 4 may represent average domain positions in the full-length protein. The same is true for the conformation of the linker loop regions, so that the probable configurations presented in Figure 4, right column, are likely to reflect the volumes most frequently populated by the linkers. Potential flexibility of the molecule may explain some systematic deviations between the experimental data from the full-length protein and the model constructed by rigid body refinement (Figure 2A, curve 4). The ab initio model in Figure 4 yielding a better fit to the data (Table I) has a somewhat bulkier appearance, which may illustrate the conformational space occupied by the flexible Btk molecule. This conformational space, however, could be formed only by extended molecules, and potential flexibility of human Btk does not question the observed linear arrangement of domains.
Our results indicate that although Btk contains the conserved SH3–SH2–kinase cassette, its regulatory mechanism might differ from that of related PTKs. In the inactive, assembled conformation of Src (stabilized by the interaction between a phosphorylated C-terminal, regulatory tyrosine binding to the SH2 domain) and Abl (non-phosphorylated and stabilized through an N-terminal myristoyl–kinase domain interaction), the SH3 and SH2 domains dock on the back of the kinase domain with the SH2–kinase linker sandwiched between the SH3 and the kinase domains (Sicheri et al., 1997; Xu et al., 1997; Nagar et al., 2003). Although the regulatory domains do not directly block access to the catalytic site, these interactions seem to favor an inactive conformation of the kinase domain. The release of these interactions either by addition of ligands of the SH3 and SH2 domains or by mutations in the SH2–kinase linker produces activation, resulting in an open conformation without any apparent domain–domain interactions. In contrast, our results indicate that in solution, Btk adopts an elongated conformation where domains are arranged in a linear manner with little or no interactions between them. This elongated shape is not likely to arise from an activated state of Btk, since we confirmed that both Tyr551 in the activation loop and Tyr223, which become phosphorylated during the activation process are not phosphorylated in our preparations. Moreover, this model is in agreement with our observation that complete deletion of the regulatory domains does not alter the intrinsic catalytic activity of the kinase domain. Similarly, three mutations in Btk (P385E, W421A and W461A) affecting the SH2–catalytic domain linker interactions, which are known to activate Src, failed to enhance kinase activity of either wild type, or a constitutively active form (E41K) of Btk in transiently transfected cells (C.M.Bäckesjö, personal communication). Moreover, when residues residing in the SH2–catalytic domain linker of wt-Btk were transferred to the Src kinase (chicken c-Src P299E and P304E) this resulted in transforming activity on fibroblasts as well as enhanced catalytic activity (Gonfloni et al., 1997).
This open conformation of Btk is likely to be a consequence of basic differences in domain composition and regulatory mechanisms controlling the activity of its catalytic domain compared with other PTKs. As mentioned before, in the assembled form of c-Src and Abl, the SH2–kinase linker region adopts a polyproline type II helix conformation and binds to the SH3 domain. This interaction is essential for keeping a regulated kinase domain (Gonfloni et al., 1997). However Btk, and other Tec kinases, contain one or two proline-rich regions adjacent to the SH3 domains. Although the limited resolution of the SAXS technique does not allow determination of whether these proline-rich regions occupy the binding groove of the SH3 domain, such conformations have been found to occur in both in Btk (Hansson et al., 2001; Laederach et al., 2002) and Itk (Andreotti et al., 1997)–another member of the Tec family–and could potentially compete with the SH2–kinase linker for binding to the SH3 domain. However, it should be cautioned that these interactions, which also include dimer formations, were only reported using protein fragments, whereas when we now study full-length proteins only monomers were found. On the other hand, a recent work reporting the crystal structure of the kinase domain of Btk (Mao et al., 2001) revealed that it could adopt an inactive conformation in the absence of regulatory domains. In this structure, Glu445 (equivalent to Src Glu310) is hydrogen bonded to Arg544, which is adjacent to Tyr551 in the activation loop. This interaction seems to have a double effect preventing the engagement of Glu445 in the catalytic centre and stabilizing the outward conformation of the C-helix. Mao and co-workers have suggested that upon phosphorylation of Tyr551 by Src family kinases, Arg544 would be more engaged in interaction with the phosphate group of phospho-Tyr551 releasing Glu445 and allowing the inward rotation of the C-helix. Such a mechanism would preclude the requirement of an assembled conformation, similar to the one seen in c-Src and Abl, for the regulation of the kinase activity as our data suggest.
