Abstract
Insulators are DNA sequences that are likely to be involved in formation of chromatin domains, functional units of gene expression in eukaryotes. Insulators can form domain boundaries and block inappropriate action of regulatory elements (such as transcriptional enhancers) in eukaryotic nuclei. Using an in vitro system supporting enhancer action over a large distance, the enhancer-blocking insulator activity has been recapitulated in a highly purified system. The insulator-like element was constructed using a sequence-specific DNA-binding protein making stable DNA loops (lac repressor). The insulation was entirely dependent on formation of a DNA loop that topologically isolates the enhancer from the promoter. This rationally designed, inducible insulator-like element recapitulates many key properties of eukaryotic insulators observed in vivo. The data suggest novel mechanisms of enhancer and insulator action.
Keywords: RNA polymerase/enhancers/initiation/insulators/transcription
Introduction
Insulators are eukaryotic DNA sequences that are defined based on two functional activities (for recent reviews see Gerasimova and Corces, 2001; West et al., 2002). The first is position-dependent enhancer-blocking activity: insulators block enhancer action only when placed between an enhancer and promoter but not upstream or downstream of an enhancer–promoter pair (Chung et al., 1993; Geyer and Corces, 1992; Kellum and Schedl, 1992). The second insulator activity is an ability to form chromatin boundaries (Ishii et al., 2002; Mutskov et al., 2002) and therefore confer position-independent transcription to transgenes stably integrated in the genome (Bonifer et al., 1990; Kellum and Schedl, 1991). Insulator activity has not been described in prokaryotes.
The mechanism of insulator action is unknown; two models were proposed to explain the enhancer-blocking activity of insulators. The ‘promoter decoy’ model suggests that insulators neutralize enhancer action by interacting with it and capturing a functionally active component of the enhancer (Geyer, 1997). The second, ‘chromatin boundary’ model suggests that insulators organize higher order chromatin structure, perhaps by formation of large chromatin loops (Schedl and Grosveld, 1995). An enhancer positioned within one chromatin loop cannot communicate with a promoter positioned within another chromatin domain. While a large body of data was obtained in support of both models (Geyer and Clark, 2002), the mechanism of insulator action remains hypothetical, in part because of the lack of an in vitro system supporting insulator action.
The mechanism of action of eukaryotic enhancers over a large distance is not well understood (see West et al., 2002 for review); this makes development of an in vitro enhancer-blocking assay extremely difficult. In contrast, enhancer-dependent promoters of Escherichia coli are well studied and highly active in vitro. Moreover, numerous data suggests that NtrC-dependent enhancer–promoter system has many properties characteristic of eukaryotic enhancer-dependent transcription, such as activation over a large distance, both upstream and downstream of the regulated promoters in an orientation-independent way (see Buck et al., 2000 for a review).
The glnAp2 promoter activated by an NtrC-dependent enhancer is one of the best-studied promoters in E.coli. In this system, NtrC activator protein binds to the enhancer and, when phosphorylated by NtrB protein kinase, forms homo-oligomers and activates transcription of the glnAp2 promoter (Porter et al., 1993; Wedel and Kustu, 1995; Wyman et al., 1997). Active enhancer-bound NtrC interacts with the RNA polymerase bound as a closed initiation complex at the promoter and stimulates transition to the open complex (Popham et al., 1989; Sasse-Dwight and Gralla, 1988). During the enhancer–promoter interaction intervening DNA is transiently looped out (Rippe et al., 1997; Su et al., 1990). More recently, it has been shown that DNA supercoiling dramatically facilitates enhancer–promoter communication over a large distance (Bondarenko et al., 2002; Liu et al., 2001).
In this work, rational design and construction of an insulator-like element having an enhancer-blocking activity have been attempted based on the detailed knowledge of the mechanism of enhancer action on the glnAp2 promoter. Topologically isolated domains (DNA loops) were formed using lac repressor tetramer that can simultaneously bind two lac operator (lacO) sites on DNA. Placing the enhancer and promoter within different topologically isolated domains on the same supercoiled plasmid DNA molecule greatly inhibits enhancer–promoter communication. Moreover, a pair of lac operators recapitulates many key properties of eukaryotic insulators observed in vivo.
