Abstract
HHR23A, a protein implicated in nucleotide excision repair, belongs to a class of proteins containing both a ubiquitin-like (Ubl) domain and one or more ubiquitin-associated (UBA) domains, suggesting a role in the ubiquitin–proteasome pathway as well. The Ubl domain binds with high affinity to the second ubiquitin-interacting motif (UIM) of the S5a subunit of the proteasome. Here we present the solution structures of the HHR23A Ubl domain, the second UIM of S5a (UIM-2), and the Ubl:S5a–UIM-2 complex. The HHR23A Ubl domain is structurally similar to ubiquitin. The S5a UIM forms an α-helix with an unexpected hairpin loop that contributes to the binding interface with Ubl. The molecular determinants of the Ubl–proteasome interaction are revealed by analysis of the structures, chemical shift mapping, mutant binding studies and sequence conservation.
Keywords: NMR/protein degradation/Rad23/S5a/Ubl
Introduction
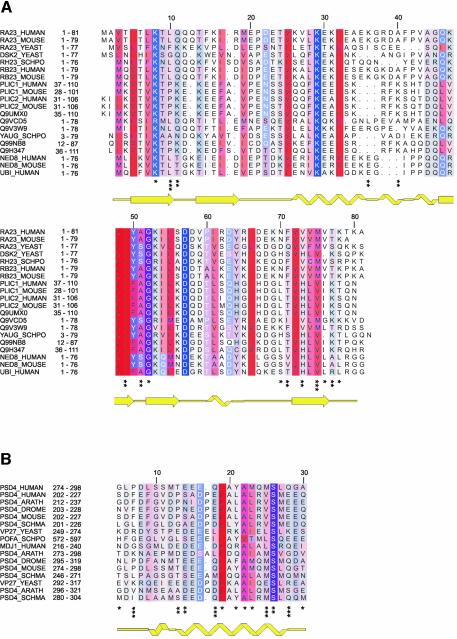
Recently, a class of proteins containing an N-terminal ubiquitin-like (Ubl) domain has been implicated in the regulation of proteolysis (Schauber et al., 1998; Hiyama et al., 1999; Kleijnen et al., 2000; Elsasser et al., 2002; Funakoshi et al., 2002; Rao and Sastry, 2002). A member of this superfamily, Saccharomyces cerevisiae Rad23 was first identified for its role in DNA repair (Haynes and Kunz, 1981). Rad23– mutants have an increased sensitivity to UV light, which can be explained by the fact that Rad23 interacts with Rad4 in the assembly of the DNA repair complex (Guzder et al., 1998). For the human homolog of Rad23, two proteins HHR23A (363 residues) and HHR23B (409 residues) exist, which share an identity of 59% on the amino acid level. Differences in function between the two homologs are unclear (Sugasawa et al., 1997). Sequence analysis of Rad23 proteins reveals a modular architecture with at least four distinct domains. The N-terminal 80 residues can be identified as a ubiquitin-like domain (Ubl), and the second and fourth domains are ubiquitin-associated domains (UBA) (Hofmann and Bucher, 1996), which flank a region identified as a binding site for the nucleotide excision repair XPC protein (Masutani et al., 1997). The UBA domains of several proteins, including those of Rad23/HHR23A, have been shown to bind to monomeric ubiquitin as well as to multiubiquitin chains, implying a regulatory function in proteasomal degradation (Ortolan et al., 2000; Bertolaet et al., 2001; Chen et al., 2001; Clarke et al., 2001; Wilkinson et al., 2001; Elder et al., 2002), although this remains controversial. Several groups have shown that multiubiquitin chains and not ubiquitin are probably the physiological target for UBA domains (Wilkinson et al., 2001; Funakoshi et al., 2002; Rao and Sastry, 2002; Raasi and Pickart, 2003). Our previous structure analysis of the UBA domains of HHR23A revealed a compact three-helix bundle with a conserved hydrophobic patch that probably represents the binding site for ubiquitin (Dieckmann et al., 1998; Withers-Ward et al., 2000; Mueller and Feigon, 2002).
The Ubl domain of HHR23A belongs to the class of type 2 Ubl domains that cannot be conjugated to target proteins and thus function as a linear non-cleavable ubiquitin fusion, in contrast to the type 1 small ubiqui tin-like modifiers such as SUMO and Nedd8. The N-terminal Ubl domain of HHR23A has been shown to bind to the polyubiquitin (polyUb) binding site in the proteasomal subunit S5a with high affinity (Hiyama et al., 1999). Since deletion of the Ubl domain is associated with increased sensitivity to UV light, a link between the proteasomal interaction and DNA repair was suggested (Schauber et al., 1998; van Laar et al., 2002). Based on the combination of UBA domains that can bind polyUb and a Ubl domain that can directly interact with the proteasome, a shuttle function for Rad23-like proteins in protein degradation was proposed (Chen and Madura, 2002; van Laar et al., 2002; Hartmann-Petersen et al., 2003). However, other proteins, e.g. 3-methyladenine DNA glycosylase and Png deglycosylating enzyme, can interact directly with the UBA domain of HHR23A (Miao et al., 2000; Suzuki et al., 2001), indicating that HHR23A/Rad23 might also have a function in ubiquitin-independent protein degradation via direct targeting of these proteins to the proteasome.
Two ubiquitin-interacting motifs (UIMs) of human S5a were originally identified in vitro as binding regions for polyUb, and were consequently named polyUb binding sites PUbS1 (UIM-1) and PUbS2 (UIM-2) (Young et al., 1998). The UIM domain was subsequently shown to be present in a wide variety of proteins in the proteasomal degradation pathway, as well as in proteins important for endocytosis, signaling adapters and several proteins that are responsible for neurological disorders (Hofmann and Falquet, 2001). The occurrence of the UIM motif in this large variety of proteins with different functions suggests that UIM-containing proteins might also regulate other functions than just proteasomal degradation. As is the case for human S5a, UIM-containing proteins often contain two or three of these motifs. The small size of the UIM (17 residues, PFAM database; http://pfam.wustl.edu) and the high level of amino acid sequence conservation suggest identical functions and binding properties at first sight. However, results from several groups indicate that multiple UIMs within one protein often exhibit different binding properties or targets (Young et al., 1998; Hiyama et al., 1999; Polo et al., 2002). Although both UIM regions of human S5a bind to polyUb in vitro, UIM-1 has a 10-fold lower affinity for polyUb chains compared with UIM-2 (Young et al., 1998). Similarly, the Ubl domain of HHR23A selectively binds with high affinity to UIM-2 of human S5a (Hiyama et al., 1999). Therefore the two UIM domains in human proteasomal subunit S5a might serve different biological functions. Chemical shift mapping of full-length S5a on the structures of the Ubl domains of hPLIC, SUMO, and a model of HHR23A along with a model α-helix of a minimal UIM-2 of S5a, have been used to generate a simple electrostatic model for binding specificity (Walters et al., 2002).
In order to provide further structural insight into the role of the modular HHR23A protein in proteasome-dependent protein degradation and DNA repair, we have used NMR to investigate the structure of the HHR23A Ubl domain and its interaction with its S5a binding site, UIM-2. Here we present the structures of the free components HHR23A Ubl and S5a263–307, as well as the first structure of a UIM bound to a member of the ubiquitin family, the Ubl:S5a263–307 complex. Both the free and the bound UIM-2 form an α-helix that is stabilized by an unexpected N-terminal hairpin turn that contributes to the binding affinity. Analysis of the protein interface based on the structures, chemical shift mapping, mutant binding studies, and sequence conservation provides insights into the specificity of the interaction between the S5a UIM-2 and the Ubl domain.