Btk belongs to the second largest group of PTKs, which includes four additional proteins in humans: Bmx, Itk, Tec and Txk and their mammalian counterparts. Our results suggest that despite the conservation of the SH3–SH2–kinase cassette Btk—and perhaps other members of the family—might have adopted a unique regulatory mechanism. Indeed, preliminary SAXS experiments performed with insect-cell-expressed full-length Bmx also indicate an extended conformation (J.A.Márquez and D.I.Svergun, unpublished). Nevertheless, this cassette might still retain some of the regulatory properties described for c-Src and Abl. For example an assembled conformation of Btk might be facilitated by sequestration of the proline-rich regions in the TH domain by interacting proteins. Such interactions might occur in the complex environment of cellular membranes, rich in signalling molecules and access to such membranes is carefully regulated by the tethering of the PH domain of Btk to transiently generated phosphoinositides. It would be interesting to know whether the SH3–SH2–kinase cassette of Btk—and Tec kinases in general—retain the ability to adopt an assembled conformation. To this end, of note is the fact that the SH3–SH2 ‘clamp’ is less conserved among Tec family kinases (Smith et al., 2001), compared with the Src and Abl families, suggesting that regulation may differ not only between different families, but also perhaps within the Tec family.
Material and methods
Protein expression and purification
Full-length human Btk, Btk-R28C, Btk-K430R and the kinase domain of Btk (aa 386–659) were cloned in pFastBac Htb (Life Technologies). All constructs were designed to contain a His6 sequence tag at the N-terminus followed by a 7 aa spacer and the tobacco etch virus (TEV) protease cleavage site. Expression was performed with Sf9 cells (Invitrogen) in suspension cultures with serum-free Sf900II medium (Life Technologies) using the fermentor facilities at KaroBio (Sweden). The proteins were purified using ion-exchange (Sepharose–Fast-Flow; Pharmacia) and nickel-affinity (Ni-NTA; Quiagen) chromatography. The preparations were transferred to a dialysis bag and TEV protease and either lambda-phage phosphatase or bacterial alkaline phosphatase was added in a 1/10 ratio. The sample was dialysed overnight against 20 mM HEPES pH 7.5, 200 mM NaCl, 2 mM DTT and 1 mM MgCl2. The digested samples were further purified by ion-exchange chromatography in a Source 15-S (Pharmacia) column. This step was introduced to remove the TEV protease, the cleaved tag and the phosphatases. For the purification of Btk.K the same procedure was followed but without the addition of protein phosphatases.
The PH–SH2 region (aa 1–394) of Btk was cloned in pBAT4 using NcoI and XhoI sites. Expression was carried out in Escherichia coli BL21-CodonPlus(DE3)-RP cells (Stratagene, USA) with 2× YT medium. The protein was purified using a three-step procedure involving cation exchange (SP-Sepharose; Amersham-Pharmacia) affinity chromatography (pY-Sepharose) and gel filtration (Superdex 75; Amersham-Pharmacia). The Btk SH3–SH2 construct (aa 212–382) was expressed in E.coli and purified as described by Nera et al. (2000).
Analytical ultracentrifugation
Protein samples were loaded in 300 µl ultracentrifugation cells at a starting concentration of 0.5 mg/ml in 20 mM HEPES pH 7.5, 200 mM NaCl and 2 mM DTT, and centrifuged at 40 000 r.p.m. in a Beckman XL-A analytical ultracentrifuge with an an-60ti Rotor. Protein concentration along the radius axis of the cells was followed at different time points during the run by monitoring absorbance at 238 nm. Sedimentation coefficients S20,w were calculated by measuring the speed of migration of the protein concentration boundary. Programs HYDRO (Garcia De La Torre et al., 1994) and HYDROPRO (Garcia De La Torre et al., 2000) were employed to compute theoretical S20,w values of the bead models and of the atomic models, respectively.
