Abstract
RNA polymerase III (Pol III) transcribes a large set of genes encoding small untranslated RNAs like tRNAs, 5S rRNA, U6 snRNA or RPR1 RNA. To get a global view of class III (Pol III-transcribed) genes, the distribution of essential components of Pol III, TFIIIC and TFIIIB was mapped across the yeast genome. During active growth, most class III genes and few additional loci were targeted by TFIIIC, TFIIIB and Pol III, indicating that they were transcriptionally active. SNR52, which encodes a snoRNA, was identified as a new class III gene. During the late growth phase, TFIIIC remained bound to most class III genes while the recruitment of Pol III and, to a lesser extent, of TFIIIB was down regulated. This study fixes a reasonable upper bound to the number of class III genes in yeast and points to a global regulation at the level of Pol III and TFIIIB recruitment.
Keywords: ChIP on chip/microarray/transcription/tRNA gene/yeast
Introduction
In eukaryotic cells, three nuclear RNA polymerases (Pol) transcribe distinct classes of genes (Sentenac, 1985). Pol I transcribes a single essential gene, the rDNA, to generate the 35S rRNA precursor. Pol II synthesizes pre-messenger RNAs and a number of small, stable RNA genes involved in mRNA splicing and in the modification of rRNAs. Pol III is essentially responsible for the synthesis of the small RNA species involved in translation like 5S rRNA and tRNAs. In yeast, Pol III was also shown, both in vivo and in vitro, to synthesize U6 spliceosomal RNA, the 7SL and the RNase P RNAs that are involved in protein secretion and in the maturation of tRNAs, respectively encoded by the SNR6, SCR1 and RPR1 genes (for a review, see White, 1998). Except for the 5S RNA gene, all known yeast class III genes (i.e. genes transcribed by RNA polymerase III) share control sequence motifs, the A and B blocks, and a run of T residues that specifies transcription termination (Willis, 1993). The transcription start site is located around 20 nucleotides upstream of the A block (see Figure 1 for a model view). In contrast, the B block is located at a variable distance downstream of the A block within the transcription unit, or exceptionally, downstream of the termination site in the case of the SNR6 gene. SNR6 and a few tRNA genes have a TATA-like element upstream of the transcription start site (Brow and Guthrie, 1990; Eschenlauer et al., 1993; Dieci et al., 2000).
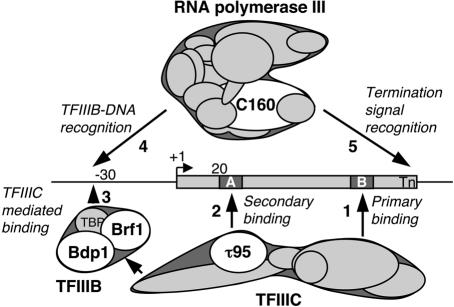
Fig. 1. Transcription complex assembly on a yeast tRNA gene. The scheme depicts the multistep pathway of transcription complex formation: promoter recognition by TFIIIC, TFIIIC-directed assembly of the initiation factor TFIIIB and recruitment of RNA Pol III (numbered arrows) (Chédin et al., 1998; Geiduschek and Kassavetis, 2001). Tagged polypeptide components used in this paper are identified by white ovals.
In vitro studies with the yeast transcription system have described the cascade of protein–DNA interactions leading to the recruitment of Pol III to its target genes (Chédin et al., 1998; Geiduschek and Kassavetis, 2001; Schramm and Hernandez, 2002). The first step in the formation of the yeast Pol III transcription initiation complex is the recognition of the A and B blocks by the multisubunit TFIIIC factor (see Figure 1 for a model). TFIIIC comprises two large protein modules, τA and τB (Marzouki et al., 1986). τB (three subunits) binds to the B block with high affinity and favors A block binding by τA (three subunits). Elegant protein–DNA crosslinking experiments have provided a coarse mapping of TFIIIC subunits over a tRNA gene and the 5S RNA gene (Bartholomew et al., 1990; Braun et al., 1992). The subunit τ95 used in the present study belongs to the τA module (Conesa et al., 1993), maps over the A block (Bartholomew et al., 1990) and influences the stability of the TFIIIC–DNA complex and start site selection (Jourdain et al., 2003). Once bound, TFIIIC directs the assembly of TFIIIB upstream of the initiation site (Kassavetis et al., 1990; Bartholomew et al., 1991). TFIIIB comprises three components, the ubiquitous TATA binding protein (TBP), Brf1 which is related to TFIIB, and Bdp1. Brf1 appears to play a key role in initiating TFIIIB assembly (Kassavetis et al., 1992) and in the subsequent Pol III recruitment and post-recruitment steps (Brun et al., 1997; Kassavetis et al., 1998). Bdp1 locks the TFIIIB–DNA complex in a highly stable form competent for the stable recruitment of Pol III over the start site (Kumar et al., 1997). The presence of the TATA box in SNR6 or some tRNA genes can bypass the requirement for TFIIIC in vitro. TATA-box directed binding of TFIIIB is mediated in that case by TBP (Margottin et al., 1991; Burnol et al., 1993b; Whitehall et al., 1995). Once formed, the TFIIIB–DNA complex suffices to recruit the 17 subunit Pol III enzyme (Kassavetis et al., 1990, 1997) via multiple interactions involving specific Pol III subunits (Bartholomew et al., 1993; Werner et al., 1993; Brun et al., 1997; Andrau et al., 1999; Ferri et al., 2000).
In yeast, the 5S RNA genes are arranged in tandem with the 35S rRNA genes. Arrays of 100–200 copies of these genes are present on the right arm of chromosome XII. In contrast, the 275 tRNA genes are dispersed in the genome (Percudani et al., 1997; Hani and Feldmann, 1998). The yeast genome has been studied in silico to predict the existence of additional class III genes, based on the presence of A and B blocks and a poly-T track within intergenic regions (Olivas et al., 1997). This study identified one additional gene with intragenic A and B blocks, RNA170, which indeed showed a dependence on the B block and Brf1 for transcription in vivo. Based on the tRNA gene paradigm, this analysis assumed that Pol III transcription would always depend on TFIIIC binding, which remains to be established, and relied on a robust conservation of A and B block elements, which is not always the case.
A complete panorama of all the yeast class III genes would be a great asset to potentially reveal new RNA species and to address a number of questions concerning the Pol III promoter elements, the general role of TFIIIC, or possible differential regulation of class III genes. In the work presented here, we have used chromatin immunoprecipitation experiments coupled to microarrays hybridization (ChIP on chip; Ren et al., 2000; Iyer et al., 2001; Lieb et al., 2001) to investigate the Pol III genome as defined as the repertory of the binding sites for the Pol III transcription machinery. The binding sites of essential components of Pol III, TFIIIC and TFIIIB were mapped across the entire yeast genome. All known class III genes and a small set of additional loci were targeted by at least one component of the Pol III transcription machinery.