Results
Structure of the ubiquitin-like domain of HHR23A
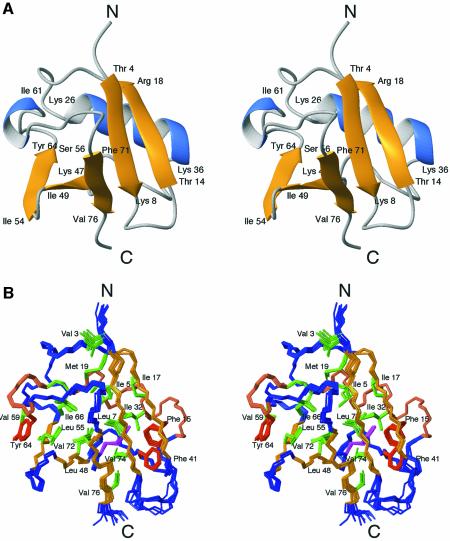
In order to understand how the Ubl domain of HHR23A (residues 1–87) specifically recognizes the S5a subunit (Hiyama et al., 1999), we determined its solution structure by NMR. One thousand six hundred and thirty Nuclear Overhauser effect (NOE)-derived distance restraints yielded a high-resolution structure with a root mean square deviation (r.m.s.d.) of 0.29 Å for the backbone and 0.90 Å for heavy atoms (residues 3–77) (Table I). Residues Thr4 to Lys78 form a compact, well defined structure exhibiting the grasp-fold observed for ubiquitin (Figure 1A and B) and ubiquitin-like proteins such as NEDD8 (Vijay-Kumar et al., 1987; Bayer et al., 1998; Rao-Naik et al., 1998; Whitby et al., 1998). The secondary structure elements are similar to those predicted by homology modeling of HHR23A Ubl using the hPLIC-2 Ubl structure (Walters et al., 2002). The N-terminus, Met1 to Val3, and the C-terminus, Thr79 to Ala87, are disordered. The structure of HHR23A Ubl contains a five-stranded β-sheet with strands β1 and β2 as well as strands β3, β4 and β5 running all-antiparallel, and with the strands β1 and β5 running parallel (Figure 1A and B). A long α-helix is located almost perpendicular to the β-sheet and both together form a compact hydrophobic core. A short 310-helix caps the hydrophobic core at one side of the β-sheet, similar to ubiquitin. Gln46 is buried inside the hydrophobic core, and its side-chain amide protons (which have an exchange lifetime in D2O of >3 months) form two hydrogen bonds with the backbone carbonyl groups of Lys29 and Phe41, thus stabilizing the conformation of the loop between the long α-helix and the strand β3. This glutamine is conserved throughout the ubiquitin family or replaced by a hydrophobic residue (Ile or Leu) (Figure 2A), which can meet the spatial requirements, consistent with its importance for the structural integrity of the fold.
Table I. Restraint and structural statistics for the structures of HHR23A Ubl and S5a263–307, and the complex of HHR23A Ubl and S5a263–307.
| |
Ubl1–78 |
S5a263–307 |
Ubl:S5a complexa |
|---|---|---|---|
| Distance restraints | |||
| Total | 1630 | 474 | 2066 |
| NOE-derived | |||
| Intra-residue | 703 | 169 | 834 |
| Sequential (|i – j| = 1) | 316 | 143 | 430 |
| Medium range (|i – j| ≤4) | 177 | 127 | 299 |
| Long range (|i – j| ≥5) | 413 | 13 | 388 |
| Intermolecular | – | – | 58 |
| Average NOE/residue | 20.6 | 10.0 | 18.2 |
| Hydrogen bonds | 21 | 22 | 67 |
| Torsion angle restraints | 39 | – | – |
| Structure statistics | |||
| R.m.s.d. from experimental distance restraints (Å) | 0.013 ± 0.0009 | 0.016 ± 0.002 | 0.019 ± 0.0008 |
| R.m.s.d. from idealized geometry | |||
| Bonds (Å) | 0.002 ± 0.0001 | 0.002 ± 0.0002 | 0.005 ± 0.0002 |
| Angles (deg.) | 0.39 ± 0.009 | 0.40 ± 0.015 | 0.60 ± 0.02 |
| Energies (kcal mol–1)b | |||
| Etotal | 101.0 ± 6.7 | 48.1 ±5.4 | 425.1 ± 19.4 |
| ENOE | 19.3 ± 2.7 | 8.8 ± 2.0 | 103.4 ± 9.3 |
| Ebonds | 6.3 ± 0.6 | 2.6 ± 0.5 | 37.8 ± 3.3 |
| Eangles | 55.4 ± 2.5 | 28.3 ± 2.1 | 178.5 ± 8.7 |
| EvdW | 15.1 ± 1.6 | 4.8 ± 1.4 | 83.9 ± 6.8 |
| Edihedral | 0.2 ± 0.2 | – | – |
| Eimproper | 4.8 ± 0.4 | 3.5 ± 0.5 | 21.5 ± 2.3 |
| R.m.s.d. from average structure (Å)c | |||
| Backbone atoms | 0.29 ± 0.08 | 1.15 ± 0.12 | 0.43 ± 0.07 |
| All heavy atoms | 0.90 ± 0.14 | 1.68 ± 0.21 | 0.93 ± 0.07 |
| Procheck analysisd | |||
| Residues in most favored region (%) | 73.0 | 81.1 | 69.7 |
| Residues in additional allowed region (%) | 21.4 | 10.0 | 23.8 |
| Residues in generously allowed region (%) | 4.2 | 7.8 | 6.0 |
| Residues in disallowed region (%) | 1.4 | 1.1 | 0.6 |
aOnly residues 270–301 of S5a were included in the structure calculation (see Materials and methods for details).
bEnergies were calculated with XPLOR 3.1 and force field parallhdg.pro version 5.0. The van der Waals energy term is a pure repulsive function, therefore vdW energies are positive.
cBackbone atoms are N, Cα and C. For the superposition and calculation of the r.m.s.ds, the following residue ranges were used: Ubl residues, 3–77; S5a263–307 residues, 278–295; complex Ubl:S5a residues, 3–77 of Ubl and residues 278–295 of S5a.
dRamachandran plot analysis was performed: free HHR23A Ubl residues 3–77, free S5a263–307 residues 278–295, complex HHR23A Ubl:S5a263–307 residues 3–77 (HHR23A Ubl) and 278–295 for S5a.
Fig. 1. Structure of the Ubl domain of HHR23A. Stereoviews of (A) a ribbon drawing and (B) an ensemble of the 11 structures of Ubl with the lowest overall and NOE energies. The boundaries of the secondary structure elements are indicated in (A). Side chains of residues contributing to the hydrophobic core are shown in green (hydrophobic), brown (aromatic) or magenta (polar).
Fig. 2. Amino acid sequence alignments of Ubl and UIM domains. (A) Amino acid sequence alignment of the Ubl domains. All proteins, with the exception of NEDD8 (NED8_HUMAN and NED8_MOUSE) and ubiquitin (UBI_HUMAN), share a similar architecture, consisting of a Ubl domain and one or more UBA domains. (B) Amino acid sequence alignment of UIMs found in a selection of proteins. The accession codes for the Swiss Prot database and amino acid numbering in the parent proteins are indicated on the left, and the secondary structures as determined for HHR23A Ubl (RA23_HUMAN) and human S5a263–307 (PSD4_HUMAN) are shown below the alignments. The degree of hydrophobicity of an amino acid position is indicated using a color ramp (red for hydrophobic residues and blue for charged amino acids). The intensity of the color is modulated by the degree of conservation of the residue position, with intense color indicating the most conserved residues. The loss of surface area for HHR23A Ubl upon binding to S5a263–307 is marked at the bottom (*≥10 Å2, **≥30 Å2, ***≥60 Å2), and for S5a263–307 upon binding to HHR23A Ubl (*≥20 Å2, **≥40 Å2, ***≥80 Å2).
All of the loops are well defined with the exception of the β1–β2 loop (Thr9 to Gln13), which is most likely due to lack of a sufficient number of restraints for these residues. The hydroxyl proton of Thr9 exhibits very slow exchange in D2O, indicating that it might be involved in a hydrogen bond, e.g. to Gln13 side-chain amide. The presence of NOEs from the side-chain hydroxyl proton of Thr9 to the amide protons of Thr9, Leu10, Gln11 and Gln12 indicate that the loop conformation is probably well defined. In ubiquitin, a threonine occupying the identical position (Thr7) forms a hydrogen bond via its hydroxyl group to both the amide and the hydroxyl group of Thr9 (Gln11 in HHR23A Ubl; Figure 2A).
