Abstract
During transcription, cellular RNA polymerases (RNAP) have to deal with numerous potential roadblocks imposed by various DNA binding proteins. Many such proteins partially or completely interrupt a single round of RNA chain elongation in vitro. Here we demonstrate that Escherichia coli RNAP can effectively read through the site-specific DNA-binding proteins in vitro and in vivo if more than one RNAP molecule is allowed to initiate from the same promoter. The anti-roadblock activity of the trailing RNAP does not require transcript cleavage activity but relies on forward translocation of roadblocked complexes. These results support a cooperation model of transcription whereby RNAP molecules behave as ‘partners’ helping one another to traverse intrinsic and extrinsic obstacles.
Keywords: protein roadblocks/RNA polymerase/transcription elongation
Introduction
In bacteria and eukaryotes DNA is organized into a condensed structure composed, in part, of histones and histone-like proteins. Despite the constant presence of such nucleoprotein complexes as well as various site-specific DNA binding proteins within intragenic regions, rapid progression of elongating RNAP is not compromised in vivo, although in vitro, protein roadblocks often impede transcription (e.g. Pavco and Steege, 1990; Izban and Luse, 1991; Reines and Mote, 1993). Eukaryotic and bacterial general elongation factors SII and GreA/GreB, respectively (Reinberg and Roeder, 1987; Borukhov et al., 1993) have been shown to stimulate transcription through site-specific DNA binding proteins (Reines and Mote, 1993; Toulmé et al., 2000). These factors function by inducing transcript cleavage in the RNAP catalytic site (Izban and Luse, 1992; Borukhov et al., 1993; Rudd et al., 1994; Orlova et al., 1995). Cleavage of the 3′ portion of the nascent transcript reactivates the elongation complex (EC) that has been backtracked for one or more nucleotides along DNA and RNA (Komissarova and Kashlev, 1997; Nudler et al., 1997). It is assumed that the repeated rounds of cleavage and re-extension of RNA in blocked EC can eventually displace the roadblock, allowing transcription to proceed through the physical barrier. More specific elongation factors have been also described that can either suppress EC backtracking (e.g. Elongin; Aso et al., 1995) or reactivate the backtracked EC by actively translocating it forward (Mfd; Park et al., 2002).
Remarkably, transcript cleavage factors, as well as Elongin and Mfd, are dispensable for cell viability under physiological conditions (Archambault et al, 1992; Selby and Sancar, 1993; Orlova et al., 1995; Yamazaki et al., 2003), suggesting that a more general transcriptional anti-arrest and anti-roadblock mechanism exists. Although specific protein complexes, such as FACT and Elongator, have been shown to facilitate RNAP II transcription to some extent on chromatin templates in vitro (Orphanides et al., 1998; Kim et al., 2002), it is unclear whether these factors are sufficient for rapid chromatin transcription in vivo by RNAP II, and also RNAP I and III. Even less clear is how various RNAPs deal with DNA-binding proteins other than histones during elongation. Con sidering the common structural organization of bacterial and eukaryotic RNAP (Ebright, 2000; Darst, 2001; Gnatt et al., 2001), it is likely that the basic anti-roadblock mechanism is also conserved in evolution.
The elongation phase of the transcription cycle often involves multiple RNAP molecules moving, one after another, along the same DNA molecule. Recently, we showed that trailing ECs directly stimulate the activity of leading ECs. Using a reconstituted transcription system from Esherichia coli, we demonstrated that trailing ECs reactivate leading backtracked ECs at pause or arrest sites by translocating them forward (Epshtein and Nudler, 2003). In the present work, we tested the hypothesis that such a cooperation mechanism between RNAP molecules also functions in overcoming elongational roadblocks. Using two different site-specific DNA-binding proteins with high affinity for their cognate sites, we demonstrate that, in both cases, trailing ECs rescue roadblocked ECs by ‘pushing’ them forward. Transcript cleavage activity is not required during this process. Taken together our in vitro and in vivo results suggest that cooperation between RNAP molecules plays an important role in rendering transcription fast and processive on protein covered (physiological) DNA.
Results and discussion
Efficient transcription through the roadblocks depends on multiple rounds of initiation
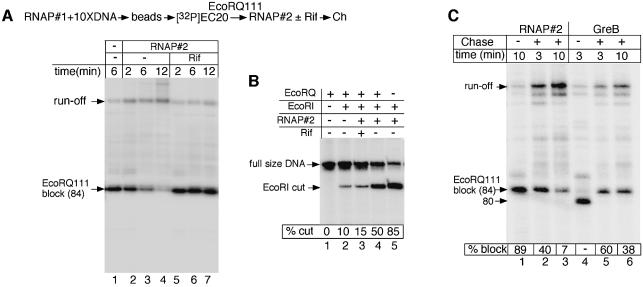
DNA-bound EcoRI restriction endonuclease (EcoRQ111), defective in DNA cleavage (Wright et al., 1989), is a formidable barrier to transcription. It halts the majority of ECs 14 ± 1 bp upstream of the EcoRI site (Pavco and Steege, 1990; Nudler et al., 1995). To monitor the effect of trailing ECs on roadblocked ECs, we utilized T7A1 promoter DNA and two kinds of E.coli RNAPs, one that carries a His6 tag at the C-terminus of the β′ subunit and another without the His tag. The non-tagged holoenzyme was purified from greA–/greB– cells and is free of transcript cleavage factors (Orlova et al., 1995). For convenience, the His-tagged enzyme is designated as 1 and non-His-tagged as 2. The experiment began with preparation of EC stalled at position +20 (EC20) using RNAP1 (Figure 1A). EC20 was labeled with [32P]CTP at three positions and immobilized on Ni2+-chelating agarose beads. Ten molar excess of the DNA template (template 1) was used to ensure that no more than one RNAP molecule initiates on a single DNA molecule. EC20 was then washed several times with high-salt (1 M KCl) and standard (100 mM KCl) transcription buffer (TB) to remove free DNA and unstable complexes, and chased to the EcoRQ111 roadblock. Almost 90% of ECs were blocked by EcoRQ111 (Figure 1A, lane 1). Next, RNAP2 was added with or without rifampicin (Rif). RNAP2 transcription allowed most of the roadblocked ECs to move through the block in 12 min of the chase reaction (Figure 1A, lanes 2–4). Rif, which inhibits promoter clearance (Campbell et al., 2001), completely abolished this effect (lanes 5–7). Transcription through EcoRQ111 was accompanied by dislocation of the block from the DNA, as evident from the loss of the restriction site protection against native EcoRI (Figure 1B).