SAXS data collection and processing
Synchrotron radiation SAXS data were collected using standard procedures on the X33 camera (Koch and Bordas, 1983; Boulin et al., 1988) (EMBL, DESY, Hamburg), using a gas detector (Gabriel and Dauvergne, 1982). All the constructs were measured for at least three protein concentrations each ranging from 1 to 10 mg/ml at the X-ray wavelength λ = 0.15 nm covering the range of momentum transfer 0.12 < s < 4.5 nm–1. To check for radiation damage, the data were collected in 15 successive 1-min frames. The data was normalized and processed, and the scattering of the buffer was subtracted using the programs PRIMUS and SAPOKO (Konarev et al., 2003). The difference curves were scaled for the solute concentration and extrapolated to infinite dilution following standard procedures (Feigin and Svergun, 1987). The maximum dimensions Dmax were estimated using the program ORTOGNOM (Svergun, 1993). The forward scattering I(0) and the radius of gyration Rg were evaluated by the Guinier approximation (Gunier, 1939) and also using the indirect transform package GNOM (Svergun, 1992), which provides the distance distribution functions p(r) of the particles. The molecular masses of the solutes were evaluated by calibration against reference solutions of bovine serum albumin.
Scattering data analysis
Low resolution models were generated by two ab initio programs. The program DAMMIN (Svergun, 1999) represents the particle as a collection of M>>1 densely packed beads inside a sphere with the diameter Dmax. Each bead belongs either to the particle or to the solvent, and the shape is described by a binary string of length M. Starting from a random string, simulated annealing is employed to search for a compact model that fits the experimental data Iexp(s) to minimize discrepancy in equation 1:
where N is the number of experimental points, c is a scaling factor and Icalc(s) and σ(sj) are the calculated intensity and the experimental error at the momentum transfer sj, respectively. The program GASBOR (Svergun et al., 2001) represents a protein by an assembly of DRs and uses simulated annealing to build a locally ‘chain-compatible’ DR-model inside the same search volume. The DR modelling is able to better account for the internal structure and generally provides more detailed models than those given by the shape determination algorithms. With each ab initio method and for each protein construct, results from at least 10 separate runs were averaged by the package DAMAVER (Volkov and Svergun, 2003). The independent models were superimposed using the program SUPCOMB (Kozin and Svergun, 2001) and analysed to select the most typical model (i.e. the model displaying the lowest deviation from the rest models) and to construct the average model representing common structural features of all the reconstructions.
Molecular modelling
The atomic models of the individual domains were first positioned interactively into the low resolution models of the appropriate constructs. This arrangement was further refined using the rigid body modelling program MASSHA (Konarev et al., 2001). The scattering from a dimeric complex consisting of two subunits A and B, where A is kept at a reference position, is described by six positional and rotational parameters of the subunit B. These parameters can be established by minimizing discrepancy (1) between the experimental and calculated scattering from the complex. Using MASSHA, rigid body modelling can be performed both in interactive and automated refinement modes. The scattering from the atomic models was calculated using the program CRYSOL (Svergun et al., 1995). The program suite CREDO (Petoukhov et al., 2002) was used to model the missing portions of the structure corresponding to the PH–SH3, SH3–SH2 (in PH–SH3–SH2 and full-length BTK constructs), SH2–kinase linker regions and C-terminus (in the full construct).
