Abstract
Protein-tyrosine kinases regulating bacterial exopolysaccharide synthesis autophosphorylate on tyrosines located in a conserved C-terminal region. So far no other substrates have been identified for these kinases. Here we demonstrate that Bacillus subtilis YwqD not only autophosphorylates at Tyr-228, but that it also phosphorylates the two UDP-glucose dehydrogenases (UDP-glucose DHs) YwqF and TuaD at a tyrosine residue. However, phosphorylation of YwqF and TuaD occurs only in the presence of the transmembrane protein YwqC. The presumed intracellular C-terminal part of YwqC (last 50 amino acids) seems to interact with the tyrosine-kinase and to allow YwqD-catalysed phosphorylation of the two UDP-glucose DHs, which are key enzymes for the synthesis of acidic polysaccharides. However, only when phosphorylated by YwqD do the two enzymes exhibit detectable UDP-glucose DH activity. Dephosphorylation of P-Tyr-YwqF and P-Tyr-TuaD by the P-Tyr-protein phosphatase YwqE switched off their UDP-glucose DH activity. YwqE, which is encoded by the fourth gene of the B.subtilis ywqCDEF operon, also dephosphorylates P-Tyr-YwqD.
Keywords: P-tyrosine phosphatase/polysaccharide synthesis/protein phosphorylation/tyrosine-kinase/UDP-glucose dehydrogenase
Introduction
In eukaryotes, protein-tyrosine kinases (PTKs) and P-Tyr-protein phosphatases (PTPs) regulate important cellular processes, such as proliferation, differentiation and oncogenesis (Fantl et al., 1993; Hunter, 1995; Schlessinger, 2000). Eukaryotic PTKs have also been reported to phosphorylate proteins of certain bacterial pathogens (Backert et al., 2001). Bacteria possess PTKs too, and in one case has the substrate of a bacterial PTK been identified. For others, only autophosphorylation at tyrosine residues in their C-terminal part has so far been reported (Grangeasse et al., 2002; Paiment et al., 2002). Bacterial PTKs differ from their eukaryotic counterparts, in that they use a Walker motif (Walker et al., 1982) as active site. In Gram-negative bacteria, these PTKs are large membrane-spanning multi-domain proteins (Grangeasse et al., 1997; Ilan et al., 1999; Doublet et al., 2002). The PTK activity is associated with the C-terminal domain located on the cytoplasmic side of the membrane (Vincent et al., 2000; Niemeyer and Becker, 2001). When synthesized separately, the C-terminal domain of the Escherichia coli PTK Wzc was soluble, but still able to autophosphorylate (Grangeasse et al., 2002). In Gram-positive bacteria, the large PTKs of Gram-negative bacteria are often split into two distinct proteins, which both have a molecular weight (MW) of ∼25 kDa, and which are encoded by genes usually located next to each other. The protein encoded by the first gene resembles the membrane-spanning and the protein encoded by the second gene the autophosphorylation domain of PTKs from Gram-negative bacteria, (Sau et al., 1997; Morona et al., 1999; Yamamoto et al., 1999).
In Gram-positive and Gram-negative bacteria, these PTKs regulate the formation of exopolysaccharides (EPS) (Morona et al., 2000; Vincent et al., 2000; Niemeyer and Becker, 2001; Wugeditsch et al., 2001; Paiment et al., 2002). Their genes are often organized in an operon together with genes implicated in capsular polysaccharide (CPS) synthesis. In E.coli, the unphosphorylated form of the autophosphorylating PTK Wzc is required for the synthesis of the EPS colanic acid (Vincent et al., 2000). However, diminished synthesis of colanic acid occurred after inactivation of not only Wzc, but also of Wzb, which catalyses the dephosphorylation of P-Wzc. In wzb mutants, probably little Wzc is present, as most of it is converted to P-Wzc, which therefore leads to slow colanic acid synthesis (Vincent et al., 2000). Phosphorylation of E.coli Wzc has also been reported to influence the assembly of group 1 CPSs (Wugeditsch et al., 2001). Among other functions, EPS production affects cell adhesion in bacterial symbionts and pathogens (Ilan et al., 1999). For example, autophosphorylation of the PTK ExoP in the soil bacterium Sinorhizobium meliloti alters the production ratio of low- and high-molecular weight succinoglycans, which affects the ability of this bacterium to invade the root nodules of its plant symbiont Medicago sativa (Niemeyer and Becker, 2001). Moreover, autophosphorylation of the PTK Cps2D slows CPS biosynthesis in the pathogen Streptococcus pneumoniae (Morona et al., 2000; Morona et al., 2003), which in turn affects the virulence of this organism. P-Cps2D is dephosphorylated by the PTP Cps2B (Bender and Yother, 2001; Morona et al., 2002), which differs from other PTPs with respect to its sequence and catalytic mechanism (Barford et al., 1998), and which has also been reported to inhibit the PTK Cps2D (Bender and Yother, 2001). PTKs of other pathogens, such as Klebsiella pneumoniae (Arakawa et al., 1995; Preneta et al., 2002) and Erwinia amylovora (Bugert and Geider, 1997), have also been reported to affect the virulence of their organisms.
Bacillus subtilis contains several genes encoding proteins with significant similarity to S.pneumoniae Cps2D (Morona et al., 2000) and the C-terminal PTK domain of E.coli Wzc (Grangeasse et al., 2002). At least one of them, YwqD, exhibits autophosphorylating PTK activity. The gene encoding this PTK is part of the ywqCDEF operon, where ywqE codes for a S.pneumoniae Cps2B-type PTP. ywqC encodes a membrane-spanning protein resembling the transmembrane domain of E.coli Wzc, and ywqF encodes a protein exhibiting significant similarity to B.subtilis TuaD (Pagni et al., 1999) and other UDP-glucose dehydrogenases (UDP-glucose DHs). Our studies concentrated on the biochemical characterization of the proteins encoded by the ywqCDEF operon, including the in vivo autophosphorylation of YwqD at Tyr-228 and the YwqC-dependent YwqD-catalysed phosphorylation of YwqF and its homologue TuaD, which activates their UDP-glucose DH function.
Results and discussion
Bacillus subtilis YwqD autophosphorylates on tyrosine
Bacillus subtilis possesses several Walker motif-containing proteins exhibiting significant similarity to the PTK domain of Wzc from E.coli (Vincent et al., 1999). Five of these proteins were purified with an N-terminal His-tag. Among these proteins, MinD is implicated in the placement of the cell division site (de Boer et al., 1988; Levin et al., 1998) and Soj in chromosome partitioning and sporulation (Sharpe and Errington, 1996; Quisel and Grossman, 2000). The three other proteins, YwqD, YveL and YlxH, are of unknown function. Autophosphorylation experiments with [γ-32P]ATP revealed that only YwqD incorporated detectable amounts of radioactivity (data not shown). To determine which amino acid becomes phosphorylated, hydrolysis of radiolabelled YwqD was carried out under acidic conditions. The main radioactive hydrolysis product comigrated with the P-Tyr standard during two-dimensional thinlayer chromatography (Figure 1A). Western blots carried out with monoclonal anti-P-Tyr antibodies confirmed that YwqD autophosphorylates on a tyrosine (Figure 1B, lane 1). Interestingly, YveL also gave a strong signal in Western blots with anti-P-Tyr antibodies (Figure 1B, lane 3), suggesting that it gets phosphorylated when expressed in E.coli. This assumption was confirmed by incubating purified YveL with alkaline phosphatase, which is known to dephosphorylate P-proteins. When YwqD or YveL were treated with alkaline phosphatase before carrying out a western blot, it diminished the intensity of both protein bands (Figure 1B, lanes 2 and 4). Phosphorylation of YveL in E.coli could be due either to autophosphorylation or trans-phosphorylation catalysed by one of the E.coli PTKs. In the first case, in vivo modifiation of YveL would prevent further in vitro autophosphorylation with [γ-32P]ATP.
