Abstract
Programmed –1 ribosomal frameshifting, involving tRNA re-pairing from an AAG codon to an AAA codon, has been reported to occur at the sequences CGA AAG and CAA AAG. In this study, using the recoding region of insertion sequence IS3, we have investigated the influence on frameshifting in Escherichia coli of the first codon of this type of motif by changing it to all other NNA codons. Two classes of NNA codons were distinguished, depending on whether they favor or limit frameshifting. Their degree of shiftiness is correlated with wobble propensity, and base 34 modification, of their decoding tRNAs. A more flexible anticodon loop very likely makes the tRNAs with extended wobble more prone to liberate the third codon base, A, for re-pairing of tRNALys in the –1 frame.
Keywords: frameshifting/hexanucleotide/insertion sequences/tRNA modification/wobble
Introduction
The expression of a minority of genes in probably all organisms involves a proportion of ribosomes shifting reading frame at specific sites. In some cases the function of this programmed event is regulatory and in others the synthesis of two products, with different C-termini, is the important consequence (Farabaugh, 1997; Atkins et al., 2001). Examples of the former often implicate +1 frameshifting, whereas many of the latter involve –1 frameshifting. Beside its role in gene expression, frameshifting elicits interest because of what it reveals about the functioning of the translational molecular machine, especially in view of the recent advances in our understanding of the ribosome at the atomic level (Ogle et al., 2001, 2002, 2003; Yusupova et al., 2001; Noller et al., 2002; Valle et al., 2002; Gao et al., 2003). In particular, frameshifting brings into light the intricacies of the relation between a tRNA, its codon and the ribosome, as illustrated by the present work, and also raises the question of the maintenance of the translational reading frame (Farabaugh and Björk, 1999; Atkins et al., 2000). All known cases of –1 ribosomal frameshifting involve dissociation of codon: anticodon pairing followed by anticodon re-pairing to mRNA at an overlapping –1 frame codon. Early work with frameshift mutant leakiness and synthetic constructs focused on low frequency dissociation and re-pairing events involving a single tRNA anticodon (Weiss et al., 1987; Gallant and Lindsley, 1992). The high frequency programmed frameshifting events involved in decoding potato virus M (Gramstat et al., 1994), bacterial insertion sequences IS3 (Sekine et al. 1994) and IS1222 (N.Mejlhede, P.Licznar, M.F.Prère, N.Wills, R.F.Gesteland, J.Atkins and O.Fayet, in preparation) and that associated with decoding the Bacillus subtilis cytidine deaminase gene (cdd) (Mejlhede et al., 1999) have been considered in these terms. However, the great majority of known programmed –1 frameshifting involves re-pairing by tandem tRNAs at heptanucleotide sequences. Tandem slippage was discovered by Jacks and Varmus (1988) in their studies on the frameshifting required for retroviral gene expression and has since been found mostly in the decoding of viruses from diverse sources and in bacterial programmed frameshifting. Searches for additional cases of frameshifting were therefore concentrated on the characteristic heptanucleotide motifs for tandem re-pairing with little attention to single re-pairing possibilities.
The frameshifting that occurs in decoding B.subtilis cdd is 16% efficient. The intrinsic level of frameshifting at its A AAG shift site is 1.5%; as originally shown in Escherichia coli, tRNALys (anticodon 3′-UUmnm5s2U-5′, where mnm5s2U is 5-methylaminomethyl-2-thiouridine) is prone to shift –1 from AAG to AAA (Weiss et al., 1989; Tsuchihashi and Brown, 1992). A Shine–Dalgarno-like sequence within the coding sequence nine bases 5′ of the shift site acts to stimulate –1 frameshifting 10.6-fold (Mejlhede et al., 1999). Analogous stimulatory effects of nearby 5′ internal Shine–Dalgarno sequences are known for tandem –1 frameshifting (Larsen et al., 1994; Rettberg et al., 1999). The identity of the codon, CGA, upstream of the AAG is crucial for high efficiency frameshifting, but not the base 5′ of it, leading to the hypothesis of a hexameric shift site (Mejlhede et al., 1999). The anticodon of the CGA-decoding tRNAArg (3′-GCI-5′) contains inosine, I. Previous studies have shown very inefficient A:I pairing in vivo (Curran, 1995; Carter et al., 1997). It was therefore suggested that apposition of the purine inosine in the anticodon with the purine A of the cdd CGA codon does not permit strong pairing and would frequently result in the liberation of the third codon base, thereby allowing re-pairing of tRNALys from AAG to AAA.
Decoding of a bacterial transposable element, insertion sequence IS1222 (Steibl and Lewecke, 1995), also uses –1 frameshifting at a CGA AAG hexamer. Frameshifting is required for synthesis of the transposase, and so for transposition, of IS1222 (N.Mejlhede, P.Licznar, M.F.Prère, N.Wills, R.F.Gesteland, J.Atkins and O.Fayet, in preparation). This recoding event, occurring at a frequency of ∼7%, is stimulated by a weaker 5′ Shine–Dalgarno sequence than in cdd, but has a 3′ stimulatory stem–loop sequence. A stimulatory 3′ stem–loop is also not unique to this type of shift site, as it is known for several cases of bacterial dual-slippage frameshift regions including that for synthesis of a DNA polymerase component encoded by the E.coli dnaX gene (Larsen et al., 1997). In another insertion sequence, IS3, an A AAG frameshift site is associated with a pseudoknot as 3′ stimulator (there is no 5′ stimulatory SD sequence) and the frequency of frameshifting was reported to be 6% (Sekine et al., 1994). In this example the two upstream nucleotides are CA, which gives a CAA AAG hexamer. However, the role in frameshift modulation of the CAA codon was not determined in the IS3 context, nor had it been tested within the IS1222 recoding signal. Possible different mechanistic consequences of the two types of sequences were examined in the current study.
The present work also determines whether NNA codons other than CGA, in the sequence NNA AAG, are decoded by tRNAs that liberate the third codon base, A, permitting realignement of tRNALys in the –1 frame. The incidence of the nucleotide 5′ of the NNA codon and the effect of the modification status of anticodon base 34 of the NNA-decoding tRNA were also analyzed. Two versions of a model for single re-pairing frameshifting are presented.