Fig. 1. Cooperation between RNAP molecules in overcoming EcoRQ111 roadblock. (A) Stimulating effect of trailing RNAP molecules on transcription through DNA-bound EcoRQ111. An EcoRI restriction site is located at position +98 from the +1 start of transcription of template 1. A step-by-step diagram of the experiment is shown at the top (see details in the text and Materials and methods). The autoradiogram displays RNA products synthesized during a single round of transcription by RNAP1. The washed roadblocked complexes were chased in the presence of RNAP2 with or without rifampicin (Rif) for the indicated time intervals (lanes 2–7). (B) Dissociation of EcoRQ111 from DNA during transcription. The auto radiogram shows end-labeled DNA template fragments (template 1) recovered from transcription reactions with RNAP1 alone (lanes 1 and 2) or together with RNAP2 (lanes 3–5). The experiment was performed as shown in (A) except that native EcoRI was also added prior to the chase reaction (lanes 2–5). The increase in EcoRI-mediated DNA cleavage (% cut) during multi-round transcription (lane 4) reflects the increased accessibility of the restriction site, i.e. dissociation of EcoRQ111. (C) Comparison of the anti-roadblock effect of RNAP2 with that of GreB. The experiment is performed as in (A) except that equal molar amounts of RNAP2 and GreB (3 pmol) were added prior to the chase reaction. The amount of the roadblocked complexes (% block) was calculated by dividing the amount of radioactivity in the ‘EcoRQ111’ band by the total radioactivity present in all readthrough bands.
To address whether transcript cleavage activity contributed to the anti-roadblock effect of trailing RNAP, we compared the ability of RNAP2 to potentiate the roadblock readthrough with that of GreB (Figure 1C). The same molar amounts (3 pmol) of RNAP2 holoenzyme and GreB were used in the experiment. Without NTPs, the transcript in the roadblocked complex remained intact after 10 min of incubation with RNAP2, thus excluding any transcript cleavage activity (Figure 1C, lane 1). Under the same conditions, GreB induced cleavage of 4 nt from the RNA 3′ terminus in ∼95% of blocked complexes (Figure 1C, lane 4). Upon addition of NTPs, RNAP1 proceeded through the block at a much faster rate in the presence of RNAP2 than GreB (Figure 1C, compare lanes 2 and 3 with 5 and 6), indicating that at least under the specified conditions, GreB has a lower anti-roadblock potential than trailing ECs.
Taken together these results indicate that transcription through the roadblock by leading RNAP relies on actively transcribed trailing RNAP molecules and does not involve transcript cleavage activity.
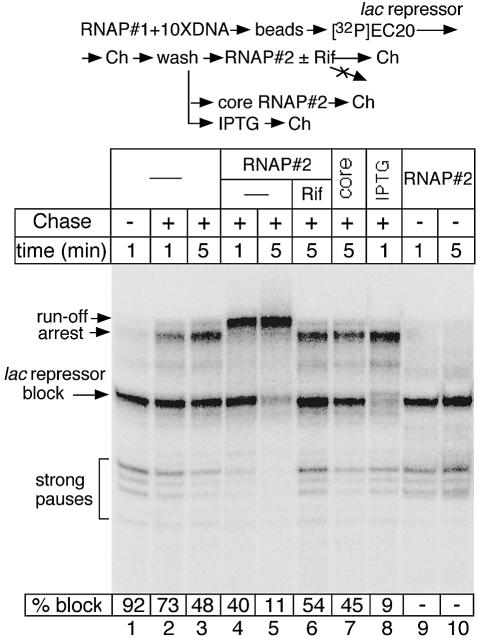
A similar study was performed using template 2 and the DNA-bound lac repressor as a different roadblock (Figure 2). Like the situation with EcoRQ111, RNAP2 markedly potentiated roadblock readthrough by RNAP1 (Figure 2, lanes 4 and 5). The stimulating effect was observed only with RNAP2 holoenzyme, not the core enzyme (Figure 2, lane 7), directly implicating transcription initiation by RNAP2 in this process. Addition of RNAP2 holoenzyme to the washed roadblocked complex lacking NTPs did not change the size of the nascent transcript (Figure 2, lanes 9 and 10), confirming the absence of any transcript cleavage activity. The release of lac repressor by IPTG immediately reactivated roadblocked ECs without RNAP2 (Figure 2, lane 8).
Fig. 2. Cooperation between RNAP molecules in overcoming the lac repressor roadblock. A step-by-step diagram of the experiment is shown at the top. The autoradiogram displays RNA products synthesized during a single round of transcription by RNAP1. The washed roadblocked complexes were chased in the presence of RNAP2 holoenzyme [with or without rifampicin (Rif)], core RNAP2 (core) or IPTG (200 µM) for the indicated time intervals. The amount of the roadblocked complexes (% block) was calculated by dividing the amount of radioactivity in the ‘lac repressor’ band by the total radioactivity present in all readthrough bands. Note that not only the roadblock band, but also specific arrest and pausing bands disappear much faster if transcription performed in the presence of RNAP2 (lanes 4 and 5).
Notably, RNAP2 transcription also stimulated RNAP1 elongation through the strong arrest site near the end of template 2, as well as several other minor arrest/pause sites. This observation exemplifies the role of cooperation between RNAP molecules in overcoming intrinsic blocks, i.e. pauses and arrests (Epshtein and Nudler, 2003), in addition to roadblocks.
