Abstract
Iron regulatory protein 1 (IRP1) is regulated through the assembly/disassembly of a [4Fe–4S] cluster, which interconverts IRP1 with cytosolic aconitase. A genetic screen to isolate Saccharomyces cerevisiae strains bearing mutations in genes required for the conversion of IRP1 to c-aconitase led to the identification of a previously uncharacterized, essential gene, which we call CFD1 (cytosolic Fe–S cluster deficient). CFD1 encodes a highly conserved, putative P-loop ATPase. A non-lethal mutation of CFD1 (cfd1-1) reduced c-aconitase specific activity in IRP1-transformed yeast by >90%, although IRP1 in these cells could be readily converted to c-aconitase in vitro upon incubation with iron alone. IRP1-transformed cfd1-1 yeast lacked EPR-detectable Fe–S clusters in c-aconitase, pointing to a defect in Fe–S cluster assembly. The specific activity of another cytosolic Fe–S protein, Leu1p, was also inhibited by >90% in cfd1-1 yeast, whereas activity of mitochondrial Fe–S proteins was not inhibited. Consistent with a cytosolic site of activity, Cfd1p was localized in the cytoplasm. To our knowledge, Cfd1p is the first cytoplasmic Fe–S cluster assembly factor described in eukaryotes.
Keywords: aconitase/ATPase/Fe–S/IRP/YIL003W/DRE3
Introduction
Fe–S proteins participate in a variety of cellular processes, including intermediary metabolism, electron transfer reactions and gene regulation. The presence of the Fe–S cluster, the type of cluster and its oxidation state are all important determinants of protein activity, and each is utilized as a control point in a number of regulated systems (for recent reviews see Johnson, 1998; Beinert and Kiley, 1999). For example, regulated assembly and disassembly of Fe–S clusters is now recognized to be central to the control of gene expression by Fe–S proteins in bacteria and animals (Beinert and Kiley, 1999). In the case of the RNA binding protein, iron regulatory protein 1 (IRP1), iron-responsive assembly and disassembly of a [4Fe–4S] cluster interconverts IRP1 with cytosolic aconitase, thereby regulating its ability to bind to iron responsive elements (IREs; Eisenstein, 2000). By binding to IREs located within the 5′ or 3′ untranslated region, IRP1 regulates the translation or stability, respectively, of mRNAs encoding a variety of proteins involved in the transport and storage of iron or intermediary metabolism (Eisenstein, 2000; Theil and Eisenstein, 2000).
The molecular mechanisms by which animal cells regulate Fe–S cluster assembly/disassembly in IRP1/c-aconitase remains to be elucidated. Low intracellular iron favors the IRE-binding form of IRP1, resulting in stronger translational repression or stabilization of target mRNAs. High intracellular iron promotes the assembly of a [4Fe–4S] cluster converting IRP1 to c-aconitase, inhibiting IRE binding and relieving translational repression or destabilizing target mRNAs (Eisenstein, 2000). Recent evidence suggests that Fe–S cluster disassembly in c-aconitase is initiated by oxidants produced via normal cellular metabolism (Brown et al., 1998, 2002). Further, oxidative stress promotes conversion of c-aconitase to IRP1 (Hentze and Kuhn, 1996) and phosphorylation of IRP1 at serine residue 138 appears to enhance the sensitivity of the Fe–S cluster of c-aconitase to oxidants (Brown et al., 1998, 2002). Taken together, these results argue that cluster disassembly in c-aconitase is a dynamic, oxidant-initiated process that proceeds through a 3Fe intermediate to apo-IRP1, the rate of disassembly being affected by metabolic activity (Eisenstein, 2000). We still know little about how apo-IRP1 is generated from oxidized, 3Fe c-aconitase, however.
Recent studies in bacteria and yeasts have led to the identification of a number of proteins implicated in the biogenesis of Fe–S clusters (for review see Lill and Kispal, 2000). Surprisingly, the Fe–S cluster assembly factors identified to date are found in the mitochondria of eukaryotic cells (Lill and Kispal, 2000). That iron-responsive Fe–S cluster assembly in IRP1 in the cytoplasm also appears to be a facile process in animal cells raises the question of what factors are involved there. Here we report the identification of an evolutionarily conserved gene that we call CFD1, which is required for cytosolic Fe–S cluster biogenesis and conversion of mammalian IRP1 to c-aconitase in yeast. Cfd1p is the first eukaryotic Fe–S cluster assembly factor that has been found to be located predominately in the cytosol.
Results
Isolation of a yeast mutant impaired for c-aconitase-dependent growth
Aconitase-deficient (aco1) yeast are auxotrophic for glutamate because of an inability to generate α-ketoglutarate through the TCA cycle (Ogur et al., 1964; Gangloff et al., 1990). We have found that animal IRP1 can rescue aco1 yeast from glutamate auxotrophy, provided that sufficient IRP1 is converted to c-aconitase (Brown et al., 1998; Narahari et al., 2000). To isolate yeast mutants with defects in the interconversion of IRP1 and c-aconitase, we relied on the requirement of c-aconitase activity for glutamate-dependent growth in IRP1-transformed aco1 yeast. Aconitase-deficient yeast transformed with an IRP1 gene on a multi-copy plasmid (0615d/IRP1) were mutagenized with ethyl methane sulfonate (EMS) and allowed to grow into individual colonies on rich (YPD) medium. (We used a high copy vector for expression of IRP1 to bias our selection against mutations in IRP1 itself.) Approximately 5 × 104 of the resulting colonies were replica-plated onto glutamate-free medium and screened for the ability to grow. In order to enhance selection of strains with mutations in genes affecting cytosolic Fe–S proteins, colonies were also replica-plated onto leucine-free medium. Leucine biosynthesis requires the activity of a cytosolic Fe–S protein Leu1p (Ryan et al., 1973; Kispal et al., 1999). As a third screen, we monitored the growth of colonies on YPD medium. Twenty-two colonies were identified that failed to grow or grew poorly on glutamate-free medium, and that showed a growth defect on leucine-free medium or YPD. Strain AR27/IRP1 showed growth defects on all three media, and its growth was particularly poor on glutamate-free medium. Growth of AR27/IRP1 on glutamate-free medium was not detectable after 4 days of incubation at 30°C (Figure 1), although very minimal growth could be detected after more than a week.
Fig. 1. IRP1-transformed AR27 mutant strain displays a severe growth defect on glutamate-free medium. Fresh overnight cultures of 0615d or AR27, each transformed with IRP1, were harvested, washed twice with sterile water, and resuspended into sterile water at an OD600 of 0.5. Ten microliters of this suspension and 10-fold serial dilutions were spotted onto SD medium lacking uracil, with (+) or without (–) glutamate. Cells were allowed to grow at 30°C for 4 days before photographing.
Analysis of c-aconitase activity in AR27/IRP1
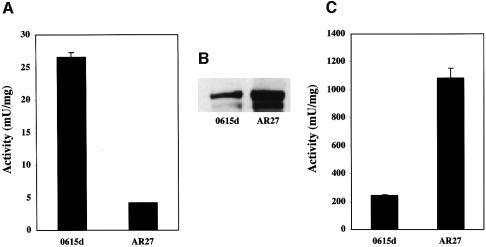
Aconitase activity in extracts prepared from AR27/IRP1 was reduced by 85% compared to this activity in extracts of 0615d/IRP1 (Figure 2A). The poor growth of AR27/IRP1 on glutamate-free medium correlated with depressed IRP1-dependent aconitase activity. This lowered c-aconitase activity could have resulted from a defect in the conversion of IRP1 to c-aconitase, from reduced stability of the c-aconitase Fe–S cluster in vivo, or from a reduction in total IRP1 protein. Results from protein immunoblot analysis exclude the latter possibility. IRP1 protein was present in AR27/IRP1 at a level higher (by ∼5-fold) than in the parental 0615d/IRP1 strain (Figure 2B). Together with the ∼6-fold reduction in c-aconitase activity, these results indicate that c-aconitase specific activity had been reduced by >95% in AR27/IRP1.