Results
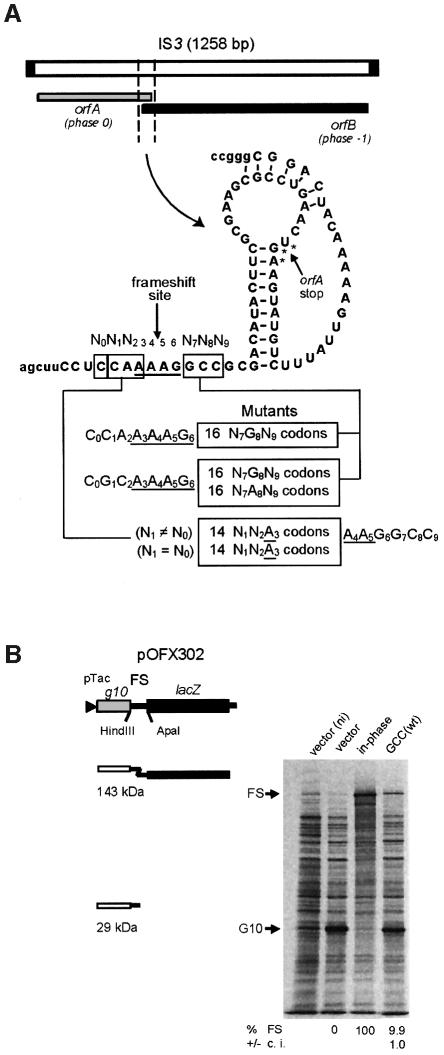
Members of the IS3 family of insertion sequences have two partially overlapping open reading frames, orfA and orfB, with –1 ribosomal frameshifting at a specific site in the overlap region yielding an OrfAB transframe protein with transposase function (Mahillon and Chandler, 1998). IS3 itself, the archetype of the family, and IS1222 have this gene organization (Figure 1A). Their orfA gene encodes a protein containing a predicted α-helix–turn–α-helix motif, as well as a leucine-zipper motif, and their orfB gene encodes a protein with a domain characteristic of retroviral integrases and IS3 family transposases (Mahillon and Chandler, 1998). So far in IS elements, the OrfB polypeptide has only been found to be important for transposition activity when fused to the OrfA protein (Polard et al., 1992). In the orfA–orfB overlap region of both IS3 and IS1222, frameshifting presumably occurs by re-alignement of one tRNALys on the A AAG sequence. To elucidate the exact role of the upstream codon in each IS, we cloned both frameshift regions into a reporter plasmid and changed the upstream codon of their respective hexamer to all 13 other N1N2A3 sense codons (diagrammed in Figure 1A). In addition, we investigated the incidence of the nucleotide on the 5′ side of the hexamer (nucleotide N0). In one set, N0 was different from N1 to prevent re-pairing of the N1N2A3-decoding tRNA, and in the other set it was identical to N1 in order to allow re-pairing of at least the third anticodon base (tRNA nucleotide 36). Since identical results were found with IS3 and IS1222, only the IS3 results are presented below. For reasons discussed in the next section, we also analyzed in the case of IS3 the effect on frameshifting of the codon 3′ to the A AAG shift site (nucleotides N7N8N9 in Figure 1A).
Fig. 1. The IS3 frameshift region, its various derivatives and the plasmid reporter system. (A) The segment of IS3 shown and the derived mutants were cloned in the pOFX302 plasmid. (B) Frameshifting efficiency was determined by protein labeling with [35S]methionine. The results obtained with two control strains (one with an in-phase construct, giving the theoretical 100% frameshifting value, and the other containing the vector plasmid, 0% frameshifting value), and with the IS3 ‘wild type’ region (wt) are shown. One culture of the vector- containing strain was labeled in the absence of IPTG (ni). The position of the product from normal translation (G10) or frameshifting (FS) is indicated. The calculated level of frameshifting and the 95% confidence interval (c.i.) are indicated below the relevant lanes. In the natural IS3 frameshift region an AUG codon (frame –1) overlapping the UGA stop codon of orfA (frame 0) is used to initiate synthesis of the OrfB protein. This AUG codon was changed to CUG in order to prevent initiation without interfering with frameshifting (Sekine et al., 1994).
To study IS3 frameshifting, the 81-nucleotide segment shown in Figure 1A was inserted between, and fused to, two genes. The end of orfA is in-frame with gene 10 of phage T7 and the beginning of orfB is in-frame with the lacZ coding sequence on a plasmid-borne construct (Figure 1B; Rettberg et al., 1999). Quantitation of the G10-OrfA′-OrfB′-LacZ transframe product (FS in Figure 1B) and G10-OrfA′ (G10 in Figure 1B) products was performed by in vivo protein pulse labeling followed by PAGE separation or by β-galactosidase assay.
Frameshifting occurs while the AAG codon is in the ribosomal A-site
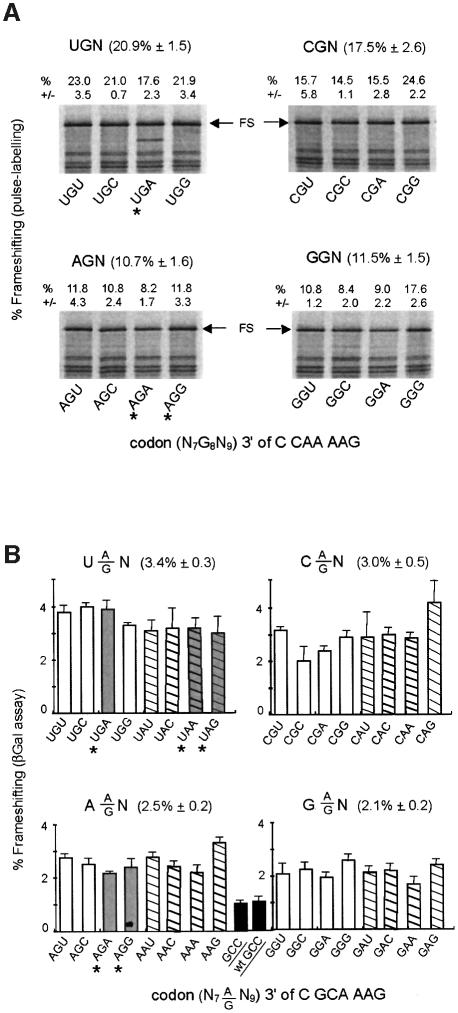
To gain evidence concerning the ribosomal site at which the frameshift occurs with the IS3 motif, the GCC codon 3′ to the C CAA AAG wild-type sequence was changed to all 16 possible N7G8N9 codons (Figure 1A); in another set of constructs, a different shift site was used, C GCA AAG, and the 3′ codon was changed to the 32 possible N7(A/G)8N9 codons. Slow-to-decode codons, especially stop codons, in the ribosomal A-site can stimulate non-programmed (i.e. low level) –1 frameshifting of peptidyl-tRNA in the P-site, if upstream re-pairing is possible (Weiss et al., 1987; Gallant and Lindsley, 1992). Consequently, if a 3′ stop or rare (e.g. AGG or AGA in E.coli) codon has a stimulatory effect, this indicates P-site slippage, whereas absence of an effect suggests that frameshifting occurred while the shifty motif was in the A-site.