Results
Specific immunoprecipitation of class III genes with TFIIIC
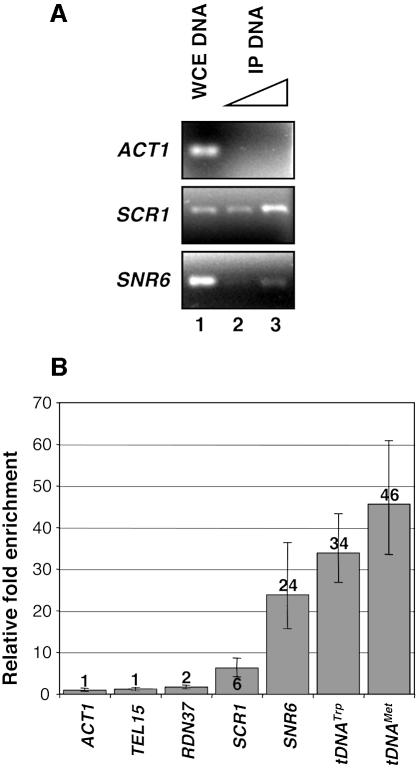
To identify the targets of the Pol III transcription apparatus, we combined chromatin immunoprecipitation and intergenic microarray hybridization. Chromatin proteins were crosslinked with formaldehyde to their target sites in vivo (Hecht and Grunstein, 1999; Hecht et al., 1999). After sonication that resulted in chromatin fragments of 250–1000 bp in size, DNA fragments that were specifically crosslinked to the myc-tagged 95 kDa subunit of TFIIIC were purified by immunoprecipitation with a monoclonal antibody directed to the epitope tag. To evaluate the selectivity of the immunoprecipitation reactions, the immunopurified DNA fragments were first analyzed by PCR amplification for the enrichment of two known class III genes, SCR1 and SNR6, that encode the RNA component of the 7SL particle and U6 snRNA, respectively (Figure 2A, lanes 2 and 3), relative to the ACT1 promoter as ACT1 (encoding actin) is transcribed by RNA polymerase II. As shown in Figure 2, SNR6 and SCR1 loci were amplified while ACT1 was not, under the same conditions (Figure 2A, compare lanes 3 and 2 with lane 1). In a second set of experiments, we quantified the specific fold enrichment of several known class III genes or non-Pol III DNA by real-time PCR quantification. The fold enrichment observed for each DNA locus was normalized to the fold enrichment observed for the ACT1 promoter. As expected, a fragment of chromosome XV telomere (TEL15) and a fragment of large rRNA 35S gene (named RDN37 in the Saccharomyces cerevisiae database) did not co-immunoprecipitate with epitope-tagged τ95 (Figure 2B, enrichment of 1 and 2 for TEL15 and RDN37, respectively). In contrast, the class III promoters of SCR1, SNR6, tDNATrp [also named tW(CCA)G1] and tDNAMet gene [tM(CAU)D] were specifically enriched, from 6- to 46-fold, in immunoprecipitated DNA as compared with the ACT1 promoter region (Figure 2B). The specific crosslinking of tagged τ95 to class III genes encouraged us to undertake a genome-wide analysis of TFIIIC binding sites.
Fig. 2. Gene-specific PCR analysis of immunoprecipitated chromatin bound to τ95-myc. (A) Primers hybridizing to the indicated promoter regions were used to amplify DNA immunoprecipitated with anti-myc antibody (lane 2 and 3) from the τ95-myc-tagged strain. DNA purified from whole-cell extract (WCE) was used as positive control (lane 1). As a Pol II transcribed gene, ACT1 is not expected to be enriched in τ95-associated chromatin, while the class III genes SCR1 and SNR6 contain TFIIIC binding sites. (B) Relative enrichment of DNA fragments, immunoprecipitated extracts, from the τ95-myc-tagged strain relative to genomic DNA from the whole-cell extract as determined by real-time PCR analysis. Enrichments levels for each gene were normalized to the level of the ACT1 promoter. The data show the mean, maximum and minimum fold enrichment from three independent experiments.
After reversal of the protein–DNA crosslinks, DNA fragments enriched by immunoprecipitation were amplified and fluorescently labeled with the Cy3 fluorophore. In parallel, a control DNA from whole-cell extract was amplified and fluorescently labeled with the Cy5 fluorophore. Genomic target loci were identified by comparative hybridization of the immunoprecipitated versus the control DNA probes to a DNA microarray harboring intergenic DNA regions. The ratio of the Cy3 to Cy5 fluorescence intensities at each site in the microarray provided a measure of the extent of binding of the bait protein to the corresponding genomic locus (for an example see Figure 4B). We used microarrays of intergenic regions to uncover all potential TFIIIC binding sites and to circumvent the problem of cross-hybridizations between conserved sequence elements (such as tRNA and Ty elements). The DNA microarray represented 6402 intergenic intervals on either side of every open reading frame, tRNA, small nuclear RNA, Ty element, solo δ, etc (see Supplementary data, available at The EMBO Journal Online). The length of the sheared crosslinked DNA (between 250 and 1000 bp; Supplementary data), the absence of sequence similarity between intergenic elements adjacent to class III genes and the small size of class III genes (the largest class III gene, SCR1, is 522 bp long; Dieci et al., 2002) was expected to allow the identification of Pol III promoters by hybridization of the crosslinked DNA fragments to intergenic elements located immediately upstream or downstream of a class III genes.
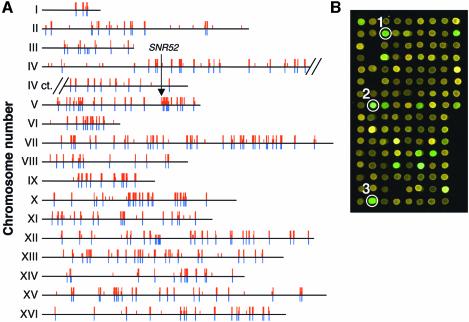
Fig. 4. Chromosomal display of genomic binding sites for τ95, Bdp1, Brf1 and the 160 kDa subunit of RNA polymerase III. (A) Map of the genomic binding sites of τ95, Brf1, Bdp1 and C160 determined by ChIP on chip analysis. Red bars correspond to the 647 intergenic regions bound by at least one of the class III transcription machinery, among which the 458 enriched by the four components together are highlighted by higher red bars. The blue bars indicate the position of the 275 annotated tRNA genes and other class III genes SNR6, RPR1, SCR1 and RDN5. An arrow points to the SNR52 gene. (B) Partial view of a microarray slide of a τ95-myc immunoprecipitation experiment. Spots corresponding to the intergenic region flanking two TATA-containing tRNA genes tP(UGG)F and tI(UAU)D (white circles 1 and 2, respectively) and a TATA-less tRNA gene tS(AGA)E (white circle 3) are highlighted. Green spots correspond to enriched intergenic regions.
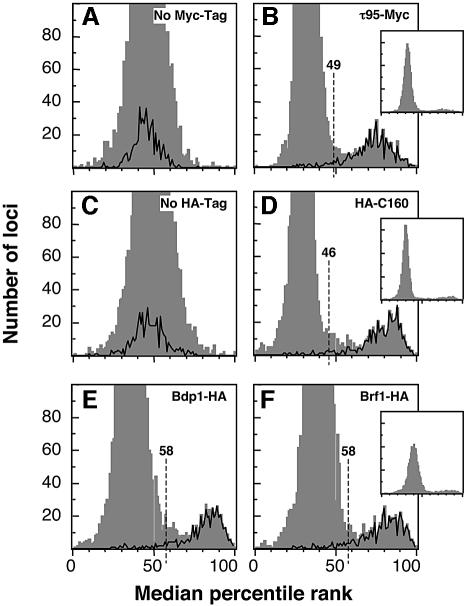
To identify the targets of the myc-tagged TFIIIC, we ranked genomic loci according to their logarithmic fluorescence ratios. The median of the percentile rank values for each locus was determined from four independent experiments. The distribution of the median percentile ranks for the control immunoprecipitation experiment with anti-myc antibodies on wild-type (untagged) cell extracts was Gaussian and thus showed no enrichment of particular genomic DNA fragments (Figure 3A, gray histogram). In contrast, the distribution of the median percentile ranks for the τ95 immunoprecipitation experiment was bimodal (Figure 3B, gray histogram), which indicates the specific enrichment of a significant number of intergenic elements.
Fig. 3. Specific enrichment of intergenic regions adjacent to class III genes. Distribution of median percentile ranks of Cy3/Cy5 fluorescence ratios are represented as gray histograms. (A) Control immunoprecipitation of untagged protein with an anti-myc antibody or (B) immunoprecipitation of myc-tagged TFIIIC. (C) Immuno precipitation of control untagged protein extract with an anti-HA antibody, (D) tagged HA-C160, (E) tagged Bdp1-HA, and (F) tagged Brf1-HA. Full-scale distributions for (B), (D) and (F) are represented in small insets (maximum y-axis is 700). The distributions of median percentile ranks of 489 flanking class III genes regions (corresponding to 271 tRNA genes, RPR1, SNR6, SCR1) are represented as black curves. The thresholds chosen to identify class III genes are indicated by dotted lines.
Remarkably, the distribution of 489 class III gene flanking regions (271 tRNA genes, RPR1, SNR6, SCR1) exactly fitted the second bulk of the global distribution of the DNA fragments immunoprecipitated with tagged TFIIIC (with the notable exception of RDN5, which encodes the 5S rRNA) whereas it was normally distributed in the mock immunoprecipitation experiments (the black curves in Figure 3A and B).