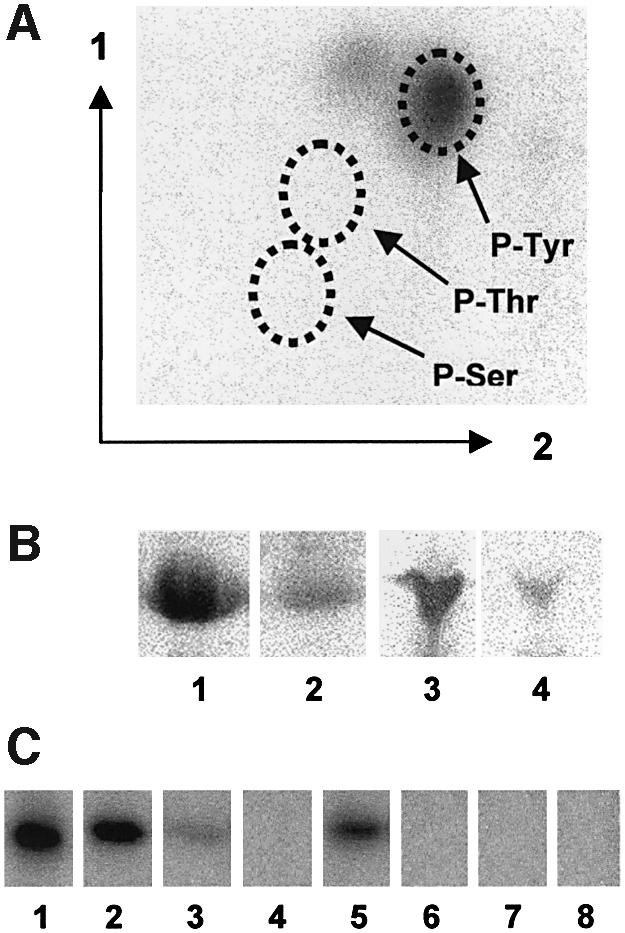
Fig. 1. Autophosphorylation of YwqD. (A) Autoradiogram obtained after two-dimensional thin layer chromatography carried out with the acid hydrolysate of [32P]Tyr-YwqD. Positions of the three P-amino acid standards were determined by ninhydrin staining and are indicated by dotted circles. (B) Western blots with anti P-Tyr antibodies and YwqD (lane 1), alkaline phosphatase-treated YwqD (lane 2), YveL (lane 3) and alkaline phosphatase-treated YveL (lane 4). (C) Autoradiogram of an SDS/polyacrylamide gel on which the following proteins autophos phorylated with [γ-32P]ATP (all at 1.5 µM) had been loaded: lane 1, YwqD; lane 2, YwqD-1; lane 3, YwqD-23; lane 4, YwqD-123; lane 5, YwqD-ΔCter; lane 6, YwqD-ΔNter; lane 7, YwqD-81; and lane 1, YwqD-83.
The sites of autophosphorylation in YwqD
In vitro phosphorylation experiments with E.coli Wzc had revealed that it autophosphorylates primarily at a C-terminal tyrosine cluster (Grangeasse et al., 2002). To test whether any of the three C-terminal tyrosines (Tyr-225, -227 and -228) of YwqD would also be autophosphoryl ated, one or several of these tyrosines were replaced with phenylalanines. In vitro autophosphorylation assays were carried out with [γ-32P]ATP and wild-type and mutant YwqDs. When Tyr-225 was altered (YwqD-1), the intensity of the radioactive signal was ∼70% of that obtained with wild-type YwqD, while the simultaneous replacement of Tyr-227 and -228 (YwqD-23) left a residual signal of only 5% compared to experiments with wild type YwqD (Figure 1C, lanes 1–3). Only replacement of all three tyrosines (YwqD-123) led to a complete loss of 32P incorporation (Figure 1C, lane 4). Wzc of E.coli also autophosphorylates on Tyr-569 located between the Walker motifs A and B (Grangeasse et al. 2002). YwqD is missing an equivalent of Tyr-569 and autophosphoryl ates only at the C-terminal tyrosine cluster. Interestingly, YwqD-123 with an artificial C-terminal extension containing a tyrosine residue was still autophosphorylated. The modification probably occurred at the additional tyrosine, suggesting that in vitro autophosphorylation is not very selective (data not shown).
To determine whether one of the three C-terminal tyrosines was preferentially phosphorylated in vivo, YwqD purified from E.coli was digested with trypsin providing a peptide (222-HSEYGYYGTK-231) containing all phosphorylatable tyrosines (underlined). Mass spectrometry (MALDI-TOF) carried out with the tryptic digest revealed that YwqD was present in mono- and non-phosphorylated form. The mono- and non-phosphorylated tryptic peptides could be separated by HPLC on a C18 reverse phase column and based on the UV elution profile we estimated that YwqD purified from E.coli contained ∼30% unphos phorylated and 70% mono-phosphorylated protein. YwqD synthesized in E.coli was found to be mainly phosphorylated at Tyr-228, as during Edman sequencing of the tryptic P-peptide no signal was detected at the step corresponding to Tyr-228. When YwqD was incubated in vitro with ATP and subsequently digested with trypsin, the di- and tri-phosphorylated peptides were obtained, which could also be separated by HPLC. Edman sequencing of the di-phosphorylated peptide revealed that Tyr-227 and -228 were modified. Phosphorylation at Tyr-225 could only be detected in the tri-phosphorylated peptide. These results suggested that phosphorylation of YwqD in E.coli occurs preferentially at Tyr-228. Equivalents of Tyr-228 of YwqD are present in Wzc of E.coli (Tyr-711) and Cps2D of S.pneumoniae (Tyr-221). However, recent mutant studies with Wzc of E.coli revealed no preferential in vivo phosphorylation at its Tyr-711 (Paiment et al., 2002). In vivo autophosphorylation has also been reported for S.pneumoniae Cps2D, but its exact phosphorylation site has not been determined (Ianelli et al., 1999; Morona et al., 2000, 2003).
YwqD autophosphorylation is intramolecular and requires Asp-81 and -83
In addition to the Walker A and B motifs, which are indispensable for the autophosphorylation of bacterial PTKs (Doublet et al., 1999; Niemeyer and Becker, 2001; Wugeditsch et al., 2001), YwqD possesses another sequence conserved in its B.subtilis paralogues YveL, YlxH, Soj and MinD [75-KKVLL(I/V)D(A/L/T/I)D-83]. The recently determined crystal structure of MinD revealed that Asp-38 and -40 of this region are implicated in binding of water molecules coordinating the magnesium ion in the Walker motif (Hayashi et al., 2001). To test whether the equivalent Asp-81 and -83 of YwqD might have the same function, we replaced the two aspartates one by one with alanine. The purifed mutant proteins YwqD-81 and YwqD-83 were devoid of autophosphorylation activity (Figure 1C, lanes 7 and 8).
Autophosphorylation of YwqD was also affected by cutting off the N- or C-terminal region. When the nine C-terminal amino acids following Tyr-228 were removed, the resulting protein YwqD-ΔCter exhibited only ∼20% of the autophosphorylation activity of wild-type YwqD (compare Figure 1C, lanes 1 and 5). Interestingly, YwqD and YveL contain a 42-amino acid N-terminal extension that is absent in Soj and MinD and is correlated with the inability of the latter proteins to autophosphorylate. Removal of this sequence prevented autophosphorylation of the resulting YwqD-ΔNter (Figure 1C, lane 6). A possible explanation for this finding was that YwqD autophosphorylation was an intermolecular reaction similar to that reported for E.coli Wzc (Grangeasse et al., 2002), and that an intact N-terminus was necessary to establish the intermolecular contacts. To test this assumption, we carried out phosphorylation experiments with YwqD-123 and either YwqD-81 or YwqD-83, which are all unable to autophosphorylate. If autophosphorylation was an intermolecular reaction, YwqD-123 was expected to phosphorylate YwqD-81 and YwqD-83. However, no phosphorylation of the two mutant proteins could be detected (data not shown). One possible explanation is that autophosphorylation of YwqD is not an intermolecular process, as reported of Wzc, but an intramolecular event.
YwqD also exhibited weak ATPase activity. Interestingly, replacing the three phosphorylatable tyrosines with phenylalanines, which completely prevented autophosphorylation, significantly stimulated the ATPase activity of YwqD. We therefore suspected that autophos phorylation might inhibit the ATPase activity of YwqD. Indeed, YwqD, which had been in vitro phosphorylated with ATP and subsequently desalted on Ni-NTA (Qiagen) and PD-10 (Pharmacia) columns, was completely devoid of ATPase activity (data not shown). Cutting off the N-terminus had no effect, whereas replacing Asp-81 and/or Asp-83 with alanines almost completely abolished the ATPase activity of YwqD.