The anti-roadblock mechanism
To investigate the mechanism of the anti-roadblock activity of trailing RNAPs, we analyzed the conformation of roadblocked ECs. Judging by its high sensitivity to GreB (Figure 3A, lanes 2 and 3), EC blocked by EcoRQ111 should be in the backtracking state (Nudler et al., 1995). Removing EcoRQ111from the DNA by washing with high-salt TB renders EC resistant to GreB (Figure 3A, lanes 4 and 5). The drastic change in GreB sensitivity in response to the roadblock and also the size of GreB cleavage products (4–5 nt) indicate that EcoRQ111 forces EC to backtrack for 4–5 nt from the position that would otherwise keep EC in the active non-backtracking state (Figure 3B). A similar EC backtracking was induced by the lac repressor roadblock in vitro (Pavco and Steege, 1990; Reines and Mote, 1993; Syroid and Capone, 1994) and in vivo (see below). These results also indicate that roadblock-induced backtracking is reversible. Addition of high-salt or IPTG not only removes EcoRQ111 or lac repressor, respectively, but also reactivates EC, i.e. the blocked complex rapidly assumes its default (active) configuration. Since trailing ECs have been shown to reactivate various spontaneously backtracked complexes by translocating them forward (Epshtein and Nudler, 2003), the present observations strongly suggest that the anti-roadblock mechanism also relies on forward translocation of the blocked complex. In other words, trailing RNAPs help the blocked complex to pass through the block by keeping it in the active state even in the presence of the roadblock. Once the blocked EC assumes its active configuration, it has a chance to move through the roadblock as soon as the latter dissociates (Figure 3B).
Fig. 3. The anti-roadblock mechanism. (A) Analysis of the conformation of the roadblocked EC. A step-by-step diagram of the experiment is shown at the top. The autoradiogram displays RNA products synthesized during a single round of transcription by RNAP1 with (lanes 2–5) or without EcoRQ111 (lane 1). All conditions are as in Figure 1. The washed roadblocked complex was treated with GreB before (lanes 2 and 3) or after washing with 1 mM KCl followed by equilibration with standard TB (lanes 4 and 5). Numbers on the left indicate the size of RNA transcripts. (B) The model for the anti-roadblock mechanism. The scheme summarizes results of Figure 1 and this figure. Trailing EC first reactivates the roadblocked EC by translocating it forward. Next, the active blocked EC has an opportunity to move through the block as soon as the latter dissociates. In addition, the local distortion of DNA and/or steric clash between EC and the block may facilitate dissociation of the block. R, EcoRQ111 roadblock. The model can be generalized for other types of blocking molecules.
This model also explains why GreB is less effective in stimulating readthrough of the roadblocked EC than a trailing EC. Unlike simple forward translocation induced by a trailing EC, the GreB-mediated assistance requires two steps: RNA cleavage and re-synthesis. While cleavage is fast, the re-synthesis step is slow, since the complex starts to pause 4–5 nt before reaching the final stoppage point in front of the roadblock. Pausing, in this case, is a result of progressive backtracking.
Trailing RNAP molecules rescue roadblocked complexes in vivo by ‘pushing’ them forward
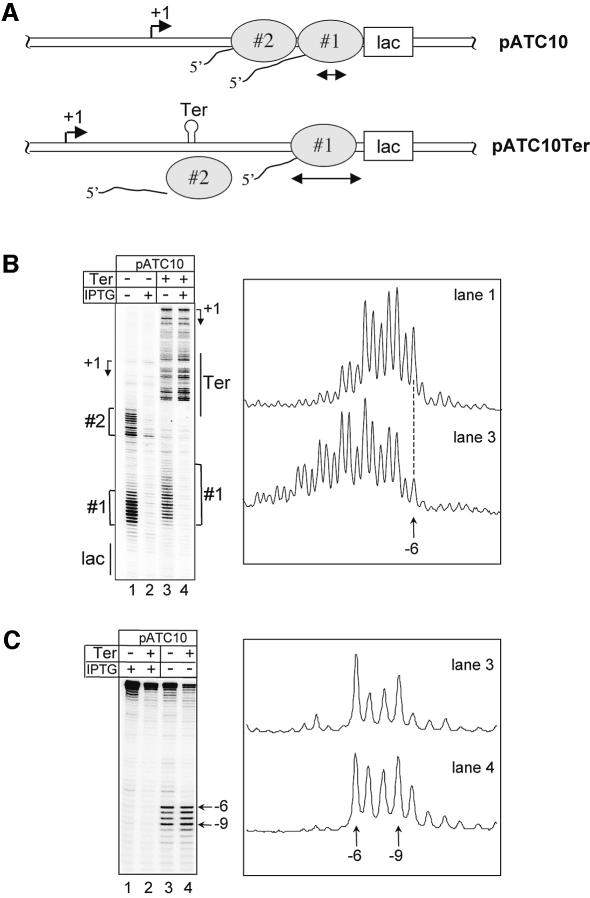
To confirm that the cooperation mechanism described above also operates in living cells, we designed an in vivo system to monitor the effect of a trailing RNAP on a roadblocked EC. In this system, E.coli RNAP that initiates transcription from a constitutive promoter within a plasmid is temporarily halted at a downstream position (at +64 relative to the transcription start site) by the lac repressor bound to its operator motif (Figure 4A). The dynamic feature of the roadblocked EC was investigated by in situ footprinting with the single-strand-specific probe, chloroacetaldehyde (CAA) and RNA 3′ end mapping with S1 nuclease. The analysis of the CAA modifications on the non-template strand of the plasmid pATC10 revealed two RNAPs transcribing in tandem along the DNA region between the promoter and the lac repressor binding site (Figure 4B, lane 1). The roadblocked EC hereafter called 1 is in close apposition to the lac repressor whereas the trailing EC2 is halted further upstream (at around +34). As previously reported (Toulmé et al., 1999, 2000), the roadblocked EC1 slides back and forth between positions –6 and –9 with accompanying Gre-mediated transcript cleavage and re-synthesis (the positions refer to the 3′ terminal nucleotide in the RNA transcript with respect to the upstream edge of the operator motif). This is revealed by the size of the apparent CAA footprint and the detection of a cluster of RNA 3′ ends between –6 and –9 (Figure 4C, lane 3).
Fig. 4. Physical interaction between RNAP molecules in vivo. (A) Schematic representation of RNAP molecules roadblocked by the lac repressor within the two plasmids, pATC10 and its terminator-containing variant pATC10Ter. (B) Primer extension analyses of the in situ CAA modifications on the non-template strand in pATC10 and pATC10Ter. The positions of the operator (lac) and the terminator (Ter) are depicted, as well as the locations of EC1 and EC2 within the two plasmids. The arrows mark the positions of the transcription start sites in each construct. The + and – IPTG indicate the presence or absence of the inducer in the growth media during the footprinting experiment. Densitometer scans of lanes 1 and 3 are shown on the right. (C) S1 mapping of the 3′ ends of the RNA transcripts produced in vivo by the roadblocked ECs from the two plasmid derivatives. Bands corresponding to positions –6 and –9 are indicated by arrows. Densitometer scans of lanes 3 and 4, obtained as in (B), are shown on the right.