Genome-wide binding of the Pol III machinery
The bimodal distribution and the specific enrichment of the class III-specific intergenic elements in the DNA sample immunoprecipitated with tagged TFIIIC demonstrated the specificity of the analysis and therefore warranted a global study of the Pol III genomic loci. We therefore extended our analysis to three other representative components of the Pol III transcription machinery: the largest subunit of Pol III, C160, and the two components of TFIIIB, Brf1 and Bdp1. Each of these protein baits were tagged with HA epitopes. As in the case of τ95, in at least two independent experiments, each protein bait enriched a collection of DNA fragments that fitted well with the known class III genes (see the bimodal distribution in Figure 3D–F), whereas no enrichment of known class III genes was observed in the mock experiment (no HA tag, Figure 3C). We defined for each experiment a threshold percentile rank above which intergenic elements formed a group that was distinct from the bulk of the distribution. Slightly different threshold percentile ranks (58th, 58th, 49th and 46th) have been selected for Bdp1, Brf1, τ95 and C160, respectively (Figure 3B and D–F), due to the better efficiency of the immunoprecipitation reactions with τ95 and C160 bait proteins. We found 647 loci with a percentile rank above the threshold value with at least one of the four protein baits. Mapping these 647 loci across the whole genome (Figure 4A, red bars) shows a striking colocalization with the known class III genes (Figure 4A, blue bars). Four hundred and fifty-eight loci were selectively enriched with each of the four protein baits (Figure 4A, highest red bars), which corresponded to 7.2l% of the 6402 intergenic regions. These 458 loci were assumed to be intergenic elements adjacent to known or unknown class III genes.
These 458 loci were designated LEPTM for loci enriched by the Pol III transcription machinery. Four hundred and forty-two LEPTM (97% of the 458 LEPTM) were located next to or at a close distance from a tRNA gene (Table I), which underscored the quality of the microarrays. Ninety-four per cent of these 442 LEPTM are adjacent to tRNA genes. The size range of the DNA fragments generated by sonication (between 250 and 1000 bp, as estimated by gel electrophoresis) can explain some rare cases of hybridization with loci located farther, 1000–1500 bp, from a tRNA gene. When a locus lay between a pair of class III genes, we considered both genes as potential transcriptional targets. Therefore, the 442 LEPTM characterized the occupancy of the four bait proteins on 259 of the 275 tRNA genes (Table I). On the 16 remaining LEPTM, two were adjacent to the SNR6 genes, one flanked the RPR1 gene and another one flanked the SCR1 gene (Table I). The impressive colocalization of the intergenic elements enriched with four components of the class III transcription apparatus with nearly all the known class III genes (259 of the 275 tRNA genes, plus SNR6, RPR1 and SCR1) strongly suggested that the remaining unassigned loci could correspond to unknown class III genes. Twelve LEPTM could not be assigned to any known class III gene and potentially encompassed, or were adjacent to, some unknown class III genes. Among these 12 LEPTM, one (distant from any known class III genes) was adjacent to the SNR52 gene that encodes the small snoRNA snR52, a methylation guide for rRNA. Six LEPTM mapped near Ty transposons, usually inserted upstream of tRNA genes (Hani and Feldmann, 1998). We found by PCR analysis that three of these Ty elements were deleted in our yeast strains (data not shown) and the corresponding loci were therefore located next to a tRNA gene. Sequence analysis of the remaining three loci upstream of authentic Ty insertions revealed partial sequence identities with other loci adjacent to tRNA genes (data not shown), which might have caused false positives by cross-hybridization. Preliminary northern blot analyses of the last five loci did not reveal the presence of RNA originating from these intergenic elements (data not shown). These negative results were surprising. However, even if one assumes that these loci were false positives, the global level of false positives (<0.1%) was remarkably low due to the stringent criteria of specific enrichment by the four protein baits. Interestingly, these criteria revealed one strong candidate class III gene, SNR52.
Table I. Summary of the 647 enriched loci.
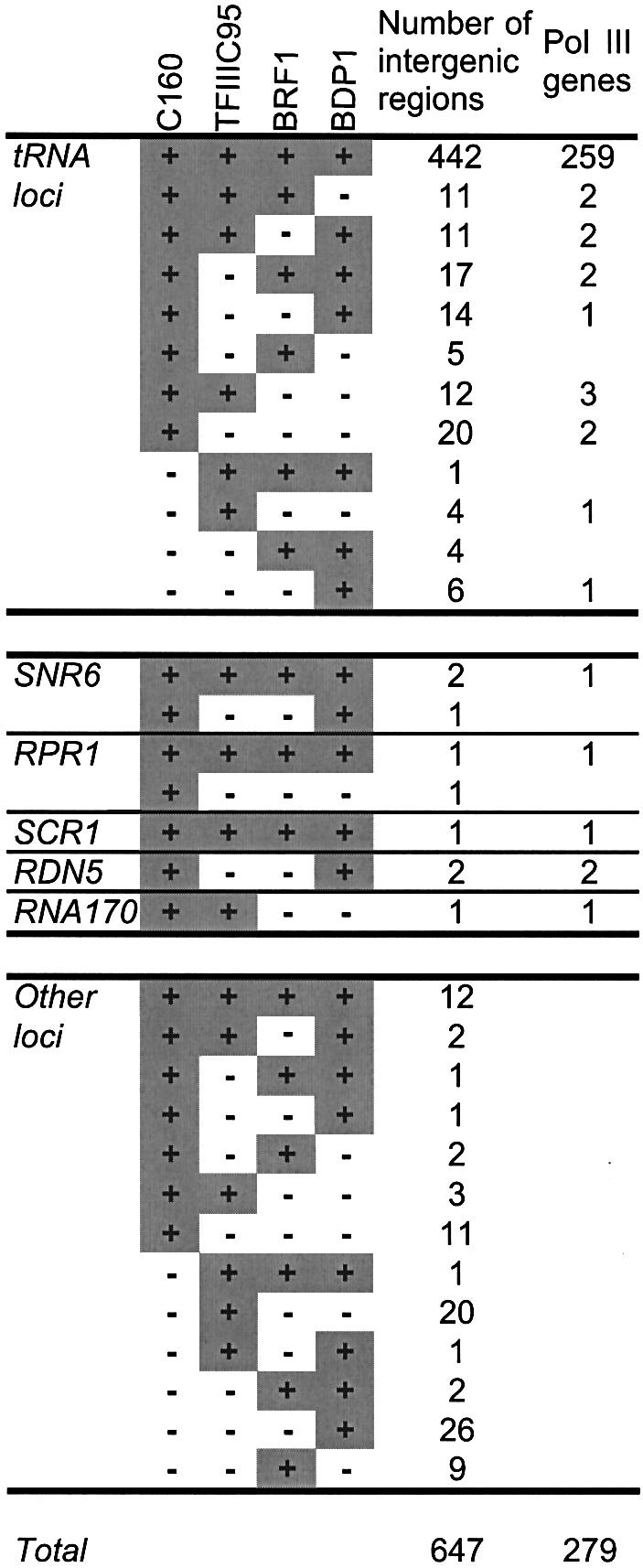
One could predict, however, that not all class III genes would be enriched by each of the four baits. For instance, the 5S RNA gene was not enriched by τ95, as mentioned above. Therefore, we further analyzed the 189 loci that were enriched by at least one of the four protein baits. Some of the 189 loci that had median percentile ranks above the threshold in one, two or three of the four immunoprecipitations could be assigned to known class III genes. Hence, 105 loci mapped within 1500 bp from a tRNA gene, adding 14 additional tRNA genes to the 259 tRNA genes already identified with the four bait proteins. In many instances, the enriched loci were not adjacent to the tRNA genes, which explains their selection by a subset of the protein baits and their low percentile rank. Note that the tD(GUC)N gene, which is considered as a pseudo gene due to a Ty insertion immediately upstream of the A box, was immunoprecipated with τ95 only. Two loci close to two tRNA genes, at a distance of <1.3 kb on the right arm of chromosome III, were not enriched by any of the protein bait. However, we did not succeed in amplifying this region by PCR analysis on S288c and YPH500 strain genomic DNA (data not shown). With these two exceptions, all the tRNA genes were targeted by at least one of the protein baits. Five loci flanked or harbored other known class III genes: RDN5 (two loci), SNR6 (one locus), RPR1 (one locus), and RNA170 (one locus). The remaining 78 loci, which could not be assigned to known class III genes (Table I), also had a relatively low median percentile rank as compared with the class III genes related loci recognized by the four protein baits (data not shown).