Fig. 2. AR27/IRP1 has reduced c-aconitase activity. Strains 0615d/IRP1 and AR27/IRP1 were grown in SD-ura medium to an OD600 of 0.8, and cells were harvested and extracts prepared as described previously (Brown et al., 1998). Extracts were used to measure endogenous c-aconitase activity (A), IRP1 protein by immunoblotting (B), or were incubated with 0.5 mM ferrous ammonium sulfate and 50 mM DTT at room temperature for 1 h to activate c-aconitase followed by aconitase assay (C). Unit activity is in µmol cis-aconitate produced per min per mg total extract protein using 20 mM isocitrate as substrate (Kennedy et al., 1983). The results of the enzyme assays are representative of three independent experiments.
We investigated whether the IRP1 expressed in strain AR27 was functional. We have found that IRP1 expressed in the parental strain 0615d could be converted to c-aconitase in vitro by incubation with iron and DTT (Figure 2C). Typically, aconitase activity was stimulated 5- to 9-fold upon incubation of 0615d/IRP1 extracts with iron (compare activity in Figure 2A–C). After incubation with iron and DTT, aconitase activity in AR27/IRP1 extracts increased to 1.09 × 103 (±67.8) mU/mg. This represents an increase of ∼250-fold over the activity in untreated AR27/IRP1 extracts (4.3 ± 0.1 mU/mg). The full level of activity achieved in AR27/IRP1 was nearly 5-fold higher than that reached in 0615d/IRP1 cell extracts (245.9 ± 2.9 mU/mg; Figure 2C), consistent with the higher amount of IRP1 protein in AR27/IRP1 (Figure 2B). These results indicate that the potential of IRP1 to function as c-aconitase was unimpaired in AR27/IRP1, consistent with the view that the consequence of the genetic deficiency in AR27 is on the assembly or maintenance of the Fe–S cluster in IRP1. We therefore call the gene responsible for this phenotype CFD1, for cytosolic Fe–S cluster deficient 1.
Cloning of CFD1 by complementation of the glutamate-dependent growth phenotype of AR27/IRP1
The mutation that caused the c-aconitase deficiency in AR27/IRP1 is recessive. To isolate the gene responsible for this cytosolic Fe–S cluster deficiency, AR27/IRP1 was transformed with a genomic library from a wild-type yeast strain and transformants were screened for restored growth on glutamate-free medium. This yielded two genomic clones containing inserts from the BET1–PAN1 intergenic region of chromosome IX (see Materials and methods for details of the library screen). A SacI–XbaI fragment isolated from one of these clones complemented AR27/IRP1 as well as the full-length genomic clone, and included only two genes, BET1 and an uncharacterized gene designated YIL003W/DRE3. Since BET1 encodes a membrane protein involved in vesicular transport between the endoplasmic reticulum and the Golgi (Newman and Ferro-Novick, 1987; Ferro-Novick et al., 1991), we chose to focus on YIL003W/DRE3, which is predicted to encode a nucleotide binding protein. YIL003W/DRE3 was PCR amplified from genomic DNA of 0615d or AR27. The PCR products were cloned into a CEN/ARS vector and used to transform AR27/IRP1. Transformation of AR27/IRP1 with YIL003W/DRE3 isolated from 0615d restored vigorous growth on glutamate-free medium, whereas the YIL003W/DRE3 gene isolated from AR27 itself did not (Figure 3). Comparison of the YIL003W/DRE3 sequence showed that the AR27 gene differed from the 0615d wild-type gene at a single nucleotide, a G→A transition at nucleotide 3 of the open reading frame (ORF). The result of this mutation was to change the AUG start codon to an AUA codon. We conclude that CFD1 and YIL003W/DRE3 are identical. Because CFD1 is a more informative name, we use this nomenclature and refer to the allele carried in AR27 as cfd1-1.
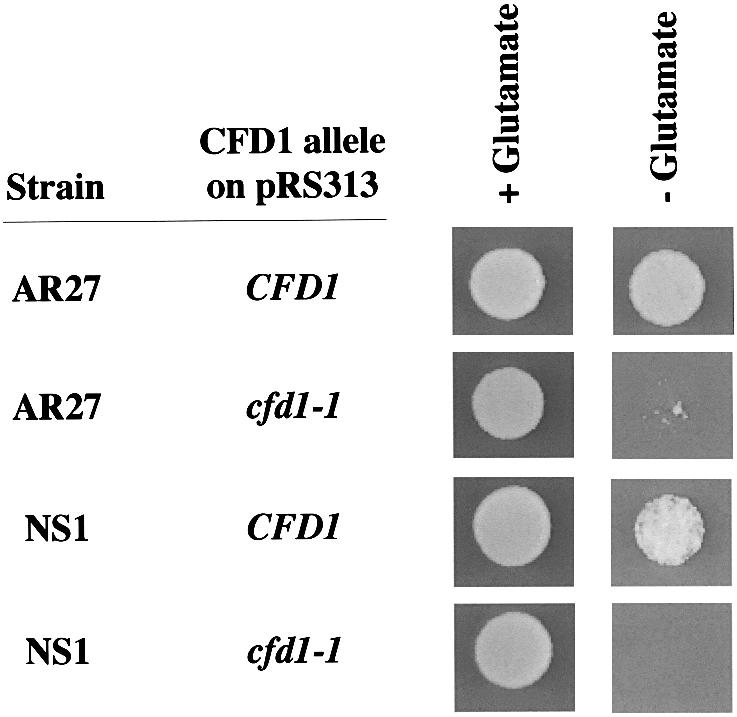
Fig. 3. Wild-type CFD1 restores glutamate-dependent growth to AR27/IRP1. Mutant strain AR27 or the cfd1-disrupted strain NS1 transformed with the indicated CFD1 allele carried on pRS313 was transformed with IRP1. Approximately 104 transformed cells were spotted in 10 µl of water onto selective SD medium, with (+) or without (–) glutamate as indicated, and allowed to grow for 4 days at 30°C.
Mutation of the predicted AUG translation start codon to an AUA in cfd1-1 should strongly inhibit synthesis of Cfd1p. Isolation of a strain carrying such a mutation in the CFD1 gene was surprising since it has been reported that disruption of YIL003W/DRE3 is lethal (Entian et al., 1999; Giaever et al., 2002). To explore the issue of lethality further, CFD1 was disrupted in 0615d. The viability of the resulting cfd1 null strain, called NS1, was dependent on ectopic expression of CFD1, confirming that CFD1 is essential in 0615d. IRP1-transformed NS1 harboring the cfd1-1 allele was viable on glutamate-supplemented media, but failed to grow vigorously on glutamate-free media, showing a similar growth restriction to AR27/IRP1 (Figure 3). NS1 carrying the wild-type CFD1 gene and transformed with IRP1 grew well on media lacking glutamate (Figure 3). Taken together, these results demonstrate that cfd1 deficiency is lethal and that the cfd1-1 allele, while sufficient for viability, is insufficient for conversion of IRP1 to c-aconitase.