Protein kinase assays
Ten microlitre protein samples (100 ng) in kinase assay buffer (10 mM HEPES pH 7.4, 200 mM NaCl, 5 mM MnCl2, 4 mM DTT, 0.5 Na3VO4, 1 mM PMSF) were mixed with an equal volume of substrate solution containing 2 µg of substrate peptide (aa 107–229 of human Btk), 20 µM ATP and 0.5 µl of [γ-32P]ATP (5000 Ci/mmol; Amersham) in kinase assay buffer and incubated at 28°C for 5, 10 and 15 min. The reaction was stopped by adding 20 µl of stop solution (1 mM EDTA, 1 mM ATP) and heating at 95°C for 5 min. The products of the reaction were separated by PAGE and autoradiographed. For quantitative assays, 10 µl of reaction sample were mixed with 10 µl of 0.2% trifluoroacetic acid (TFA) and combined with 20 µl of a 50% (V/V) C-18 resin slurry (Poros 50 R2; PerSeptive Biosystems). The mixture was centrifuged and the pellet was washed three times with 0.1% TFA to remove the unincorporated radioactivity. The radioactivity in the pellets was measured with a liquid scintillation counter. For the determination of the apparent Km values for ATP, initial reaction velocities were estimated at ATP concentrations ranging from 100 nM to 1 mM.
Acknowledgments
Acknowledgements
The authors thank M.H.J.Koch for the support at the X33 beamline. Matti Saraste died during the initial stages of this work. We all miss him deeply. This work was supported by the Swedish Cancer Fund, the Swedish Science Council, the Wallenberg Foundation, European Union grants B104-CT98-0142 and QLRT-2000-01395. D.S. and M.P. acknowledge support from the International Association for the Promotion of Cooperation with Scientists from the Independent States of the Former Soviet Union, Grant 00–243 and from the EU SPINE contract QLG2-CT-2002-00988. J.A.M. is thankful to the Fundación Ramón Areces (Madrid).
References
- Amrein K.E., Takacs,B., Stieger,M., Molnos,J., Flint,N.A. and Burn,P. (1995) Purification and characterization of recombinant human p50csk protein-tyrosine kinase from an Escherichia coli expression system overproducing the bacterial chaperones GroES and GroEL. Proc. Natl Acad. Sci. USA, 92, 1048–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreotti A.H., Bunnell,S.C., Feng,S., Berg,L.J. and Schreiber,S.L. (1997) Regulatory intramolecular association in a tyrosine kinase of the Tec family. Nature, 385, 93–97. [DOI] [PubMed] [Google Scholar]
- Backesjo C.M., Vargas,L., Superti-Furga,G. and Smith,C.I. (2002) Phosphorylation of Bruton’s tyrosine kinase by c-Abl. Biochem. Biophys. Res. Commun., 299, 510–515. [DOI] [PubMed] [Google Scholar]
- Bajpai U.D., Zhang,K., Teutsch,M., Sen,R. and Wortis,H.H. (2000) Bruton’s tyrosine kinase links the B cell receptor to nuclear factor κB activation. J. Exp. Med., 191, 1735–1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baraldi E., Carugo,K.D., Hyvonen,M., Surdo,P.L., Riley,A.M., Potter,B.V., O’Brien,R., Ladbury,J.E. and Saraste,M. (1999) Structure of the PH domain from Bruton’s tyrosine kinase in complex with inositol 1,3,4,5-tetrakisphosphate. Struct. Fold Des., 7, 449–460. [DOI] [PubMed] [Google Scholar]
- Boulin C.J., Kempf,R., Gabriel,A. and Koch,M.H.J. (1988) Data acquisition systems for linear and area X-ray detectors using delay line readout. Nucl. Instrum. Methods A, 269, 312–320. [Google Scholar]
- Cariappa A. and Pillai,S. (2002) Antigen-dependent B-cell development. Curr. Opin. Immunol., 14, 241–249. [DOI] [PubMed] [Google Scholar]
- Feigin L.A. and Svergun,D.I. (1987) In G.W.Taylor (ed.), Structure Analysis by Small-Angle X-ray and Neutron Scattering. Plenum Press, New York. [Google Scholar]