SNR52 is transcribed by RNA polymerase III
We have focused our study on the intergenic region adjacent to the SNR52 gene (Figure 4). SNR52 has been previously identified by a computational screen as a snoRNA gene required for 2′-O-methylation of rRNA (Lowe and Eddy, 1999). To confirm that the SNR52 gene was transcribed by Pol III in vivo, we analyzed the transcription profile of this gene by northern blot analysis in a mutant strain harboring a thermosensitive mutation rpc160-112 in the catalytic site of the Pol III largest subunit (Dieci et al., 1995). Total RNA was extracted from wild-type and mutant cells grown at permissive temperature (24°C) or 3 h after a temperature shift at 37°C, fractioned by electrophoresis, transferred onto a membrane and hybridized with various probes (Figure 5). While the level of control spliceosomal U4 RNA synthesized by RNA polymerase II was unaffected by the temperature shift (Figure 5, lanes 1–4), that of 5′- and 3′-processed but unspliced precursor of tRNAPro was drastically decreased in mutant cells, as expected for a Pol III transcript (Figure 5, compare lanes 2 and 3, upper panel). The transcription defect was not apparent at the level of mature tRNAPro due to the stability of tRNAs (Figure 5, compare lanes 2 and 3, lower panel). Similarly, the precursor RPR1 RNA was undetectable and the mature RPR1 RNA level was reduced in the mutant cells grown at 37°C (Figure 5, compare lanes 2 and 3). The level of unspliced and mature RPR1 RNA was reduced even at permissive temperature (24°C) in the mutant cells although the same amount of total RNA was analyzed in these northern blot experiments (Figure 5, compare lanes 1 and 4). Similarly, the level of snR52 was reduced in the rpc160-112 cells grown at permissive temperature (Figure 5, compare lanes 1 and 4) and became nearly undetectable at the restrictive temperature (Figure 5, lane 3).
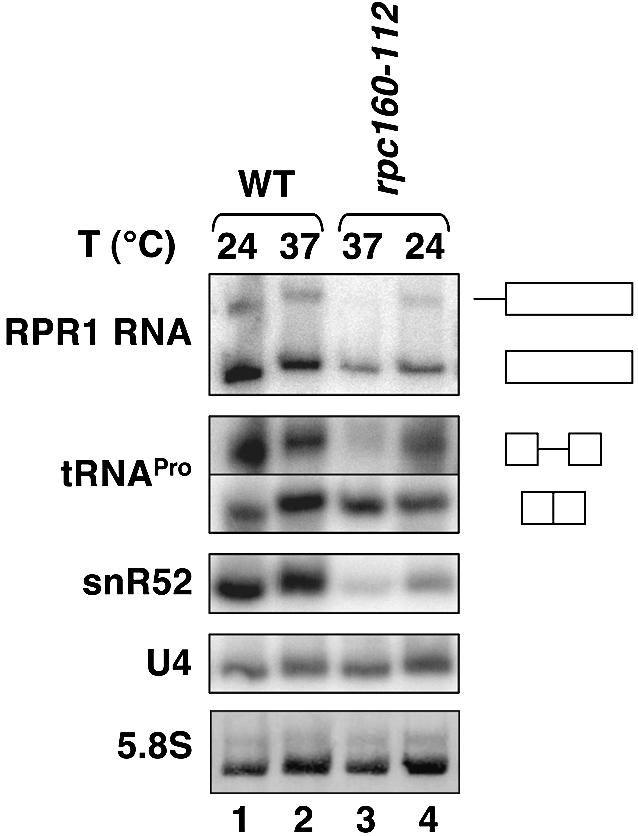
Fig. 5. Northern blot analysis of snR52 from wild-type and rpc160-112 cells. Wild-type (lanes 1 and 2) or rpc160-112 (lanes 3 and 4) cells were grown at 24°C (lanes 1 and 4) or shifted for 3 h at 37°C (lanes 2 and 3). RNA extraction, gel electrophoresis, electrophoretic transfer on nylon membrane, and hybridization with DNA probe were carried out as described in Materials and methods. The RNAs probes are indicated on the left. The precursor and mature RPR1 RNA, the 5′- and 3′-processed, unspliced precursor and mature tRNAPro are indicated on the right. As a loading control, the bottom panel shows a post-transfer methylene blue staining of 5.8S RNA on the membrane.
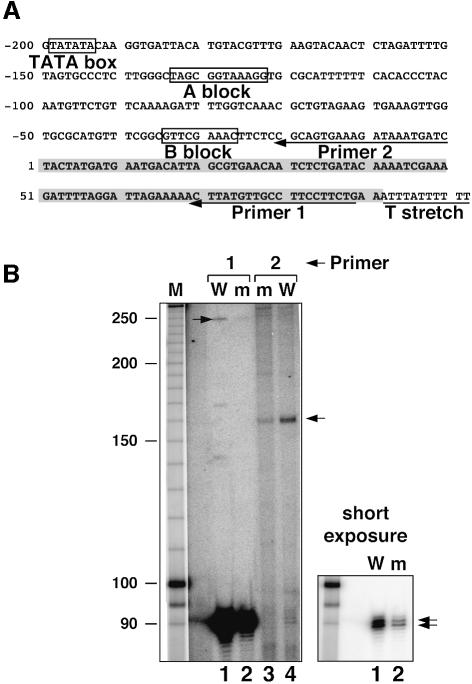
Interestingly, two transcripts of ∼90 (Figure 5) and ∼250 (data not shown) nucleotides were revealed by this northern blot analysis of the SNR52 gene product. The size of the most-abundant RNA was in good agreement with the predicted size of snR52 (92 bases long; see Figure 5; Lowe and Eddy, 1999). The second RNA band was present at a very low level (about 0.5% of the ∼90 nucleotide transcript) in the wild-type strain and was undetectable in the mutant strain (data not shown). To investigate whether this longer RNA band was a precursor of snR52, we performed a primer extension analysis using two probes. The first one (Primer 1, located 3 nucleotides from the 3′-end; see Figure 6A) will hybridize with the snR52 RNA (Lowe and Eddy, 1999) and the other one, located just upstream of the predicted mature snR52 5′-end (Primer 2; see Figure 6A), should hybridize with a putative precursor. Two DNA fragments of 89 and 90 bases long were observed with the 3′ probe and therefore corresponded to the predicted snR52 sequence (Lowe and Eddy, 1999). A second DNA fragment of ∼250 bases, 160 bases longer than the predicted sequence, was synthesized with the same primer (Figure 6B). PhosphorImager analysis indicated that this large fragment was ∼1000-fold less abundant than the 89–90 base fragments. The 90 and 250 base DNA fragments corresponded well to the small and large snR52 transcripts observed by northern blot. Finally, a DNA fragment of 160 bases was synthesized by extension of the second primer located just upstream of the predicted sequence (Figure 6B, lanes 3 and 4). These results suggested that snR52 is synthesized as a precursor RNA with a leader sequence of 160 bases that is efficiently cleaved to generate the 92–93 base mature snR52. We therefore looked for promoter elements in this putative leader sequence. Indeed, sequence analysis revealed a well-conserved A block element (one mismatch to the tRNA gene consensus) ∼26 bp downstream of the predicted 5′-end of the transcript, a perfect B block 87 bp downstream, and a TATA-like element ∼33 bp upstream of the predicted start region (see Figure 6A). In addition, as expected, the snR52 gene ends with a stretch of T residues, typical for Pol III termination sequences. Furthermore, a Ty1 LTR scar of a past transposition event is present about 110 bp upstream of the predicted start region, in good agreement with the known colocalization of retrotransposons with class III genes (Hani and Feldmann, 1998; Sandmeyer, 1998).
Fig. 6. SNR52 gene and RNA transcripts. (A) SNR52 gene sequence shows canonical class III promoter elements (boxed) in a leader sequence and a putative Pol III termination signal (underlined T stretch). The sequence of mature snR52 is labeled in gray (Lowe and Eddy, 1999); arrows underline the sequence of Primers 1 and 2 used for primer extension. (B) Primer extension analysis of SNR52 snoRNA and its precursor. Total RNA (3 µg) from YPH500 (W; lanes 1 and 4) or rpc160-112 mutant cells (m; lanes 2 and 3) grown at 24°C was reverse transcribed using Primer 1 (lanes 1 and 2) or Primer 2 (lanes 3 and 4).