Figure 2A and B shows the results of the analysis carried out on the wild-type and mutant IS3 signals, respectively. Interestingly, in this set of 48 N7(A/G)8N9 constructs the absolute level of frameshifting varies with the 3′ context. In Figure 2A, for example, there is a 2-fold difference between GGA and GGC or a 3-fold factor between GGC and CGG. In Figure 2B, β-galactosidase assay was used to measure frameshifting. Even if levels of frameshifting thus measured are lower than with the pulse-labeling method (see Materials and methods), significant differences also exist between constructs (e.g 2.5-fold between GAA and CAG). In a recent study we observed a similar effect of the 3′ context with the four heptameric X XXA AAG dual slippage motifs (Bertrand et al., 2002). The statistical analysis of nearly 200 mutants showed that the first nucleotide after the motif has the primary effect on frameshifting, with, in order of decreasing efficiency, U > C > A > G. Our interpretation was that when the AAG slippery codon enter the A-site, there is a competition between standard decoding and –1 frameshifting the outcome of which could be in part determined by the stacking of the next nucleotide of the message on the AAG codon–anticodon helix. Purines, having a higher stacking potential than pyrimidines, would therefore tend to limit frameshifting (see discussion in Bertrand et al., 2002). Comparison of the mean value after grouping of the constructs according to the identity of the first nucleotide of the 3′ codon (Figure 2) demonstrates that, for the IS3 shift site also, pyrimidines in this position generally results in a higher level of frameshifting than when it is a purine. However, exceptions to the rule do exist (e.g. CGG or GGG in Figure 2A are higher than their three relatives), suggesting that the second and third 3′ nucleotides can also influence frameshifting.
Fig. 2. Effect on frameshifting of variants of the codon 3′ of the IS3 shift site. The mutations indicated in Figure 1A were introduced on the 3′side (nucleotides N7N8N9) of the A AAG shift site and the modified frameshift regions were cloned into the pOFX302 reporter plasmid. (A) Summary of the results obtained by pulse-labeling of C CAA AAG N7G8N9 constructs. (B) Results obtained by performing β-galactosidase assays with C GCA AAG N7(G/A)8N9 constructs. The value for both motifs with GCC as 3′ codon is also given (wt stands for C CAA AAG).
Whatever the real cause(s) of the observed 3′ context effect is, and more importantly in view of the initial question concerning the position of the AAG-decoding tRNA, the results presented in Figure 2 show clearly that the levels of frameshifting do not significantly vary between the eight constructs with a 3′ stop, or rare, codon (marked with an asterisk in Figure 2A and B) and most of their related sense codon constructs. There is no increase in frameshifting caused by the presence of a 3′ stop or rare codon. Therefore, in the context of the IS3 recoding region, frameshifting of tRNALys from AAG to AAA is most likely initiated while AAG is in the A-site.
First codon of the hexanucleotide shift site
tRNALys re-pairing to mRNA at a cognate codon requires that the last base of the previous codon be A. Previous partial mutational analysis of the cdd signal suggested a strong influence on recoding of the identity of the NNA codon, with CGA apparently being the most shift prone (Mejlhede et al., 1999). To investigate this question more systematically, CAA was substituted in the IS3 recoding region by all other NNA codons except for UAA and UGA that would be in-frame stop codons. The 5′ nucleotide, N0, was also changed as indicated to preclude its involvement in Watson–Crick pairing with a tRNA attempting to re-pair to mRNA at the overlapping –1 frame codon. The results presented in Figure 3A show clearly that the level of frameshifting is strongly influenced by the identity of the first two nucleotides of the NNA AAG hexamer and that several codons are equal to, or better, than CGA. The 14 NNA codons can be separated into two classes, the ones that lead to ‘low’ frameshifting and those that give ‘high’ frameshifting. Within each class, there is a notable amount of variation indicative of an extra layer of idiosyncratic behavior. For example, GUA and AGA are respectively remarkably higher and lower than the others. Codons UCA and ACA first appeared as intermediate, but not overlapping with any in the low category (>95% confidence level).
Fig. 3. Effect on frameshifting of N0N1N2A variants: correlation with wobble properties and modification of base 34 of the N1N2A-decoding tRNA (A and B), and variants where N0 is identical to N1 (C). In (A) and (C), frameshift efficiencies were measured by quantitation of [35S]methionine labeled products. The error bars indicate the 95% confidence interval. In (B), the sequences of the anticodons of the E.coli NNA-decoding tRNAs including modifications of base 34 are shown. The modifications are abbreviated as follows: 5-methylaminomethyluridine (mnm5U), 5-methylaminomethyl-2-thiouridine (mnm5s2U), 5-caboxymethylaminomethyluridine (cmnm5U), 5-methoxyuridine (mo5U), uridine-5-oxyacetic acid (cmo5U), inosine (I) and lysidine (k2C). Three anticodon sequences are not from E.coli but from Bacillus subtilis (B.s.) or Mycoplasma capricolum (M.c.) as indicated.
The decoding properties of the wobble base (Crick, 1966; Yokoyama and Nishimura, 1995) of the cognate tRNAs for NNA codons are given in Figure 3B. With the exception of GGA, the NNA codons which give a low level of frameshifting belong to split codon boxes. These codons are decoded by tRNAs with a ‘restricted’ wobble capacity, i.e. they have a 3′-N36N35U34-5′ anticodon (except tRNAIle, which has a 3′-UAC-5′ anticodon) that read NNA and NNG codons only (or AUA only for tRNAIle). In contrast, the NNA codons associated with high frameshifting come from four-codon family boxes. Their respective tRNAs also have a 3′-N36N35U34-5′ anticodon, except tRNAArg 3′-GCI-5′, but can read three codons, those ending with A, G and U (or C, U and A for tRNAArg 3′-GCI-5′). All the NNA-specific tRNAs have a modified anticodon base U34 (or C34 to k2C in tRNAIle, or A34 to I in tRNAArg) and the type of U34 modification is clearly correlated with ‘low’ or ‘high’ shiftiness (Figure 3B). The tRNAs which have an xm5 type modification (i.e. mnm5, cmnm5 and mnm5s2) restrict frameshifting, whereas those having a modification of the xo5 type allow more frameshifting to occur.
Possible upstream re-pairing for tRNANNA
A second set of 14 constructs was generated by changing the 5′ N0 nucleotide to one identical to the first of the NNA codon. The consequences in terms of re-pairing in the –1 frame vary according to each tRNA/N0N1N2A3 pair (Figure 3C). In four cases a consensus heptameric X XXY YYZ site for tandem slippage is generated, allowing cognate (A AAA AAG, G GGA AAG) or near cognate (C CCA AAG and U UUA AAG) interaction of P-site tRNANNA in the –1 phase. Accordingly, frameshifting is greatly stimulated from 6- to nearly 50-fold. For the 10 other NNA codons, the outcome is variable. In two cases, G GAA and C CAA, there is a 4-fold stimulation perhaps related to Watson–Crick pairing of tRNA bases 34 and 36 with the –1 frame codon (middle base 35 would form either an acceptable U·G pair or a less favorable U·C pair). Two others have a 2-fold increase (A ACA and C CGA) and the six remaining cases are not affected; for all eight, Watson–Crick pairing is limited to the interaction between N0 and anticodon base 36. From this we conclude that providing tRNANNA with an opportunity to re-pair in a cognate or near-cognate manner in the –1 phase increases frameshifting efficiency. Restricting pairing to the first position, N0, of the codon in the new frame has no, or only a marginal, positive effect on frameshifting which must then proceed via re-pairing of the AAG-decoding tRNA only.