Structure of the UIM of the proteasome subunit S5a
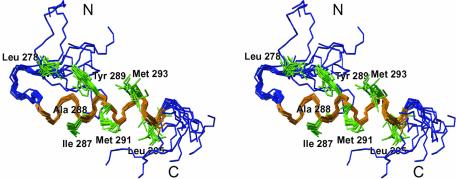
Human S5a contains two UIMs comprised of residues 210–227 (UIM-1) and 281–298 (UIM-2), which form two conserved hydrophobic stretches (Figure 2B). Secondary structure predictions as well as NMR studies of UIM-1 (PUbS1) of human S5a propose an α-helical conformation (Young et al., 1998; Hofmann and Falquet, 2001). A high resolution structure of UIM-2 of human S5a was determined using 13C-,15N-labeled S5a263–307, and standard double and triple resonance experiments (Figure 3; Table I). Structure calculations using 452 NOE-derived distance restraints yielded a well defined structure with an r.m.s.d. of 1.2 Å for the backbone atoms of residues Leu278 to Leu295 (Figure 3). Residues Glu283 to Leu295 form a single stable amphipathic α-helix. There were no indications of dimerization or oligomer assembly to stabilize this single helix. The N-terminal residues Met263 to Asp277 and the C-terminal residues Gln296 to Asp307 are not well defined by the NMR data, which likely reflects inherent disorder. Leu275, Pro276, Leu278 and Tyr289 form a small hydrophobic core, which leads to the formation of a hairpin structure for Leu278 to Thr282 (Figure 3). This hydrophobic core is well defined by 13 long-range NOEs. The hairpin structure is probably required to induce helix formation and to stabilize the helix starting from Glu283. The high degree of conservation for the negatively charged residues adjacent to the N-terminal end of the α-helix (position 10–12; Figure 2B) might be necessary due to the rules for N-capping of α-helices (Richardson and Richardson, 1988).
Fig. 3. Structure of human S5a UIM-2 (Met263 to Asp307). An ensemble of 10 structures is shown. The α-helix (E283 to L295) is colored orange, and the side chains of hydrophobic residues P277 to L295 are green.
Mapping the interaction sites for S5a263–307 and the Ubl domain of HHR23A
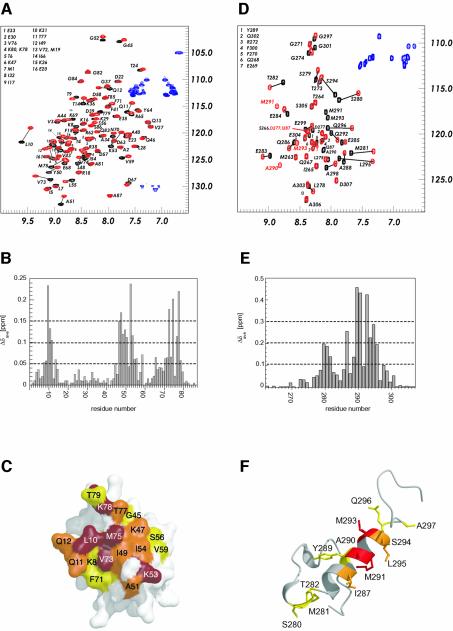
In order to map the binding sites of UIM-2 of S5a on the HHR23A Ubl domain, NMR chemical shift perturbation experiments were performed. 1H-15N HSQC spectra of 15N-labeled HHR23A Ubl were acquired as a function of added unlabeled S5a263–307. Additional small signals appeared upon addition of S5a263–307, indicating that the complex is in slow exchange on the NMR timescale. At an equimolar ratio of HHR23A Ubl and S5a peptide, only one set of correlations for the Ubl protein was observed (Figure 4A), clearly indicating that the stoichiometry of the interaction of HHR23A Ubl with S5a UIM-2 is 1:1. The results were mapped onto the structure of HHR23A Ubl (Figure 4B and C). The binding epitope is located on a hydrophobic patch formed by the five-stranded β-sheet. Based on the chemical shift mapping, the hydrophobic residues located on strands β1 (Leu10), β3 (Lys47, Leu48, Ile 9), β4 (Lys53, Ile54, Leu55), β5 (Val73 and Met75) and on the connecting loops (Gly45, Ala51, Ser56, Val59) as well as the non-conserved β1–β2 loop (Gln11 and Gln12) and the C-terminus (Lys78), are predicted to be involved in the binding to S5a263–307 (Figure 4B and C). Lys53 is the equivalent to Lys48 in ubiquitin, the major site for ubiquitin conjugation. The results for the mapping of the S5a263–307 binding site on HHR23A Ubl are in full agreement with those reported by Walters et al. (2002) using full-length S5a, which indicates that the isolated UIM-2 interacts with HHR23A Ubl using the same residues.
Fig. 4. Mapping the interaction sites for S5a263–307 and the Ubl domain of HHR23A. (A) 1H-15N HSQC spectra of 15N-labeled HHR23A Ubl in free (black contours) and bound (1:1 Ubl:S5a263–307) conformation (red contours). The contours of the side-chain amide protons of Asp and Glu residues in (A) and (D) are shown in blue. (B) Graph representing the changes in chemical shifts of the amide nitrogens and protons of HHR23A Ubl upon addition of an equimolar amount of S5a263–307. (C) Surface representation of HHR23A Ubl, color-coded by the changes in chemical shift upon binding to S5a263–307. Residues exhibiting a change in chemical shift of >0.15 p.p.m. are shown in red, 0.1 < x < 0.15 p.p.m. in orange, and 0.05 < x < 0.1 p.p.m. in yellow. (D) 1H-15N HSQC spectra of 15N-labeled S5a263–307 in free (black contours) and bound (1:1 Ubl:S5a263–307) conformation (red contours). (E) Graph representing the changes in chemical shifts of the amide nitrogens and protons of S5a263–307 upon addition of an equimolar amount of HHR23A Ubl. (F) Ribbon representation of the S5a263–307 color-coded by the changes in chemical shift upon binding HHR23A Ubl. Residues exhibiting a change in chemical shift of >0.3 p.p.m. are shown in red, 0.2 < x < 0.3 p.p.m. in orange, and 0.1 < x < 0.2 in yellow.
We also performed the reverse mapping study using 15N-labeled S5a263–307 peptide and unlabeled HHR23A Ubl protein in order to determine the binding site of the Ubl domain on S5a UIM-2. At an equimolar ratio of both components, a complete set of correlations for the backbone (47 correlations) and side-chain amides of S5a263–307 was observed, confirming that the stoichiometry of the interaction is indeed 1:1 (Figure 4D). The addition of HHR23A Ubl to the S5a sample leads to a large change in the chemical shifts of >25 residues, with the greatest changes >1 p.p.m. in the proton and ∼2 p.p.m. in the nitrogen dimension of the HSQC spectra. All residues of S5a263–307 that show changes in their chemical shifts upon addition of HHR23A Ubl are located in the α-helix or at the flanking sequences close to the helix (Figure 4E and F). The residues in the conserved hydrophobic patch Ile287-Ala288-Tyr289-Ala290-Met291 (the LALAL motif) are in the center of the binding epitope (Figure 4F). Ala290, Met291 and Met293 exhibit the largest change in chemical shift. In addition to the latter residues, which that are clustered within two turns of the helix, three residues (Ser280, Met281 and Thr282) that precede the conserved acidic amino acids in the first turn of the α-helix also exhibit smaller, but significant, changes in their chemical shifts. Leu278 and Ser279 in the hairpin are also affected by the binding to HHR23A Ubl, although their changes in chemical shift are below the significance level chosen here.