The C-terminal domain of YwqC allows YwqD to phosphorylate proteins fused to YwqC
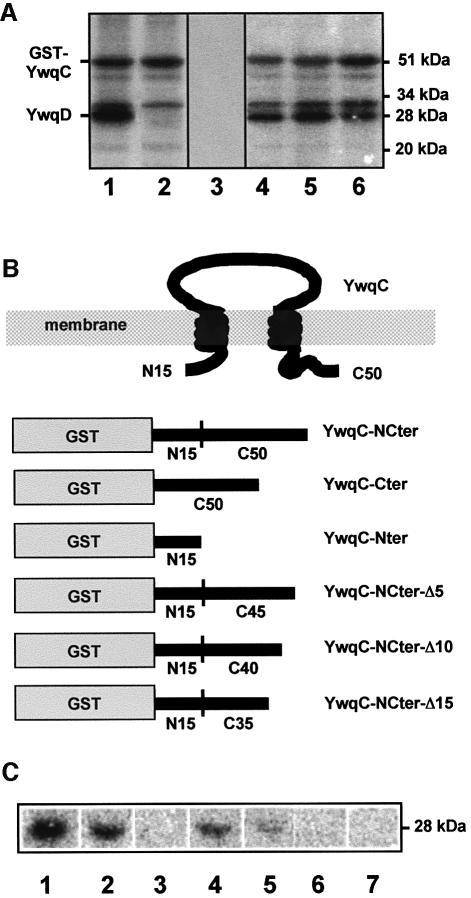
YwqC and YwqD resemble the transmembrane and the C-terminal PTK domain of E.coli Wzc, respectively. When the two Wzc domains were synthesized separately, the PTK domain could not phosphorylate the N-terminal domain (Grangeasse et al., 2002). However, it could not be excluded that the intact protein could autophosphorylate its N-terminal domain. We therefore tested whether YwqC could function as substrate for YwqD. Since YwqC and the autophosphorylating YwqD have a similar MW, YwqC was fused to glutathione transferase (GST) to better detect its possible phosphorylation. The membrane-associated hybrid protein was indeed phosphorylated with [γ-32P]ATP and YwqD (Figure 2A, lane 1). Purified [32P]Tyr-YwqD (free of [γ-32P]ATP) could not transfer its phosphoryl group to GST–YwqC (data not shown). It was therefore not surprising that GST–YwqC was also phosphorylated with YwqD-123 (Figure 2A, lane 2), confirming that the kinase activity of YwqD does not depend on its autophosphorylation. The mutant proteins YwqD-81, YwqD-83 and YwqD-ΔNter, which cannot autophosphorylate, could also not phosphorylate GST–YwqC (data not shown). Phosphorylation of GST–YwqC by YwqD-123 occurred at a tyrosine, as was demonstrated by acid hydrolysis/two-dimensional thin layer chromatography and anti-P-Tyr antibodies (data not shown). YwqC contains two tyrosines, which were replaced by alanines providing the mutant proteins YwqC-1, YwqC-2 and YwqC-12. Surprisingly, all three mutant GST–YwqC fusions were still phosphorylated by YwqD (Figure 2A, lanes 4–6). After cleaving [32P]GST–YwqC with thrombin it became obvious that not YwqC but GST was phosphorylated at a tyrosine, although GST without YwqC was not phosphorylated by YwqD-123 (Figure 2A, lane 3). To test whether YwqC would also mediate phosphorylation of other proteins, it was fused to the maltose binding protein (MBP). Phosphorylation experiments with MBP–YwqC provided results identical to those obtained with GST–YwqC (data not shown), suggesting that YwqC specifically interacts with YwqD thereby allowing tyrosine phosphorylation of the fusion proteins.
Fig. 2. Effect of YwqC on the protein kinase activity of YwqD. (A) Autoradiograms of SDS/polyacrylamide gels on which reaction mixtures containing [γ-32P]ATP and the following proteins had been separated (YwqD and YwqD-123, 1 µM; GST–YwqC and GST, 5 µM): lane 1, YwqD and GST–YwqC; lane 2, YwqD-123 and GST–YwqC; lane 3, YwqD-123 and GST; lane 4, YwqD and YwqC-1; lane 5, YwqD and YwqC-2; and lane 6, YwqD and YwqC-12. (B) Presumed YwqC topology and schematic presentation of the GST fusions with YwqC or various parts of it. YwqC was predicted to contain two transmembrane regions with the 15 N-terminal and 50 C-terminal amino acids oriented towards the cytoplasm. (C) Autoradiograms of SDS–polyacrylamide gels on which reaction mixtures containing [γ-32P]ATP and the following proteins had been separated (YwqD-123 and all forms of GST–YwqC, 1.5 µM): YwqD-123 and either GST–YwqC-NCter (lane 1), GST–YwqC-Cter (lane 2), GST–YwqC-Nter (lane 3), GST–YwqC-NCter-Δ5 (lane 4), GST–YwqC-NCter-Δ10 (lane 5) and GST–YwqC-NCter-Δ15 (lane 6). Lane 7, control sample with [γ-32P]ATP and GST–YwqC-NCter.
Web-based algorithms (http://www.expasy.org/tools/) predicted that YwqC contained two transmembrane helices and that the 15 N-terminal and 50 C-terminal amino acids were exposed to the cytosol and the major central region to the periplasm (Figure 2B). If this prediction was correct, only the C- and/or N-terminal parts of YwqC could interact with the cytosolic YwqD and allow phosphorylation of GST and MBP. We therefore fused the first 15 and last 50 amino acids of YwqC (YwqC-NCter) to GST (Figure 2B) and MBP. GST–YwqC-NCter was not modified by [γ-32P]ATP (Figure 2C, lane 7), while phosphorylation occurred when YwqD-123 was also present (Figure 2C, lane 1). Identical results were obtained with MBP–YwqC-NCter. To pinpoint the minimal region able to interact with YwqD, a series of deletions were introduced into GST–YwqC-NCter (Figure 2B). GST–YwqC-Cter (missing the N-terminus of YwqC) was phosphorylated at about half the rate of GST–YwqC-NCter, whereas cutting off the C-terminus prevented phosphorylation of the resulting GST–YwqC-Nter (Figure 2C, lanes 2 and 3). A 5-amino acid deletion at the C-terminus of GST–YwqC-NCter reduced the radioactive signal by 70% and a 10-amino acid deletion by 95%. Finally, after cutting off the 15 C-terminal amino acids, phosphorylation of the hybrid protein could no longer be detected (Figure 2C, lanes 4–6). In conclusion, the last 50 amino acids of YwqC were sufficient to allow YwqD-catalysed phosphorylation of GST and MBP. The additional presence of the N-terminal 15 amino acids stimulated GST phosphorylation ∼2-fold. However, these 15 amino acids alone could not promote GST phosphorylation, suggesting that they act as spacers, improving the interaction between YwqD and the fusion protein.
YwqC allows YwqD-catalysed phosphorylation of UDP-glucose DHs
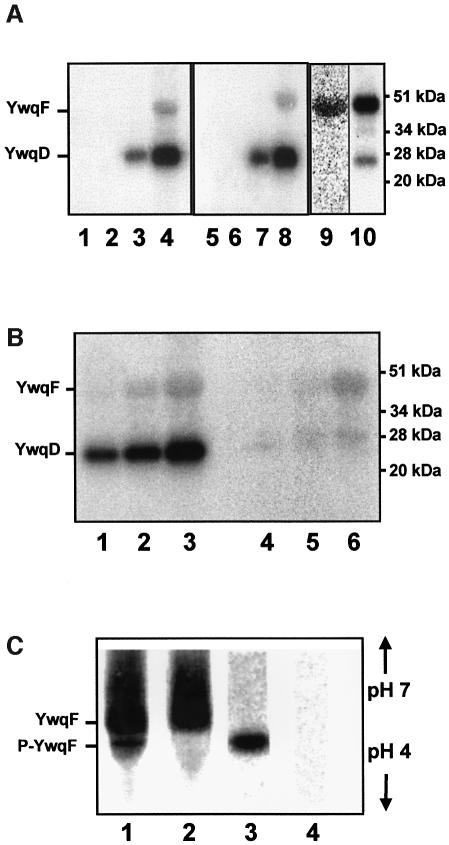
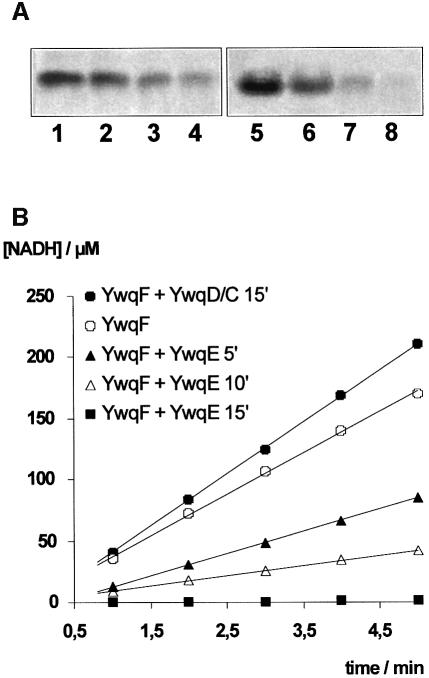
YwqF, which is encoded by the fourth gene of the ywqCDEF operon, exhibits significant homology to the B.subtilis UDP-glucose DH TuaD (Pagni et al., 1999). To test whether YwqF and its homologue TuaD were substrates for YwqD, both proteins were synthesized with a His-tag and purified. No phosphorylation of YwqF and TuaD was observed when they were incubated with [γ-32P]ATP and YwqD (Figure 3A, lanes 3 and 7). However, when GST–YwqC-NCter was included in the assay mixture, YwqF and TuaD were phosphorylated (Figure 3A, lanes 4 and 8, respectively). YwqF and TuaD were also phosphorylated when GST–YwqC was used instead of GST–YwqC-NCter (data not shown). In both proteins, phosphorylation occurred at a tyrosine, as was demonstrated by acid hydrolysis/two-dimensional thin layer chromatography and with anti P-Tyr antibodies (data not shown). UDP-glucose DHs are the second class of substrates identified for bacterial PTKs, the first being heat shock-related proteins (Klein et al., 2003). The rate of YwqF phosphorylation by YwqD under the employed experimental conditions was calculated as 3 nM/min, while YwqD autophosphorylates more rapidly at 24 nM/min (Figure 3B, lanes 1–3). When YwqD-123 was used instead of wild-type YwqD, the rate of YwqF phosphorylation remained essentially the same (Figure 3B, compare lanes 4–6 to lanes 1–3), confirming that autophosphoryl ation of YwqD is not necessary for its protein kinase activity. Interestingly, YwqF already gave a strong signal in western blots with anti P-Tyr antibodies before in vitro phosphorylation with YwqD/GST–YwqC, suggesting that YwqF had been phosphorylated by an E.coli PTK. In agreement with this assumption, the PTK domain of E.coli Wzc (Grangeasse et al., 2002) could effectively phosphorylate B.subtilis YwqF (Figure 3A, lane 10). Moreover, radiolabelled [33P]Tyr-YwqF was obtained when E.coli cells synthesizing YwqF were grown in [33P]phosphate-containing medium (Figure 3A, lane 9). When [33P]Tyr-YwqF was analysed by isoelectric focusing, a major non-radioactive protein band corresponding to YwqF and a minor slightly more acidic band (Figure 3C, lane 1), which migrated to the same position as the radioactive band (Figure 3C, lane 3), were observed. When incubated with alkaline phosphatase or the PTP YwqE (see below) before isoelectric focusing, the minor protein band as well as the radioactive band disappeared (Figure 3C, lanes 2 and 4). These results suggested that ∼5% of YwqF purified from E.coli cells is present as P-Tyr-YwqF.