- Fluckiger A.C. et al. (1998) Btk/Tec kinases regulate sustained increases in intracellular Ca2+ following B-cell receptor activation. EMBO J., 17, 1973–1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel A. and Dauvergne,F. (1982) The localization method used at EMBL. Nucl. Instrum. Methods, 201, 223–224. [Google Scholar]
- Garcia De La Torre J., Navarro,S., Lopez Martinez,M.C., Diaz,F.G. and Lopez Cascales,J. (1994) A computer software for the prediction of hydrodynamic properties of macromolecules. Biophys. J., 67, 530–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia De La Torre J., Huertas,M.L. and Carrasco,B. (2000) Calculation of hydrodynamic properties of globular proteins from their atomic-level structure. Biophys. J., 78, 719–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonfloni S., Williams,J.C., Hattula,K., Weijland,A., Wierenga,R.K. and Superti-Furga,G. (1997) The role of the linker between the SH2 domain and catalytic domain in the regulation and function of Src. EMBO J., 16, 7261–7271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonfloni S., Frischknecht,F., Way,M. and Superti-Furga,G. (1999) Leucine 255 of Src couples intramolecular interactions to inhibition of catalysis. Nat. Struct. Biol., 6, 760–764. [DOI] [PubMed] [Google Scholar]
- Guex N. and Peitsch,M.C. (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis, 18, 2714–2723. [DOI] [PubMed] [Google Scholar]
- Gunier A. (1939) La diffarction des rayons X aux tres petits angles; application a l’etude de phenomenes ultramicroscopiques. Ann. Phys. (Paris), 12, 161–237. [Google Scholar]
- Hansson H., Mattsson,P.T., Allard,P., Haapaniemi,P., Vihinen,M., Smith,C.I. and Hard,T. (1998) Solution structure of the SH3 domain from Bruton’s tyrosine kinase. Biochemistry, 37, 2912–2924. [DOI] [PubMed] [Google Scholar]
- Hansson H., Smith,C.I. and Hard,T. (2001) Both proline-rich sequences in the TH region of Bruton’s tyrosine kinase stabilize intermolecular interactions with the SH3 domain. FEBS Lett., 508, 11–15. [DOI] [PubMed] [Google Scholar]
- Hantschel O., Nagar,B., Guettler,S., Kretzschmar,J., Dorey,K., Kuriyan,J. and Superti-Furga,G. (2003) A myristoyl/phosphotyrosine switch regulates c-Abl. Cell, 112, 845–857. [DOI] [PubMed] [Google Scholar]
- Harrison S.C. (2003) Variation on an Src-like theme. Cell, 112, 737–740. [DOI] [PubMed] [Google Scholar]
- Hyvonen M. and Saraste,M. (1997) Structure of the PH domain and Btk motif from Bruton’s tyrosine kinase: molecular explanations for X-linked agammaglobulinaemia. EMBO J., 16, 3396–3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S.W. et al. (2001) PKCβ modulates antigen receptor signaling via regulation of Btk membrane localization. EMBO J., 20, 5692–5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch M.H.J. and Bordas,J. (1983) X-ray diffraction and scattering on disordered systems using synchrotron radiation. Nucl. Instrum. Methods, 208, 461–469. [Google Scholar]
- Konarev P.V., Petouknov,M.V. and Svergun,D.I. (2001) MASSHA—a graphic sytem for rigid body modelling of macromolecular complexes against solution scattering data. J. Appl. Crystallogr., 34, 527–532. [Google Scholar]
- Konarev P.V., Volkov,V.V., Sokolova,A.V., Koch,M.H.J. and Svergun,D.I. (2003) PRIMUS—a Windows-PC based system for small-angle scattering data analysis. J. Appl. Crystallogr., in press. [Google Scholar]
- Kozin M.B. and Svergun,D.I. (2001) Automated matching of high- and low-resolution structural models. J. Appl. Crystallogr., 34, 33–41. [Google Scholar]
- Kozin M.B., Volkov,V.V. and Svergun,D.I. (1997) ASSA—a program for the three-dimensional rendering in solution scattering from biopolymers. J. Appl. Crystallogr., 30, 811–815. [Google Scholar]