Role of tRNA base U34 modification
The correlation between base U34 modification and frameshifting propensity, as well as many data suggesting that U34 modification may contribute to the wobble property of the tRNAs, prompted us to investigate the effect of mutations affecting specifically the xo5 or the xm5 modification. Inactivation of the aroD gene prevents the formation of cmo5U (Björk, 1995). Inactivation of mnmA and mnmE, respectively, precludes replacement of o2 by s2 and insertion of the mnm5 group (Björk, 1995). Two subsets of the NNA AAG constructs, three ‘low’ and three ‘high’ frameshifters, were tested in the three modification-deficient mutants (Figure 4); in the chosen constructs, the N0 nucleotide does not allow upstream re-pairing. The aroD mutation did not appear to have any significant effect on frameshifting modulation by the two classes of NNA codons. This indicates that the cmo5 modification is not what makes the tRNAs with extended wobbling more shift prone. In contrast, the mnmE and, more clearly, the mnmA mutations led to reduced frameshifting frequency, especially in the case of the ‘high’ frameshifting NNA codons, for which there is a 2- to 4-fold reduction. With these two mutants, the modification deficiencies affect not only the ‘low’ frameshifting NNA-decoding tRNAs, but also the downstream tRNALys, the one that shifts from the 0 to the –1 frame. So in the case of ‘high’ frameshifting NNA codons, the assay in the mnm mutants reveals the importance of modification for the frameshifting capacity of tRNALys. Obviously, both mnmA and mnmE alter this capacity, the former more than the latter.
Fig. 4. Frameshifting in modification-deficient mutants. Six NNA mutants (three low frameshifters and three high) were introduced into isogenic wild-type, aroD, mnmA and mnmE strains. Frameshifting was estimated by measuring the β-galactosidase activity of the resulting strains (in the absence of IPTG). The construct in which g10 and lacZ are in the same frame (in-phase construct in Figure 1) served to define the 100% reference activity and a construct in which the A AAG sequence was mutated to a non-slippery one (G AAA) was used to determine the background level.
Discussion
P-site pairing maintained or irreversibly disrupted in hexanucleotide shifting
Influence of a stop codon (or rare codon) on –1 frameshifting is evidence that disruption of codon:anticodon base-pairing and re-pairing in a new frame occurs in the P-site. The observed lack of influence of a stop or rare codon placed immediately 3′ of the CAA AAG or GCA AAG hexanucleotide shift site is interpreted to mean that CAA or GCA, and by extension any other NNA codon, is in the ribosomal P-site and AAG is in the A-site when frameshifting occurs. This is comparable to classical heptanucleotide frameshifting on X XXY YYZ sequences, where the XXY and YYZ 0 frame codons are in the P- and A-site, respectively (Jacks et al., 1988; Weiss et al., 1989; Harger et al., 2002).
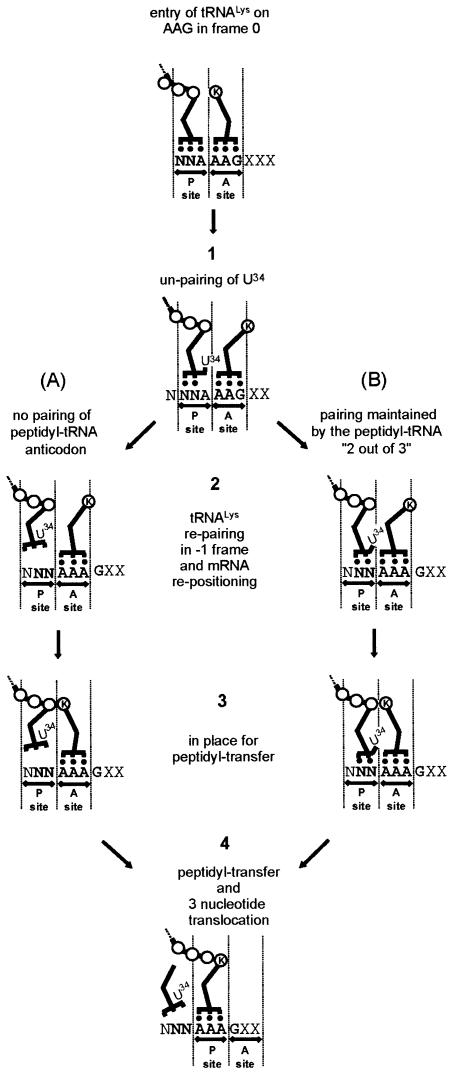
An interesting feature of the results is the involvement of hexanucleotide, rather than heptanucleotide, shift sites for the lesser, but still significantly efficient, –1 frameshifting studied. This hexanucleotide frameshifting likely involves the same mRNA movement as in tandem slippage in –1 heptanucleotide frameshifting (Jacks et al., 1988; Weiss et al., 1989; Harger et al., 2002). The difference is that in hexanucleotide frameshifting there is no re-pairing of the P-site tRNA to mRNA. In one model of hexanucleotide frameshifting, outlined in Figure 5A, there is dissociation of pairing in the P-site without re-pairing to mRNA. Lack of involvement of P-site re-pairing is quite plausible, since peptidyl-transfer can sometimes be carried out, in vitro and in vivo, in the absence of codon–anticodon interaction (Yusupova et al., 1986; Atkins et al., 2001; A.J.Herr, N.M.Wills, C.Nelson, R.F.Gesteland and J.F.Atkins, in preparation). Our data show that –1 frameshifting is more efficient when there is limited P-site re-pairing potential on non-standard heptamers (e.g. C CAA AAG or G GAA AAG; see Figure 3), provided that two conditions are met: N0 and N1 have to be identical and at least one other Watson–Crick pair exists between the tRNA and the –1 frame N0N1N2 codon.
Fig. 5. Models for frameshifting on hexameric motifs. The N1N2A3 AAG hexamer is normally read as N1N2A3 and A4A5G6 in frame 0 (top). Base 34 of the N1N2A3-decoding tRNA disengages from pairing with A3 (stage 1). A-site tRNALys re-pairs on the –1 frame A3A4A5 codon with re-positioning of the mRNA (stage 2). In (A), the P-site anticodon irreversibly dissociates from the mRNA whereas in (B), anticodon bases 36 and 35 maintain pairing. Accommodation is then completed (stage 3), bringing the two tRNAs and the mRNA in the configuration seen in crystallized complexes (Yusupova et al., 2001) allowing peptidyl-transfer and standard three-base translocation establishing the change in frame (stage 4).