HHR23A Ubl binds the human S5a UIM-2 domain across the five-stranded β-sheet
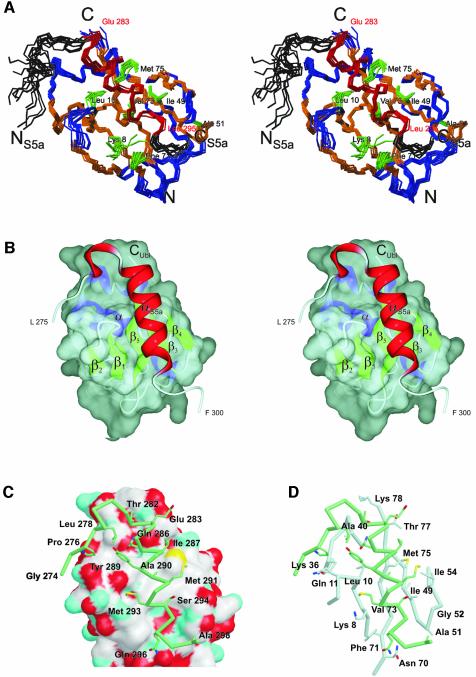
In order to understand the molecular basis for the interaction of UIM-2 of S5a263–307 and the Ubl domain of HHR23A, we determined the NMR solution structure of the complex (Figure 5A). Complete assignments for the backbone as well as the side-chain atoms of both protein domains in the complex were obtained by using NMR samples of 13C-,15N-labeled HHR23A Ubl in complex with unlabeled S5a263–307 peptide and vice versa. Whereas for HHR23A Ubl the absence of larger structural rearrangements upon binding to S5a was concluded from the fact that the chemical shifts for all residues far from the binding interface remained unchanged, this conclusion could not be drawn a priori for S5a263–307. The majority of the residues of S5a263–307 that form the helix exhibit drastic changes in their chemical shift in the bound conformation. However, almost all medium-range NOEs (of the type HαiHβi+3) defining the α-helix of S5a263–307 in its unbound conformation were also observed in the bound conformation. In addition, the NOEs between Leu278 and Tyr289 are also present for S5a263–307 when bound to HHR23A Ubl, indicating that the hairpin loop preceding the α-helix is retained in the structure of the complex. The protein–peptide interactions were defined by 58 unambiguous, unique intermolecular NOEs. Most NOEs observed were between the residues in the α-helix of S5a263–307, i.e. residues Gln286 to Gln296, and residues of HHR23A Ubl whose side chains are located on strands β1, β3, β4 and β5. The orientation and location of the α-helical S5a peptide could be determined by several ‘key interactions’: Tyr289 of S5a shows several NOEs to Leu10 of HHR23A Ubl, and Ile287 Hα exhibits NOEs to Met75 and Ile49 of HHR23A Ubl. The largest number of intermolecular NOEs was assigned between Met291 of S5a and Ile49 of HHR23A Ubl, which are in close proximity and form the center of the interface (Figure 5B and C). For the C-terminal end of the S5a helix, NOEs were identified between Leu295 of S5a and Ala51 and Gly52 in the β3–β4 loop, as well as Ile49, of HHR23A Ubl.
Fig. 5. Structure of the HHR23A Ubl:S5a UIM-2 complex. (A) Stereoviews of an ensemble of 11 structures. The backbone atoms of HHR23A Ubl (V3-T77) are shown in blue and orange (α-helices, β-strands), and the backbone atoms of S5a (L275-G297) are shown in black and red (α-helix). The side chains of the residues of HHR23A Ubl that form the center of the hydrophobic binding interface are shown in green and marked accordingly. (B) Representation of the binding interface between Ubl and S5a. A van der Waals surface is shown for HHR23A Ubl, along with the secondary structure elements and a ribbon representation for S5a. (C) The van der Waals surface of HHR23A Ubl color coded by atoms (white for C, cyan for N, red for O, yellow for S) and Cα trace (thick tube) for S5a (L275 to F300). The side chains of S5a UIM-2 that interact with HHR23A Ubl according to Figure 2B are shown. V76 (not shown) binds via main-chain atoms; I54 contributes ∼10 Å2 surface area. I54 contributes to binding affinity (Table II) probably by maintaining the side-chain conformation of I49. The conserved LALAL motif (here I287AYAM291) of the UIM is in direct contact with the hydrophobic patch on the surface of HHR23A Ubl, which consists mainly of K8, L10, I49, F71, V73 and M75. (D) Stick representation of interacting residues of UIM-2 (green) and Ubl (gray). Side chains of Ubl that are buried upon complex formation (see Figure 2A) are shown. The backbone Cα trace for residues 6–13, 34–41, 46–56 and 69–78 is presented as a bold tube; UIM-2 residues are the same as in (C).
S5a263–307 binds near the center of the five-stranded β-sheet of HHR23A Ubl, with its hairpin turn lying in a small groove near the N-terminal end of the α-helix of the Ubl (Figure 5A and B). The amphipatic α-helix of S5a263–307 runs almost perfectly in parallel and on top of strand β5 of HHR23A Ubl, with its hydrophobic side facing the surface of the β-sheet. The S5a α-helix is flanked by strand β1 and the β1–β2 loop on one side, and strand β3 and the β3–β4 loop on the other side. The conserved acidic residues at the N-terminal end of the α-helix of S5a (Glu283 to Glu285), which form a patch of negative electrostatic potential, are in close proximity to the positively charged residues Lys78 and Lys80 at the C-terminus of HHR23A Ubl (Figure 5C). The conserved LALAL (I287AYAM291) motif on S5a UIM-2 is in the center of the binding interface. Ile287 of S5a is located in a pocket formed by Met75 and Thr77 of Ubl, and Tyr289 of S5a is in contact with Leu10 of Ubl. Ala290 is in the center of the hydrophobic surface patch of HHR23A Ubl, comprised of residues Leu10, Ile49, Val73 and Met75, and Met291 of S5a is in direct contact with Ile49, Ile54 and Met75 of HHR23A Ubl (Figure 5C). The N-terminal tail also interacts with the Ubl, although the position of the residues of S5a preceding Leu278 is not well defined. NOEs between Leu278 and β1–β2 loop residues, as well as between Ser280 of S5a and Lys36 and Ala40 of HHR23A Ubl, show that the hairpin is located at the N-terminal end of the long α-helix of the Ubl domain. In three of 10 structure models, the N-terminal tail binds to Ubl like a paper clip, with Pro276 located in the hydrophobic groove formed by Thr9, Lys36 and Phe41 of HHR23A Ubl. For the other seven selected structure models of the complex, the N-terminal tail of S5a does not run through this hydrophobic groove but is close to the residues in the β1–β2 loop of HHR23A Ubl. Consequently, in these structure models, the N-terminal tail of S5a UIM-2 points away from the surface of the Ubl domain (Figure 5A and C). Additional NOEs between Pro276 of S5a and residues of HHR23A Ubl to resolve this ambiguity have not been identified so far; however, residues 273–275 of S5a exhibit changes in their chemical shift (Figure 4) which could not be explained if those residues were not close to the surface of HHR23A Ubl. The conformation of the β1–β2 loop is well defined in the structure of the complex due to additional van der Waals contacts and intermolecular NOEs with S5a. The binding interface for S5a is preformed on the surface of HHR23A Ubl, and no significant structural rearrangements are required for the binding.
Although most of the interacting residues in the complex interface are hydrophobic, two polar residues, Gln286 and Ser294, located at the N- and C-terminal ends of the α-helix of S5a263–307, become buried upon binding to HHR23A Ubl. Ser294 is almost completely conserved among the UIM family. The high degree of conservation of this serine might be due to spatial constraints in the binding interface. Mutation of this serine to an alanine resulted in disruption of the binding to polyUb (Young et al., 1998). Based on the structure, possible hydrogen bonds can be formed between the Ser294 hydroxyl and the backbone of HHR23A Ubl, e.g. Ile49 carbonyl and Gly52 amide. Gln286 is not absolutely conserved throughout the UIM family, but a polar residue of the type Asp, Glu, Asn or Gln often occupies this position. The structure of the complex reveals that the side chain of Gln286 may form a hydrogen bond with the Val76 carbonyl of HHR23A Ubl.