Rescue of c-aconitase activity by wild-type CFD1
Since CFD1 rescued AR27/IRP1 from its growth restriction on glutamate-free medium, it was of interest to determine whether it also restored c-aconitase activity. Cell extracts were prepared from AR27/IRP1 that carried an extra-chromosomal copy of wild-type CFD1 or the cfd1-1 allele. Extracts of AR27/IRP1 transformed with wild-type CFD1 showed a nearly 9-fold increase in aconitase activity over that seen in AR27/IRP1 transformed with empty vector (Table I). Moreover, the presence of wild-type CFD1 raised aconitase activity to a level close to that seen in 0615d/IRP1. Transformation of AR27/IRP1 with cfd1-1 did not significantly enhance c-aconitase activity (Table I). The presence of the cfd1-1 allele also did not depress c-aconitase activity in 0615d/IRP1, consistent with its recessive phenotype (Table I). Routinely, a very slight increase in c-aconitase activity was seen in cfd1-1-transformed AR27/IRP1 compared to cells transformed with empty vector. We believe that this is an effect of gene dosage, since the cfd1-1 allele should encode a functional protein but at a much lower expression level than the wild-type gene. Similarly, CFD1-dependent conversion of IRP1 to c-aconitase was observed in the NS1 strain (Table I). These results demonstrate that the non-lethal mutation in the cfd1-1 allele impairs the ability of yeast to convert IRP1 to c-aconitase, most likely through an inhibition of cytosolic Fe–S cluster assembly.
Table I. Cytosolic aconitase activity in IRP1-transformed yeast strains.
| Strain |
CFD1 allele on pRS313 |
Aconitase activity (mU/mg) |
| 0615d | None | 11.3 ± 1.0 |
| AR27 | None | 1.0 ± 0.3 |
| AR27 | CFD1 | 8.8 ± 0.4 |
| AR27 | cfd1-1 | 1.5 ± 0.2 |
| 0615d | cfd1-1 | 11.0 ± 0.4 |
| NS1-16 | cfd1-1 | 0.1 ± 0.1 |
| NS1-16 | CFD1 | 3.1 ± 0.1 |
| NS1-18 | cfd1-1 | 0.4 ± 0.1 |
| NS1-18 | CFD1 | 3.9 ± 0.4 |
Yeast, transformed with IRP1 and CFD1 as indicated, were grown to mid-log phase in selective SD medium, harvested, and extracts were prepared for aconitase assay. NS1-16 and NS1-18 are two independently isolated cfd1Δ strains.
Characteristics of the protein encoded by CFD1 and mutational analysis
CFD1 encodes a predicted 31 922 Da Mr protein that is highly conserved and shares sequence homology with the family of P-loop ATPases (Leipe et al., 2002). The Mrp and MinD proteins of bacteria are closely related, as are the NifH Fe-protein and the ArsA ATPase. Figure 4 shows an alignment of yeast Cfd1p with human and mouse Cfd1p (also called NuBP2), and with bacterial Mrp and MinD proteins. Structural features found in the bacterial cell division protein MinD, whose crystal structure has been solved (Cordell and Lowe, 2001; Hayashi et al., 2001; Sakai et al., 2001), are aligned below the MinD sequence. The predicted nucleotide binding elements in Cfd1p, particularly the Walker P-loop and Walker B motif (Walker et al., 1982), comprise the most highly conserved regions (Figure 4). Unique to Cfd1p is a CX2CX2C motif that is conserved in all Cfd1p homologs, and which aligns with a region that forms a surface exposed loop in MinD (Figure 4; Cordell and Lowe, 2001; Sakai et al., 2001). In Cfd1p, the CX2CX2C motif is part of a large insertion, relative to the MinD sequence (Figure 4).
Fig. 4. Comparative alignment of yeast Cfd1p. Yeast Cfd1p (Sc Cfd1p) was aligned with human Cfd1p (Hu Cfd1p), mouse Cfd1p (Mu Cfd1p), Salmonella enterica serovar typhimurium ApbC (St Mrp) and Archaeoglobus fulgidis MinD (Af MinD) using CLUSTAL_X. The human and mouse proteins are also called NuBP2 (Nakashima et al., 1999). Dark shading in the alignment show identities and light shading shows conserved residues. Residues that form the Walker P-loop, Switch I and Switch II (Walker B motif) elements, which comprise important nucleotide binding and intramolecular signaling domains in P-loop ATPases, are indicated beneath the alignment (Cordell and Lowe, 2001; Leipe et al., 2002). The location of the CX2CX2C motif (residues 201–207) is also indicated beneath the alignment as CXXCXXC. Regions that form β-strands (black rectangles) or α-helices (gray rectangles) in the MinD protein are aligned below its sequence (Cordell and Lowe, 2001).
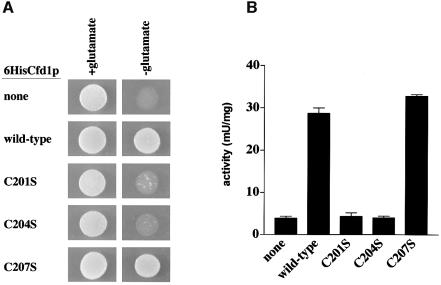
The CX2CX2C motif in Cfd1p holds interest as a potential metal binding domain. To investigate the importance of this element to Cfd1p function, each of the cysteine residues was independently changed to serine in a CFD1 fusion gene encoding an N-terminal His6 tagged Cfd1p (see Materials and methods); the N-terminal His6 tag did not alter function (Figure 5). The resulting mutant genes were tested for their ability to rescue the glutamate-dependent growth defect of AR27/IRP1. While CFD1C207S supported growth, CFD1C201S and CFD1C204S were defective, showing no visible improvement in growth compared to cells transformed with empty vector (Figure 5A). Analysis of c-aconitase activity in these yeast gave results consistent with the growth data. Wild-type CFD1 and CFD1C207S restored c-aconitase activity in AR27/IRP1, while CFD1C201S and CFD1C204S did not (Figure 5B). These results strongly implicate the CX2CX2C motif, and especially the first two cysteine residues, as important to Cfd1p function.
Fig. 5. Cys201 and Cys204 of the CX2CX2C element are required for Cfd1p function. AR27/IRP1 was transformed with empty vector (none) or with vector carrying His6-tagged wild-type, CFD1C201S, CFD1C204S or CFD1C207S fusion genes (see Materials and methods). (A) Glutamate-dependent growth. Approximately 104 cells were spotted in 10 µl onto media with or without glutamate, as indicated. Photographs were taken after 4 days of growth at 30°C. (B) Cytosolic aconitase activity. Aconitase assays were performed as described in the legend to Figure 2A using extracts from logarithmically growing AR27/IRP1 transformed with the indicated CFD1 genes.
EPR analysis of the Fe–S cluster of c-aconitase in vivo
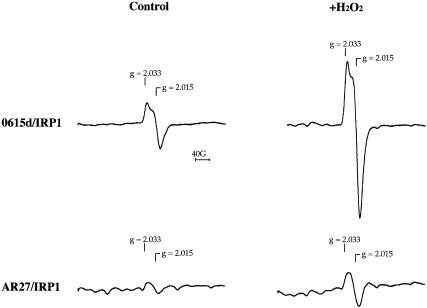
Cfd1p could play a role in Fe–S cluster assembly or disassembly in cytosolic Fe–S proteins. To investigate in what process Cfd1p might function, we subjected IRP1-transformed yeast to EPR analysis. A [3Fe–4S] cluster, representing a stable breakdown intermediate of the [4Fe–4S] cluster is present in ∼20% of c-aconitase in logarithmically growing 0615d/IRP1, and this form can be detected in intact cells by EPR (Brown et al., 2002). More than 90% of the c-aconitase can be converted to the 3Fe form by treatment of cells with H2O2 prior to EPR analysis. If the defect associated with cfd1-1 affects Fe–S cluster disassembly, an increase in the 3Fe EPR signal obtained from cells should be evident. On the other hand, if the cfd1-1 defect precludes Fe–S cluster assembly in the first place, no 3Fe EPR signal would be expected. When subjected to whole-cell EPR, AR27/IRP1 failed to give a detectable 3Fe signal for c-aconitase, and a typical 3Fe c-aconitase signal did not appear even following treatment with 0.6 mM H2O2 (Figure 6). Note that the overlapping, weak, signal seen in the subtracted spectrum of H2O2-treated AR27/IRP1 is distinct from the 3Fe c-aconitase signal. As previously reported, 0615d/IRP1 gave an easily detectable 3Fe signal, with the characteristic peaks at g values of 2.015 and 2.033 (Kennedy et al., 1992; Brown et al., 2002). This signal was enhanced ∼5-fold by treatment of cells with 0.6 mM H2O2 (Figure 6), as predicted if 20% of c-aconitase was in the 3Fe form and 80% was in the 4Fe form prior to treatment with H2O2. These results demonstrate that little 4Fe c-aconitase exists in AR27 cells and strongly support the conclusion that Cfd1p plays an important role in the assembly of Fe–S clusters in the cytoplasm.