An alternative model for hexanucleotide –1 frameshifting is that P-site pairing is partially maintained, detachment of only anticodon base 34 from the third codon base is involved allowing tRNALys to re-pair in the –1 frame (Figure 5B). Retention of codon pairing by P-site anticodon bases 36 and 35 would require not only a change of the relative positions of anticodon bases 35 and 34, but also a large change in position within the P-site of the whole anticodon to permit pairing between anticodon base 36 of A-site tRNALys and what was the third codon base of the P-site. It is unlikely that tRNALys initially pairs in the –1 frame with AAA in the sequence A AAG since the AAG lysine codon is required for efficient frameshifting.
A-site tRNA
Both models require re-pairing of the A-site tRNA in the –1 frame. The frame change could then happen before and/or after GTP hydrolysis and EF-Tu release, after codon recognition, during the second-half of the ‘initial selection’ steps or at the onset of the following ‘accommodation’ step, as defined by Rodnina and Wintermeyer (2001). It possibly occurs after the correct codon–anticodon interaction in the A-site triggers adoption of a ‘closed’ conformation by the 30S subunit (Ogle et al., 2002, 2003). Once accepted-in, the A-site tRNA, still in the A/T hybrid state, with or without EF-Tu attached, must have more leeway: it can disengage, re-pair in the –1 frame and stay there, especially in the case of E.coli tRNALys, which has a stronger interaction with AAA than with AAG (Lustig et al., 1981; Yokoyama and Nishimura, 1995). Re-adjustment by one nucleotide of the mRNA position, brought along by the 3′ pseudoknot (Plant et al., 2003), probably occurs at that time (stage 2 in Figure 5). The A-site tRNA eventually moves to reach the A/A state; this requires a large movement (56 Å) of its CCA end and the repositioning by 9 Å of anticodon base 34, accompanied by a rotation around the phosphodiester bond between the P and A codons (Noller et al., 2002). Locking in the P/P and A/A states of the two tRNAs and their anticodons probably makes the change in frame irreversible. There is, then, a kink in the message between the P and A codons (stage 3 in Figure 5) and the phosphate group of base 1401 of 16S RNA is wedged between the last and the first bases of each codon (Ogle et al., 2001; Yusupova et al., 2001). What is unusual here is that the P-site codon should contain at most two paired bases. Re-pairing of the A-site tRNA in the –1 frame and mRNA movement resets the reading frame. Peptidyl transfer can now take place and be followed by a normal three-base translocation (stage 4 in Figure 5).
This model of re-alignment of aminoacyl tRNALys occurring prior to translocation derives from the one originally proposed for tandem –1 slippage (Jacks et al., 1988) and very recently refined and justified by Harger et al. (2002). However, for both tandem –1 slippage (Weiss et al., 1989; Atkins and Gesteland, 1995) and the P-site pairing model presented here, another proposal invokes slippage after transpeptidation, and perhaps during translocation resulting effectively in a two-base translocation. Arguments against this version were presented in detail by Harger et al. (2002). What we would like to add, in view of the three-dimensional model for tRNAs movements outlined by Noller et al. (2002), is that directly after transpeptidation the tRNAs are still in the same ‘locked’ configuration, and are therefore unlikely to change frame. Translocation per se is, together with peptidyl-transfer, the fundamental function of the ribosome, and as such it probably is the most constrained one. Again it is difficult to envision the tRNAs and the mRNA being loose at this critical stage. The advantage of frameshifting occurring at the accommodation step is that it does not tamper with the strict three-base translocation mechanism.
P-site tRNA
In both versions of the model, anticodon base 34 of the P-site tRNA has the ability (to different extents depending on the tRNA) of un-pairing from the third codon base. In the version where P-site pairing is irreversibly disrupted, there is no necessary tRNA distortion. However, in the version where anticodon bases 35 and 36 maintain pairing, disruption of the anticodon base 34 interaction is most likely due to its flipping out of the anticodon stack. tRNAs are known to possess a large degree of structural flexibility. Recent studies by cryo-electron microscopy suggest that the aminoacyl-tRNA could participate actively in the accommodation step via conformational changes in its anticodon arm (Stark et al., 2002; Valle et al., 2002). At a more refined level, molecular dynamics studies indicates that the anticodon loop (as well as the acceptor arm) is potentially a region with a large amplitude mobility (Matsumoto et al., 1999). NMR analyses of synthetic anticodon regions derived from a few tRNAs give an even more precise idea of the degree of mobility of individual nucleotides in the anticodon loop and of the effect of modification of bases 34 and 37 in particular (Clore et al., 1984; Schweisguth and Moore, 1997; Sundaram et al., 2000; Cabello-Villegas et al., 2002). Even if most, in their fully modified form, adopt, in solution, a classical 3′-stacked loop configuration, with a U-turn between nucleotide 33 and 34, anticodon bases 34, 35 and 36 are still fairly mobile. In one case, E.coli tRNAPhe, the loop is reduced to bases 34 to 36 even when base 37 is modified as it is in vivo, suggesting that some anticodon loops may adopt, in solution, a conformation differing from the classical one (Cabello-Villegas et al., 2002). However, two missing pseudouridine modifications (U32 and U39) in the analyzed anticodon stem–loop may be in part responsible for this unorthodox configuration. In the case of tRNALys, absence of modification also lead to a pseudo tri-loop anticodon, and addition of the modifications (t6A37, mnm5s2U34, ψU39) bring the structure to the standard 7-nucleotide loop, which, however, remains flexible (Durant and Davis, 1999; Sundaram et al., 2000). It therefore appears that the s2 and mnm5 modifications increase the rigidity of the anticodon loop, in particular by strongly shifting the ribose conformation toward the C3′-endo form, and thus allowing reading of A- and G-ending codons only (restricted wobble). In contrast the cmo5, and related modification of U34, have been proposed to tilt the balance in a more moderate manner. The C2′-endo form predominates but the C3′-endo form is also present and the end result is more flexibility. This allows interaction of U34 with G- or U-ending codons (extended wobble), when in the C2′-endo configuration, and also with A-ending codons, when in the C3′-endo form (Yokoyama et al., 1985; Yokoyama and Nishimura, 1995). Thus these data, linking anticodon base flexibility and wobble capacity, are in agreement with our finding that there are two types tRNAs as judged by their effect on frameshifting on NNA AAG hexamers. We can now re-formulate our conclusion and say that tRNAs with a more flexible base 34 (xmo5U in six cases and I in one; see Figure 3) are more frameshift-promoting than those with less flexibility at that position (xm5U in six cases and k2C in one). At the molecular level, the C2′/C3′-endo interconversion may be what temporarily brings base 34 out of pairing with the third base of NNA codons (or what causes all three bases of tRNANNA anticodon to disengage, according to the alternate scenario). To conclude, it appears that tRNAs anticodons are not extremely rigid and that there are probably large differences among them from that point of view. Such flexibility may well allow transitory un-pairing of base 34 (and perhaps of bases 35 and 36 also), especially since there is not a close monitoring of the codon–anticodon interaction in the P-site (Ogle et al., 2001).