Fig. 3. YwqD-catalysed phosphorylation of YwqF and TuaD. Protein concentrations in (A) and (B) were as follows: YwqD, YwqD-123 and GST–YwqC-NCter, 1.5 µM; YwqF and TuaD, 10 µM. (A) Autoradiograms of SDS–polyacrylamide gels on which reaction mixtures containing [γ-32P]ATP and the following proteins had been separated: lane 1, YwqF; lane 2, YwqF and GST–YwqC-NCter; lane 3, YwqD and YwqF; lane 4, YwqD, GST–YwqC-NCter and YwqF; lane 5, TuaD; lane 6, TuaD and GST–YwqC-NCter; lane 7, YwqD and TuaD; lane 8, YwqD, GST–YwqC-NCter and TuaD; and lane 10, YwqF and the cytoplasmic PTK domain of E.coli Wzc (Wzccyto). Lane 9, YwqF (3 µg) purified from E.coli cells grown in [33P]phosphate- containing medium (exposure time 60 h). (B) Comparison of the rates of YwqD- and YwqD-123-catalysed YwqF phosphorylation. Autoradiograms of SDS–polyacrylamide gels on which reaction mixtures containing [γ-32P]ATP, YwqF, GST–YwqC-NCter and either YwqD (lanes 1–3) or YwqD-123 (lanes 4–6) were separated. Incubations were carried out for 5, 10 and 15 min. The slower migrating bands correspond to YwqF or TuaD and the faster migrating bands to YwqD and/or GST–YwqC-NCter [to Wzccyto in (A), lane 10]. (C) YwqF purified from E.coli grown in [33P]phosphate-containing medium was submitted to isoelectric focusing without (lanes 1 and 3) or with prior YwqE-treatment (lanes 2 and 4). The strips were stained with Coomassie blue (lanes 1 and 2) and submitted to autoradiography (lanes 3 and 4).
YwqE, a tyrosine phosphatase switching off the UDP-glucose DH activity of phosphorylated YwqF and TuaD
YwqE encoded by the third gene of the ywqCDEF operon exhibits similarity to the recently described S.pneumoniae PTP Cps2B (Bender and Yother, 2001; Morona et al., 2002). To test whether YwqE also functions as PTP, samples of 32P-labelled YwqD, YwqF and TuaD were prepared, which had been desalted to remove [γ-32P]ATP to prevent re-phosphorylation. YwqE indeed functions as PTP, as it could dephosphorylate autophosphorylated YwqD (Figure 4A, lanes 1–4), P-Tyr-YwqF (Figure 4A, lanes 5–8) and P-Tyr-TuaD (data not shown). These three P-proteins were also dephosphorylated by B.subtilis YwlE, which exhibits similarity to Wzb, an E.coli low MW PTP (Vincent et al., 1999). However, YwlE was clearly less active than YwqE (data not shown) and the physiological significance of this ‘cross’ reactivity remains questionable. Escherichia coli only possesses YwlE- and no Cps2B-like PTPs, which might partly be responsible for the accumulation of P-Tyr-YwqD in E.coli.
Fig. 4. Phosphorylation of YwqF affects its UDP-glucose DH activity. (A) Autoradiography of SDS/polyacrylamide gels on which 3 µM [32P]YwqD (lanes 1–4) or 10 µM [32P]YwqF (lanes 5–8) incubated for 0, 5, 10 and 15 min with 1.5 µM YwqE had been separated. (B) UDP-glucose DH assays with different forms of YwqF. The UDP-glucose DH activity of YwqF purified from E.coli (open circles), which contains ∼5% P-Tyr-YwqF (Figure 3C), diminished when it was incubated with YwqE for 5 (filled triangles), 10 (open triangles) and 15 min (filled squares), and slightly increased when incubated with YwqD, GST–YwqC-NCter, 100 µM ATP and 5 mM MgCl2 (filled circles).
Similar to TuaD (Pagni et al., 1999), YwqF also seems to function as specific UDP-glucose DH, as YwqF purified from E.coli was capable of oxidizing UDP-glucose (Figure 4B, open circles), but not UDP-galactose (data not shown). Incubating YwqF with YwqD, YwqC-NCter and 100 µM ATP slightly increased its UDP-glucose DH activity. In contrast, when YwqF was incubated for increasing time periods with YwqE (or alkaline phosphatase) before carrying out the UDP-glucose DH assay, its activity decreased and finally completely disappeared (Figure 4B). When [32P]YwqF was treated for 15 min with YwqE under identical conditions, it was completely dephosphorylated (Figure 4A, lane 8), suggesting that only the 5% P-Tyr-YwqF present in YwqF purified from E.coli (Figure 3C) is active as UDP-glucose DH. In agreement with this assumption, the YwqF fraction not retained on an anti-P-Tyr antibody-carrying agarose column exhibited very low UDP-glucose DH activity (data not shown), as probably most of the active P-Tyr-YwqF was bound to the column. UDP-glucose DH assays carried out with TuaD purified from E.coli and phosphorylation and dephosphorylation experiments with YwqD/YwqC-NCter and YwqE, respectively, provided results identical to those obtained with YwqF, confirming that TuaD is also phosphorylated in E.coli (data not shown).
The UDP-glucose DH activity of B.subtilis is modified by YwqD and YwqE
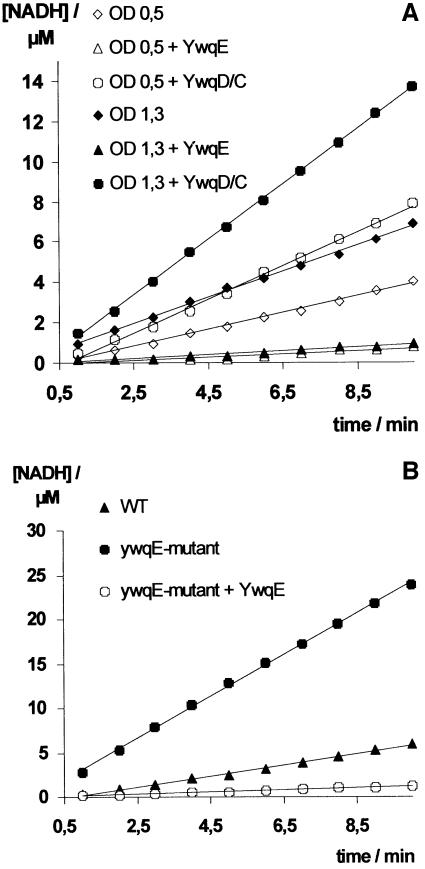
To test whether the UDP-glucose DHs TuaD and YwqF are also phosphorylated in B.subtilis, we measured the UDP-glucose DH activity in cell extracts prepared from B.subtilis wild-type and a ywqE mutant and also purified His-tagged YwqF from B.subtilis. Cell extracts were prepared as described by Pagni et al. (1999) after growth to exponential (OD600 = 0.5) or to the beginning of stationary phase (OD600 = 1.3). The UDP-glucose DH activity detected in extracts of B.subtilis wild-type cells harvested during exponential growth (Figure 5A, open diamonds) was ∼2-fold increased when the cell extracts were incubated with ATP, YwqD and YwqC (Figure 5A, open circles). In contrast, when the cell extract was treated for 30 min with YwqE, the UDP-glucose DH activity disappeared almost completely (Figure 5A, open triangles). Similar results were obtained with extracts prepared from B.subtilis cells harvested at the onset of stationary growth, except that the UDP-glucose DH activity was ∼2-fold higher than in cells grown to exponential phase (Figure 5A). By using a pEJ9-derived plasmid we could overproduce His-tagged YwqF in B.subtilis. YwqF purified from B.subtilis also exhibited UDP-glucose DH activity, which was ∼2-fold stimulated when incubated with ATP, GST–YwqC and YwqD, but almost completely disappeared when it was treated with YwqE (data not shown). We also inactivated the B.subtilis ywqE gene by disrupting it with pMUTIN2 (Vagner et al., 1998). Interestingly, when grown in LB medium without IPTG, the resulting strain YWQE1 exhibited a 4-fold higher UDP-glucose DH activity than the wild type strain. In the absence of IPTG, the ywqF gene, which in YWQE1 is under control of the spac promoter, is only very weakly expressed (Vagner et al., 1998), as pMUTIN2 carries the E.coli lacI gene. The observed increase in UDP-glucose DH activity is therefore likely to be due to enhanced phosphorylation of TuaD. When cell extracts of YWQE1 were incubated with YwqE, the UDP-glucose DH activity disappeared almost completely (Figure 5B).