To investigate the potential effect of the trailing EC2 on the roadblocked EC1, we constructed a plasmid derivative (pATC10Ter) in which the trp transcription terminator was inserted between the promoter and the lac operator upstream from the position occupied by EC2 in pATC10. The rational of this design is that the relatively high efficiency of the terminator in vivo (80–85%) should dramatically decrease the flow of RNAPs transcribing downstream from the termination point. Under the steady state conditions of transcription, the operator-bound lac repressor is expected to impede RNAP progression only for a certain period of time, which is determined by the repressor concentration and operator affinity. Thus, in the context of a low level of RNAPs bypassing the terminator, the roadblocked EC1 may have enough time to escape the repressor impediment before a trailing EC2 arrives behind. As shown in Figure 4B (lanes 3 and 4), the CAA footprinting of pATC10Ter revealed strong reactivity of the DNA region between the promoter and the termination site, in both the absence and in the presence of IPTG. This reflects the overall transcriptional activity in the region, with a possible queuing effect due to RNAP pausing at the terminator before RNA release (Gusarov and Nudler, 1999). In this case, the CAA footprint in the absence of IPTG revealed a single roadblocked EC, which is isolated between the terminator and the operator (Figure 4B, lane 3). The footprint is significantly different from the one observed for EC1 in the construct lacking the terminator (compare lanes 1 and 3 and the corresponding scans in Figure 4B). Obviously, the isolated EC1 in pATC10Ter backtracks over a longer distance, since new CAA reactive sites are detected upstream and the reactivity of the downstream margin of the footprint (around position –6) is strongly decreased. The retreat of isolated EC1 is also clearly seen with the S1 mapping experiments where the cluster of RNA 3′ ends is now shifted toward more upstream positions (compare the scans of lanes 3 and 4 in Figure 4C).
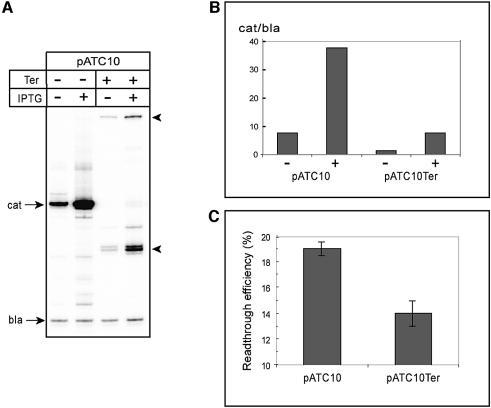
In the context of the back and forth movements of the roadblocked EC, the efficiency with which RNAP will read through the operator motif, following repressor dissociation, depends upon whether the enzyme is able to shift forward rapidly before the repressor rebinds DNA. Therefore, since the trailing RNAP restricts backtracking of the leading EC, this physical constrain should have an effect on the transcriptional readthrough. This hypothesis was tested by measuring the amounts of downstream cat mRNA produced with the two plasmids in vivo. A 32P end-labeled oligonucleotide complementary to the early region of the cat message was used to quantitate mRNA by primer extension with reverse transcriptase. A second primer was also included to detect the plasmid-encoded β-lactamase transcript (bla), for normalization. As shown in Figure 5A and B, a fraction of the RNAPs transcribed beyond the operator motif under repressing conditions on both constructs, yet the level of readthrough was particularly low in the case of the terminator-containing plasmid. To deduce the roadblock effect, we normalized the level of cat mRNA produced under repressing conditions to that obtained under inducing conditions. This analysis (Figure 5C) revealed a significant in vivo cooperation between two RNAPs in overcoming the roadblock, with a 30% readthrough reduction for the terminator-containing plasmid as compared with pATC10. This effect is independent of Gre factors, since the same stimulation by the trailing RNAP was observed when the two plasmids were analyzed in greA–/greB– cells (A.R.Rahmouni and E.Nudler, unpublished observations). Altogether, these results provide a clear view of the dynamic interactions between two RNAPs moving one after another along a transcription unit in vivo. They also highlight a transcription regulation mechanism by which transcribing RNAPs may help each other to traverse protein roadblocks inside the cell.
Fig. 5. Anti-roadblock activity by the trailing EC in vivo. (A) Comparison of the transcriptional readthrough within plasmids pATC10 and pATC10Ter. Autoradiogram of the extension products obtained after in vitro reverse transcription of the RNAs with 32P primers hybridizing to either cat or bla transcripts. Note, that the reverse transcriptase does not read efficiently across the terminator hairpin. As a result, the extension reaction with the terminator-containing RNAs generates two products (indicated by arrowheads), the runoff at +1 of the transcript, and additional products resulting from the extension up to the base of the terminator hairpin stem. (B) Quantification of the primer extension products from (A) using STORM PhosphorImager. The value plotted in each lane indicates the level of cat mRNA relative to the internal control bla transcript. (C) Determination of the readthrough efficiency within the two plasmids. The readthrough was calculated using the following formula: [(cat/bla transcripts)–IPTG/(cat/bla transcripts)+IPTG] × 100. The error bars reflect the standard deviation from the mean of three independent experiments.
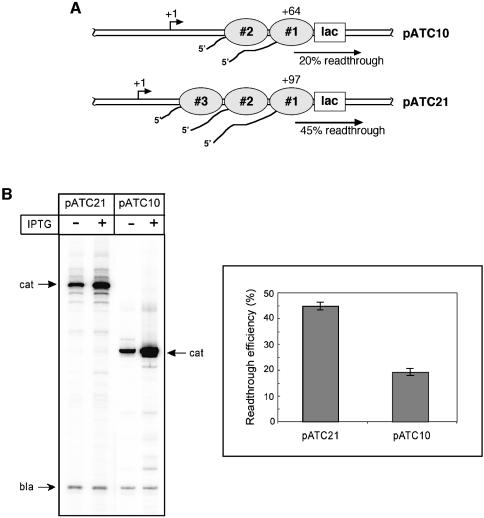
The cumulative anti-roadblock effect by trailing RNAP molecules
Since each active EC exerts ∼20 pN of force (Wang et al., 1998; Davenport et al., 2000), the cooperation mechanism described above is expected to be cumulative, i.e. the efficiency of the anti-roadblock process should depend on a number of RNAPs trailing behind the blocked EC. This hypothesis was tested in vivo by comparing the efficiency of the lac repressor readthrough between two plasmids, pATC10 and its derivative pATC21. The latter construct has an additional 33 bp between the promoter and lac operator, so that it could accommodate at least one extra EC transcribing behind the roadblocked EC (Figure 6A). As depicted in Figure 6B, the ability to load more RNAPs on the same DNA molecule by increasing the distance between the promoter and the roadblock has a marked effect on transcriptional readthrough, with the efficiency rising from 20 to 45%. A similar distance effect was reported previously for T7 RNAP halted by the lac repressor in vitro (Lopez et al., 1998). These results argue that in the anti-roadblock mechanism, every additional RNAP contributes to the forward ‘push’.