Modification of tRNA base 34
The apparent correlation between frameshifting, wobbling and the modification pattern of base U34 (Figure 3) led us to examine frameshifting in modification-deficient mutants, with the hope it would provide a new window for assessing the function of modification at that position in the anticodon (Figure 4). This hope was only partly fulfilled. Absence of the cmo5 modification did not change the incidence on frameshifting of the relevant NNA-tRNAs. This means that the flexibility of base U34 is still the same without the cmo5 group. The result was not entirely unexpected in view of the higher mobility displayed by that nucleotide when it is non-modified. This higher mobility expands wobbling further, since unmodified U34 recognizes codons ending with any nucleotide, at least in vitro (Yokoyama and Nishimura, 1995). Thus, the cmo5 modification is not the cause of U34 mobility, it probably limits it to prevent pairing with C; it rather is, like frameshifting stimulation, a consequence of structural properties of the anticodon region shared by one class of NNA-decoding tRNAs (Grosjean et al., 1996). In contrast, absence of either the s2 or mnm5 modifications had a negative effect on frameshifting on the most efficient NNA AAG motifs. There, it was the A-site tRNALys that was affected by the mutation. A known effect of s2, and of mnm5 to a lesser extent, is to favor pairing of tRNALys on AAA over AAG by increasing the rigidity of the anticodon (Yokoyama et al., 1985; Yokoyama and Nishimura, 1995). In the absence of one or other modification, anticodon base 34 is more flexible, allowing easier adjustment for proper pairing with G. This makes re-pairing from AAG to AAA energetically less advantageous and therefore frameshifting becomes less frequent.
The finding of significant levels of frameshifting at multiple hexanucleotide sequences has substantial relevance for ongoing searches to discover where programmed frameshifting is utilized for gene expression. While utilization by the IS elements mentioned above provide some initial examples, the generality of this form of recoding remains to be determined.
Materials and methods
Bacterial strains and growth conditions
The E.coli K12 strain JS238 [MC1061, araD Δ(ara leu) galU galK hsdS rpsL Δ(lacIOPZYA)X74 malP::lacIQ srlC::Tn10 recA1] was used for all cloning experiments.
Strains with mutations in tRNA modification genes were provided by Professor G.Björk: TH194 (aroD+, mnmA+, mnmE+), GRB2162 (aroD), TH193 (mnmA) and TH99 (mnmE) (Urbonavicius et al., 2001). These strains were transformed with plasmid pAP2-lacIQ (P.Polard, unpublished) before introduction of the various pOFX302-based frameshift constructions. This plasmid, being based on a p15A replicon, is compatible with pBR322 derivatives and carries a kanamycin resistance gene as well as the lacIQ gene, which ensure a tight control of the Tac promoter carried by pOFX302. Bacteria were grown in LB medium (Sambrook et al., 1989) or, for protein labeling, in MOPS medium (Neidhardt et al., 1974) supplemented with glucose (0.5%), thiamine (2 mg/l) and all amino acids at 50 µg/ml each (except methionine, tryptophan and tyrosine). Rambach agar plates (Merck) were used to identify clones expressing β-galactosidase. Ampicillin (40 µg/ml) plus oxacillin (200 µg/ml), and kanamycin (25 µg/ml) were added when necessary.
DNA techniques and quantitation of radioactive macromolecules
Plasmid DNA was prepared using the Qiaprep or Qiagen-tip100 systems as indicated by the supplier (Qiagen). Restriction enzymes, T4 polynucleotide kinase and T4 DNA ligase were from New England Biolabs. AmpliTaq DNA polymerase and the Amplicycle sequencing kit were from Applera. Cloning, transformation, agarose gel electrophoresis, and sequencing gels were carried out according to standard procedures (Sambrook et al., 1989). Radioactive products ([γ-33P]ATP for DNA sequencing and [35S]methionine for in vivo protein labeling) were obtained from Amersham. The Fuji X BAS1000 phosphoimager and the PCBas software were used for the quantitative analysis of electrophoresis gels in which 35S-labeled proteins were separated.
Plasmids constructions
Mutants of the IS3 (or IS1222) frameshift region were cloned into the reporter plasmid pOFX302 described by Rettberg et al. (1999). In one set the second and third nucleotides of the C CAA AAG sequence containing the IS3 shift site was changed to all possible sequences (except TA and TG, to avoid in-frame stop codon); the first base was changed to G when the second was C. In a second set of 14 constructions, the first base was made identical to the second. Two control plasmids were also generated, in one the C CAA AAG was changed to C CAG AAA, to prevent frameshifting (0% frameshifting construct) and in the other a base was added to C CAA AAA G in order to set g10 and lacZ in the same phase (100% frameshifting construct).
Measurement of frameshifting frequency
Frameshifting frequency was determined by in vivo protein pulse labeling with [35S]methionine on four independent clones for each construct, following a previously described protocol (Rettberg et al., 1999; Bertrand et al., 2002). To calculate the frequency of frameshifting, the fraction of the total radioactivity present in the relevant band was divided by the corresponding value obtained for the in-phase control. Precision was assessed by calculation of the 95% confidence interval.
In some experiments frameshifting was estimated by measuring β-galactosidase activity. For each strain, four to eight tubes containing 0.5 ml of LB (supplemented with kanamycin, ampicillin and oxacillin) were inoculated with independent clones and incubated overnight at 37°C. After a 1/5 dilution in LB, the absorbance at 600 nm of each culture was measured on 125 µl in a 96 flat-bottomed wells microplate (optical path of 0.38 cm) with a spectramax 340PC spectrophotometer (Molecular Devices). The diluted cultures (0.5 ml) were adjusted to 1× Z* buffer [Z buffer from Miller (1992), supplemented with 0.005% SDS, 1 mg/ml BSA and 10 mM DTT instead of β-mercaptoethanol] and treated for 10 min at 0°C with 10 µl of CHCL3. Assays were prepared in 96-well microplates. A volume of extract depending on the activity was completed to 200 µl with Z* buffer and 50 µl of 4 mg/ml ONPG were added. Absorbance was read at 420 nm each minute over a 30 min period with a Spectramax 340PC spectrophotometer. In order to be directly comparable to those obtained with the classical protocol of Miller (1992), our specific activities were calculated for a volume of extract of 125 µl and for an OD600 of 1. As in the pulse-labeling experiments, the in-phase control served as 100% reference and precision was assessed by determining the 95% confidence interval.
Note that absolute levels of frameshifting measured that way are about nine times lower than those obtained by pulse-labeling of the same constructs. This is likely due to underestimation of the 100% value in the case of pulse-labeling (induction of the strong pTac promoter probably saturates the protein synthesis capacity). However, both methods gave identical results in terms of relative activities of the various constructs.