Fig. 6. IRP1-transformed cfd1-1 yeast lack [3Fe–4S] c-aconitase. Strains 0615d and AR27 were transformed with IRP1 and transformants were grown to mid-log phase in selective SD medium, harvested, and prepared for whole-cell EPR as described in Materials and methods. Non-transformed yeast were grown in SD-complete medium. Where indicated, cells were exposed to 0.6 mM H2O2 for 15 min prior to harvest. Spectra shown are the difference spectra resulting after subtracting non-transformed cell spectra from IRP1-transformed cell spectra.
Effect of the cfd1-1 mutation on the activity of other Fe–S proteins
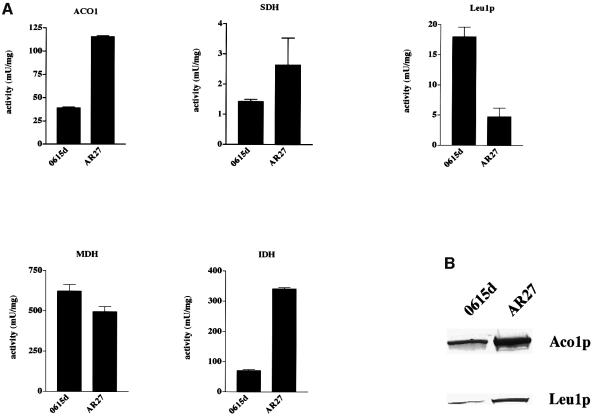
We examined the effect of the cfd1-1 mutation on other Fe–S cluster proteins, including cytosolic Leu1p and mitochondrial succinate dehydrogenase (SDH) and aconitase (Aco1p; the yeast mitochondrial ACO1 gene was transformed into 0615d and AR27 for these experiments). Like IRP1-dependent aconitase activity, Leu1p activity was reduced >4-fold in the AR27/ACO1 strain compared with 0615d/ACO1 (Figure 7A). A similar difference in Leu1p activity was seen in extracts from the NS1 strains, with NS1/cfd1-1 having only 25% of the Leu1p activity of NS1/CFD1 (data not shown). Analysis of Leu1p protein in these strains showed that AR27/ACO1 had ∼4-fold more Leu1p than did 0615d/ACO1 (Figure 7B), indicating a >90% reduction in the specific activity of Leu1p in AR27/ACO1 cells.
Fig. 7. Inhibition caused by the cfd1-1 mutation is specific to cytosolic Fe–S proteins. (A) Strains 0615d and AR27 were transformed with ACO1 and transformants were grown to mid-log phase in selective SD medium, harvested, and extracts were prepared for assay of aconitase (ACO1), succinate dehydrogenase (SDH), Leu1p, malate dehydrogenase (MDH) and NAD+-dependent isocitrate dehydrogenase (IDH) as described in Materials and methods. (B) Protein immunoblotting performed with 50 µg of total extract protein using antiserum specific for mitochondrial aconitase (Aco1p) or Leu1p.
In contrast, the activities of mitochondrial Fe–S enzymes were not diminished in AR27/ACO1 relative to 0615d/ACO1 (Figure 7A). The elevation in aconitase activity seen in AR27/ACO1 may be due, at least in part, to higher Aco1p in these yeast compared with 0615d/ACO1 (Figure 7B). While SDH activity was also modestly enhanced, this may be a more general consequence of the cfd1-1 mutation on mitochondrial oxidative metabolism. For example, the activity of NAD+-dependent isocitrate dehydrogenase (IDH), an enzyme that lacks a Fe–S cluster, was also increased (Figure 7A). We are currently investigating the basis of this phenomenon. In any case, these results demonstrate the selective effect of the cfd1-1 mutation on cytosolic Fe–S proteins, and suggest that the lethal phenotype associated with CFD1 deletion may relate to this localized function.
Subcellular localization of Cfd1p
The Fe–S cluster assembly factors that have been identified to date in eukaryotes are found predominantly in mitochondria, with only minor fractions of a few being found in other subcellular compartments. To determine the intracellular location of Cfd1p, subcellular fractions were prepared from log phase yeast and analyzed for the presence of Cfd1p along with various organelle marker proteins (see Materials and methods). Cfd1p was abundant in fractions containing cytoplasm (S100 and N1 in Figure 8; the N1 fraction contains residual soluble material from the cytoplasm, which floats on the Nycodenz gradient) and was present to a lesser extent in the high-speed pellet from cytoplasm (P100) and in the fraction containing light organelles and membranes (N2). Based on the percentage of each subcellular fraction that was loaded onto the gel in this experiment, we estimate that ≥90% of Cfd1p fractionated with cytosolic S100 (see Figure 8 legend). Importantly, Cfd1p was absent from fractions that were rich in mitochondria (M, Figure 8). The presence of the mitochondrial proteins Aco1p and porin in the mitochondrial fraction, and their absence from cytosolic S100 and N1 fractions (Figure 8) confirms the efficacy of this fractionation regimen. Further, the small amount of porin in the P100 fraction, and its absence from the S100, indicates that minimal breakage of mitochondria occurred during the cell fractionation procedure. As expected, IRP1 was present exclusively in fractions containing cytoplasm (Figure 8), and Leu1p also showed cytoplasmic localization in these experiments (not shown). The similar fractionation patterns for Cfd1p and IRP1 and the lack of co-fractionation of Cfd1p with porin and Aco1p provide strong evidence that Cfd1p is localized in the cytoplasm and is not associated with mitochondria.
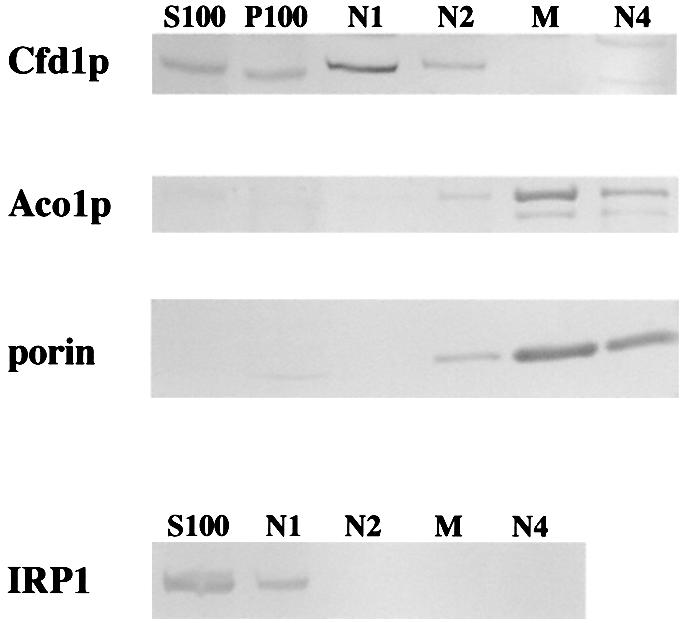
Fig. 8. Cfd1p localizes to the cytoplasm. Mid-log phase yeast were subjected to subcellular fractionation as described in Materials and methods. The presence of individual proteins in each fraction was assessed by protein immunoblotting. S100, soluble cytoplasmic material after 100 000 g centrifugation; P100, material pelleted from crude cytoplasmic fraction by 100 000 g centrifugation; N1, floating material from Nycodenz gradient; N2, light membrane vesicles; M, mitochondria-enriched fraction; N4, large aggregates and heavy membrane vesicles. The fractionation patterns shown are from NS1/CFD1 (Cfd1p), 0615d/myc tagged-IRP1 (IRP1) and AR27/ACO1 transformed with 6HisCFD1 (Aco1p and porin; note that 6HisCfd1p showed a similar fractionation pattern to native Cfd1p). Relative to the fraction of S100 material loaded, ∼10-fold more material from P100, N1 and N2, and 100-fold more material from M and N4 were loaded. The results shown are representative of five independent experiments.