Fig. 6. Transcriptional readthrough in vivo as a function of a distance between the promoter and roadblock. (A) Schematic comparison of the pATC10 and pATC21 constructs and their capacity for ECs. (B) The amount of cat mRNA produced by the plasmids was determined by the reverse transcriptase assay, as in Figure 5B. The efficiencies of transcriptional readthrough were calculated and compared as in Figure 5C. The error bars reflect the standard deviation from the mean of three independent experiments.
Physiological implications of the cooperation mechanism of transcription
Considering the presence of numerous specific and non-specific DNA binding proteins that could potentially impede transcription in vivo, the ability of cellular RNAPs to transcribe DNA at the rate of 20–80 nt/s (Vogel and Jensen, 1994; Reines et al., 1996; Uptain et al., 1997) is remarkable. Although, this general property of RNAP must be essential for efficient transcription of chromosomal DNA, its mechanism is poorly understood. There are many examples of bacterial and eukaryotic RNAP being able to transcribe beyond a potential roadblock in natural systems. For example, in E.coli, DnaA protein bound to its box within the dnaA coding region does not repress transcription of the gene (Perez-Roger et al., 1995). RNAP I initiated from the upstream promoter freely reads through downstream promoter-bound transcription initiation factor (TIF) by disrupting the TIF–DNA complex (Bateman and Paule, 1988). RNAP III proceeds without impediment through TFIIIC, which is bound downstream of the promoter (Bardeleben et al., 1994; Matsuzaki et al., 1994). Most significantly, transcription through multiple nucleosomes by RNAP I and II, or histone-like proteins by bacterial RNAP, seems to occur without any delay in vivo. However, the ability of RNAP II to transcribe through nucleosomes is severely compromised in a reconstituted system and inversely proportional to the number of nucleosomes bound to template DNA (Izban and Luse, 1991; Chang and Luse, 1997). Although factors such as IIS, IIF, FACT and Elongator have been shown to relieve the nucleosome block in vitro (Chang and Luse, 1997; Orphanides et al., 1998; Kim et al., 2002), the effect was only partial if compared with transcription in vivo.
Intuitively, the anti-roadblock mechanism of transcription should rely on some common strategy, since various RNAPs have to deal with many obstructive proteins that bind DNA in different ways and with different affinity. In the present study, we compared the ability of an E.coli RNAP molecule alone or in the presence of other RNAP molecules trailing behind to read through two different roadblocks in vitro and in vivo. The results clearly demonstrate that in both cases cooperation between RNAP molecules facilitates transcription through the blocks. In the case of EcoRQ111, the cooperation was virtually absolute, i.e. without trailing molecules, the first round of transcription was almost completely and permanently blocked (Figure 1; data not shown). EcoRQ111 has an exceptionally high affinity for its site (Kd = 10–15 M; Wright et al., 1989), suggesting that this mechanism would work even better with other less avid DNA binding molecules. Our results with the lac repressor (Kd =10–13 M; Barkley, 1981) are consistent with this notion (Figure 2). The analysis of the conformation of EC blocked by either EcoRQ111 or lac repressor indicates that in both cases EC was in the backtracking (inactive) state, suggesting that backtracking may be a common outcome of the collision between EC and a roadblock. The observation that the distance between the RNA 3′ terminus and the rear edge of the roadblock (lac repressor and EcoRQ111) is shorter than expected from the footprint of halted EC (Deuschle et al., 1990; Pavco and Steege, 1990; Reines and Mote, 1993; Syroid and Capone, 1994) supports this model. Thus, there must be at least two consecutive steps in the process of passing a transcriptional roadblock: (i) EC reactivation and (ii) relocation and/or dissociation of the roadblock. Cooperation between RNAP molecules serves to increase the rate of the first step, i.e. reactivation of EC via its forward translocation. The second step, which is primarily determined by the nature of the roadblock–DNA contacts, may be also affected by the first step due to local distortion of the DNA and/or steric clash between two proteins. The same mechanism may work to facilitating transcription through nucleosomes. Indeed, the increased TFIIS sensitivity of the EC blocked by a nucleosome (Izban and Luse, 1992) suggests that pausing due to nucleosome-induced backtracking of RNAP II is the rate-limiting step in chromatin transcription. Thus, preventing backtracking by trailing ECs may contribute to relief of the nucleosomal block.
The anti-roadblock mechanism that relies on RNAP–RNAP cooperation fits the criteria of a general mechanism of transcription. It does not require transcript cleavage activity, which is consistent with the dispensability of the transcript cleavage factors (TFIIS or GreA/GreB) in vivo under physiological conditions (Archambault et al, 1992; Orlova et al., 1995), and should only depend on the density of elongating RNAP molecules (Epshtein and Nudler, 2003). In other words, the efficiency of this mechanism should be directly proportional to the transcriptional activity of a gene and a number of ECs trailing behind. Experimental evidence supports this model. We detected strong blockage of elongation by the lac repressor in vivo when it was initiated from the relatively weak his promoter, which is located at a short distance from the lac operator site (Figure 4). The readthrough increased with increasing distance between the his promoter and lac operator since additional ECs could be accommodated on the same DNA molecule and exert their cumulative ‘pushing’ effect (Figure 6). Furthermore, much higher readthrough levels were observed even at two or three consecutive lac operator sites, if transcription was driven from the very strong T7 A1 promoter located at a relatively long distance from the operator (S.Borukhov, unpublished results). The difference in the promoter activity and its location relative to the roadblock may also explain the previously observed broad variations in the abilities of RNAP to read through the lac repressor in vivo (Horowitz and Platt, 1982; Sellitti et al., 1987) and also EcoRQ111 in vitro (Pavco and Steege, 1991). Moreover, we have demonstrated recently that the rate and efficiency of transcription elongation in living E.coli cells is directly proportional to the transcriptional initiation rate (Epshtein and Nudler, 2003). A similar correlation was observed with RNAP II transcription in various eukaryotic cells (Yankulov et al., 1994). In this regard, one can argue that the highly active genes (e.g. rRNA or heat shock genes) should have the least problems with protein roadblocks or numerous intrinsic pauses and arrests associated with EC backtracking. This explains, at least in part, the highest elongational rate observed with these genes in vivo (Giardina and Lis, 1993; Condon et al., 1995). Alternatively, many genes (particularly in eukaryotes) are constantly weak, having only a few RNAP molecules engaged in transcription. We believe that various stimulating elongation factors are needed in such cases to compensate for lack of efficient cooperation between RNAP molecules.