Conserved hydrophobic residues of the Ubl and the UIM are required for recognition
The importance of several hydrophobic residues of HHR23A Ubl for the binding of S5a263–307 was also studied by mutagenesis and binding analysis using surface plasmon resonance methodology (BIAcore). The analysis of the binding kinetics revealed very fast association and dissociation rates for the binding of HHR23A Ubl to S5a263–307. Binding affinity constants were therefore deduced from equilibrium binding experiments, yielding a dissociation constant of ∼13 µM for the interaction of the wild-type proteins (Table II). Four residues of HHR23A Ubl were mutated: Ile49, Ile54 and Phe71 to alanine, and Thr77 to serine. Exchange of any one of the three hydrophobic residues, Ile49, Ile54 or Phe71, with alanine resulted in a modest reduction of the binding affinity, with a 4.5-fold decrease for Ile49Ala, ∼3.7-fold for Phe71Ala, and a 3.1-fold decrease for Ile54Ala (Table II). Ile49 of HHR23A Ubl is located in the center of the binding interface and shares ∼45 Å2 surface area between Ubl and S5a peptide. Phe71 exhibits about the same amount of interface area (45 Å2); the contribution to the binding energy is slightly smaller, probably due to the non-central location at the C-terminal end of the α-helix of S5a UIM-2 (Figure 5C). Substitution of Thr77 to a serine does not alter the binding affinity between HHR23A Ubl and S5a263–307, showing that hydrophobic contacts between the C-terminus of HHR23A Ubl and the N-terminal region of the helix of S5a263–307 do not contribute to the binding energy. The modest decreases in binding affinity for the other mutations are consistent with the largely hydrophobic nature of the interactions in this part of the interface.
Table II. Binding affinity for the interaction of HHR23A UBL and S5a263–307.
| Protein | Binding affinity × 10–6 M | Standard deviation × 0–6 M |
|---|---|---|
| Wild type | 12.9 | ± 1.0 |
| I49A | 60.2 | ± 3.6 |
| I54A | 40.0 | ± 2.5 |
| F71A | 47.9 | ± 3.7 |
| T77S | 15.2 | ± 1.1 |
Discussion
Rad23 Ubl domains have a ubiquitin fold with a unique α-helical turn
In this study we determined a high-resolution structure of the N-terminal Ubl domain of HHR23A. The structure is very similar to the overall fold of ubiquitin and other ubiquitin-like domain proteins (Vijay-Kumar et al., 1987; Bayer et al., 1998; Rao-Naik et al., 1998; Whitby et al., 1998; Walters et al., 2002). However, one structural feature seems to be unique among the Ubl domains of Rad23 from different species. The loop between the β2 strand and the α-helix varies in length, with Ubl domains of Rad23 having an insertion of three amino acids. Because of the extended loop, a type IV α-helical turn is formed by the residues Arg38 to Phe41 (Figure 1C). This insertion and hence the short α-helix is not found in either ubiquitin (Vijay-Kumar et al., 1987), NEDD8 (Whitby et al., 1998) or the Ubl domain of PLIC-2 (Walters et al., 2002), which also bind to S5a (Figure 2). Since residues Lys36 and Ala40 interact directly with S5a UIM-2, this unique structural feature may contribute to the specificity of S5a UIM-2 for HHR23A Ubl. The side chain of Phe41 is oriented into the hydrophobic core and has numerous van der Waals contacts with residues at the C-terminal end of the helix (Ile32), and strands β1 (Thr9), β2 (Phe15) and β5 (Val74). The succeeding four-residue turn (Pro42 to Gln45) is, however, also present in ubiquitin (Pro37 to Asp40), as well as NEDD8 (Pro37 to Gln40) and PLIC-2 Ubl (Gln68 to Gln71).
The second UIM of S5a has a conserved hairpin turn
In this study we report the first experimental structure of a full-length UIM domain, i.e. UIM-2 of human S5a (residues 263–307). As predicted by secondary structure programs (Hofmann and Falquet, 2001) and NMR studies (Shekhtman and Cowburn, 2002), the UIM forms a long α-helix. However, structure analysis revealed that the UIM-2 of S5a has a short hairpin loop at the N-terminal end of the long α-helix. This hairpin loop is formed by a small hydrophobic core that contributes to stabilization of the α-helix. Based on the structure of S5a263–307, it appears that a functional UIM is longer than described by the PFAM database (residues 281–298 for UIM-2 of human S5a) and that it should be extended by at least five residues at the N-terminus to include the residues of the hairpin loop. If all residues showing changes in their chemical shift upon binding to HHR23A Ubl are considered, the UIM from the PFAM database would have to be extended by 11 residues at the N-terminus and by three residues at the C-terminus according to the current definition.
Our hypothesis that the N-terminal sequence in front of the α-helix of the UIM is important for structure and/or binding to ubiquitin as well as to Ubl domains is also supported by the observation that binding to the PUbS1 (UIM-1) and PUbS2 (UIM-2) sequences of human S5a were dependent on the length of the respective N-terminus (Young et al., 1998). The residue pair Leu278-Tyr289, which is important for the formation of the small hydrophobic core, is conserved in the UIM-2 of S5a from different species. For UIM-1 of human S5a, the two amino acids flip positions, with the aromatic amino acid occupying the position of Leu278 and a leucine substituting Tyr289 in the α-helix (Figure 2B). However, in UIM-1 of S5a proteins, an additional residue (usually a proline) is inserted in the conserved stretch of negatively charged residues. Homology modeling based on our structure data suggests that this insertion would disrupt the helical turn formed by the three acidic residues and would consequently lead to a one-turn shorter α-helix. In addition, the backbone conformation of the hairpin loop would have to change to allow the formation of the hydrophobic interactions between the residues equivalent to Leu278 and Tyr289. Alternatively, the hydrophobic core might not be present for these UIM domains, and hence the hairpin observed for UIM-2 of S5a could be a unique structural feature of the C-terminal UIMs of S5a.
Very recently, a crystal structure of a 20 amino acid peptide (residues 301–320) from the UIM-2 of Vps27p (yeast vacuolar protein sorting) (corresponding to residues 281–301 of S5a) was reported (Fisher et al., 2003). For all but the two N-terminal residues, the peptide forms an amphipathic α-helix, which unexpectedly tetramerizes as a left-handed, antiparallel four-helix bundle. Although Fisher et al. discuss the possible biological relevance of this tetramerization, they also present analytical ultracentrifugation studies showing that the peptide is a monomer in solution, consistent with our results for S5a263–307. Interestingly, in the UIM domains of Vps27 and the human homolog Hrs, a hydrophobic residue is located at a similar distance from the hydrophobic patch as Leu278 in human S5a UIM-2, thus allowing the formation of a hairpin loop as found in S5a UIM-2. Therefore the hairpin loop might be a common structural feature of other UIM domains, besides those of S5a. The presence of a hairpin loop would likely disrupt the tetramerization found in the crystal structure of the minimal UIM.
Specificity of the second UIM of S5a for the Ubl of HHR23A
Chemical shift mapping of S5a UIM-2 on HHR23A Ubl and vice versa demonstrated that the two domains form a tight complex with 1:1 stoichiometry. HHR23A Ubl and S5a263–307 interact with preformed binding surfaces, with no large changes for the backbone conformations of either protein. The interacting surfaces determined by chemical shift mapping provided additional constraints for the orientation and location of S5a on the β-sheet surface of the HHR23A Ubl revealed in the three-dimensional (3D) structure. As discussed, S5a UIM-2 binds on top of, and parallel to, the central strand β5, with its hairpin loop interacting near the N-terminal end of the long α-helix. The binding sites for S5a or the yeast homolog Rpn10 on type-2 Ubl proteins like PLIC-2 and Parkin, respectively, have been mapped or modeled previously (Walters et al., 2002; Sakata et al., 2003), showing that the location of the binding epitopes is retained and that the binding mechanism for the UIMs to Ubl domains are probably very similar.
Analysis of the complex structure reveals that a large, mainly hydrophobic surface area (∼700 Å2) between the two molecules becomes buried upon binding of the S5a263–307 peptide. The surface area provided by the α-helix (residues Glu283 to Gln296) of S5a263–307 amounts to only 490 Å2, suggesting that the preceding hairpin structure contributes to the affinity and specificity of the protein–peptide interaction. This confirms the observation of Young et al. (1998) that the amino acid sequences at the N-terminus of the PUbS1 (UIM-1) and PUbS2 (UIM-2) in human S5a is contributing to high-affinity binding of polyUb. Consistent with this, Fisher et al. (2003) have also reported that the addition of five to nine extra residues on the N- and C-termini of minimal UIMs of Hrs and Stam 1 resulted in 2- to 7-fold increased binding affinities to ubiquitin.