Down regulation of Pol III and TFIIIB recruitment in stationary-phase cultures
Yeast Pol III transcription decreases markedly during late growth phase (Sethy et al., 1995; Clarke et al., 1996). We analyzed the occupancy of class III genes by τ95, Bdp1 and C160 during exponential, late-exponential and stationary growth phases to investigate on a genomic scale the basis of this regulation and whether all the genes would be regulated in a coordinated manner (see Materials and methods). To compare the three growth stages, we determined for each of 419 LEPTM the fold enrichment of the late-exponential and stationary phase relative to the exponential phase (Figure 7). The data were subjected to hierarchical clustering and visualized with TreeView (see Supplementary data). With the C160 bait, the 419 LEPTM were found to be uniformly and markedly less enriched during stationary phase relative to the exponential phase (Figure 7, compare lanes 9 with lanes 3 and 6). This low occupancy of class III genes by RNA polymerase III in stationary phase is in keeping with the general decrease in Pol III transcription. Most of the 419 LEPTM were also significantly less enriched in stationary phase with the Bdp1 bait. However, the decrease of Bdp1 binding was less pronounced than with the C160 bait (Figure 7, compare lane 6 with lane 9). Notably, some intergenic elements (59 of the 419 LEPTM analyzed) were equally enriched in stationary and exponential-phase cells. Most remarkably, with the τ95 bait, the LEPTM were either equally enriched in stationary and exponential phases (249 LEPTM) or their enrichment was much-less affected in stationary phase than in the case of C160 or Bdp1 (Figure 7, compare lane 3 with lanes 9 and 6). We noted that an intergenic element adjacent to RPR1 was one of eight loci that were equally occupied in exponential and stationary phases by the two transcription factors TFIIIC and TFIIIB, whereas Pol III binding was much decreased; there were also cases (51 LEPTM) where TFIIIC (τ95) binding decreased, whereas TFIIIB (Bdp1) binding did not (Figure 7, lower part). These results suggest that, in stationary-phase cells, Pol III transcription is globally regulated at the level of TFIIIB and Pol III recruitment while TFIIIC remains mostly bound to the genes.
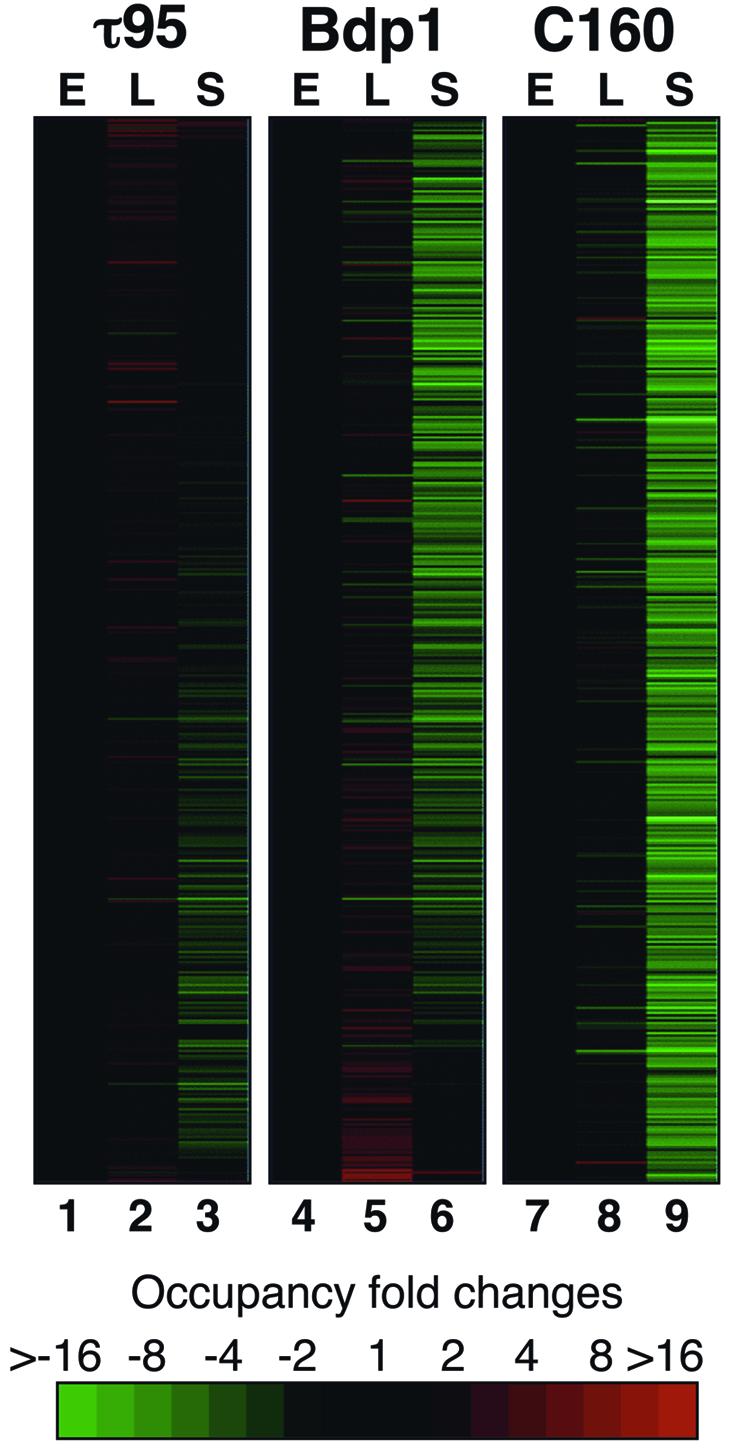
Fig. 7. Occupancy profiles of loci enriched by the Pol III transcription machinery (LEPTM) at different growth phases. Four hundred and nineteen LEPTM arranged in rows are classified according to their enrichment fold changes from exponential (columns E, normalized to 1) to late exponential (L) and stationary (S) growth phases for τ95, Bdp1 and C160 bait proteins. As indicated on the color scale, black means no changes whereas green and red indicate decreased and increased DNA binding levels, respectively.
Discussion
We have mapped the genomic binding sites for four components of the Pol III transcription apparatus which are each essential for TFIIIC, TFIIIB or Pol III activity. Based on the enrichment of intergenic regions co-immunopurified with epitope-tagged proteins, the colocalization of the four proteins targeted 94% of the tRNA genes and identified a new class III gene, SNR52. All the known class III genes and a relatively small set of additional loci were targeted by at least one component of the Pol III transcription machinery, which suggests an upper bound to the number of class III genes in S.cerevisiae.
The four components selected for this study are the largest subunit of Pol III (C160), two components of TFIIIB (Brf1 and Bdp1), and τ95, a TFIIIC subunit involved in A block binding. All four are part of multiprotein complexes and although none was shown to bind DNA by itself in vitro, each of them is thought to contact DNA during pre-initiation or transcription complex formation (Huet et al., 1997; Colbert et al., 1998; Persinger et al., 1999; Shah et al., 1999; Geiduschek and Kassavetis, 2001; Juo et al., 2003). Therefore, crosslinking of τ95 to genomic DNA can be considered as a signature for TFIIIC binding, crosslinking of Brf1, which initiates TFIIIB assembly, and Bdp1 that locks TFIIIB–DNA complex in its highly stable form both witness TFIIIB binding, and crosslinking of C160 indicates transcription complex formation or elongation. In the clearest case where the four components colocalized (i.e. enrich the same intergenic regions), we infer that the genes were transcribed by Pol III in a TFIIIC- and TFIIIB-dependent manner. This was the case for 259 tRNA genes (out of 275) and three additional class III genes SNR6, RPR1 and SCR1. It is interesting to note that the vast majority of the redundant tRNA genes were found to be transcriptionally active under active growth conditions (exponential phase in rich medium). Together with the observation that there is a good correlation between tRNA gene redundancy (copy number) and the overall amino acid composition of yeast proteins, this result confirms the assumption that intracellular levels of tRNA in growing yeast cells are mainly determined by gene copy number (Percudani et al., 1997).