Results
Designing an insulator-like element
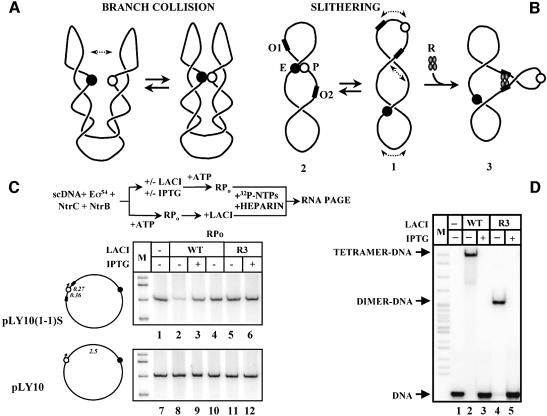
Design of a regulatory element having enhancer-blocking activity was based on the knowledge of the mechanism of action of the NtrC-dependent enhancer over a large distance. DNA supercoiling greatly (∼50-fold) facilitates enhancer–promoter communication; as a result, the enhancer works over short (0.11 kb) and large (2.5 kb) distances with similar efficiencies (Liu et al., 2001). Two models were proposed to explain the role of DNA supercoiling in facilitating communication between two DNA regions: ‘slithering’ (sliding of intertwined DNA double helixes) and ‘branch collision’ (Figure 1A and B; Chirico and Langowski, 1996; Vologodskii and Cozzarelli, 1996). Based on computer simulations of Brownian dynamics of supercoiled DNA it has been proposed that slithering is the predominant mechanism facilitating enhancer–promoter communication over distances up to 10 kb (Huang et al., 2001). Since plasmids used in this work have 2.5 or 3.3 kb enhancer–promoter spacing, an insulator-like element was designed based on the slithering model of enhancer action. This model suggests that placing the enhancer and promoter on topologically isolated DNA loops within the same supercoiled plasmid would prevent enhancer–promoter communication because sliding of DNA double helixes will only be possible within each domain, but not between the domains (Figures 1B and 6B).
Fig. 1. The enhancer cannot cis-activate the glnAp2 promoter localized within a topologically isolated DNA loop. (A) The branch collision mechanism. This mechanism was proposed to explain the effect of DNA supercoiling on the rate of communication between DNA regions on supercoiled DNA. Frequent collisions between branches formed on supercoiled DNA could facilitate communication between the enhancer and promoter localized on different branches of the same DNA molecule. The enhancer and promoter are indicated by black and white circles, respectively. (B) LacI-induced DNA loop formation topologically isolates the enhancer from the promoter. When spaced by 2.5 kb, the enhancer (E) and promoter (P) probably communicate by slithering on supercoiled DNA (structures 1 and 2). The slithering model suggests that intertwined DNA helixes can slide relative to each other on supercoiled DNA at a high rate; sliding greatly increases the probability of enhancer–promoter collision. LacI tetramer (R) can simultaneously bind two lac operators positioned upstream and downstream of the promoter (O1 and O2), form a DNA loop (structure 3), and thus could topologically isolate the enhancer from the promoter. (C) LacI-induced DNA loop formation greatly inhibits enhancer-dependent transcription. The experimental strategy is outlined at the top. Plasmid containing two lac operators localized 0.36 kb upstream and 0.27 kb downstream of the promoter [pLY10(1-1)S, upper panel] and control plasmid not containing lac operators (pLY10, lower panel) were transcribed in the presence or in the absence of wt LacI or the R3 mutant. R3 mutant binds DNA well but cannot form DNA loops. In some cases, LacI was added after formation of the open complexes (lanes 4 and 10). The distances between key regulatory elements are shown in italics. RPo, open initiation complex. Other designations are as in (B). M, labeled pBR322-MspI markers. Note that, although LacI can present a weak road-block to transcript elongation (Oehler et al., 1990), the block was not detected on the templates used in this work. (D) LacI and the R3 mutant LacI are quantitatively bound to their binding sites. Analysis by native PAGE. Aliquots of transcription reactions described in (C), lanes 2, 3, 5 and 6, containing LacI or R3, respectively, but not containing plasmid DNA were incubated in the presence of labeled double-stranded oligonucleotide containing an ‘ideal’ lac operator. Mobilities of corresponding complexes in the gel are indicated.