Acknowledgments
Acknowledgements
We are very grateful to Isabelle Canal for expert technical help, to Darrell Davis, Jan Neuhard, Jim McCloskey, Frode Engbæk, Glenn Björk and especially to Pavel Baranov for discussions, sharing of unpublished results and strains. N.M. appreciates an EMBO short-term fellowship for her work in Toulouse. This work was supported by the Centre National de la Recherche Scientifique, the Université Paul Sabatier of Toulouse, by a grant to O.F. from the Ministère de l’Education Nationale, de la Recherche et de la Technologie (programme de recherche fondamentale en microbiologie et maladies infectieuses et parasitaires), a Department of Energy grant to R.F.G. (DEFG03-99ER62732) and an NIH grant to J.F.A.(R01-GM48152).
Note added in proof
An important relevant paper has just been published [Napthine,S., Vidakovic,M., Ginary,R., Namy,O. and Brierley,I. (2003) Prokaryotic-style frameshifting in a plant translation system: conservation of an unusual single-tRNA slippage event. EMBO J., 22, 3941–3950].
References
- Atkins J.F. and Gesteland,R.F. (1995) Discontinuous triplet decoding with or without re-pairing by peptidyl tRNA. In Söll,D. and RajBhandary,U.L. (eds), tRNA: Structure, Biosynthesis and Function. ASM Press, Washington, DC, pp. 471–490. [Google Scholar]
- Atkins J.F., Herr,A., Massire,C., O’Connor,M., Ivanov,I. and Gesteland,R.F. (2000) Poking a hole in the sanctity of the triplet code: inferences for framing. In Garrett,R.A., Douthwaite,S.R., Liljas,A., Matheson,A.T., Moore,P.B. and Noller,H.F. (eds), The Ribosome: Structure, Function, Antibiotics and Cellular Interactions. ASM Press, Washington, DC, pp. 369–383. [Google Scholar]
- Atkins J.F. et al. (2001) Over-riding standard decoding: implication of recoding for ribosome function and enrichment of gene expression. In Cold Spring Harbor Symposium on Quantitative Biology, Vol. LXVI, The Ribosome. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 217–232. [DOI] [PubMed] [Google Scholar]
- Bertrand C., Prère,M.F., Gesteland,R.F., Atkins,J.F. and Fayet,O. (2002) Influence of the stacking potential of the base 3′ of tandem shift codons on –1 ribosomal frameshifting used for gene expression. RNA, 8, 16–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björk G.R. (1995) Biosynthesis and function of modified nucleosides in tRNA. In Söll,D. and RajBhandary,U.L. (eds), tRNA: Structure, Biosynthesis and Function. ASM Press, Washington, DC, pp. 165–205. [Google Scholar]
- Brierley I., Jenner,A.J. and Inglis,S.C. (1992) Mutational analysis of the “slippery-sequence” component of a coronavirus ribosomal frameshifting signal. J. Mol. Biol., 227, 463–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabello-Villegas J., Winkler,M.E. and Nikonowicz,E.P. (2002) Solution conformations of unmodified and A(37)N(6)-dimethylallyl modified anticodon stem–loops of Escherichia coli tRNA(Phe). J. Mol. Biol., 319, 1015–1034. [DOI] [PubMed] [Google Scholar]
- Carter R.J., Baeyens,K.J., SantaLucia,J., Turner,D.H. and Holbrook,S.R. (1997) The crystal structure of an RNA oligomer incorporating tandem adenosine-inosine mismatches. Nucleic Acids Res., 25, 4117–4122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler M. and Fayet,O. (1993) Translational frameshifting in the control of transposition in bacteria. Mol. Microbiol., 7, 497–503. [DOI] [PubMed] [Google Scholar]
- Clore G.M., Gronenborn,A.M., Piper,E.A., McLaughlin,L.W., Graeser,E. and van Boom,J.H. (1984) The solution structure of a RNA pentadecamer comprising the anticodon loop and stem of yeast tRNAPhe. A 500 MHz 1H-n.m.r. study. Biochem. J., 221, 737–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crick F.H.C. (1966) Codon–anticodon pairing: the wobble hypothesis. J. Mol. Biol., 19, 548–545. [DOI] [PubMed] [Google Scholar]
- Curran J.F. (1995) Decoding with the A:I wobble pair is inefficient. Nucleic Acids Res., 23, 683–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durant P.C. and Davis,D.R. (1999) Stabilization of the anticodon stem-loop of tRNALys,3 by an A+–C base-pair and by pseudouridine. J. Mol. Biol., 285, 115–131. [DOI] [PubMed] [Google Scholar]
- Farabaugh P.J. (1997) Programmed Alternative Reading of the Genetic Code. R.G.Landes Company, Austin, TX. [Google Scholar]
- Farabaugh P.J. and Björk,G.R. (1999) How translational accuracy influences reading frame maintenance. EMBO J., 18, 1427–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallant J.A. and Lindsley,D. (1992) Leftward ribosome frameshifting at a hungry codon. J. Mol. Biol., 223, 31–40. [DOI] [PubMed] [Google Scholar]
- Gao H. et al. (2003) Study of the structural dynamics of the E. coli 70S ribosome using real-space refinement. Cell, 113, 789–801. [DOI] [PubMed] [Google Scholar]
- Gramstat A., Prüfer,D. and Rohde,W. (1994) The nucleic acid-binding zinc finger protein of potato virus M is translated by internal initiation as well as by ribosomal frameshifting involving a novel mechanism of P-site slippage. Nucleic Acids Res., 22, 3911–3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosjean H., Edqvist,J., Straby,K.B. and Giegé,R. (1996) Enzymatic formation of modified nucleosides in tRNA: dependence on tRNA architecture. J. Mol. Biol., 255, 67–85. [DOI] [PubMed] [Google Scholar]
- Harger J.W., Meskauskas,A. and Dinman,J.D. (2002) An “integrated model” of programmed ribosomal frameshifting. Trends Biochem. Sci., 27, 448–454. [DOI] [PubMed] [Google Scholar]
- Jacks T., Madhani,H.D., Masiarz,F.R. and Varmus,H.E. (1988) Signals for ribosomal frameshifting in the Rous Sarcoma Virus gag-pol region. Cell, 55, 447–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen B., Wills,N.M., Gesteland,R.F. and Atkins,J.F. (1994) rRNA–mRNA base pairing stimulates a programmed –1 ribosomal frameshift. J. Bacteriol., 176, 6842–6851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen B., Gesteland,R.F. and Atkins,J.F. (1997) Structural probing and mutagenic analysis of the stem–loop required for Escherichia coli dnaX ribosomal frameshifting: Programmed efficiency of 50%. J. Mol. Biol., 271, 47–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lustig F., Elias,P., Axberg,T., Samuelsson,T., Tittawella,I. and Lagerkvist,U. (1981) Codon reading and translational error. Reading of the glutamine and lysine codons during protein synthesis in vitro. J. Biol. Chem., 256, 2635–2643. [PubMed] [Google Scholar]