- Laederach A., Cradic,K.W., Brazin,K.N., Zamoon,J., Fulton,D.B., Huang,X.Y. and Andreotti,A.H. (2002) Competing modes of self-association in the regulatory domains of Bruton’s tyrosine kinase: intramolecular contact versus asymmetric homodimerization. Protein Sci., 11, 36–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan S., Fargnoli,J., Burkhardt,A.L., Kut,S.A., Saouaf,S.J. and Bolen,J.B. (1995) Src family protein tyrosine kinases induce autoactivation of Bruton’s tyrosine kinase. Mol. Cell. Biol., 15, 5304–5311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao C., Zhou,M. and Uckun,F.M. (2001) Crystal structure of Bruton’s tyrosine kinase domain suggests a novel pathway for activation and provides insights into the molecular basis of X-linked agammaglobulinemia. J. Biol. Chem., 276, 41435–41443. [DOI] [PubMed] [Google Scholar]
- Nagar B., Hantschel,O., Young,M.A., Scheffzek,K., Veach,D., Bornmann,W., Clarkson,B., Superti-Furga,G. and Kuriyan,J. (2003) Structural basis for the autoinhibition of c-Abl tyrosine kinase. Cell, 112, 859–871. [DOI] [PubMed] [Google Scholar]
- Neet K. and Hunter,T. (1996) Vertebrate non-receptor protein-tyrosine kinase families. Genes Cells, 1, 147–169. [DOI] [PubMed] [Google Scholar]
- Nera K.P., Brockmann,E., Vihinen,M., Smith,C.I. and Mattsson,P.T. (2000) Rational design and purification of human Bruton’s tyrosine kinase SH3–SH2 protein for structure–function studies. Protein Expr. Purif., 20, 365–371. [DOI] [PubMed] [Google Scholar]
- Park H., Wahl,M.I., Afar,D.E., Turck,C.W., Rawlings,D.J., Tam,C., Scharenberg,A.M., Kinet,J.P. and Witte,O.N. (1996) Regulation of Btk function by a major autophosphorylation site within the SH3 domain. Immunity, 4, 515–525. [DOI] [PubMed] [Google Scholar]
- Pawson T. and Nash,P. (2003) Assembly of cell regulatory systems through protein interaction domains. Science, 300, 445–452. [DOI] [PubMed] [Google Scholar]
- Petoukhov M.V., Eady,N.A., Brown,K.A. and Svergun,D.I. (2002) Addition of missing loops and domains to protein models by x-ray solution scattering. Biophys. J., 83, 3113–3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petro J.B., Rahman,S.M., Ballard,D.W. and Khan,W.N. (2000) Bruton’s tyrosine kinase is required for activation of IκB kinase and nuclear factor κB in response to B cell receptor engagement. J. Exp. Med., 191, 1745–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y. and Kung,H.J. (2000) Signaling network of the Btk family kinases. Oncogene, 19, 5651–5661. [DOI] [PubMed] [Google Scholar]
- Rameh L.E. et al. (1997) A comparative analysis of the phosphoinositide binding specificity of pleckstrin homology domains. J. Biol. Chem., 272, 22059–22066. [DOI] [PubMed] [Google Scholar]
- Rawlings D.J. et al. (1993) Mutation of unique region of Bruton’s tyrosine kinase in immunodeficient XID mice. Science, 261, 358–361. [DOI] [PubMed] [Google Scholar]
- Rawlings D.J., Scharenberg,A.M., Park,H., Wahl,M.I., Lin,S., Kato,R.M., Fluckiger,A.C., Witte,O.N. and Kinet,J.P. (1996) Activation of BTK by a phosphorylation mechanism initiated by SRC family kinases. Science, 271, 822–825. [DOI] [PubMed] [Google Scholar]
- Robinson D.R., Wu,Y.M. and Lin,S.F. (2000) The protein tyrosine kinase family of the human genome. Oncogene, 19, 5548–5557. [DOI] [PubMed] [Google Scholar]
- Salim K. et al. (1996) Distinct specificity in the recognition of phosphoinositides by the pleckstrin homology domains of dynamin and Bruton’s tyrosine kinase. EMBO J., 15, 6241–6250. [PMC free article] [PubMed] [Google Scholar]