Discussion
We have utilized the yeast Saccharomyces cerevisiae as a model organism to investigate the process of Fe–S cluster assembly/disassembly, focusing on this dynamic process in the interconversion of IRP1 and c-aconitase. In IRP1-transformed aco1 yeast, conversion of IRP1 to c-aconitase is essential for growth on glutamate-free medium. Using this trait as the basis for a genetic screen, we identified CFD1, a previously uncharacterized gene in yeast, to play a crucial role in cytosolic Fe–S cluster assembly. Recent studies, primarily in yeast, have identified a number of eukaryotic Fe–S cluster assembly factors (Knight et al., 1998; Land and Rouault, 1998; Garland et al., 1999; Kispal et al., 1999; Li et al., 1999; Schilke et al., 1999; Jensen and Culotta, 2000; Kaut et al., 2000; Pelzer et al., 2000; Tong and Rouault, 2000; Lange et al., 2001; Voisine et al., 2001; Muhlenhoff et al., 2002; Sipos et al., 2002). These factors are localized predominantly in mitochondria and they comprise a cluster assembly machinery that is conserved in prokaryotes (Zheng et al., 1998; Muhlenhoff and Lill, 2000). Cfd1p is the first cytosolic Fe–S cluster assembly factor to be identified.
Cfd1p shares sequence homology with the large family of P-loop ATPases and falls into the subfamily of P-loop ATPases that include NifH, the Fe-protein in nitrogenase (Dean et al., 1993; Leipe et al., 2002). NifH is itself an Fe–S protein; however, the cluster-coordinating residues of NifH are not conserved in Cfd1p. Cfd1p is even more closely related to the bacterial Mrp proteins, which have recently been shown to play a role in Fe–S cluster assembly (Figure 4; Skovran and Downs, 2003). Therefore, a role for P-loop ATPases in Fe–S cluster biogenesis seems to have been conserved through evolution. Interestingly, a homolog of Cfd1p in yeast, called Nbp35, is localized primarily in the nucleus (Vitale et al., 1996), raising the possibility that it could function in Fe–S cluster assembly there. That eukaryotes may have evolved unique Fe–S cluster assembly machineries for distinct subcellular compartments underscores the importance of Fe–S proteins in varied aspects of cell physiology.
Mutations in genes encoding several of the mitochondrial cluster assembly factors have been found to inhibit the biogenesis of clusters in both mitochondrial and cytosolic Fe–S proteins, prompting the proposal that all Fe–S cluster assembly occurs in mitochondria (Lill and Kispal, 2000). This has led to the view that preassembled Fe–S clusters are exported from mitochondria for insertion into cytosolic proteins. At this time, CFD1 is one of three genes identified genetically that affect cytosolic Fe–S proteins exclusively. The other two genes, ATM1 and ERV1, encode a mitochondrial inner-membrane ABC-type transporter and a mitochondrial inter-membrane space sulfhydryl oxidase, respectively. These proteins are thought to act in the export of Fe–S clusters and/or cluster components from the mitochondria to cytosol (Kispal et al., 1999; Lange et al., 2001). Since Cfd1p does not stably associate with mitochondria, it does not appear likely that it is directly involved in the export of cluster components from this organelle. Interestingly, a small percentage of Cfd1p fractionated with other subcellular organelles and membranes. At the present time, it is not clear whether this small fraction of Cfd1p is associated specifically with a subcellular compartment or that it is the result of cytosolic contamination of these fractions. In either case, the amount of Cfd1p in membrane-enriched fractions represents <10% of the total Cfd1p in yeast cells, with the remaining ≥90% of Cfd1p being a soluble cytoplasmic protein.
The ability to convert IRP1 to c-aconitase in extracts of cfd1-1 yeast with iron alone leaves open the question of what is required from mitochondria for cytosolic Fe–S cluster assembly. We favor the view that Cfd1p functions to make iron available for cytosolic Fe–S cluster assembly, whether the iron is exported from mitochondria or acquired from other cellular pools. An intriguing possibility is that Cfd1p is an iron chaperone, ferrying iron for cytosolic Fe–S cluster biogenesis. We speculate that the CX2CX2C motif in Cfd1p is a metal binding site for such a process. Alignment of the Cfd1p amino acid sequence with that of homologous proteins for which structural information is available predicts that the CX2CX2C motif is surface exposed (Figure 4). By analogy to known metallochaperones (Arnesano et al., 2002), we expect that the metal binding element of an iron chaperone would be at or near the protein surface. That a change of either of the first two cysteine residues of the CX2CX2C motif to serine impaired Cfd1p activity is also consistent with a metal binding role for this motif.
Cytosolic aconitase in IRP1-transformed 0615d exists in both active [4Fe–4S] (80%) and inactive [3Fe–4S] forms (20%; Brown et al., 2002). The 3Fe form of the enzyme is a relatively long-lived intermediate in the oxidant-initiated Fe–S cluster disassembly process, and can be detected in cells by EPR (Brown et al., 2002). The cfd1-1 strain lacks any detectable 3Fe c-aconitase when transformed with IRP1. This suggests that very little [4Fe–4S] c-aconitase is ever formed in IRP1-transformed cfd1-1 yeast and provides strong support for the view that Cfd1p is essential for de novo assembly of Fe–S clusters in the cytosol.
The ability to convert IRP1 to c-aconitase in vitro by incubation with iron alone suggests that IRP1 expressed in cfd1-1 yeast already contains bound sulfur. One possibility is that sulfide can be held in IRP1 as protein-bound polysulfide (Cys-SSxS-Cys). Apo-mitochondrial aconitase generated via oxidant-mediated Fe–S cluster disassembly retains sulfide, apparently as Cys-SSxS-Cys (Kennedy and Beinert, 1988). It has been suggested that the formation of polysulfides in mitochondrial aconitase might keep the apo-protein in a structure in which the Fe–S cluster can be efficiently reconstituted with iron alone (Kennedy and Beinert, 1988). Given that Fe–S cluster assembly in IRP1 is acutely responsive to cellular iron status, we believe that IRP1 behaves similarly to mitochondrial aconitase and can form polysulfide bonds in the absence of iron; thereby being ready to accept excess iron in the cytoplasm. By making iron available for cluster assembly, Cfd1p might play a key role in the regulation of IRP1 in animals. Nevertheless, our results support the view that Cfd1p plays a critical and indispensable role in cytosolic Fe–S cluster assembly.