Fig. 5. (A) UDP-glucose DH activity of B.subtilis extracts prepared from exponential (open symbols) or stationary phase cells (filled symbols). Diamonds, triangles and circles represent untreated, YwqE- dephosphorylated and YwqD-phosphorylated extracts, respectively. (B) UDP-glucose DH activity in B.subtilis YWQE1 (filled circles) compared with that of the wild type strain 168 (filled triangles). Preincubation with YwqE strongly diminished the UDP-glucose DH activity activity of YWQE1 (open circles).
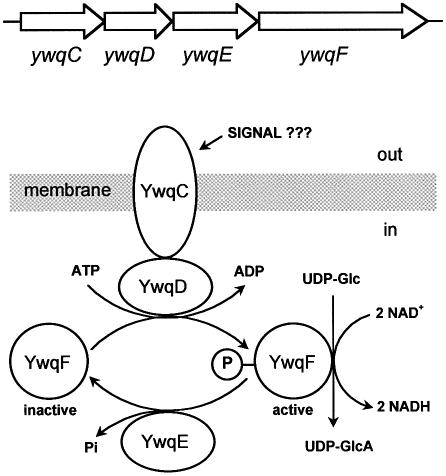
Proposed model of YwqD function
Our results suggest that the PTK YwqD can bind to the membrane-spanning YwqC. This interaction probably allows YwqD to recognize its substrates YwqF and TuaD (Figure 6). Alternatively, YwqC might also bind YwqF and TuaD and thus bring them into contact with YwqD. Although other modes of YwqC function can be envisaged, it is tempting to assume that it receives an external signal regulating its capacity to interact with YwqD and/or YwqF and TuaD. In Gram-negative bacteria, the homologues of YwqC and YwqD are fused to a single protein, and the proposed signal transfer mechanism therefore resembles that of eukaryotic receptor-binding PTKs. The proposed external signal is expected to be related to EPS synthesis, as mutations affecting YwqD-like PTKs were found to alter EPS synthesis in bacteria (Morona et al., 2000, 2003; Wugeditsch et al., 2001). In B.subtilis, the YwqC/YwqD-mediated signal transduction mechanism probably allows this organism to regulate the synthesis of EPSs with a high content of glucuronic acid, as P-YwqF and P-TuaD function as UDP-glucose DHs, while dephosphorylation by YwqE leads to a loss of this activity. The implication of TuaD in B.subtilis teichuronic acid synthesis has been established (Pagni et al., 1999). The UDP-glucose DH Ugd of E.coli has also been shown to be phosphorylated by its PTK Wzc (Grangeasse et al., 2003). The role of autophosphorylation at the C-terminal tyrosine cluster of YwqD is not clear, as it had no significant effect on the PTK activity.
Fig. 6. Proposed role of the proteins encoded by the B.subtilis ywqCDEF operon. Interaction of the transmembrane modulator YwqC with YwqD allows the PTK to phosphorylate its substrate YwqF (or TuaD) and to switch ‘on’ its UDP-glucose DH activity. The PTP YwqE switches ‘off’ the UDP-glucose DH activity by dephosphorylating P-YwqF (or P-TuaD). The predicted localization of YwqC in the membrane suggests an external signal probably affecting the kinase/modulator interaction.
Materials and methods
PCR amplification and cloning experiments
The genes minD, soj, ylxH, yveK, yveL, ywlE, ywqC, ywqD, ywqE, ywqF and tuaD were PCR-amplified by using genomic DNA of B.subtilis 168 as template and specific primers, to which appropriate restriction sites had been added (Table I). The PCR products of yveK, ywlE and ywqC were inserted into pGEX-2T (Pharmacia), all others into pQE-30 (Qiagen). The ywqC gene was also inserted into pMAL-c2x (NEB). For overexpression in B.subtilis, ywqF was inserted into the shuttle vector pEJ9 cut with BamHI and SphI. The absence of PCR-introduced mutations was verified by DNA sequencing. The plasmids with the various insertions were used to transform E.coli NM522 (B.subtilis 168 for pEJ9) and transformants were selected on LB medium containing appropriate antibiotics.
Table I. PCR primers used in this study for amplification and mutagenesis with the introduced restriction site or mutation indicated on the righta.
| Amplification primers | ||
| YwqD+ | GTGGGATCCATGGCGCTTAGAAAAAACAG | BamHI |
| YwqD– | GGCACGCTGCAGGTTATTTATTTTTGC | PstI |
| YveL+ | GTGGGATCCATGATCTTTAGAAAAAAGAAA | BamHI |
| YveL– | GACATAAGCTTTACAAAAACTAGTAGG | HindIII |
| YlxH+ | GAGGGATCCATGCAGATGAACAGATATGAC | BamHI |
| YlxH– | CGAGTCGACTTAAGCCCTCCTCATT | SalI |
| Soj+ | GTAGGTGGATCCATGGGAAAAATCATAGCA | BamHI |
| Soj– | TTCCAAAGCTTTTAGCCATTCGCAGCCAC | HindIII |
| MinD+ | GGAAGGATCCATGGGTGAGGCTATCGTAAT | BamHI |
| MinD– | GATTCTCTGCAGTTAAGATCTTACTCCG | PstI |
| YwqC+ | GGAGGATCCATGGGAGAATCTACAAGCTTAAAAG | BamHI |
| YwqC– | CTGGAATTCCTAAGCGCCAAAATGTCCAC | EcoRI |
| YveK+ | GGCTGGGATCCATGAATACATGAATGAGAA | BamHI |
| YveK– | CGAATTCTCACTCGCTTCACTCCCCG | EcoRI |
| YwlE+ | GTGGGATCCATGGATATTATTTTTGTCTGTACTGGAAAT | BamHI |
| YwlE– | TCTGAATTCTTATCTACGGTCTTTTTTCAGCTGTTTTGCC | EcoRI |
| YwqE+ | CGCGGATCCATGATCGATATTCACTGTCACATTCTTCCC | BamHI |
| YwqE– | AAACTGCAGTCAAAAAAAACCAAACAATTTTCTTCTTTTGACCGG | PstI |
| YwqF+ | CGCGGATCCATGAATATAACAGTCATCGGAACAGGCTATGTCGGT | BamHI |
| YwqF– | ACGCGTCGACCATTGAATTGCACCTGACGGAACAACCGCCTTCC | SalI |
| YwqF Sph– | ACATGCATGCTCACTATTGAATTGCACCTGA | SphI |
| TuaD+ | CCGGATCCATGAAAAAAATAGCTGTCATTGGAACAGG | BamHI |
| TuaD– | CCGTCGACTTATAAATTGACGCTTCCCAAGTCTTTAGCC | SalI |
| Xyl+ | GCGAATTCGATCAGCGATATCCACTTC | EcoRI |
| Xyl– | GCCGAAGCTTCAGATGCATTTTATTTCATATAG | HindIII |
| Mutagenic primers for ywqD | ||
| CT | GGCTGCAGTTAGTAATATCCGTATTCAGAGTGC | aa 229–237 deleted |
| Y1 | GGCTGCAGGTTATTTATTTTTGCATGAAATTGTCCTTGGTTCCGTAATATCCGAATTCAGAGTGC | Y225→F |
| Y23 | GGCTGCAGGTTATTTATTTTTGCATGAAATTGTCCTTGGTTCCGAAAAATCCGTATTCAGAGTGC | Y227, 228→F |
| Y123 | GGCTGCAGGTTATTTATTTTTGCATGAAATTGTCCTTGGTTCCGAAAAATCCGAATTCAGAGTGC | Y225, 227, 228→F |
| D81F | GCTCCTGATTGCTGCTGATTTGCGAAAACCAAC | D81→A |
| D81R | GCAAATCAGCAGCAATCAGGAGCACTTTTTTTCC | D81→A |
| D83F | GATTGATGCTGCTTTGCGAAAACCAACAGTACA | D83→A |
| D83R | GTTTTCGCAAAGCAGCATCAATCAGGAGCACTT | D83→A |
| NT | GGGGATCCATGCAAATGAAATCAGTCATGATTACATCGGCT | aa 1–42 deleted |
| Mutagenic primers for ywqC | ||
| Y44F | AGTTGAATTTTCAGCGATGGGTGTAAGTGCGAAGAA | Y44→A |
| Y44R | CTTACACCCATCGCTGAAAATTCAACTCAGATTCTG | Y44→A |
| Y77F | GATGACATTGGCCGTATTGATTAACTGAAGATT | Y77→A |
| Y44R | ATCAATACGGCCAATGCTCATCATTAAGAGCCCT | Y77→A |
| NCterF | GGAGGATCCATGGGAGAATCTACAAGCTTAAAAGAGATATTATCAACGTTAACATGGATAATACGATCAAATCGG | aa 16–198 deleted |
| NCterR | CTGGAATTCCTAAGCGCCAAAATGTCCAC | aa 16–198 deleted |
| NCter-Δ15F | GGAGGATCCATGATAATACGATCAAATCGG | aa 1–15 deleted |
| NCter-Δ5R | CCGAATTCCTAACTCCCCGTTTTTTCAGATTGGAAACCC | aa 243–248 deleted |
| NCter-Δ10R | CCGAATTCCTACAGATTGGAAACCCTGTAAGTCTTTGC | aa 238–248 deleted |
| NCter-Δ15R | CCGAATTCCTAGTAAGGTCTTTGCTGGTTTTCTGCTCATTC | aa 233–248 deleted |
| Primers for insertional mutagenesis of ywqE | ||
| Emutin+ | CCCAAGCTTAGTTAAACAAACGTTTAATCAAAG | EcoRI |
| Emutin– | CGGGATCCTCACATCTGAGGCTACGAAATGG | BamHI |
aNewly introduced restriction sites and mutated codons are in bold type.