- Scharenberg A.M. et al. (1998) Phosphatidylinositol-3,4,5-trisphosphate (PtdIns-3,4,5-P3)/Tec kinase-dependent calcium signaling pathway: a target for SHIP-mediated inhibitory signals. EMBO J., 17, 1961–1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sicheri F., Moarefi,I. and Kuriyan,J. (1997) Crystal structure of the Src family tyrosine kinase Hck. Nature, 385, 602–609. [DOI] [PubMed] [Google Scholar]
- Smith C.I. et al. (1994a) Expression of Bruton’s agammaglobulinemia tyrosine kinase gene, BTK, is selectively down-regulated in T lymphocytes and plasma cells. J. Immunol., 152, 557–565. [PubMed] [Google Scholar]
- Smith C.I., Islam,K.B., Vorechovsky,I., Olerup,O., Wallin,E., Rabbani,H., Baskin,B. and Hammarstrom,L. (1994b) X-linked agammaglobulinemia and other immunoglobulin deficiencies. Immunol. Rev., 138, 159–183. [DOI] [PubMed] [Google Scholar]
- Smith C.I., Islam,T.C., Mattsson,P.T., Mohamed,A.J., Nore,B.F. and Vihinen,M. (2001) The Tec family of cytoplasmic tyrosine kinases: mammalian Btk, Bmx, Itk, Tec, Txk and homologs in other species. BioEssays, 23, 436–446. [DOI] [PubMed] [Google Scholar]
- Svergun D.I. (1992) Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr., 25, 495–503. [Google Scholar]
- Svergun D.I. (1993) A direct indirect method of small-angle scattering data treatment. J. Appl. Crystallogr., 26, 258–267. [Google Scholar]
- Svergun D.I. (1999) Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys. J., 76, 2879–2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svergun D.I., Barberato,C. and Koch,M.H.J. (1995) CRYSOL—a program to evaluate X-ray scattering of biological macromolecules from atomic coodinates. J. Appl. Crystallogr., 28, 768–773. [Google Scholar]
- Svergun D.I., Petoukhov,M.V. and Koch,M.H. (2001) Determination of domain structure of proteins from X-ray solution scattering. Biophys. J., 80, 2946–2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata M. and Kurosaki,T. (1996) A role for Bruton’s tyrosine kinase in B cell antigen receptor-mediated activation of phospholipase C-γ2. J. Exp. Med., 184, 31–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J.D., Sideras,P., Smith,C.I., Vorechovsky,I., Chapman,V. and Paul,W.E. (1993) Colocalization of X-linked agammaglobulinemia and X-linked immunodeficiency genes. Science, 261, 355–358. [DOI] [PubMed] [Google Scholar]
- Vargas L., Nore,B.F., Berglof,A., Heinonen,J.E., Mattsson,P.T., Smith,C.I. and Mohamed,A.J. (2002) Functional interaction of caveolin-1 with Bruton’s tyrosine kinase and Bmx. J. Biol. Chem., 277, 9351–9357. [DOI] [PubMed] [Google Scholar]
- Vihinen M., Nilsson,L. and Smith,C.I. (1994) Tec homology (TH) adjacent to the PH domain. FEBS Lett., 350, 263–265. [DOI] [PubMed] [Google Scholar]
- Volkov V.V. and Svergun,D.I. (2003) Uniqueness of ab initio shape determination in small angle scattering. J. Appl. Crystallogr., 36, 860–864. [Google Scholar]
- Watanabe D., Hashimoto,S., Ishiai,M., Matsushita,M., Baba,Y., Kishimoto,T., Kurosaki,T. and Tsukada,S. (2001) Four tyrosine residues in phospholipase C-γ2, identified as Btk-dependent phosphorylation sites, are required for B cell antigen receptor-coupled calcium signaling. J. Biol. Chem., 276, 38595–38601. [DOI] [PubMed] [Google Scholar]
- Weijland A., Williams,J.C., Neubauer,G., Courtneidge,S.A., Wierenga,R.K. and Superti-Furga,G. (1997) Src regulated by C-terminal phosphorylation is monomeric. Proc. Natl Acad. Sci. USA, 94, 3590–3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W., Harrison,S.C. and Eck,M.J. (1997) Three-dimensional structure of the tyrosine kinase c-Src. Nature, 385, 595–602. [DOI] [PubMed] [Google Scholar]