The structure of the complex provides insight into the differences in binding of Ubl domains to UIM-1 and UIM-2 of S5a. The hydrophobic residues Ile287, Tyr289 and Met291 in UIM-2 are substituted for leucine residues in UIM-1. Modeling of the complex of Ubl:UIM-1 shows that these exchanges do not alter the size and geometry of the UIM binding interface appreciably. The high specificity of HHR23A Ubl for the UIM-2 of human S5a versus UIM-1 is probably due to Pro214 inserted in the helix of the UIM-1. As discussed above, this proline would cause the first helical turn (Glu283 to Gln286) to be absent or kinked, and therefore the interaction of the LALAL motif with the hydrophobic surface patch of Ubl would be weakened. In addition, the hairpin conformation is most likely affected, which would lead to a reduction in binding affinity. The second UIM of Vps27p has a proline in the same position as found in S5a UIM-1 (while Vps27p UIM-1 lacks a proline). Our suggestion, based on modeling, that this proline would disrupt the helical conformation, resulting in an N-terminal shortened α-helix for the UIM, is confirmed by the crystal structure of the Vps27p-2 UIM (Fisher et al., 2003). Here the beginning of the α-helix is induced by the proline, but is one turn shorter than the one in S5a UIM-2. In yeast, in which S5a (S.cerevisiae) does not contain a second UIM similar to human S5a, RAD23A does not interact with the proteasome via the S5a subunit, but binds to the S2 subunit (RPN1) instead (Elsasser et al., 2002).
Differences in the binding of ubiquitin and ubiquitin-like proteins to the proteasome subunit S5a
Previous NMR (Shekhtman and Cowburn, 2002) and Biacore (Raiborg et al., 2002) experiments have shown that ubiquitin binds to the Hrs UIM with a KD of ∼300 µM, ∼30-fold more weakly than Ubl binds to UIM-2 of S5a. A comparison of the binding epitopes of monomeric ubiquitin and HHR23A Ubl reveals possible explanations for the large difference in binding affinity for S5a. Amino acid exchanges in the binding epitope are mainly located on strand β5, i.e. Phe71→Thr66, Val73→His68 and Met75→Val70. Gln11 in the β1–β2 loop is replaced by Thr9 in ubiquitin, and the four-residue turn of HHR23A Ubl, Arg38 to Phe41, which is in close contact to the hairpin of S5a263–307 and interacts with the hairpin loop of S5a UIM-2, is absent in ubiquitin. Modeling the complex of ubiquitin and UIM-2 of S5a reveals that the substitution of Val73 of HHR23A Ubl for His68 of ubiquitin might be responsible for the lower binding affinity. The larger side chain of His68 would not allow the tight interactions of the hydrophobic IAYAM motif to form with the hydrophobic patch on the surface of ubiquitin, as observed for HHR23A Ubl. In addition, if the buried histidine is positively charged this could also contribute to a decrease in binding affinity through electrostatic repulsion. Surprisingly, the Ubl domain of PLIC-2, which binds to S5a with high affinity, also has a histidine at the position of Val73 (Walters et al., 2002). However, no experimental dissociation constant has been reported, and the binding region of S5a interacting with PLIC-2, i.e. UIM-1 or UIM-2 of human S5a, has not been defined. Finally, we note that Ala288, which is completely conserved in S5a and is located in the LALAL motif, has no contacts to HHR23AUbl, and we speculate that this residue is involved in binding to polyubiquitin.
A conserved binding surface on Ubl and ubiquitin for interacting protein domains?
Several groups have proposed that proteins with a modular architecture similar to Rad23 might function as shuttle carriers targeting polyubiquitinated substrates to the proteasome, utilizing the UBA domains to bind the substrate and the Ubl domain to interact with the regulatory subunit at the proteasome (Chen and Madura, 2002; Funakoshi et al., 2002; van Laar et al., 2002). At first sight, the low apparent binding affinity (KD ∼10 µM) of HHR23A Ubl for S5a seems to contradict this function, since tetraubiquitin (tetraUb) binds the proteasome with an affinity of <0.2 µM (Thrower et al., 2000), and competition of HHR23A Ubl for the binding to the proteasome would therefore be limited in the presence of polyubiquitinated substrate. However, the dissociation constant determined here represents only the isolated interaction between the Ubl domain of HHR23A and the UIM-2 of human S5a, whereas the affinity of polyUb chains was determined for complete 26S proteasome particles. Sequences outside the Ubl domain of full-length HHR23A might contribute to the binding to complete 26S proteasome particles and, in addition, binding to full-length S5a in the structural framework of complete proteasome particles could increase the affinity.
Identical residue positions of HHR23A Ubl and polyUb appear to be important for the binding to the UIMs of S5a, i.e. Leu10/Leu8, Ile49/Ile44 and Met75/Val70 (HHR23A/Ub) (Beal et al., 1996). The binding epitope of ubiquitin/polyUb for UIM of S5a and other proteins as determined by mutagenesis and NMR chemical shift mapping (Beal et al., 1996, 1998; Shekhtman and Cowburn, 2002; Walters et al., 2002) is remarkably similar to the binding epitope found for the complex of HHR23A Ubl and S5a263–307. This suggests that even if the binding mechanisms are different, Ubl and ubiquitin will compete for binding sites on S5a. Other protein modules have also recently been found to bind to ubiquitin and promote monoubiquitination, including the UBA domains of HHR23A and CUE (coupling of ubiquitin conjugation to ER degradation). After this work was submitted, two papers describing complexes formed between CUE domains and ubiquitin were reported (Kang et al., 2003). The interacting surface of ubiquitin found in the solution structure of yeast CUE2 (Kang et al., 2003; Prag et al., 2003) corresponds closely to that found for Ubl with S5a in this study. Furthermore, the interacting surface on CUE2, which is structurally homologous to the UBA domain (Dieckmann et al., 1998; Withers-Ward et al., 2000; Mueller and Feigon, 2002), corresponds to the hydrophobic patch predicted to be the interacting surface for binding to ubiquitin (Mueller and Feigon, 2002) and confirmed by chemical shift mapping (T.D.Mueller and J.Feigon, unpublished). While these protein modules have similar interacting surfaces (α-helices for UIM, UBA and CUE, and β-sheet for ubiquitin and Ubl), their affinities and specificities for particular domains vary. More detailed structural and functional studies will be needed to sort out how these domains cooperate and compete to promote monoubiquitination and control protein degradation during the cell cycle. The structure of the full-length UIM, S5a UIM-2, presented in this study shows that the tertiary structure deviates from the simple α-helical model used so far. The hairpin structure located at the N-terminus of the amphipathic α-helix is important for the specific recognition and binding to HHR23A Ubl. The complex of S5a UIM-2 with HHR23A Ubl presented here provides the first molecular details of how the proteasome interacts with Ubl domains, as well as insight into how ubiquitin and Ubl domains are specifically recognized.
Materials and methods
Sample preparation
The genes encoding residues 1–87 of the Ubl domain of HHR23A and residues 263–307 of the human S5a subunit of the 26S proteasome were cloned into a pGEX-2T expression system (Pharmacia). Expression and purification were performed according to manufacturer’s recommendations. The final Ubl and S5a proteins each contained an additional Gly-Ser dipeptide at the N-terminus, resulting from the thrombin cleavage site. Protein homogeneity and purity were checked by SDS–PAGE and analytical reverse phase HPLC (C8-column). Uniformly 15N-labeled and 13C-,15N-labeled proteins were obtained by growing cells in M9 minimal medium containing [15N]ammonium chloride and [13C6]glucose as sole nitrogen and carbon sources.
NMR spectroscopy
NMR experiments were performed at 27°C on a Bruker DRX-500 or DRX-600 instrument equipped with a triple-resonance/triple-axis gradient probe. All NMR experiments were processed using the software XWINNMR and analyzed with AURELIA (Bruker). Backbone atom chemical shifts of Ubl and S5a263–307 were assigned from four triple-resonance experiments, CBCA(CO)NH, CBCANH, HBHA(CO)NH and HBHANH (Cavanagh et al., 1996). Side-chain assignments were obtained from two triple-resonance experiments, CC(CO)NH-TOCSY and HC(C)(CO)NH-TOCSY, and two 13C-edited experiments, HCCH-COSY and HCCH-TOCSY (Cavanagh et al., 1996). NOEs for distance restraints were assigned using 13C-edited 3D NOESY-HSQC experiments in either 95% H2O/5% 2H2O or in 100% 2H2O. Additional restraints were obtained from two 15N-edited 3D experiments, i.e. a 15N-HSQC NOESY and 15N-HSQC-NOE-HSQC, and 2D NOESY experiments with different τm. NOE information from these spectra was classified as weak (≥5 Å), medium (≥4 Å) or strong (≥3 Å) on the basis of their intensities. For the assignment of the chemical shifts of the complex of HHR23A Ubl and S5a, samples containing only one component in labeled form were used to acquire a full set of triple-resonance and 13C-filtered experiments. A series of 2D filtered/edited experiments were acquired (Peterson et al., 2003), that allow for the exclusive selection of either intramolecular or the intermolecular NOEs.