Oligonucleotide-directed mutagenesis
To construct the various ywqC and ywqD alleles and fusions, PCR-based mutagenesis was performed with appropriate primers (Table I) and plasmids containing the desired template gene. Mutations introduced into ywqD led to the deletion of either the 42 N-terminal amino acids (ywqD-ΔNT) or the nine C-terminal amino acids (ywqD-ΔCT), or to the replacement of Tyr-225 (ywqD-1), Tyr-227 and -228 (ywqD-23) or Tyr-225, -227 and -228 (ywqD-123) with phenylalnine or Asp-81 (ywqD-D81) or Asp-83 (ywqD-D83) with alanine. Mutations introduced into ywqC led to the replacement of Tyr-44 (ywqC-1), Tyr-77 (ywqC-2) or Tyr-44 and -77 (ywqC-12) with alanine. To construct a fused ywqC allele encoding only the 15 N-and 50 C-terminal amino acids (Figure 2B), ywqC-containing pGEX-2T was cut with AciI prior to PCR amplification, thus minimizing the probability of amplifying the full-length gene. The resulting PCR product ywqC-NCter was cloned into pGEX-2T and pMAL-c2x. The ywqC-NCter-containing pGEX-2T served as PCR template to delete the sequence encoding either the N-terminus (ywqC-Cter), the complete C-terminus (ywqC-Nter) or the last 5, 10 or 15 amino acids of the C-terminus (ywqC-NCter-Δ5, ywqC-NCter-Δ10 and ywqC-NCter-Δ15, respectively) (Figure 2B).
Construction of pEJ9, a vector for the synthesis of His-tagged proteins in B.subtilis
To construct plasmid pEJ9, a 0.24 kb fragment containing the B.subtilis xylA promoter and EcoRI and HindIII restriction sites was PCR- amplified with chromosomal DNA. After restriction with EcoRI and HindIII it was ligated to a DNA fragment, which was obtained by hybridizing in TE buffer the two reverse-complementary oligo nucleotides 5′-AGCTTAAAGGAAGGTTTCATATGCACCATCACCATCACCATGGATCCAGATCTGCA-3′ and 3′-ATTTCCTTCCAAAGTATACGTGGTAGTGGTAGTGGTACCTAGGTCTAGACGTACGC-5′ and which contained a sticky end for ligation to HindIII sites (bold letters), the ribosome binding site and the start codon of B.subtilis pckA (in italics) followed by six codons for histidine and the restriction sites BamHI, BglII and SphI (underlined). The resulting linear 0.3 kb fragment was ligated to the shuttle vector pDG1730 (Guerout-Fleury et al., 1996) cut with EcoRI and BamHI. When this construct was used to transform an E.coli strain, the gap at the SphI–BamHI ligation site was filled in, as was confirmed by DNA sequencing (5′-CTGCATGCGATCCGA-3′), thus retaining the SphI site of the 0.3 kb fragment, but not the BamHI site of the vector. Expression of genes inserted between the BamHI and BglII or SphI sites (on the 0.3 kb fragment) in the resulting plasmid pEJ9 can be induced by xylose, leading to the synthesis of proteins carrying a His6-tag at the N-terminus.
Protein synthesis and purification
YwqE (30 kDa), TuaD (51 kDa) and YwqF (49 kDa) carrying an N-terminal (His6)-extension were synthesized in E.coli strain M15[pREP4] by following the standard protocol of Qiagen. The ∼27 kDa proteins MinD, Soj, YlxH, YveL and YwqD and its mutants also contained an N-terminal His-tag, but they were less soluble and strain M15[pREP4-groESL] (Amrein et al., 1995) was therefore used for their synthesis. YveK, YwlE, YwqC, YwqC-NCter (28 kDa), YwqC-Cter, YwqC-Nter, YwqC-NCter-Δ5, YwqC-NCter-Δ10 and YwqC-NCter-Δ15 were all synthesized as N-terminal GST-fusions in E.coli strain BL21(DE23) by following the protocol of Pharmacia. YwqC and YwqC-NCter were also synthesized as MBP fusions. The two ywqC alleles were inserted into pMAL-c2x and the resulting plasmids were used to transform E.coli strain BL21(DE23). Bacillus subtilis strain 168 was transformed with ywqF-containing pEJ9, as described by Agnostopulos and Spizizen (1961), and transformants were grown in LB medium containing 0.5% xylose and 5 µg/ml kanamycin. Cells were harvested in the late exponential phase (OD600 = 0.8), treated for 30 min with 1 mg/ml lysozyme and broken by sonication.
His-tagged proteins synthesized in E.coli or B.subtilis were purified on Ni-NTA columns by following the protocol of Qiagen. We only replaced the phosphate buffer with 50 mM Tris–HCl pH 7.4. Soluble GST-tagged proteins were purified on glutathione sepharose™4B columns by following the standard protocol of Pharmacia. Proteins were desalted on PD-10 columns and aliquots were stored at –80°C in a buffer containing 50 mM Tris–HCl pH 7.4, 100 mM NaCl, 25% glycerol, 1 mM β-mercaptoethanol and 0.5 mM PMSF. When indicated, the GST-tag was removed by incubating 1 mg of the fusion protein with 10 U of thrombin for 6 h at 22°C. MBP-tagged YwqC-NCter was purified on an amylose column as recommended by NEB. Cells containing insoluble MBP- or GST-tagged membrane-integrated YwqC were incubated with 1 mg/ml lysozyme for 15 min at 37°C and subsequently sonicated. Unbroken cells were removed by a 5 min centrifugation at 5000 g, and membrane fractions containing the tagged proteins were spun down by centrifugation at 100 000 g for 1 h, washed twice with 50 mM Tris–HCl pH 7.4, 10 mM DTT, 0.5 mM PMSF and stored at –80°C.
Protein phosphorylation and dephosphorylation experiments
Protein phosphorylation reactions were performed with 1.5 µM PTK, 10 µM target protein (YwqF or TuaD) and 1.5 µM YwqC-NCter unless stated otherwise. In addition to the appropriate proteins, a typical 40 µl reaction mixture contained 50 µM [γ-32P]ATP (20 µCi/mmol), 5 mM MgCl2 and 50 mM Tris–HCl pH 7.4. After a 15 min incubation at 37°C, the reactions were stopped by adding sample buffer for SDS–PAGE and heating at 100°C for 5 min. Proteins were separated by electrophoresis on denaturing 0.1% SDS/15% polyacrylamide gels, which were subsequently treated with boiling 0.5 M HCl for 10 min. After drying the gels, radioactive bands were visualized with a STORM Phosphoimager (Molecular Dynamics). Exposure time was 16 h, except for 33P-labelled YwqF purified from E.coli, which was exposed for 60 h.
For dephosphorylation experiments, 32P-labelled P-Tyr proteins were desalted on Ni-NTA and PD-10 columns to remove [γ-32P]ATP before 1.5 µM YwqE was added. The reaction mixtures were incubated for 5, 10 or 15 min at 37°C before dephosphorylation was stopped by adding sample buffer for SDS–PAGE and heating at 100°C for 5 min. PAGE and autoradiography were carried out as described above.