Materials and methods
Media, strains and growth conditions
The 0615d yeast strain (MATa, ura3-52, trp1-Δ63, his3-Δ200, aco1-1, ade2, IDP2up) was used throughout this study and has been described elsewhere (Brown et al., 1998; Narahari et al., 2000). Yeasts were routinely transformed with plasmid DNA using the lithium acetate method (Ito et al., 1983). Transformed yeast cells were grown at 30°C in minimal medium supplemented with 2% dextrose (SD; Sherman, 1991) and lacking specific nutrients as necessary for selection and maintenance of specific plasmids. For testing aconitase function in vivo, glutamate was omitted from this medium. Non-transformed yeast were grown in rich YPD medium containing 2% dextrose, or in SD-complete medium, as indicated (Sherman, 1991). URA3-containing plasmids were shuffled out of ura3 yeast by selection on SD-complete medium supplemented with 5-fluoroorotic acid (5-FOA; Sikorski and Boeke, 1991). Yeast were transformed with IRP1 carried on 2 µm plasmids, either pYADFRP (Narahari et al., 2000) or pYESHIS-IRP. These plasmids are identical except that the URA3 gene marker on pYADFRP is replaced with a HIS3 gene marker on pYESHIS-IRP. The level of IRP1 expression obtained with these plasmids in any particular yeast strain was indistinguishable.
Mutagenesis
IRP1-transformed 0615d was grown overnight in SD medium lacking uracil (SD-ura) to an OD600 of 0.8–1.0. Cells were pelleted, washed and treated with EMS as described elsewhere (Lawrence, 1991). EMS treatment gave ∼40–50% viability. Mutagenized cells were plated onto YPD solid medium and cells were allowed to grow at 30°C until colonies appeared. Cells were then replica-plated onto SD-glu and SD-leu media. Colonies that failed to grow or which grew poorly on SD-glu medium, and which grew poorly on SD-leu or YPD media were picked and tested directly for growth on glutamate-free SD-ura medium. AR27/IRP1 was picked because it displayed a severe growth defect on glutamate-free medium, and showed growth defects on leucine-free medium and YPD.
Library screening
AR27/IRP1 was transformed with a wild-type yeast genomic library cloned in YCp50 (average insert size equal 12 kb; kindly provided by Andy Dancis, University of Pennsylvania). To identify cells that had been transformed with library clones that could complement the strain’s glutamate-dependent growth restriction, ∼105 transformants were plated onto SD medium lacking uracil, histidine and glutamate and incubated at 30°C. Three hundred colonies appeared within 5 days of growth. A large majority of these colonies had acquired a genomic clone containing the mitochondrial aconitase gene, ACO1, which could also rescue AR27 from glutamate auxotrophy. To identify clones that had picked up the wild-type complement to the relevant mutant gene in AR27, the 300 colonies were replica-plated onto glutamate-free medium supplemented with the cell-impermeant iron chelator ferrozine. Addition of cell-impermeant iron chelators to glutamate-free medium strongly inhibits growth of yeast that are dependent on IRP1 for aconitase activity, whereas ACO1+ yeast are much more resistant to this inhibition (Narahari et al., 2000). Two transformants were highly sensitive to ferrozine. These transformants each carried a genomic fragment from the BET1–PAN1 region of yeast chromosome IX, and were subjected to further analysis as described in Results.
PCR cloning of CFD1, gene disruption and site-directed mutagenesis
To clone the CFD1 gene, yeast genomic DNA was prepared as previously described (Narahari et al., 2000) and used for amplification of CFD1 genes by the polymerase chain reaction (PCR) using the Expand High Fidelity PCR System (Roche), according to the manufacturer’s recommendation. The CFD1 gene plus 456 bp upstream and 168 bp downstream of the ORF was amplified using the following primer set: 3F, 5′-ATGGAAGAAAGAGCCGTGAA-3′ (5′ end); 3R, 5′-CACTTGAAGAACATGCTGCA-3′ (3′ end). This yielded the 1506 bp CFD1 and cfd1-1 genes used for complementation studies described in the text. A 940 bp fragment comprising the CFD1 or cfd1-1 ORF alone was amplified using the following primers: 2F, 5′-AGCACCCAAAACTAATACGAC-3′ (nucleotides –35 to –15 relative to the ORF); 2R, 5′-ACGTACATGCATAGGAGAGGA-3′ (nucleotides +905 to +885 relative to the ORF).
The ORF extends from nucleotide 1 to 882. The DNA fragments resulting from PCR amplifications were initially cloned into pGEM using the pGEM-T Easy Vector System I (Promega) following the manufacturer’s recommendation. A total of six wild-type or six mutant PCR clones (four 940 bp and two 1506 bp products each) were sequenced at the UIC DNA Sequencing Facility. For transformation into yeast, wild-type or mutant 1506 bp CFD1 genes were cloned into pRS313 or pRS316 (Sikorski and Hieter, 1989). Three independent wild-type or mutant PCR clones of CFD1 were tested in complementation assays.
The chromosomal CFD1 gene was disrupted in strain 0615d by one-step gene disruption (Rothstein, 1991). TRP1 was cloned between two unique ClaI sites within the 940 bp CFD1 ORF. The resulting cfd1::TRP1 gene was amplified by PCR using the 2F and 2R primer set. This PCR product was used to transform 0615d, which also had an extra copy of wild-type CFD1 carried on pRS316 (pRS316-CFD1). TRP+ transformants were tested for growth on medium containing 5-FOA to identify cells in which the chromosomal CFD1 gene had been disrupted. Since the CFD1 gene is essential in yeast, transformants with a disruption of the chromosomal CFD1 gene would not grow on media containing 5-FOA because loss of pRS316-CFD1 would be lethal. Disruption of the chromosomal copy of the CFD1 gene was further confirmed by PCR using primers specific for the chromosomal gene. The resulting strain was called NS1/CFD1. To replace the pRS316-CFD1 plasmid in disrupted strains with plasmids carrying either wild-type CFD1 or cfd1-1, NS1/CFD1 was transformed with these genes on a HIS3 plasmid and HIS+ transformants were plated onto media containing 5-FOA. Colonies that grew up on this medium were collected and transformed with IRP1 for further study.
6HisCfd1p was generated by subcloning the CFD1 ORF between the NdeI and EcoRI sites in pET-28a (Novagen). This placed the CFD1 ORF downstream of and in-frame with the sequence encoding a His6 polypeptide. For expression in yeast, an XbaI–SalI fragment containing the 6HisCFD1 fusion gene was subcloned downstream of the yeast ADH1 promoter on pRS313, generating p6HisCFD1. All site-directed mutagenesis was performed using p6HisCFD1 and the Transformer Site-directed Mutagenesis kit (Clontech) following the manufacturer’s protocol, and was confirmed by sequencing.
Whole-cell EPR analysis
Whole-cell EPR analysis was performed as described elsewhere (Brown et al., 2002) with a Bruker ELEXSYS series spectrometer at a temperature of 10–11K using standard methods. Conditions of spectroscopy were as follows: microwave power, 1 mW; microwave frequency, 9.45 GHz; modulation amplitude, 10G; modulation frequency, 100 kHz; time constant, 0.082 s; scanning time, 83 s. Each spectrum represents an average of four scans. Subtraction of one spectrum from another was performed using software supplied with the spectrometer. Where indicated, yeast were treated with H2O2 for 15 min in growth media prior to harvest for EPR analysis.
Enzyme assays and protein analysis
Preparation of yeast cytoplasmic extract, aconitase assay and protein immunoblotting were performed as described elsewhere (Brown et al., 1998; Narahari et al., 2000). Leu1p assays were performed using freshly prepared extracts and citraconic acid as substrate as described elsewhere (Kohlhaw, 1988). SDH, IDH and MDH enzyme assays were performed on freshly prepared whole-cell extracts (SDH) or mitochondrial extracts (IDH and MDH) following published methods (Cook and Sanwal, 1969; Williamson and Corkey, 1969; Robinson et al., 1991). All enzyme assays were carried out in triplicate and are representative of at least two independent experiments. When comparing enzyme activities between wild-type and mutant strains, yeast cultures were grown on the same day to an OD600 of 0.7–0.8.