Materials and methods
DNA templates and proteins
Templates 1 and 2 were generated by PCR-directed mutagenesis from ‘template 1’ (Nudler et al., 1995) using Deep Vent DNA polymerase (New England Biolabs) and synthetic DNA oligos (IDT). They have the identical T7 A1 promoter and initial transcribed sequence up to position +36 (from the +1 start of the transcription). The transcribed sequence of template 1: atcgagagggccacggcgaatagccaaccccaatcgaacagccatcatcctcagtattcaggtagctgttgagcctggggcggtagcgtgcttttttcgaattcacttaatggtaatctcgc. The EcoRI site is underlined. The transcribed sequence of template 2: atcgagagggccacggcgaatagccaaccccaatcgacaccggatccccgggttgtgtgaaattgttatccgctcacaattccacacatgatacg. The lac operator site is underlined. Both templates were purified from a low-melting agarose gel and diluted in TE buffer to a concentration ∼1 pmol/µl. His6-tagged RNAP (1) and GreA/GreB-free RNAP (2) and its core were purified as described previously (Orlova et al., 1995; Nudler et al., 1996). GreB was isolated from GreA–/GreB– E.coli AD8571 strain carrying pMO1.4 plasmid as described by Koulich et al. (1997). EcoRQ111 was a gift from Dr P.Modrich (Duke University). The lac repressor was purified from MG1665 E.coli cells harboring a placIq plasmid (Gusarov and Nudler, 2001). T4 polynucleotide kinase and EcoRI were from New England Biolabs.
In vitro transcription
Two to 5 pmol RNAP were mixed with 10× molar excess of DNA in 20 µl of TB (100 mM KCl, 10 mM MgCl2, 10 mM Tris–HCl pH 7.2) for 5 min at 37°C followed by addition of ApUpC (10 µM), GTP and ATP (25 µM) for 7 min. Next, 5 µl TB-equilibrated Ni2+-chelating agarose beads (Qiagen) were added for 5 min at room temperature followed by washing with 2× 1.5 ml of TB1000 (1 M KCl) and 2× 1.5 ml of TB. To produce EC20, ATP, GTP (5 µM) and 1 µl of [α-32P]CTP were added for 5 min at room temperature followed by unlabeled CTP (5 µM) for a further 2 min. Beads were washed five times with TB. Probes were treated with roadblocking proteins as described below. RNAP2 holoenzyme (3 pmol), RNAP2 core (3 pmol), GreB (3 pmol), RNAP2 holoenzyme preincubated with rifampicin (100 µM, 5 min at 37°C) were added at 37°C as indicated in each figure. All chase reactions were performed with 100 µM NTPs and quenched by an equal volume of stop solution (8 M urea, 20 mM EDTA, 1× TBE, 0.25% bromphenol blue, 0.25% xylene cianol). The products were separated by electrophoresis in 12% sequencing PAGE (8 M urea). Relative amounts of [32P]RNA and DNA species were determined using a PhosphorImager and software from Molecular Dynamics.
EcoRQ111 mutant protein was diluted immediately prior to use with ice-cold 20 mM KPO4 pH 7.4, 0.2 M KCl, 0.2 mM DTT, 1 mM EDTA. EcoRQ111 (2 pmol) was added to the TB-washed EC20 for 1 min at 37°C. RNAP2 (2 pmol) was added together with 100 µM NTPs with or without rifampicin for the indicated intervals at 37°C. To remove EcoRQ111, EC was washed once with TB1000 (1 min) followed by several washes with TB. The completeness of EcoRQ111 removal was verified by a control demonstrating 100% readthrough from the upstream position. For the experiment of Figure 1B, Template 1 was obtained by PCR using non-phosphorylated (left) and 5′-phosphorylated (right) primers to produce a 5′-OH group in the non-template strand for subsequent enzymatic phosphorylation. DNA was labeled by T4 polynucleotide kinase (25 U) and [γ-32P]ATP for 10 min at room temperature in TB. Ten units of EcoRI per reaction were added for 3 min at 37°C. The lac repressor was added to the final concentration 0.5 µM for 5 min at 25°C prior to the chase reaction.
In vivo studies
The pATC10 plasmid was constructed as previously described using pKK232-8 as the starting vector (Toulmé et al., 1999). It contains a constitutive promoter that drives transcription through an (ATC/TAG)10 repeat upstream of the operator motif and followed by the cat gene. The (ATC/TAG)10 repeat with the downstream operator are inserted at position +40 relative to the transcription start site with the ATC sequence on the non-template strand. pATC21 is the same as pATC10 except that it has 21 (ATC/TAG) repeats, which increase the distance between the promoter and the lac operator site by 33 bp. Its construction was reported previously (Toulmé et al., 1999). The plasmid pATC10Ter was obtained by inserting the trp transcription terminator as a 36mer duplex oligonucleotide into the filled-in BamHI site in pATC10. The plasmids were maintained in E.coli strain SU1675 (Toulmé et al., 1999). To have a high yield of intracellular lac repressor, the cells were co-transformed with a pACYC177 derivative (pAC177IQ) that harbors the lacIQ gene. The two plasmids were maintained within the cell by double selection in ampicillin (100 µg/ml) and kanamycin (30 µg/ml). The in situ DNA footprinting with CAA and the subsequent analyses of the modifications by primer extension with Klenow fragment of DNA polymerase I as well as total RNA extraction and 3′ end mapping with S1 nuclease protection experiments were carried out as previously reported (Toulmé et al., 1999, 2000). Densitometer scans were obtained on a Molecular Dynamics PhosphorImager using ImageQuant software version 5.1 for data processing. Quantitative analyses of the cat transcripts by primer extension with reverse transcriptase were also performed exactly as described previously (Toulmé et al., 2000). In these experiments, a primer that hybridizes to the plasmid encoded β-lactamase (bla) transcript was also included for normalization.