Acid hydrolysis and thin layer chromatography
32P-labelled proteins were separated by SDS–PAGE and subsequently electroblotted (300 mA for 1 h) onto PVDF membranes by using a 10% methanol solution containing 25 mM Tris and 192 mM glycine pH 8.3 as transfer buffer. Transferred proteins were detected by staining with Ponceau-S (Aebersold et al., 1987). Stained radioactive protein bands were excised and treated with 6 M HCl at 110°C for 1.75 h. The hydrolysate was lyophilized and resuspended in 20 µl of water containing 1 mg/ml of P-Ser, P-Thr and P-Tyr standards. The samples were separated by two-dimensional thin layer chromatography on cellulose sheets (MERCK). Isobutyric acid and 0.5 M NH4OH (at a ratio of 5:3, v/v) was used as solvent for the first dimension (12 h) and 2-propanol, HCl and water (at a ratio of 7:1.5:1.5, v/v/v) for the second dimension (8 h). The dried plates were stained with ninhydrin to reveal the positions of the P-amino acid standards, whereas the radioactive hydrolysis products were visualized with a STORM Phosphoimager.
Identification of P-peptides by MALDI-TOF mass spectrometry and Edman sequencing
Peptides were obtained by incubating 400 pmol of YwqD with trypsin (Roche) at an enzyme-to-substrate ratio of 1:20 (w/w) in 40 µl of 50 mM ammonium bicarbonate pH 8.0 for 16 h at 37°C. Tryptic peptides were loaded on a 250 × 2.1 mm Vydac C18 reversed phase column (300 Å pore size) and eluted with a 90 min linear gradient of 0–70% acetonitrile in 0.1% trifluoroacetic acid. The flow rate was 0.3 ml/min. Fractions were collected manually based on the absorbance profile at 215 nm. An aliquot of each HPLC fraction was concentrated 5- to 10-fold under vacuum before 1 µl of the concentrate was mixed with 1 µl of a saturated solution of α-cyano-4-hydroxycinnamic acid in 0.3% trifluoroacetic acid/50% acetonitrile. The premix was deposited on a standard stainless steel probe and allowed to air dry. MALDI analysis was carried out on a Voyager-DE STR MALDI mass spectrometer equipped with a 337 nm laser source (Perseptive Biosystems). Spectra were recorded in both positive-ion reflectron and positive-ion linear mode, with a 20 kV acceleration voltage in the ion source. External calibration was obtained with a mixture of six reference peptides covering the m/z 900–3700 Da range. To assure a reliable identification of P-peptides, aliquots of HPLC fractions assumed to contain a P-peptide were treated with alkaline phophatase (1 U/µl, Roche) on the MALDI target as described by Larsen et al. (2001) before they were analysed. P-peptides were present when the signal of the alkaline phosphatase-treated sample was diminished by (n × 80) Da compared to the untreated peptide, with n being the number of phosphate groups in the P-peptide. HPLC fractions containing P-peptides were sequenced using an Applied Biosystems model 473 sequencer. P-Tyr residues were identified by the lack of a signal.
ATPase assays
The ATPase activity of YwqD was tested in reaction mixtures containing 0.75 µM protein, 50 µM [γ-32P]ATP (20 µCi/mmol), 5 mM MgCl2 and 50 mM Tris–HCl pH 7.4. After 10 min incubation at 37°C, the products were separated by thin layer chromatography on polyethyleneimine cellulose sheets (Macherey-Nagel) using 0.3 M potassium phosphate buffer pH 7.4 as solvent. Radioactive signals on the dried sheets were visualized with a STORM Phosphoimager.
Western blotting
Proteins were electroblotted (300 mA for 1 h) from SDS–PAGE gels onto PVDF membranes by using a 10% methanol solution containing 25 mM Tris and 192 mM glycine pH 8.3 as transfer buffer. The PVDF membranes were subsequently treated for 16 h with blocking buffer (25 mM Tris and 192 mM glycine pH 8.3, 0.05% Tween-20, 3% BSA) and incubated for 1 h with anti-P-Tyr antibody-peroxidase conjugates (Sigma) in blocking buffer (1:40 000 dilution). After three 10 min washes with 0.05% Tween-20 in 25 mM Tris and 192 mM glycine pH 8.3, P-Tyr-containing proteins were revealed with the AEC-staining kit (Sigma). Dephosphorylation of P-proteins immobilized on a PVDF membrane was performed by including 1 U/ml alkaline phosphatase in the blocking buffer.
In vivo labeling of YwqF with [33P]phosphate
Escherichia coli cells harbouring the plasmid to overproduce YwqF were grown in 20 ml ampicillin-containing LB medium. When the OD600 approached 0.3, 25 µCi [33P]phosphate was added per ml of culture. Thirty minutes later, ywqF expression was induced with 0.5 mM IPTG. After 5 h induction, the cells were centrifuged, chilled on ice, broken by vortexing with glass beads and cell debris was removed by centrifugation. 6×His-YwqF present in the supernatant was purified on a Ni-NTA column and aliquots were separated by electrophoresis on a 0.1% SDS/15% polyacrylamide gel that was subsequently treated with boiling 0.5 M HCl. Radioactive protein bands were detected as described above.
Isoelectric focusing
A sample of 15 µg of [33P]Tyr-YwqF purified from E.coli was either directly loaded on Immobiline DryStrips pH 4–7 (Amersham Biosciences) or first treated for 30 min with 1.5 µg of YwqE. Isolectric focusing was carried out on an IPGphor apparatus from Pharmacia Biotech using the program recommended by the supplier. To visualize protein bands, the strips were stained with Coomassie Blue and dried. Radioactive signals were detected by autoradiography. YwqF (theoretical pI of 5.0) was clearly separated from YwqE (theoretical pI of 6.6).
Inactivation of B.subtilis ywqE
A 400-bp DNA fragment corresponding to the central part of ywqE was integrated in pMUTIN2 (Vagner et al., 1998) cut with EcoRI and BamHI. Bacillus subtilis strain 168 was transformed with the resulting plasmid (Agnostopulos and Spizizen, 1961) and a transformant (YWQE1) was isolated on LB plates containing 0.3 µg/ml erythromycin. Disruption of ywqE in this strain was verified by PCR amplification using genomic DNA and pMUTIN2-specific internal primers.
UDP-glucose DH assay
UDP-glucose DH activity of purified enzymes or in cell extracts was measured by using a spectrophotometric assay described by Pagni et al. (1999). We only replaced the glycylglycine/NaOH buffer pH 9.2 with Tris–HCl buffer pH 8.8. The tests were performed with 2.5 mM UDP-glucose and 5 mM NAD+ and either 10 µM YwqF or 10 µM TuaD purified from E.coli, 10 µM YwqF purified from B.subtilis or dialysed extracts prepared from various B.subtilis strains (Pagni et al., 1999). Assay mixtures with cell extracts contained ∼0.5 mg protein. To measure the effect of dephosphorylation or phosphorylation on their UDP-glucose DH activity, YwqF or TuaD were treated for 30 min (if not stated otherwise) with either YwqE or YwqD/GST–YwqC-NCter in the presence of 100 µM ATP and 5 mM MgCl2.
Acknowledgments
Acknowledgements
We wish to thank Dina Petranovic for critically reading the manuscript. This research was supported by the CNRS, the INRA and the INA-PG. I.M. received fellowships from the French Government and the INRA.