Subcellular fractionation of yeast was performed following the procedure of Diekert et al. (2001). Crude, whole-cell extracts were centrifuged at 12 000 g for 10 min at 4°C to separate mitochondria and other organelles (P12) from the crude cytoplasmic fraction (S12). The S12 was separated into S100 and P100 by centrifugation at 100 000 g for 2 h at 4°C. Mitochondria and other membrane vesicles in the P12 were separated by Nycodenz density gradient and four fractions were collected containing residual soluble material (N1), light membrane vesicles (N2), mitochondria (M) and aggregated and heavy membrane vesicles (N4). All fractions generated through this protocol were analyzed for the presence of Cfd1p and other marker proteins. Cfd1p was detected with polyclonal anti-peptide antibody raised against KLH-conjugated DKHEQPQIESPK found in the Cfd1p sequence. The IRP1 used for this analysis was a functional fusion with a myc epitope tag at the C-terminus and was detected with an anti-myc epitope monoclonal antibody (Sigma). The outer mitochondrial membrane protein porin was detected with a monoclonal antibody (Molecular Probes). Aco1p and Leu1p were detected with polyclonal antiserum specific to each protein.
Acknowledgments
Acknowledgements
The authors thank David Ucker for critical reading of the manuscript and Violeta Chavez for technical assistance. We also thank Andy Dancis for the generous gift of the yeast genomic library and Aco1p antiserum, Roland Lill for the gift of Leu1p antiserum and acknowledge the assistance of Bob Lee in the Protein Research Laboratory of the UIC Research Resources Center in anti-Cfd1p antibody production. This work was supported by grant DK47281 from the National Institutes of Health to W.E.W. We acknowledge the use of the facilities of the National Biomedical ESR Center at the Medical College of Wisconsin, which was supported by National Institutes of Health Grant RR01008.
References
- Arnesano F., Banci,L., Bertini,I., Ciofi-Baffoni,S., Molteni,E., Huffman,D.L. and O’Halloran,T.V. (2002) Metallochaperones and metal-transporting ATPases: a comparative analysis of sequences and structures. Genome Res., 12, 255–271. [DOI] [PubMed] [Google Scholar]
- Beinert H. and Kiley,P.J. (1999) Fe–S proteins in sensing and regulatory functions. Curr. Opin. Chem. Biol., 3, 152–157. [DOI] [PubMed] [Google Scholar]
- Brown N.M., Anderson,S.A., Steffen,D.W., Carpenter,T.B., Kennedy,M.C., Walden,W.E. and Eisenstein,R.S. (1998) Novel role of phosphorylation in Fe–S cluster stability revealed by phospho mimetic mutations at Ser-138 of iron regulatory protein 1. Proc. Natl Acad. Sci. USA, 95, 15235–15240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown N.M., Kennedy,M.C., Antholine,W.E., Eisenstein,R.S. and Walden,W.E. (2002) Detection of a [3Fe–4S] cluster intermediate of cytosolic aconitase in yeast expressing iron regulatory protein 1. Insights into the mechanism of Fe–S cluster cycling. J. Biol. Chem., 277, 7246–7254. [DOI] [PubMed] [Google Scholar]
- Cook R.A. and Sanwal,B.D. (1969) Isocitrate dehydrogenase (NAD-specific) from Neurospora crassa. Methods Enzymol., 13, 42–47. [Google Scholar]
- Cordell S.C. and Lowe,J. (2001) Crystal structure of the bacterial cell division regulator MinD. FEBS Lett., 492, 160–165. [DOI] [PubMed] [Google Scholar]
- Dean D.R., Bolin,J.T. and Zheng,L. (1993) Nitrogenase metalloclusters: structures, organization and synthesis. J. Bacteriol., 175, 6737–6744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diekert K., de Kroon,A.I., Kispal,G. and Lill,R. (2001) Isolation and subfractionation of mitochondria from the yeast Saccharomyces cerevisiae. Methods Cell Biol., 65, 37–51. [DOI] [PubMed] [Google Scholar]
- Eisenstein R.S. (2000) Iron regulatory proteins and the molecular control of mammalian iron metabolism. Annu. Rev. Nutr., 20, 627–662. [DOI] [PubMed] [Google Scholar]
- Entian K.D. et al. (1999) Functional analysis of 150 deletion mutants in Saccharomyces cerevisiae by a systematic approach. Mol. Gen. Genet., 262, 683–702. [DOI] [PubMed] [Google Scholar]
- Ferro-Novick S., Newman,A.P., Groesch,M., Ruohola,H., Rossi,G., Graf,J. and Shim,J. (1991) An analysis of BET1, BET2 and BOS1. Three factors mediating ER to Golgi transport in yeast. Cell Biophys., 19, 25–33. [DOI] [PubMed] [Google Scholar]
- Gangloff S.P., Marguet,D. and Lauquin,G.J. (1990) Molecular cloning of the yeast mitochondrial aconitase gene (ACO1) and evidence of a synergistic regulation of expression by glucose plus glutamate. Mol. Cell. Biol., 10, 3551–3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garland S.A., Hoff,K., Vickery,L.E. and Culotta,V.C. (1999) Saccharomyces cerevisiae ISU1 and ISU2: members of a well-conserved gene family for iron-sulfur cluster assembly. J. Mol. Biol., 294, 897–907. [DOI] [PubMed] [Google Scholar]
- Giaever G. et al. (2002) Functional profiling of the Saccharomyces cerevisiae genome. Nature, 418, 387–391. [DOI] [PubMed] [Google Scholar]
- Hayashi I., Oyama,T. and Morikawa,K. (2001) Structural and functional studies of MinD ATPase: implications for the molecular recognition of the bacterial cell division apparatus. EMBO J., 20, 1819–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentze M.W. and Kuhn,L.C. (1996) Molecular control of vertebrate iron metabolism: mRNA-based regulatory circuits operated by iron, nitric oxide and oxidative stress. Proc. Natl Acad. Sci. USA, 93, 8175–8182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Fukuda,Y., Murata,K. and Kimura,A. (1983) Transformation of intact yeast cells treated with alkali cations. J. Bacteriol., 153, 163–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen L.T. and Culotta,V.C. (2000) Role of Saccharomyces cerevisiae ISA1 and ISA2 in iron homeostasis. Mol. Cell. Biol., 20, 3918–3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson M.K. (1998) Iron-sulfur proteins: new roles for old clusters. Curr. Opin. Chem. Biol., 2, 173–181. [DOI] [PubMed] [Google Scholar]
- Kaut A., Lange,H., Diekert,K., Kispal,G. and Lill,R. (2000) Isa1p is a component of the mitochondrial machinery for maturation of cellular iron-sulfur proteins and requires conserved cysteine residues for function. J. Biol. Chem., 275, 15955–15961. [DOI] [PubMed] [Google Scholar]
- Kennedy M.C. and Beinert,H. (1988) The state of cluster SH and S2- of aconitase during cluster interconversions and removal. A convenient preparation of apoenzyme. J. Biol. Chem., 263, 8194–8198. [PubMed] [Google Scholar]
- Kennedy M.C., Emptage,M.H., Dreyer,J.L. and Beinert,H. (1983) The role of iron in the activation–inactivation of aconitase. J. Biol. Chem., 258, 11098–11105. [PubMed] [Google Scholar]