Structure calculations
The program XPLOR3.1 was used for calculation of the structures of HHR23A Ubl, S5a263–307, and the Ubl:S5a complex. NOE distance restraints were treated as point-to-point distances with a force constant of 50 kcal/mol/Å2 (1 cal = 4.184 J), using a simulated annealing protocol. For the structure calculation of the complex, only the residues 270–301 of S5a were used. The flexible and undefined termini of the S5a263–307 structure led to a low convergence rate in the structure calculation, and we therefore included only the residues that showed changes in their chemical shifts in the titration study. Intramolecular distance restraints were applied with a higher force constant of 100 kcal/mol/Å2 compared with the intermolecular distance restraints (50 kcal/mol/Å2) to maintain the structures of the two components during the complex calculation. About 10% (complex, 30%) of the calculated structures were then selected for analysis on the basis of lowest overall energy and of the lowest NOE energy. None of the selected structures exhibits NOE violations >0.25 Å. The quality of the structures was assessed using the programs PROCHECK and Quanta98.
NMR chemical shift mapping
NMR samples for titration studies contained 0.25–0.5 mM 15N-labeled Ubl or S5a263–307. Unlabeled Ubl or S5a263–307 was added stepwise (0.2 equivalents per step) up to a final ratio of 1:1. Changes in chemical shift or the appearance of additional signals were monitored by acquiring 2D 1H-15N HSQC experiments. The addition of further unlabeled Ubl or S5a263–307 beyond a 1:1 ratio did not lead to further changes in the spectrum. Chemical shift mapping was determined using the average chemical shift change of the amide proton and nitrogen Δδave [Δδave = 0.5x (|Δδ (1H)|+(0.125x|Δδ (15N)|)].
Mutagenesis and BIAcore binding experiments
Mutagenesis for HHR23A Ubl was performed using Stratagene’s QuickChange method. The S5a peptide was covalently linked to the surface of a CM5 Chip (BIAcore) using amino-coupling via 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide hydrochloride and N-hydroxysuccinimide immobilizing between 150 and 1000 resonance units. The glutathione S-transferase (GST)–S5a fusion protein was used, since the S5a itself does not contain primary amino groups other than the N-terminus. Wild-type and mutant Ubl proteins were then perfused at 5–20 µl/min flow rate using HBS-N Buffer (BIAcore). Equilibrium binding constants were determined from steady-state analysis. Five to six different concentrations were used for the Ubl variants, and all binding experiments were repeated at least two times for statistical analysis. Analysis of the binding experiments was performed using the BIAevalution software package.
Coordinate deposition
The atomic coordinates for the structures of HHR23A Ubl, human S5a263–307 and the 1:1 complex of HHR23A Ubl and S5a263–307 have been deposited in the Protein Data Bank (accession codes 1P98, 1P9C and 1P9D, respectively).
Acknowledgments
Acknowledgements
We thank Robert Peterson for help with the filtered/edited NMR experiments, My Sam for assistance in the structure determination of the HHR23A Ubl domain, and Evan Feinstein for help with figure preparation. The work was supported by National Institutes of Health grant AI43190 to I.S.Y.Chen and J.F.
References
- Bayer P., Arndt,A., Metzger,S., Mahajan,R., Melchior,F., Jaenicke,R. and Becker,J. (1998) Structure determination of the small ubiquitin-related modifier SUMO-1. J. Mol. Biol., 280, 275–286. [DOI] [PubMed] [Google Scholar]
- Beal R., Deveraux,Q., Xia,G., Rechsteiner,M. and Pickart,C. (1996) Surface hydrophobic residues of multiubiquitin chains essential for proteolytic targeting. Proc. Natl Acad. Sci. USA, 93, 861–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beal R.E., Toscano-Cantaffa,D., Young,P., Rechsteiner,M. and Pickart,C.M. (1998) The hydrophobic effect contributes to polyubiquitin chain recognition. Biochemistry, 37, 2925–2934. [DOI] [PubMed] [Google Scholar]
- Bertolaet B.L., Clarke,D.J., Wolff,M., Watson,M.H., Henze,M., Divita,G. and Reed,S.I. (2001) UBA domains of DNA damage-inducible proteins interact with ubiquitin. Nat. Struct. Biol., 8, 417–422. [DOI] [PubMed] [Google Scholar]
- Cavanagh J., Palmer,A.G., Fairbrothers,W. and Skelton,N. (1996) Protein NMR Spectroscopy: Principles and Practice. Academic Press, San Diego, CA. [Google Scholar]
- Chen L. and Madura,K. (2002) Rad23 promotes the targeting of proteolytic substrates to the proteasome. Mol. Cell. Biol., 22, 4902–4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Shinde,U., Ortolan,T.G. and Madura,K. (2001) Ubiquitin-associated (UBA) domains in Rad23 bind ubiquitin and promote inhibition of multi-ubiquitin chain assembly. EMBO Rep., 2, 933–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke D.J., Mondesert,G., Segal,M., Bertolaet,B.L., Jensen,S., Wolff,M., Henze,M. and Reed,S.I. (2001) Dosage suppressors of pds1 implicate ubiquitin-associated domains in checkpoint control. Mol. Cell. Biol., 21, 1997–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieckmann T., Withers-Ward,E.S., Jarosinski,M.A., Liu,C.F., Chen,I.S. and Feigon,J. (1998) Structure of a human DNA repair protein UBA domain that interacts with HIV-1 Vpr. Nat. Struct. Biol., 5, 1042–1047. [DOI] [PubMed] [Google Scholar]
- Elder R.T., Song,X.Q., Chen,M., Hopkins,K.M., Lieberman,H.B. and Zhao,Y. (2002) Involvement of rhp23, a Schizosaccharomyces pombe homolog of the human HHR23A and Saccharomyces cerevisiae RAD23 nucleotide excision repair genes, in cell cycle control and protein ubiquitination. Nucleic Acids Res., 30, 581–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsasser S. et al. (2002) Proteasome subunit Rpn1 binds ubiquitin-like protein domains. Nat. Cell Biol., 4, 725–730. [DOI] [PubMed] [Google Scholar]
- Fisher R.D., Wang,B., Alam,S.L., Higginson,D.S., Robinson,H., Sundquist,W.I. and Hill,C.P. (2003) Structure and ubiquitin binding of the ubiquitin-interacting motif. J. Biol. Chem., 278, 28976–28984. [DOI] [PubMed] [Google Scholar]
- Funakoshi M., Sasaki,T., Nishimoto,T. and Kobayashi,H. (2002) Budding yeast Dsk2p is a polyubiquitin-binding protein that can interact with the proteasome. Proc. Natl Acad. Sci. USA, 99, 745–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzder S.N., Sung,P., Prakash,L. and Prakash,S. (1998) Affinity of yeast nucleotide excision repair factor 2, consisting of the Rad4 and Rad23 proteins, for ultraviolet damaged DNA. J. Biol. Chem., 273, 31541–31546. [DOI] [PubMed] [Google Scholar]
- Hartmann-Petersen R., Seeger,M. and Gordon,C. (2003) Transferring substrates to the 26S proteasome. Trends Biochem Sci., 28, 26–31. [DOI] [PubMed] [Google Scholar]
- Haynes R.H. and Kunz,B.A. (1981) DNA repair and mutagenesis in yeast. In Strathern,J.N., Jones,E.W. and Broach,J.R. (eds), The Molecular Biology of the Yeast Saccharomyces. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY, pp. 371–414. [Google Scholar]