References
- Agnosotpulos C., and Spizizen,J. (1961) Requirements for trans formation in Bacillus subtilis. J. Bacteriol., 81, 741–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aebersold R.H., Leavitt,J., Saavedra,R.A., Hood,L.E. and Ken,S.B. (1987) Internal amino acid sequence analysis of proteins separated by one- or two-dimensional gel electrophoresis after in situ protease digestion on nitrocellulose. Proc. Natl Acad. Sci. USA, 84, 6970–6974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amrein K.E., Takacs,B., Stieger,M., Molnos,J., Flint,N.A. and Burn,P. (1995) Purification and characterization of recombinant human p50csk protein-tyrosine kinase from an Escherichia coli expression system overproducing the bacterial chaperones GroES and GroEL. Proc. Natl Acad. Sci. USA, 92, 1048–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arakawa Y., Wacharotayankun,R., Nagatsuka,T., Ito,H., Kato,N. and Ohta, M. (1995) Genomic organization of the Klebsiella pneumoniae cps region responsible for serotype K2 capsular polysaccharide synthesis in the virulent strain Chedid. J. Bacteriol., 177, 1788–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backert S., Moese,S., Selbach,M., Brinkmann,V. and Meyer,T.F. (2001) Phosphorylation of tyrosine 972 of the Helicobacter pylori CagA protein is essential for induction of a scattering phenotype in gastric epithelial cells. Mol. Microbiol., 42, 631–644. [DOI] [PubMed] [Google Scholar]
- Barford D., Das,A.K. and Egloff,M.P. (1998) The structure and mechanism of protein phosphatases: insights into catalysis and regulation. Annu. Rev. Biophys. Biomol. Struct., 27, 133–164. [DOI] [PubMed] [Google Scholar]
- Bender M.H. and Yother,J. (2001) CpsB is a modulator of capsule-associated tyrosine kinase activity in Streptococcus pneumoniae. J. Biol. Chem., 276, 47966–47974. [DOI] [PubMed] [Google Scholar]
- Bugert P. and Geider,K. (1997) Characterization of the amsI gene product as a low molecular weight acid phosphatase controlling exopolysaccharide synthesis of Erwinia amylovora. FEBS Lett., 400, 252–256. [DOI] [PubMed] [Google Scholar]
- de Boer P.A., Crossley,R.E. and Rothfield,L.I. (1988) Isolation and properties of minB, a complex genetic locus involved in correct placement of the division site in Escherichia coli. J. Bacteriol., 170, 2106–2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doublet P., Vincent,C., Grangeasse,C., Cozzone,A.J. and Duclos,B. (1999) On the binding of ATP to the autophosphorylating protein, Ptk, of the bacterium Acinetobacter johnsonii. FEBS Lett., 445, 137–143. [DOI] [PubMed] [Google Scholar]
- Doublet P., Grangeasse,C., Obadia,B., Vaganay,E. and Cozzone,A.J. (2002) Structural organization of the protein-tyrosine autokinase Wzc within Escherichia coli cells. J. Biol. Chem., 277, 37339–37348. [DOI] [PubMed] [Google Scholar]
- Fantl W.J., Johnson,D.E. and Williams,L.T. (1993) Signalling by receptor tyrosine kinases. Annu. Rev. Biochem., 62, 453–481. [DOI] [PubMed] [Google Scholar]
- Grangeasse C., Doublet,P., Vaganay,E., Vincent,C., Deleage,G., Duclos,B. and Cozzone,A.J. (1997) Characterization of a bacterial gene encoding an autophosphorylating protein tyrosine kinase. Gene, 204, 259–265. [DOI] [PubMed] [Google Scholar]
- Grangeasse C., Doublet,P. and Cozzone,A.J. (2002) Tyrosine phosphorylation of protein kinase Wzc from Escherichia coli K12 occurs through a two-step process. J. Biol. Chem., 277, 7127–7135. [DOI] [PubMed] [Google Scholar]
- Grangeasse C., Obadia,B., Mijakovic,I., Deutscher,J., Cozzone,A.J. and Doublet,P. (2003) Autophosphorylation of the Escherichia coli protein kinase Wzc regulates tyrosine phosphorylation of Ugd, a UDP-glucosedehydrogenase. J. Biol. Chem., 278, Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- Guerout-Fleury A.M., Frandsen,N. and Stragier,P. (1996) Plasmids for ectopic integration in Bacillus subtilis. Gene, 180, 57–61. [DOI] [PubMed] [Google Scholar]
- Hayashi I., Oyama,T. and Morikawa,K. (2001) Structural and functional studies of MinD ATPase: implications for the molecular recognition of the bacterial cell division apparatus. EMBO J., 20, 1819–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter T. (1995) Protein kinases and phosphatases: the yin and yang of protein phosphorylation and signaling. Cell, 80, 225–236. [DOI] [PubMed] [Google Scholar]
- Iannelli F., Pearce,B.J. and Pozzi,G. (1999) The type 2 capsule locus of Streptococcus pneumoniae. J. Bacteriol., 181, 2652–2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilan O., Bloch,Y., Frankel,G., Ullrich,H., Geider,K. and Rosenshine,I. (1999) Protein tyrosine kinases in bacterial pathogens are associated with virulence and production of exopolysaccharide. EMBO J., 18, 3241–3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein G., Dartigalongue,C. and Raina,S. (2003) Phosphorylation-mediated regulation of heat shock response in Escherichia coli. Mol. Microbiol., 48, 269–285. [DOI] [PubMed] [Google Scholar]
- Larsen M.R., Sørensen,G.L., Fey,S.J., Larsen,P.M. and Roepstorff,P. (2001) Phospho-proteomics: evaluation of the use of enzymatic de-phosphorylation and differential mass spectrometric peptide mass mapping for site specific phosphorylation assignment in proteins separated by gel electrophoresis. Proteomics, 1, 223–238. [DOI] [PubMed] [Google Scholar]
- Levin P.A., Shim,J.J. and Grossman,A.D. (1998) Effect of minCD on FtsZ ring position and polar septation in Bacillus subtilis. J. Bacteriol., 180, 6048–6051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morona J.K., Morona,R. and Paton,J.C. (1999) Analysis of the 5′ portion of the type 19A capsule locus identifies two classes of cpsC, cpsD, and cpsE genes in Streptococcus pneumoniae. J. Bacteriol., 181, 3599–3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morona J.K., Paton,J.C., Miller,D.C. and Morona,R. (2000) Tyrosine phosphorylation of CpsD negatively regulates capsular polysaccharide biosynthesis in Streptococcus pneumoniae. Mol. Microbiol., 35, 1431–1442. [DOI] [PubMed] [Google Scholar]
- Morona J.K., Morona,R., Miller,D.C. and Paton,J.C. (2002) Streptococcus pneumoniae capsule biosynthesis protein CpsB is a novel manganese-dependent phosphotyrosine-protein phosphatase. J. Bacteriol., 184, 577–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morona J.K., Morona,R., Miller,D.C. and Paton,J.C. (2003) Mutational analysis of the carboxy-terminal (YGX)(4) repeat domain of CpsD, an autophosphorylating tyrosine kinase required for capsule biosynthesis in Streptococcus pneumoniae. J. Bacteriol., 185, 3009–3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemeyer D. and Becker,A. (2001) The molecular weight distribution of succinoglycan produced by Sinorhizobium meliloti is influenced by specific tyrosine phosphorylation and ATPase activity of the cytoplasmic domain of the ExoP protein. J. Bacteriol., 183, 5163–5170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagni M., Lazarevic,V., Soldo,B. and Karamata,D. (1999) Assay for UDPglucose 6-dehydrogenase in phosphate-starved cells: gene tuaD of Bacillus subtilis 168 encodes the UDPglucose 6-dehydrogenase involved in teichuronic acid synthesis. Microbiology, 145, 1049–1053. [DOI] [PubMed] [Google Scholar]
- Paiment A., Hocking,J. and Whitfield,C. (2002) Impact of Phosphorylation of Specific Residues in the Tyrosine Autokinase, Wzc, on Its Activity in Assembly of Group 1 Capsules in Escherichia coli. J. Bacteriol., 184, 6437–6447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preneta R., Jarraud,S., Vincent,C., Doublet,P., Duclos,B., Etienne,J. and Cozzone,A.J. (2002) Isolation and characterization of a protein-tyrosine kinase and a phosphotyrosine-protein phosphatase from Klebsiella pneumoniae. Comp. Biochem. Physiol., 131B, 103–112. [DOI] [PubMed] [Google Scholar]
- Quisel J.D. and Grossman,A.D. (2000) Control of sporulation gene expression in Bacillus subtilis by the chromosome partitioning proteins Soj (ParA) and Spo0J (ParB). J. Bacteriol., 182, 3446–3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sau S., Bhasin,N., Wann,E.R., Lee,J.C., Foster,T.J. and Lee,C.Y. (1997) The Staphylococcus aureus allelic genetic loci for serotype 5 and 8 capsule expression contain the type-specific genes flanked by common genes. Microbiology, 143, 2395–2405. [DOI] [PubMed] [Google Scholar]
- Schlessinger J. (2000) Cell signaling by receptor tyrosine kinases. Cell, 103, 211–225. [DOI] [PubMed] [Google Scholar]
- Sharpe M.E. and Errington,J. (1996) The Bacillus subtilis soj-spo0J locus is required for a centromere-like function involved in prespore chromosome partitioning. Mol. Microbiol., 21, 501–509. [DOI] [PubMed] [Google Scholar]
- Vagner V., Dervyn,E., and Ehrlich,S.D. (1998) A vector for systematic gene inactivation in Bacillus subtilis. Microbiology, 144, 3097–3104. [DOI] [PubMed] [Google Scholar]
- Vincent C., Doublet,P., Grangeasse,C., Vaganay,E., Cozzone,A.J. and Duclos,B. (1999) Cells of Escherichia coli contain a protein-tyrosine kinase, Wzc, and a phosphotyrosine-protein phosphatase, Wzb. J. Bacteriol., 181, 3472–3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent C., Duclos,B., Grangeasse,C., Vaganay,E., Riberty,M., Cozzone,A.J. and Doublet,P. (2000) Relationship between exopolysaccharide production and protein-tyrosine phosphorylation in gram-negative bacteria. J. Mol. Biol., 304, 311–321. [DOI] [PubMed] [Google Scholar]
- Walker J.E., Saraste,M., Runswick,M.J. and Gay,N.J. (1982) Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J., 1, 945–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wugeditsch T., Paiment,A., Hocking,J., Drummelsmith,J., Forrester,C. and Whitfield,C. (2001) Phosphorylation of Wzc, a tyrosine autokinase, is essential for assembly of group 1 capsular polysaccharides in Escherichia coli. J. Biol. Chem., 276, 2361–2371. [DOI] [PubMed] [Google Scholar]
- Yamamoto S., Miyake,K., Koike,Y., Watanabe,M., Machida,Y., Ohta,M. and Ijima,S. (1999) Molecular characterization of type-specific capsular polysaccharide biosynthesis genes of Streptococcus agalactiae type Ia. J. Bacteriol., 181, 5176–5184. [DOI] [PMC free article] [PubMed] [Google Scholar]