- Kennedy M.C., Mende-Mueller,L., Blondin,G.A. and Beinert,H. (1992) Purification and characterization of cytosolic aconitase from beef liver and its relationship to the iron-responsive element binding protein. Proc. Natl Acad. Sci. USA, 89, 11730–11734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kispal G., Csere,P., Prohl,C. and Lill,R. (1999) The mitochondrial proteins Atm1p and Nfs1p are essential for biogenesis of cytosolic Fe/S proteins. EMBO J., 18, 3981–3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight S.A., Sepuri,N.B., Pain,D. and Dancis,A. (1998) Mt-Hsp70 homolog, Ssc2p, required for maturation of yeast frataxin and mitochondrial iron homeostasis. J. Biol. Chem., 273, 18389–18393. [DOI] [PubMed] [Google Scholar]
- Kohlhaw G.B. (1988) Isopropylmalate dehydratase from yeast. Methods Enzymol., 166, 423–429. [DOI] [PubMed] [Google Scholar]
- Land T. and Rouault,T.A. (1998) Targeting of a human iron-sulfur cluster assembly enzyme, nifs, to different subcellular compartments is regulated through alternative AUG utilization. Mol. Cell, 2, 807–815. [DOI] [PubMed] [Google Scholar]
- Lange H., Lisowsky,T., Gerber,J., Muhlenhoff,U., Kispal,G. and Lill,R. (2001) An essential function of the mitochondrial sulfhydryl oxidase Erv1p/ALR in the maturation of cytosolic Fe/S proteins. EMBO Rep., 2, 715–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence C.W. (1991) Classical mutagenesis techniques. Methods Enzymol., 194, 273–281. [DOI] [PubMed] [Google Scholar]
- Leipe D.D., Wolf,Y.I., Koonin,E.V. and Aravind,L. (2002) Classification and evolution of P-loop GTPases and related ATPases. J. Mol. Biol., 317, 41–72. [DOI] [PubMed] [Google Scholar]
- Li J., Kogan,M., Knight,S.A., Pain,D. and Dancis,A. (1999) Yeast mitochondrial protein, Nfs1p, coordinately regulates iron-sulfur cluster proteins, cellular iron uptake and iron distribution. J. Biol. Chem., 274, 33025–33034. [DOI] [PubMed] [Google Scholar]
- Lill R. and Kispal,G. (2000) Maturation of cellular Fe–S proteins: an essential function of mitochondria. Trends Biochem. Sci., 25, 352–356. [DOI] [PubMed] [Google Scholar]
- Muhlenhoff U. and Lill,R. (2000) Biogenesis of iron-sulfur proteins in eukaryotes: a novel task of mitochondria that is inherited from bacteria. Biochim. Biophys. Acta, 1459, 370–382. [DOI] [PubMed] [Google Scholar]
- Muhlenhoff U., Richhardt,N., Gerber,J. and Lill,R. (2002) Characterization of iron-sulfur protein assembly in isolated mitochondria. A requirement for ATP, NADH and reduced iron. J. Biol. Chem., 277, 29810–29816. [DOI] [PubMed] [Google Scholar]
- Nakashima H., Grahovac,M.J., Mazzarella,R., Fujiwara,H., Kitchen,J.R., Threat,T.A. and Ko,M.S. (1999) Two novel mouse genes—Nubp2, mapped to the t-complex on chromosome 17 and Nubp1, mapped to chromosome 16—establish a new gene family of nucleotide-binding proteins in eukaryotes. Genomics, 60, 152–160. [DOI] [PubMed] [Google Scholar]
- Narahari J., Ma,R., Wang,M. and Walden,W.E. (2000) The aconitase function of iron regulatory protein 1. Genetic studies in yeast implicate its role in iron-mediated redox regulation. J. Biol. Chem., 275, 16227–16234. [DOI] [PubMed] [Google Scholar]
- Newman A.P. and Ferro-Novick,S. (1987) Characterization of new mutants in the early part of the yeast secretory pathway isolated by a [3H]mannose suicide selection. J. Cell Biol., 105, 1587–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogur M., Coker,L. and Ogur,S. (1964) Glutamate auxotrophs in Saccharomyces 1. I. The biochemical lesion in the glt-1 mutants-2. Biochem. Biophys. Res. Commun., 14, 193–197. [DOI] [PubMed] [Google Scholar]
- Pelzer W., Muhlenhoff,U., Diekert,K., Siegmund,K., Kispal,G. and Lill,R. (2000) Mitochondrial Isa2p plays a crucial role in the maturation of cellular iron-sulfur proteins. FEBS Lett., 476, 134–139. [DOI] [PubMed] [Google Scholar]
- Robinson K.M., von Kieckebusch-Guck,A. and Lemire,B.D. (1991) Isolation and characterization of a Saccharomyces cerevisiae mutant disrupted for the succinate dehydrogenase flavoprotein subunit. J. Biol. Chem., 266, 21347–21350. [PubMed] [Google Scholar]
- Rothstein R. (1991) Targeting, disruption, replacement and allele rescue: integrative DNA transformation in yeast. Methods Enzymol., 194, 281–301. [DOI] [PubMed] [Google Scholar]
- Ryan E.D., Tracy,J.W. and Kohlhaw,G.B. (1973) Subcellular localization of the leucine biosynthetic enzymes in yeast. J. Bacteriol., 116, 222–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai N., Yao,M., Itou,H., Watanabe,N., Yumoto,F., Tanokura,M. and Tanaka,I. (2001) The three-dimensional structure of septum site-determining protein MinD from Pyrococcus horikoshii OT3 in complex with Mg-ADP. Structure, 9, 817–826. [DOI] [PubMed] [Google Scholar]
- Schilke B., Voisine,C., Beinert,H. and Craig,E. (1999) Evidence for a conserved system for iron metabolism in the mitochondria of Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 96, 10206–10211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F. (1991) Getting started with yeast. Methods Enzymol., 194, 3–21. [DOI] [PubMed] [Google Scholar]
- Sikorski R.S. and Boeke,J.D. (1991) In vitro mutagenesis and plasmid shuffling: from cloned gene to mutant yeast. Methods Enzymol., 194, 302–318. [DOI] [PubMed] [Google Scholar]
- Sikorski R.S. and Hieter,P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics, 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipos K., Lange,H., Fekete,Z., Ullmann,P., Lill,R. and Kispal,G. (2002) Maturation of cytosolic iron-sulfur proteins requires glutathione. J. Biol. Chem., 277, 26944–26949. [DOI] [PubMed] [Google Scholar]
- Skovran E. and Downs,D.M. (2003) Lack of the ApbC or ApbE protein results in a defect in Fe–S cluster metabolism in Salmonella enterica serovar typhimurium. J. Bacteriol., 185, 98–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theil E.C. and Eisenstein,R.S. (2000) Combinatorial mRNA regulation: iron regulatory proteins and iso-iron-responsive elements (Iso-IREs). J. Biol. Chem., 275, 40659–40662. [DOI] [PubMed] [Google Scholar]
- Tong W.H. and Rouault,T. (2000) Distinct iron-sulfur cluster assembly complexes exist in the cytosol and mitochondria of human cells. EMBO J., 19, 5692–5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitale G., Fabre,E. and Hurt,E.C. (1996) NBP35 encodes an essential and evolutionary conserved protein in Saccharomyces cerevisiae with homology to a superfamily of bacterial ATPases. Gene, 178, 97–106. [DOI] [PubMed] [Google Scholar]
- Voisine C., Cheng,Y.C., Ohlson,M., Schilke,B., Hoff,K., Beinert,H., Marszalek,J. and Craig,E.A. (2001) Jac1, a mitochondrial J-type chaperone, is involved in the biogenesis of Fe/S clusters in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 98, 1483–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker J.E., Saraste,M., Runswick,M.J. and Gay,N.J. (1982) Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J., 1, 945–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson J.R. and Corkey,B.E. (1969) Assays of intermediates of the citric acid cycle and related compounds by fluorometric enzyme methods. Methods Enzymol., 13, 434–513. [DOI] [PubMed] [Google Scholar]
- Zheng L., Cash,V.L., Flint,D.H. and Dean,D.R. (1998) Assembly of iron-sulfur clusters. Identification of an iscSUA-hscBA-fdx gene cluster from Azotobacter vinelandii. J. Biol. Chem., 273, 13264–13272. [DOI] [PubMed] [Google Scholar]