Using the same criteria, six other loci targeted by the four components were strong class III gene candidates. Among them is SNR52, which encodes an snoRNA, snR52, required for 2′-O-methylation of rRNA (Lowe and Eddy, 1999). The observation that snR52 RNA level was severely reduced in a Pol III mutant provided further evidence that SNR52 is a class III gene. Sequence analysis revealed the presence of a T-rich termination site at the end of the gene and well conserved A and B blocks upstream of the snR52 sequence. This promoter configuration is reminiscent of that of the RPR1 gene that encodes the RNA subunit of RNase P (Lee et al., 1991a,b). RPR1 gene harbors a transcribed leader sequence with internal A and B block elements recognized by TFIIIC. Similarly, northern blotting and primer-extension experiments revealed a mature form of snR52 of the expected length (90 nt) and a minor version of ∼250 nt that encompasses the putative A and a B block elements. The very low level of the long RNA species suggests a very efficient maturation process. Despite the presence of tRNA gene-like promoter elements, the 5′-leader regions of RPR1 and SNR52 cannot be folded into a cloverleaf structure characteristic of tRNAs, and did not share other sequence motifs. It would be interesting to know whether the precursors of these two RNA species are processed by the same mechanism. As the function of snR52 is to serve as a methylation guide for rRNA (Lowe and Eddy, 1999), the activity of Pol III is therefore important for rRNA modification. This observation is reminiscent of studies that reported some cross-talks between transcription of class III genes and rRNA processing (Hermann-Le Denmat et al., 1994; Dechampesme et al., 1999; Briand et al., 2001).
Northern blot experiments on the five remaining loci targeted by the four protein baits did not reveal any detectable RNA species. These negative results are intriguing. Formally, they do not exclude that these loci recruit the Pol III machinery unproductively, or that the transcripts are much-less abundant than usual class III RNAs and remained undetected. Alternatively, these few loci could be false positives. Decreasing the criteria stringency was nevertheless warranted by the observation that some bona fide class III genes were detectably targeted by only one, two or three of the four components (see Table I). Considering the 189 loci that were enriched with one, two or three protein baits, 105 mapped within 1500 bp from a tRNA gene and therefore were assigned to these genes since they mapped within the limit of resolution of the method. This assignment was reasonably validated by the recovery of 14 missing tRNA genes that were not targeted by the four components together (259 out of 275 tRNA genes). The decision on stringent thresholds was in part responsible for the incomplete recovery of all tRNA genes by the four protein baits but had the advantage of reducing false positives to a minimum. Therefore, one can expect that new class III genes will be identified among the 78 still unassigned loci that were selected by a subset of the transcription components. These 78 loci, which are scattered in the genome, appear to be an upper bound to the number of undiscovered class III genes in yeast. This upper bound may have to be reconsidered since additional intergenic (inter-ORF) regions were recently identified by sequence comparison of close yeast species (Kellis et al., 2003). Considering that the 110/189 ratio of bona fide class III genes among the loci enriched by three or less baits parallels the ratio of seven class III genes among the 12 loci selected by the four components (that include five false positives), one may suspect, however, that the number of new class III genes yet to be discovered will be very limited.
A few tRNA genes did not bind TFIIIC detectably, which raises the question of the general requirement for TFIIIC in Pol III transcription. Some tRNA genes and SNR6 have a TATA box that directs TFIIIC-independent transcription in vitro (Dieci et al., 2000). TFIIIB assembly in that case is initiated by TBP binding (Whitehall et al., 1995). However, TFIIIC was shown to be required for SNR6 transcription in vivo (Burnol et al., 1993b; Gerlach et al., 1995) and in vitro on chromatin templates (Burnol et al., 1993a; Eschenlauer et al., 1993). In the present experiments, the four tRNA genes with a TATA box (Dieci et al., 2000) were found to be targeted by TFIIIC (see Figure 4B). The lack of significant binding of τ95 to the 5S RNA genes was unexpected since there is also a clear requirement for TFIIIC for 5S RNA transcription in vitro and in vivo (Lefebvre et al., 1994). On 5S RNA gene, the oriented binding of TFIIIC is directed by DNA-bound TFIIIA. No sequence-specific binding site for TFIIIC alone has been detected (Braun et al., 1989; Challice and Segall, 1989). It is possible that TFIIIA binding may have reduced τ95 crosslinking efficiency under the threshold value. Therefore, in yeast, until a counter example is clearly identified, TFIIIC appears to be a general transcription factor for class III genes. The above considerations concerning TFIIIC apply to Brf1 and Bdp1. There are cases of bona fide class III genes that bound only one of these TFIIIB components or none of them (RDN5 and RNA170; Table I). However, these genes are transcribed in vitro by a TFIIIB-dependent mechanism and the in vivo synthesis of RNA170 is affected in a brf1 mutant strain (Kassavetis et al., 1990; Olivas et al., 1997). The discrepancies illustrate the complexity of interpreting negative results and are likely related to our selection of stringent thresholds. On the other hand, beside a role in transcription, the possibility remains that unproductive binding of some Pol III transcription component may have some local regulatory role. This could well be the case for TFIIIC, which can influence the local chromatin state (Marsolier et al., 1995).
The ChIP on chip technique provides a global, though indirect, analysis of the activation and transcription status of all the class III genes, which would be difficult to reach by northern blot or priming extension analysis due to the number of genes and the redundancy of tRNA genes. This technique also allows a more precise evaluation of the in vivo DNA occupancy by the different components of the transcription machinery (TFIIIB, TFIIIC and Pol III) than in vivo footprinting. The present analysis revealed that Pol III and, to a lesser level Bdp1, dissociated during stationary phase whereas TFIIIC mainly remained bound. Early genome footprinting experiments revealed a near complete Dnase I protection of the upstream regions of the SUP53 tRNA3leu, from –40 to +15 in exponentially growing cells and from –40 to –10 in stationary phase cultures (Huibregtse and Engelke, 1989). Interpreted in terms of TFIIIB and Pol III binding (Kassavetis et al., 1990; Bartholomew et al., 1993), these observations suggest the loss of Pol III but the persistence of TFIIIB binding in stationary phase. The present analysis confirms and extends the loss of Pol III to all class III genes but suggests instead a global drop of TFIIIB binding on the majority of the genes. It is interesting that, while the assembly of Pol III transcription complexes goes stepwise from TFIIIC binding to TFIIIB assembly then Pol III recruitment, the tightness of the regulation appears to be in reverse order, essentially at the level of Pol III recruitment, less so on TFIIIB binding and not detectably on TFIIIC binding. It is probably of interest for the cells to maintain a partial level of gene activation with bound TFIIIC and some TFIIIB binding in order to rapidly resume transcription when growth conditions improve.
Materials and methods
Yeast strains, growth conditions and construction of epitope-tagged strains
The 3HA-C160 (MW671) strain has been described elsewhere (Dieci et al., 1995). All strains were derived from YPH499 or YPH500 strains (Sikorski and Hieter, 1989). 3 HA- or 13 myc-tagged strains were constructed as described previously Longtine et al. (1998). The epitope-coding sequence was inserted between the last codon and the stop codon of the target ORF. The proteins Brf1, Bdp1 and τ95 were tagged, respectively, by 3 HA-, 3 HA- and 13 myc-epitope (in the MW4034, MW4035 and yOH1 strains, respectively). No growth defect was detected for the Bdp1-HA- and τ95-myc-tagged strains. The Brf1-HA-tagged strain grew normally at 24°C, but its growth was reduced at 30°C compared with the wild-type strain and stopped at 37°C. Thus, it was grown at 24°C instead of 30°C for the other strains. Expression of HA- and myc-tagged proteins was verified by western blot analysis using the 12CA5 (anti-HA) and 9E10 (anti-myc) mouse monoclonal antibodies (both a gift from the DRIP, CEA/Saclay). The wild-type and rpc160-112 strains used in northern blot and primer-extension experiments are YPH500 and MW670, respectively (Sikorski and Hieter, 1989; Dieci et al., 1995).
To analyze the binding of the Pol III transcription machinery at different growth phases, cells from the τ95-myc-, Bdp1-HA- or C160-HA-tagged strains were harvested from YPD medium at ∼0.7 OD600 (exponential phase), ∼4.8 OD600 (late exponential phase) and ∼10 OD600 (stationary phase).
Northern blotting and primer extension
RNA extraction and northern blot analysis were performed as previously described (Burnol et al., 1993b) using body-labeled DNA fragments encompassing the sequence of the probed RNA. Quantifications were performed using PhosphorImager apparatus and software (Amersham Pharmacia).