Acknowledgments
Acknowledgements
This work was supported in part by a grant from the French AIDS research program ANRS (A.R.R.), and NIH grants GM54098 (S.B.) and GM58750 (E.N.).
References
- Archambault J., Lacroute,F., Ruet,A. and Friesen,J.D. (1992) Genetic interaction between transcription elongation factor TFIIS and RNA polymerase II. Mol. Cell. Biol., 12, 4142–4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aso T., Lane,W.S., Conaway,J.W. and Conaway,R.C. (1995) Elongin (SIII): a multisubunit regulator of elongation by RNA polymerase II. Science, 269, 1439–1443. [DOI] [PubMed] [Google Scholar]
- Bardeleben C., Kassavetis,G.A. and Geiduschek,E.P. (1994) Encounters of Saccharomyces cerevisiae RNA polymerase III with its transcription factors during RNA chain elongation. J. Mol. Biol., 235, 1193–1205. [DOI] [PubMed] [Google Scholar]
- Barkley M.D. (1981) Salt dependence of the kinetics of the lac repressor–operator interaction: role of nonoperator deoxyribonucleic acid in the association reaction. Biochemistry, 20, 3833–3842. [DOI] [PubMed] [Google Scholar]
- Bateman E. and Paule,M.R. (1988) Promoter occlusion during ribosomal RNA transcription. Cell, 54, 985–992. [DOI] [PubMed] [Google Scholar]
- Borukhov S., Sagitov,V. and Goldfarb,A. (1993) Transcript cleavage factors from E. coli. Cell, 72, 459–466. [DOI] [PubMed] [Google Scholar]
- Campbell E.A., Korzheva,N., Mustaev,A., Murakami,K., Nair,S., Goldfarb,A. and Darst,S.A. (2001) Structural mechanism for rifampicin inhibition of bacterial RNA polymerase. Cell, 104, 901–912. [DOI] [PubMed] [Google Scholar]
- Chang C.H. and Luse,D.S. (1997) The H3/H4 tetramer blocks transcript elongation by RNA polymerase II in vitro. J. Biol. Chem., 272, 23427–23434. [DOI] [PubMed] [Google Scholar]
- Condon C., Squires,C. and Squires,C.L. (1995) Control of rRNA transcription in Escherichia coli. Microbiol. Rev., 59, 623–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darst S.A. (2001) Bacterial RNA polymerase. Curr. Opin. Struct. Biol., 11, 155–162. [DOI] [PubMed] [Google Scholar]
- Davenport R.J., Wuite,G.J, Landick R and Bustamante,C. (2000) Single-molecule study of transcriptional pausing and arrest by E. coli RNA polymerase. Science, 287, 2497–2500. [DOI] [PubMed] [Google Scholar]
- Deuschle U., Hipskind R.A. and Bujard,H. (1990) RNA polymerase II transcription blocked by Escherichia coli lac repressor. Science, 248, 480–483. [DOI] [PubMed] [Google Scholar]
- Ebright R.H. (2000) RNA polymerase: structural similarities between bacterial RNA polymerase and eukaryotic RNA polymerase II. J. Mol. Biol., 304, 687–698. [DOI] [PubMed] [Google Scholar]
- Epshtein V. and Nudler,E. (2003) Cooperation between RNA polymerase molecules in transcription elongation. Science, 300, 801–805. [DOI] [PubMed] [Google Scholar]
- Giardina C. and Lis,J.T. (1993) Polymerase processivity and termination on Drosophila heat shock genes. J. Biol. Chem., 268, 23806–23811. [PubMed] [Google Scholar]
- Gnatt A.L., Cramer,P., Fu,J., Bushnell,D.A and Kornberg,R.D. (2001) Structural basis of transcription: an RNA polymerase II elongation complex at 3.3 Å resolution. Science, 292, 1876–1882. [DOI] [PubMed] [Google Scholar]
- Gusarov I. and Nudler,E. (1999) The mechanism of intrinsic transcription termination. Mol. Cell, 3, 495–504. [DOI] [PubMed] [Google Scholar]
- Gusarov I. and Nudler,E. (2001) Control of intrinsic transcription termination by N and NusA: the basic mechanisms. Cell, 107, 437–449. [DOI] [PubMed] [Google Scholar]
- Horowitz H. and Platt,T. (1982) Regulation of transcription from tandem and convergent promoters. Nucleic Acids Res., 10, 5447–5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izban M.G. and Luse,D.S. (1991) Transcription on nucleosomal templates by RNA polymerase II in vitro: inhibition of elongation with enhancement of sequence-specific pausing. Genes Dev., 5, 683–696. [DOI] [PubMed] [Google Scholar]
- Izban M.G. and Luse,D.S. (1992) The RNA polymerase II ternary complex cleaves the nascent transcript in a 3′–5′ direction in the presence of elongation factor SII. Genes Dev., 6, 1342–1356. [DOI] [PubMed] [Google Scholar]
- Kim J.H., Lane W.S. and Reinberg,D. (2002) Human Elongator facilitates RNA polymerase II transcription through chromatin. Proc. Natl Acad. Sci. USA, 99, 1241–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komissarova N. and Kashlev,M. (1997) RNA polymerase switches between inactivated and activated states by translocating back and forth along the DNA and the RNA. J. Biol. Chem., 272, 15329–15338. [DOI] [PubMed] [Google Scholar]
- Koulich D., Orlova,M., Malhotra,A., Sali,A., Darst,S.A. and Borukhov,S. (1997) Domain organization of Escherichia coli transcript cleavage factors GreA and GreB. J. Biol. Chem., 272, 7201–7210. [DOI] [PubMed] [Google Scholar]
- Lopez P.J, Guillerez,J., Sousa,R. and Dreyfus,M. (1998) On the mechanism of inhibition of phage T7 RNA polymerase by lac repressor. J. Mol. Biol., 276, 861–875. [DOI] [PubMed] [Google Scholar]
- Matsuzaki H., Kassavetis,G.A. and Geiduschek,E.P. (1994) Analysis of RNA chain elongation and termination by Saccharomyces cerevisiae RNA polymerase III. J. Mol. Biol., 235, 1173–1192. [DOI] [PubMed] [Google Scholar]