- Hiyama H., Yokoi,M., Masutani,C., Sugasawa,K., Maekawa,T., Tanaka,K., Hoeijmakers,J.H. and Hanaoka,F. (1999) Interaction of hHR23 with S5a. The ubiquitin-like domain of hHR23 mediates interaction with S5a subunit of 26S proteasome. J. Biol. Chem., 274, 28019–28025. [DOI] [PubMed] [Google Scholar]
- Hofmann K. and Bucher,P. (1996) The UBA domain: a sequence motif present in multiple enzyme classes of the ubiquitination pathway. Trends Biochem. Sci., 21, 172–173. [PubMed] [Google Scholar]
- Hofmann K. and Falquet,L. (2001) A ubiquitin-interacting motif conserved in components of the proteasomal and lysosomal protein degradation systems. Trends Biochem. Sci., 26, 347–350. [DOI] [PubMed] [Google Scholar]
- Kang R.S., Daniels,C.M., Francis,S.A., Shih,S.C., Salerno,W.J., Hicke,L. and Radhakrishnan,I. (2003) Solution structure of a CUE-ubiquitin complex reveals a conserved mode of ubiquitin binding. Cell, 113, 621–630. [DOI] [PubMed] [Google Scholar]
- Kleijnen M.F., Shih,A.H., Zhou,P., Kumar,S., Soccio,R.E., Kedersha,N.L., Gill,G. and Howley,P.M. (2000) The hPLIC proteins may provide a link between the ubiquitination machinery and the proteasome. Mol. Cell, 6, 409–419. [DOI] [PubMed] [Google Scholar]
- Masutani C. et al. (1997) Identification and characterization of XPC-binding domain of hHR23B. Mol. Cell. Biol., 17, 6915–6923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao F., Bouziane,M., Dammann,R., Masutani,C., Hanaoka,F., Pfeifer,G. and O’Connor,T.R. (2000) 3-Methyladenine-DNA glycosylase (MPG protein) interacts with human RAD23 proteins. J. Biol. Chem., 275, 28433–28438. [DOI] [PubMed] [Google Scholar]
- Mueller T.D. and Feigon,J. (2002) Solution structures of UBA domains reveal a conserved hydrophobic surface for protein–protein interactions. J. Mol. Biol., 319, 1243–1255. [DOI] [PubMed] [Google Scholar]
- Ortolan T.G., Tongaonkar,P., Lambertson,D., Chen,L., Schauber,C. and Madura,K. (2000) The DNA repair protein rad23 is a negative regulator of multi-ubiquitin chain assembly. Nat. Cell Biol., 2, 601–608. [DOI] [PubMed] [Google Scholar]
- Peterson R.D., Theimer,C.A., Wu,H. and Feigon,J. (2003) New applications of 2D filtered/edited NOESY for assignment and structure elucidation of RNA and RNA–protein complexes. J. Biomol. NMR, in press. [DOI] [PubMed] [Google Scholar]
- Polo S., Sigismund,S., Faretta,M., Guidi,M., Capua,M.R., Bossi,G., Chen,H., De Camilli,P. and Di Fiore,P.P. (2002) A single motif responsible for ubiquitin recognition and monoubiquitination in endocytic proteins. Nature, 416, 451–455. [DOI] [PubMed] [Google Scholar]
- Prag G., Misra,S., Jones,E.A., Ghirlando,R., Davies,B.A., Horazdovsky,B.F. and Hurley,J.H. (2003) Mechanism of ubiquitin recognition by the CUE domain of Vps9p. Cell, 113, 609–620. [DOI] [PubMed] [Google Scholar]
- Raasi S. and Pickart,C.M. (2003) Rad23 UBA domains inhibit 26S proteasome-catalyzed proteolysis by sequestering lysine 48-linked polyubiquitin chains. J. Biol. Chem., 278, 8951–8959. [DOI] [PubMed] [Google Scholar]
- Raiborg C., Bache,K.G., Gillooly,D.J., Madshus,I.H., Stang,E. and Stenmark,H. (2002) Hrs sorts ubiquitinated proteins into clathrin-coated microdomains of early endosomes. Nat. Cell Biol., 4, 394–398. [DOI] [PubMed] [Google Scholar]
- Rao H. and Sastry,A. (2002) Recognition of specific ubiquitin conjugates is important for the proteolytic functions of the UBA domain proteins Dsk2 and Rad23. J. Biol. Chem., 277, 11691–11695. [DOI] [PubMed] [Google Scholar]
- Rao-Naik C., de la Cruz,W., Laplaza,J.M., Tan,S., Callis,J. and Fisher,A.J. (1998) The rub family of ubiquitin-like proteins. Crystal structure of Arabidopsis rub1 and expression of multiple rubs in Arabidopsis. J. Biol. Chem., 273, 34976–34982. [DOI] [PubMed] [Google Scholar]
- Richardson J.S. and Richardson,D.C. (1988) Amino acid preferences for specific locations at the ends of alpha helices. Science, 240, 1648–1652. [DOI] [PubMed] [Google Scholar]
- Sakata E. et al. (2003) Parkin binds the Rpn10 subunit of 26S proteasomes through its ubiquitin-like domain. EMBO Rep., 4, 301–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauber C., Chen,L., Tongaonkar,P., Vega,I., Lambertson,D., Potts,W. and Madura,K. (1998) Rad23 links DNA repair to the ubiquitin/proteasome pathway. Nature, 391, 715–718. [DOI] [PubMed] [Google Scholar]
- Shekhtman A. and Cowburn,D. (2002) A ubiquitin-interacting motif from Hrs binds to and occludes the ubiquitin surface necessary for polyubiquitination in monoubiquitinated proteins. Biochem. Biophys. Res. Commun., 296, 1222–1227. [DOI] [PubMed] [Google Scholar]
- Sugasawa K. et al. (1997) Two human homologs of Rad23 are functionally interchangeable in complex formation and stimulation of XPC repair activity. Mol. Cell. Biol., 17, 6924–6931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T., Park,H., Kwofie,M.A. and Lennarz,W.J. (2001) Rad23 provides a link between the Png1 deglycosylating enzyme and the 26S proteasome in yeast. J. Biol. Chem., 276, 21601–21607. [DOI] [PubMed] [Google Scholar]
- Thrower J.S., Hoffman,L., Rechsteiner,M. and Pickart,C.M. (2000) Recognition of the polyubiquitin proteolytic signal. EMBO J., 19, 94–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Laar T., van der Eb,A.J. and Terleth,C. (2002) A role for Rad23 proteins in 26S proteasome-dependent protein degradation? Mutat. Res., 499, 53–61. [DOI] [PubMed] [Google Scholar]
- Vijay-Kumar S., Bugg,C.E. and Cook,W.J. (1987) Structure of ubiquitin refined at 1.8 Å resolution. J. Mol. Biol., 194, 531–544. [DOI] [PubMed] [Google Scholar]
- Walters K.J., Kleijnen,M.F., Goh,A.M., Wagner,G. and Howley,P.M. (2002) Structural studies of the interaction between ubiquitin family proteins and proteasome subunit S5a. Biochemistry, 41, 1767–1777. [DOI] [PubMed] [Google Scholar]
- Whitby F.G., Xia,G., Pickart,C.M. and Hill,C.P. (1998) Crystal structure of the human ubiquitin-like protein NEDD8 and interactions with ubiquitin pathway enzymes. J. Biol. Chem., 273, 34983–34991. [DOI] [PubMed] [Google Scholar]
- Wilkinson C.R., Seeger,M., Hartmann-Petersen,R., Stone,M., Wallace,M., Semple,C. and Gordon,C. (2001) Proteins containing the UBA domain are able to bind to multi-ubiquitin chains. Nat. Cell Biol., 3, 939–943. [DOI] [PubMed] [Google Scholar]
- Withers-Ward E.S., Mueller,T.D., Chen,I.S. and Feigon,J. (2000) Biochemical and structural analysis of the interaction between the UBA(2) domain of the DNA repair protein HHR23A and HIV-1 Vpr. Biochemistry, 39, 14103–14112. [DOI] [PubMed] [Google Scholar]
- Young P., Deveraux,Q., Beal,R.E., Pickart,C.M. and Rechsteiner,M. (1998) Characterization of two polyubiquitin binding sites in the 26S protease subunit 5a. J. Biol. Chem., 273, 5461–5467. [DOI] [PubMed] [Google Scholar]