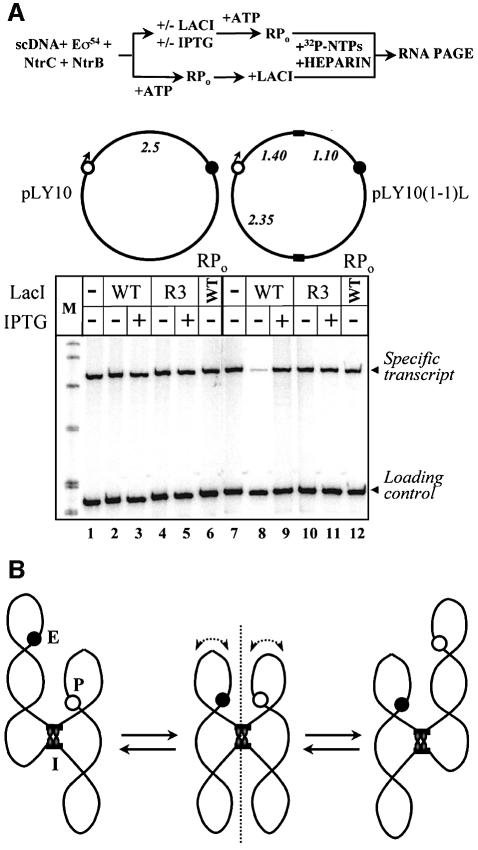
Fig. 6. Two lac operators can inhibit enhancer-dependent transcription over a large distance. (A) Two lac operators positioned 3.5 kb from each other can inhibit enhancer-dependent transcription. Plasmid containing two lac operators localized 3.7 kb from each other and at least 1.4 kb from the promoter [pLY10(1-1)L] and control plasmid not containing lac operators (pLY10) were transcribed in the presence or in the absence of LacI or the R3 mutant (see diagram at the top). A single-round assay was used. In some cases, LacI was added after formation of the open complexes (lanes 6 and 12). Labeled transcripts were analyzed in a denaturing gel. Labeled DNA fragment was added to all samples immediately after termination of transcription (loading control). Other designations are as in Figure 1C. (B) Slithering between two topologically isolated domains is impossible. Designations are as in Figure 1B. Binding of LacI tetramer to two lac operators results in separation of plasmid DNA into two topological domains. Slithering is only possible within each domain but not between them. Placing promoter and enhancer in topologically isolated domains prevents communication between them by slithering on supercoiled DNA.
To form stable DNA loops, a well-studied DNA looping protein (lac repressor, LacI) was used. LacI is a homotetramer that has two DNA-binding surfaces (Barkley et al., 1975; O’Gorman et al., 1980; Whitson et al., 1987) and is capable of binding two lac operators in vivo and in vitro, both on linear and supercoiled DNA (Hsieh et al., 1987; Kramer et al., 1987, 1988). The promoter was surrounded by two ‘ideal’ lac operators (Simons et al., 1984; Figure 1B). Lac operators were positioned far upstream and downstream the promoter (0.36 and 0.27 kb from the promoter, respectively) to avoid inhibition of transcription due to binding of LacI to promoter DNA (Borowiec et al., 1987; Choy and Adhya, 1992; Flashner and Gralla, 1988; Oehler et al., 1990). It was expected that at optimal concentration, LacI tetramer would bind both operators simultaneously, forming a stable DNA loop that would topologically isolate the promoter from the enhancer and thus prevent enhancer–promoter communication.