- Nudler E., Kashlev,M., Nikiforov,V. and Goldfarb,A. (1995) Coupling between transcription termination and RNA polymerase inchworming. Cell, 81, 351–357. [DOI] [PubMed] [Google Scholar]
- Nudler E., Avetissova,E., Markovtsov,V. and Goldfarb,A. (1996) Transcription processivity: protein-DNA interactions holding together the elongation complex. Science, 273, 211–217. [DOI] [PubMed] [Google Scholar]
- Nudler E., Mustaev,A., Lukhtanov,E. and Goldfarb,A. (1997) The RNA–DNA hybrid maintains the register of transcription by preventing backtracking of RNA polymerase. Cell, 89, 33–41. [DOI] [PubMed] [Google Scholar]
- Orlova M., Newlands,J., Das,A., Goldfarb,A. and Borukhov,S. (1995) Intrinsic transcript cleavage activity of RNA polymerase. Proc. Natl Acad. Sci. USA, 92, 4596–4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orphanides G., LeRoy,G., Chang,C.H., Luse,D.S. and Reinberg,D. (1998) FACT, a factor that facilitates transcript elongation through nucleosomes. Cell, 92, 105–116. [DOI] [PubMed] [Google Scholar]
- Park J.S., Marr,M.T. and Roberts,J.W. (2002) E. coli transcription repair coupling factor (Mfd protein) rescues arrested complexes by promoting forward translocation. Cell, 109, 757–767. [DOI] [PubMed] [Google Scholar]
- Pavco P.A. and Steege,D.A. (1990) Elongation by Escherichia coli RNA polymerase is blocked in vitro by a site-specific DNA binding protein. J. Biol. Chem., 265, 9960–9969. [PubMed] [Google Scholar]
- Pavco P.A. and Steege,D.A. (1991) Characterization of elongating T7 and SP6 RNA polymerases and their response to a roadblock generated by a site-specific DNA binding protein. Nucleic Acids Res., 19, 4639–4646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Roger I., Macian,F. and Armengod,M.E. (1995) Transcription termination in the Escherichia coli dnaA gene is not mediated by the internal DnaA box. J. Bacteriol., 177, 1896–1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinberg D. and Roeder,R.G. (1987) Factors involved in specific transcription by mammalian RNA polymerase II. Transcription factor IIS stimulates elongation of RNA chains. J. Biol. Chem., 262, 3331–3337. [PubMed] [Google Scholar]
- Reines D. and Mote,J.,Jr (1993) Elongation factor SII-dependent transcription by RNA polymerase II through a sequence-specific DNA-binding protein. Proc. Natl Acad. Sci. USA, 90, 1917–1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reines D., Conaway,J.W. and Conaway,R.C. (1996) The RNA polymerase II general elongation factors. Trends Biochem. Sci., 21, 351–355. [PMC free article] [PubMed] [Google Scholar]
- Rudd M.D., Izban,M.G. and Luse,D.S. (1994) The active site of RNA polymerase II participates in transcript cleavage within arrested ternary complexes. Proc. Natl Acad. Sci. USA, 91, 8057–8061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selby C.P. and Sancar,A. (1993) Molecular mechanism of transcription–repair coupling. Science, 260, 53–58. [DOI] [PubMed] [Google Scholar]
- Sellitti M.A., Pavco,P.A. and Steege,D.A. (1987) lac repressor blocks in vivo transcription of lac control region DNA. Proc. Natl Acad. Sci. USA, 84, 3199–3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syroid D.E. and Capone,J.P. (1994) RNA chain elongation and termination by mammalian RNA polymerase III. Analysis of tRNA gene transcription by imposing a reversible factor-mediated block to elongation using a sequence-specific DNA binding protein. J. Mol. Biol., 244, 482–493. [DOI] [PubMed] [Google Scholar]
- Toulmé F., Guerin,M., Robichon,N., Leng,M. and Rahmouni,A.R. (1999) In vivo evidence for back and forth oscillations of the transcription elongation complex. EMBO J., 18, 5052–5060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toulmé F., Mosrin-Huaman,C., Sparkowski,J., Das,A., Leng,M. and Rahmouni,A.R. (2000) GreA and GreB proteins revive backtracked RNA polymerase in vivo by promoting transcript trimming. EMBO J., 19, 6853–6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uptain S.M., Kane,C.M. and Chamberlin,M.J. (1997) Basic mechanisms of transcript elongation and its regulation. Annu. Rev. Biochem., 66, 117–172. [DOI] [PubMed] [Google Scholar]
- Vogel U. and Jensen,K.F. (1994) The RNA chain elongation rate in Escherichia coli depends on the growth rate. J. Bacteriol., 176, 2807–2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M.D., Schnitzer,M.J., Yin,H., Landick,R., Gelles,J. and Block,S.M. (1998) Force and velocity measured for single molecules of RNA polymerase. Science, 282, 902–907. [DOI] [PubMed] [Google Scholar]
- Wright D.J., King,K. and Modrich,P. (1989) The negative charge of Glu-111 is required to activate the cleavage center of EcoRI endonuclease. J. Biol. Chem., 264, 11816–11821. [PubMed] [Google Scholar]
- Yamazaki K., Aso,T., Ohnishi,Y., Ohno,M., Tamura,K., Shuin,T., Kitajima.S. and Nakabeppu,Y. (2003) Mammalian Elongin A is not essential for cell viability but is required for proper cell cycle progression with limited alteration of gene expression. J. Biol. Chem., 278, 13585–13589. [DOI] [PubMed] [Google Scholar]
- Yankulov K., Blau,J., Purton,T., Roberts,S. and Bentley,D.L. (1994) Transcriptional elongation by RNA polymerase II is stimulated by transactivators. Cell, 77, 749–759. [DOI] [PubMed] [Google Scholar]