Primer extension was performed using 3 µg of RNA as described previously (Prioleau et al., 1994). Yeast cells (25 ml cultures) were grown in YPD medium at 30°C to an OD of ∼0.7. After two washes with sterile water, total yeast RNA was purified (Burnol et al., 1993b) and primer extensions were performed with oligonucleotide primers specific for the snR52 mature sequence or the putative leader sequence of snR52. The sequences of the oligonucleotides used are as follows: Primer 1, 5′-CAGAAGGAAGGCAACATAAGT-3′; Primer 2: 5′-GATCATTTATC TTTCACTGCGG-3′.
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We would like to thank Pascale Voisin and Amandine Pitaval for technical support. We are grateful to Pierre Thuriaux for helpful discussions, to Christian Marck for help in class III promoter elements analysis and to Giorgio Dieci (University of Parma) for kindly providing unpublished data. We wish to thank Jean-Pierre Bachellerie (CNRS, Université Paul-Sabatier, Toulouse) for stimulating comments and information on snR52. This work was supported in part by a PRFMMIP grant from the Ministère de la Recherche et de la Technologie, a grant from HFSPO and from the Association pour la recherche contre le cancer (ARC).
References
- Andrau J.-C., Sentenac,A. and Werner,M. (1999) Mutagenesis of yeast TFIIIB70 reveals C-terminal residues critical for interaction with TBP and C34. J. Mol. Biol., 288, 511–520. [DOI] [PubMed] [Google Scholar]
- Bartholomew B., Kassavetis,G.A., Braun,B.B. and Geiduschek. (1990) The subunit structure of Saccharomyces cerevisiae transcription factor IIIC probed with a novel photocrosslinking reagent. EMBO J., 9, 2197–2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartholomew B., Kassavetis,G.A. and Geiduschek,E.P. (1991) Two components of Saccharomyces cerevisiae transcription factor IIIB (TFIIIB) are stereospecifically located upstream of a tRNA gene and interact with the second-largest subunit of TFIIIC. Mol. Cell. Biol., 11, 5181–5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartholomew B., Durkovich,D., Kassavetis,G.A. and Geiduschek,E.P. (1993) Orientation and topography of RNA polymerase III in transcription complexes. Mol. Cell. Biol., 13, 942–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun B.R., Riggs,D.L., Kassavetis,G.A. and Geiduschek,E.P. (1989) Multiple states of protein–DNA interaction in the assembly of transcription complexes on Saccharomyces cerevisiae 5S ribosomal RNA genes. Proc. Natl Acad. Sci. USA, 86, 2530–2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun B.R., Bartholomew,B., Kassavetis,G.A. and Geiduschek,E.P. (1992) Topography of transcription factor complexes on the Saccharomyces cerevisiae 5S RNA gene. J. Mol. Biol., 228, 1063–1077. [DOI] [PubMed] [Google Scholar]
- Briand J.F., Navarro,F., Gadal,O. and Thuriaux,P. (2001) Cross talk between tRNA and rRNA synthesis in Saccharomyces cerevisiae. Mol. Cell. Biol., 21, 189–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brow D.A. and Guthrie,C. (1990) Transcription of a yeast U6 snRNA gene requires a polymerase III promoter element in a novel position. Genes Dev., 4, 1345–1356. [DOI] [PubMed] [Google Scholar]
- Brun I., Sentenac,A. and Werner,M. (1997) Dual role of the C34 subunit of RNA polymerase III in transcription initiation. EMBO J., 16, 5730–5741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnol A.-F., Margottin,F., Huet,J., Almouzni,G., Prioleau,M.-N., Méchali,M. and Sentenac,A. (1993a) TFIIIC relieves repression of U6 snRNA transcription by chromatin. Nature, 362, 475–477. [DOI] [PubMed] [Google Scholar]
- Burnol A.-F., Margottin,F., Schultz,P., Marsolier,M.-C., Oudet,P. and Sentenac,A. (1993b) Basal promoter and enhancer element of yeast U6 snRNA gene. J. Mol. Biol., 233, 644–658. [DOI] [PubMed] [Google Scholar]
- Challice J.M. and Segall,J. (1989) Transcription of the 5S rRNA gene of Saccharomyces cerevisiae requires a promoter element at +1 and a 14-base pair internal control region. J. Biol. Chem., 264, 20060–20067. [PubMed] [Google Scholar]
- Chédin S., Ferri,L., Peyroche,G., Andrau,J.-C., Jourdain,S., Lefebvre,O., Werner,M., Carles,C. and Sentenac,A. (1998) The Yeast RNA Polymerase III Transcription Machinery. A Paradigm for Eukaryotic Gene Activation. Vol. 63. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 381–389. [DOI] [PubMed] [Google Scholar]
- Clarke E.M., Peterson,C.L., Brainard,A.V. and Riggs,D.L. (1996) Regulation of the RNA polymerase I and III transcription systems in response to growth conditions. J. Biol. Chem., 271, 22189–22195. [DOI] [PubMed] [Google Scholar]
- Colbert T., Lee,S., Schimmack,G. and Hahn,S. (1998) Architecture of protein and DNA contacts within the TFIIIB–DNA complex. Mol. Cell. Biol., 18, 1682–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conesa C., Swanson,R., Schultz,P., Oudet,P. and Sentenac,A. (1993) On the subunit composition, stoichiometry and phosphorylation of the yeast transcription factor TFIIC/τ. J. Biol. Chem., 268, 18047–18052. [PubMed] [Google Scholar]
- Dechampesme A.M., Koroleva,O., Leger-Silvestre,I., Gas,N. and Camier,S. (1999) Assembly of 5S ribosomal RNA is required at a specific step of the pre-rRNA processing pathway. J. Cell Biol., 145, 1369–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieci G., Hermann-Le Denmat,S., Lukhtanov,E., Thuriaux,P., Werner,M. and Sentenac,A. (1995) A universally conserved region of the largest subunit participates in the active site of RNA polymerase III. EMBO J., 14, 3766–3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieci G., Percudani,R., Giuliodori,S., Bottarelli,L. and Ottonello,S. (2000) TFIIIC-independent in vitro transcription of yeast tRNA genes. J. Mol. Biol., 299, 601–613. [DOI] [PubMed] [Google Scholar]
- Dieci G., Giuliodori,S., Catellani,M., Percudani,R. and Ottonello,S. (2002) Intragenic promoter adaptation and facilitated RNA polymerase III recycling in the transcription of SCR1, the 7SL RNA gene of Saccharomyces cerevisiae. J. Biol. Chem., 277, 6903–6914. [DOI] [PubMed] [Google Scholar]
- Eschenlauer J.B., Kaiser,M.W., Gerlach,V.L. and Brow,D.A. (1993) Architecture of a yeast U6 RNA gene promoter. Mol. Cell. Biol., 13, 3015–3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferri M.L., Peyroche,G., Siaut,M., Lefebvre,O., Carles,C., Conesa,C. and Sentenac,A. (2000) A novel subunit of yeast RNA polymerase III interacts with the TFIIB-related domain of TFIIIB70. Mol. Cell. Biol., 20, 488–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiduschek E.P. and Kassavetis,G.A. (2001) The RNA polymerase III transcription apparatus. J. Mol. Biol., 310, 1–26. [DOI] [PubMed] [Google Scholar]
- Gerlach V.L., Whitehall,S.K., Geiduschek,E.P. and Brow,D.A. (1995) TFIIIB placement on a yeast U6 RNA gene in vivo is directed primarily by TFIIIC rather than by sequence-specific DNA contacts. Mol. Cell. Biol., 15, 1455–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hani J. and Feldmann,H. (1998) tRNA genes and retroelement in the yeast genome. Nucleic Acids Res., 26, 689–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht A. and Grunstein,M. (1999) Mapping DNA interaction sites of chromosomal proteins using immunoprecipitation and polymerase chain reaction. Methods Enzymol., 304, 399–414. [DOI] [PubMed] [Google Scholar]
- Hecht A., Strahl-Bolsinger,S. and Grunstein,M. (1999) Mapping DNA interaction sites of chromosomal proteins. Crosslinking studies in yeast. Methods Mol. Biol., 119, 469–479. [DOI] [PubMed] [Google Scholar]