- Mahillon J. and Chandler,M. (1998) Insertion sequences. Microbiol. Mol. Biol. Rev., 62, 725–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto A., Tomimoto,M. and Go,N. (1999) Dynamical structure of transfer RNA studied by normal mode analysis. Eur. Biophys. J., 28, 369–379. [DOI] [PubMed] [Google Scholar]
- Mejlhede N., Atkins,J.F. and Neuhard,J. (1999) Ribosomal frameshifting during decoding of Bacillus subtilis cdd occurs at the sequence CGA AAG. J. Bacteriol., 181, 2930–2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J.H. (1992) A Short Course in Bacterial Genetics: A Laboratory Manual and Handbook for Escherichia coli and Related Bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Neidhardt F.C., Bloch,P.L. and Smith,D.F. (1974) Culture medium for enterobacteria. J. Bacteriol., 119, 736–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noller H.F., Yusupov,M.M., Yusupova,G.Z., Baucom,A. and Cate,J.H. (2002) Translocation of tRNA during protein synthesis. FEBS Lett., 514, 11–16. [DOI] [PubMed] [Google Scholar]
- Ogle J.M., Brodersen,D.E., Clemons,W.M.,Jr, Tarry,M.J., Carter,A.P. and Ramakrishnan,V. (2001) Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science, 292, 897–902. [DOI] [PubMed] [Google Scholar]
- Ogle J.M., Murphy,F.K., Tarry,M.J. and Ramakrishnan,V. (2002) Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell, 111, 721–732. [DOI] [PubMed] [Google Scholar]
- Ogle J.M., Carter,A.P. and Ramakrishnan,V. (2003) Insights into the decoding mechanism from recent ribosome structures. Trends Biochem. Sci., 28, 259–266. [DOI] [PubMed] [Google Scholar]
- Plant E.P., Jacobs,K.L., Harger,J.W., Meskauskas,A., Jacobs,J.L., Baxter,J.L., Petrov,A.N. and Dinman,J.D. (2003) The 9-Å solution: How mRNA pseudoknots promote efficient programmed –1 ribosomal frameshifting. RNA, 9, 168–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polard P., Prère,M.F., Chandler,M. and Fayet,O. (1991) Programmed translational frameshifting and initiation at an AUU codon in gene expression of bacterial insertion sequence IS911. J. Mol. Biol., 222, 465–477. [DOI] [PubMed] [Google Scholar]
- Polard P., Prère,M.F., Fayet,O. and Chandler,M. (1992) Transposase-induced excision and circularization of the bacterial insertion sequence IS911. EMBO J., 11, 5079–5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rettberg C.C., Prère,M.F., Gesteland,R.F., Atkins,J.F. and Fayet,O. (1999) A three-way junction and constituent stem–loops as the stimulator for programmed –1 frameshifting in bacterial insertion sequence IS911. J. Mol. Biol., 286, 1365–1378. [DOI] [PubMed] [Google Scholar]
- Rodnina M. and Wintermeyer,W. (2001) Fidelity of aminoacyl-tRNA selection on the ribosome: kinetic and structural mechanisms. Annu. Rev. Biochem., 70, 415–435. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Schweisguth D.C. and Moore,P.B. (1997) On the conformation of the anticodon loops of initiator and elongator methionine tRNAs. J. Mol. Biol., 267, 505–519. [DOI] [PubMed] [Google Scholar]
- Sekine Y., Eisaki,N. and Ohtsubo,E. (1994) Translational control in production of transposase and in transposition of insertion sequence IS3. J. Mol. Biol., 235, 1406–1420. [DOI] [PubMed] [Google Scholar]
- Stark H., Rodnina,M.V., Wieden,H.J., Zemlin,F., Wintermeyer,W. and van Heel,M. (2002) Ribosome interactions of aminoacyl-tRNA and elongation factor Tu in the codon-recognition complex. Nat. Struct. Biol., 9, 849–854. [DOI] [PubMed] [Google Scholar]
- Steibl H. and Lewecke,F. (1995) IS1222: analysis and distribution of a new insertion sequence in Enterobacter agglomerans 339. Gene, 156, 37–42. [DOI] [PubMed] [Google Scholar]
- Sundaram M., Durant,P.C. and Davis,D. (2000) Hypermodified nucleosides in the anticodon of tRNALys stabilize a canonical U-turn structure. Biochemistry, 39, 12575–12584. [DOI] [PubMed] [Google Scholar]
- Tsuchihashi Z. and Brown,P.O. (1992) Sequence requirements for efficient translational frameshifting in the Escherichia coli dnaX gene and the role of an unstable interaction between tRNALys and an AAG lysine codon. Genes Dev., 6, 511–519. [DOI] [PubMed] [Google Scholar]
- Urbonavicius J., Qian,Q., Durand,J.M., Hagervall,T.G. and Björk,G.R. (2001) Improvement of reading frame maintenance is a common function for several tRNA modifications. EMBO J., 20, 4863–4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valle M., Sengupta,J., Swami,N.K., Grassucci,R.A., Burkhardt,N., Nierhaus,K.H., Agrawal,R.K. and Frank,J. (2002) Cryo-EM reveals an active role for aminoacyl-tRNA in the accommodation process. EMBO J., 21, 3557–3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss R.B., Dunn,D.M., Atkins,J.F. and Gesteland,R.F. (1987) Slippery runs, shifty stops, backward steps and forward hops: –2, –1, +1, +2, +5 and +6 ribosomal frameshifting. In Cold Spring Harbor Symposium on Quantitative Biology, Vol. LII. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 687–693. [DOI] [PubMed] [Google Scholar]
- Weiss R.B., Dunn,D.M., Shuh,M., Atkins,J.F. and Gesteland,R.F. (1989) E. coli ribosomes re-phase on retroviral frameshift signals at rates ranging from 2 to 50 percent. New Biol., 1, 159–169. [PubMed] [Google Scholar]
- Yokoyama S. and Nishimura,S. (1995) Modified nucleosides and codon recognition. In Söll,D. and RajBhandary,U.L. (eds), tRNA: Structure, Biosynthesis and Function. ASM Press, Washington, DC, pp. 207–223. [Google Scholar]
- Yokoyama S., Watanabe,T., Murod,K., Ishikura,H., Yamaizumi,Z., Nishimura,S. and Miyazawa,T. (1985) Molecular mechanism of codon recognition by tRNA species with modified uridine in the first position of the anticodon. Proc. Natl Acad. Sci. USA, 82, 4905–4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusupova G.Z., Belitsina,N.V. and Spirin,A.S. (1986) Template-free ribosomal synthesis of polypeptides from aminoacyl-tRNA. Polyphenylalanine synthesis from phenylalanine-tRNALys. FEBS Lett., 206, 142–146. [DOI] [PubMed] [Google Scholar]
- Yusupova G.Z., Yusupov,M.M., Cate,J.H. and Noller,H.F. (2001) The path of messenger RNA through the ribosome. Cell, 106, 233–241. [DOI] [PubMed] [Google Scholar]