- Hermann-Le Denmat S., Werner,M., Sentenac,A. and Thuriaux,P. (1994) Suppression of yeast RNA polymerase III mutations by FHL1, a gene coding for a fork head protein involved in rRNA processing. Mol. Cell. Biol., 14, 2905–2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huet J., Conesa,C., Carles,C. and Sentenac,A. (1997) A cryptic DNA binding domain at the C-terminus of TFIIIB70 affects formation, stability and function of preinitiation complexes. J. Biol. Chem., 272, 18341–18349. [DOI] [PubMed] [Google Scholar]
- Huibregtse J.M. and Engelke,D.R. (1989) Genomic footprinting of a yeast tRNA gene reveals stable complexes over the 5′-flanking region. Mol. Cell. Biol., 9, 3244–3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer V.R., Horak,C.E., Scafe,C.S., Botstein,D., Snyder,M. and Brown,P.O. (2001) Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature, 409, 533–538. [DOI] [PubMed] [Google Scholar]
- Jourdain S., Acker,J., Ducrot,C., Sentenac,A. and Lefebvre,O. (2003) The tau95 subunit of yeast TFIIIC influences upstream and downstream functions of TFIIIC.DNA complexes. J. Biol. Chem., 278, 10450–10457. [DOI] [PubMed] [Google Scholar]
- Juo Z.S., Kassavetis,G.A., Wang,J., Geiduschek,E.P. and Sigler,P.B. (2003) Crystal structure of a transcription factor IIIB core interface ternary complex. Nature, 422, 534–539. [DOI] [PubMed] [Google Scholar]
- Kassavetis G.A., Braun,B.R., Nguyen,L.H. and Geiduschek,E.P. (1990) S. cerevisiae TFIIIB is the transcription initiation factor proper of RNA polymerase III, while TFIIIA and TFIIIC are assembly factors. Cell, 60, 247–257. [DOI] [PubMed] [Google Scholar]
- Kassavetis G.A., Joazeiro,C.A.P., Pisano,M., Geiduschek,E.P., Colbert,T., Hahn,S. and Blanco,J.A. (1992) The role of the TATA-binding protein in the assembly and function of the multisubunit yeast RNA polymerase III transcription factor, TFIIIB. Cell, 71, 1055–1064. [DOI] [PubMed] [Google Scholar]
- Kassavetis G.A., Barbeleben,C., Kumar,A., Ramirez,E. and Geiduschek,E.P. (1997) Domains of the Brf component of RNA polymerase III transcription factor III (TFIIIB): functions in assembly of TFIIIB–DNA complexes and recruitment of RNA polymerase to the promoter. Mol. Cell. Biol., 17, 5299–5306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassavetis G.A., Kumar,A., Letts,G.A. and Geiduschek,E.P. (1998) A post-recruitment function for the RNA polymerase III transcription-initiation factor IIIB. Proc. Natl Acad. Sci. USA, 95, 9196–9201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellis M., Patterson,N., Endrizzi,M., Birren,B. and Lander,E.S. (2003) Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature, 423, 241–254. [DOI] [PubMed] [Google Scholar]
- Kumar A., Kassavetis,G.A., Geiduschek,E.P., Hambalko,M. and Brent,C.J. (1997) Functional dissection of the B” component of RNA polymerase III transcription factor IIIB: a scaffolding protein with multiple roles in assembly and initiation of transcription. Mol. Cell. Biol., 17, 1868–1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.-Y., Evans,C.F. and Engelke,D.R. (1991a) Expression of RNase P RNA in Saccharomyces cerevisiae is controlled by an unusual RNA polymerase III promoter. Proc. Natl Acad. Sci. USA, 88, 6986–6990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.Y., Rohlman,C.E., Molony,L.A. and Engelke,D.R. (1991b) Characterization of RPR1, an essential gene encoding the RNA component of Saccharomyces cerevisiae nuclear RNase P. Mol. Cell. Biol., 11, 721–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre O., Rüth,J. and Sentenac,A. (1994) A mutation in the largest subunit of yeast TFIIIC affects tRNA and 5S RNA synthesis. J. Biol. Chem., 269, 23374–23381. [PubMed] [Google Scholar]
- Lieb J.D., Liu,X., Botstein,D. and Brown,P.O. (2001) Promoter-specific binding of Rap1 revealed by genome-wide maps of protein–DNA association. Nat. Genet., 28, 327–334. [DOI] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie,A.,III, Demarini,D.J., Shah,N.G., Wach,A., Brachat,A., Philipsen,P. and Pringle,J.R. (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast, 14, 953–961. [DOI] [PubMed] [Google Scholar]
- Lowe T.M. and Eddy,S.R. (1999) A computational screen for methylation guide snoRNAs in yeast. Science, 283, 1168–1171. [DOI] [PubMed] [Google Scholar]
- Margottin F., Dujardin,G., Gérard,M., Egly,J.-M., Huet,J. and Sentenac,A. (1991) Participation of the TATA factor in transcription of the yeast U6 gene by RNA polymerase C. Science, 251, 424–426. [DOI] [PubMed] [Google Scholar]
- Marsolier M.-C., Tanaka,S., Livingstone-Zatchej,M., Grunstein,M., Thoma,F. and Sentenac,A. (1995) Reciprocal interferences between nucleosomal organization and transcriptional activation of the yeast SNR6 gene. Genes Dev., 9, 410–422. [DOI] [PubMed] [Google Scholar]
- Marzouki N., Camier,S., Ruet,A., Moenne,A. and Sentenac,A. (1986) Selective proteolysis defines two DNA binding domains in yeast transcription factor tau. Nature, 323, 176–178. [DOI] [PubMed] [Google Scholar]
- Olivas W.M., Muhlrad,D. and Parker,R. (1997) Analysis of the yeast genome: identification of new non-coding and small ORF-containing RNAs. Nucleic Acids Res., 25, 4619–4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percudani R., Pavesi,A. and Ottonello,S. (1997) Transfer RNA gene redundancy and translational selection in Saccharomyces cerevisiae. J. Mol. Biol., 268, 322–330. [DOI] [PubMed] [Google Scholar]
- Persinger J., Sengupta,S.M. and Bartholomew,B. (1999) Spatial organization of the core region of yeast TFIIIB–DNA complexes. Mol. Cell. Biol., 19, 5218–5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prioleau M.-N., Huet,J., Sentenac,A. and Méchali,M. (1994) Competition between chromatin and transcription complex assembly regulates gene expression during early development. Cell, 77, 439–449. [DOI] [PubMed] [Google Scholar]
- Ren B. et al. (2000) Genome-wide location and function of DNA binding proteins. Science, 290, 2306–2309. [DOI] [PubMed] [Google Scholar]
- Sandmeyer S. (1998) Targeting transposition: at home in the genome. Genome Res., 8, 416–418. [DOI] [PubMed] [Google Scholar]
- Schramm L. and Hernandez,N. (2002) Recruitment of RNA polymerase III to its target promoters. Genes Dev., 16, 2593–2620. [DOI] [PubMed] [Google Scholar]
- Sentenac A. (1985) Eukaryotic RNA polymerases. CRC Crit. Rev., 18, 31–91. [DOI] [PubMed] [Google Scholar]
- Sethy I., Moir,R.D., Librizzi,M. and Willis,I.M. (1995) In vitro evidence for growth regulation of tRNA gene transcription in yeast: a role for transcription factor (TF) IIIB70 and TFIIIC. J. Biol. Chem., 270, 28463–28470. [DOI] [PubMed] [Google Scholar]
- Shah S.M., Kumar,A., Geiduschek,E.P. and Kassavetis,G.A. (1999) Alignment of the B” subunit of RNA polymerase III transcription factor IIIB in its promoter complex. J. Biol. Chem., 274, 28736–28744. [DOI] [PubMed] [Google Scholar]
- Sikorski R.S. and Hieter,P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics, 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner M., Chaussivert,N., Willis,I.M. and Sentenac,A. (1993) Interaction between a complex of RNA polymerase III subunits and the 70-kDa component of transcription factor IIIB. J. Biol. Chem., 268, 20721–20724. [PubMed] [Google Scholar]
- White R.J. (1998) RNA Polymerase III Transcription. Springer-Verlag, Berlin, Germany. [Google Scholar]
- Whitehall S.K., Kassavetis,G.A. and Geiduschek,E.P. (1995) The symmetry of the yeast U6 RNA gene’s TATA box and the orientation of the TATA-binding protein in yeast TFIIIB. Genes Dev., 9, 2974–2985. [DOI] [PubMed] [Google Scholar]
- Willis I.M. (1993) RNA polymerase III: genes, factors and transcriptional specificity. Eur. J. Biochem., 212, 1–11. [DOI] [PubMed] [Google Scholar]