DNA loop formation topologically isolates the enhancer from the promoter
All plasmid constructs used in this work contain the glnAp2 promoter and NtrC-dependent enhancer positioned 2.5 or 3.3 kb downstream of the promoter. Activity of the promoter entirely depends on the presence of the enhancer in cis (Liu et al., 2001). Transcription of the pLY10(1-1)S plasmid containing two LacI-binding sites positioned 0.36 kb upstream and 0.27 kb downstream of the glnAp2 promoter, respectively, is strongly (5- to 10-fold) inhibited by LacI in a single-round transcription assay (Figure 1C, lanes 1 and 2). Addition of IPTG (an inducer of LacI which strongly inhibits its binding to the operator) almost completely reverses the inhibition (lane 3). There is no inhibition when the repressor is added after formation of the open initiation complex (RPo) suggesting that the repressor acts before this step (lane 4). Moreover, R3, a mutant LacI that binds DNA equally well but cannot form the tetramer [it can only form dimers (Chen et al., 1994)] and therefore cannot form the DNA loop, does not have any effect on transcription (lanes 5 and 6). Neither repressor affects transcription of the control plasmid (not containing lac operators, lanes 7–12).
In a control experiment, aliquots of transcription reactions containing LacI and R3 (but not containing plasmid templates) were incubated with labeled double-stranded oligonucleotide containing ‘perfect’ lacO and analyzed in a native gel (Figure 1D). The oligonucleotide was present at the same molar amount as the plasmids in the experiment described in Figure 1C. As expected, only wt repressor forms the tetramer (R3 forms dimers, compare lanes 2 and 4), and addition of IPTG completely reverses binding of LacI and R3 to DNA (lanes 3 and 5). The data suggest that in the absence of IPTG, both LacI and R3 quantitatively occupy lac operators, but only wt repressor can form the DNA loop and inhibit transcription. Thus, quantitative DNA binding by R3 is not sufficient to inhibit transcription.
Taken together, the data suggest that LacI can strongly and specifically inhibit transcription initiation when its binding sites are positioned over more than 250 bp from the promoter. The novel type of inhibition over a distance requires the ability of the repressor to form the DNA loop; DNA binding per se is not sufficient for inhibition. These features of the repression make lac operators formally similar to eukaryotic ‘insulators’ blocking enhancer action over a large distance (see Introduction).
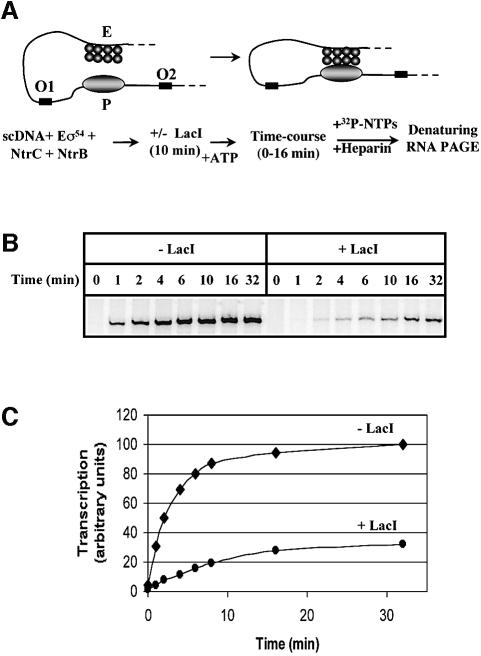
According to the slithering model of enhancer action (Figure 1B), LacI-mediated formation of the DNA loop should decrease the rate of enhancer–promoter communication. The rates of enhancer–promoter communication in the presence and in the absence of LacI were compared (Figure 2). First, all components of the transcription reaction were pre-incubated with the templates in the absence of ATP. Under these conditions, both RNA polymerase and NtrC bind to the promoter and the enhancer, respectively, but cannot functionally communicate with each other, and the polymerase cannot form the open complex (Buck and Cannon, 1992; Popham et al., 1989; Sasse-Dwight and Gralla, 1988). Enhancer–promoter communication was induced by adding ATP to the reaction to induce NtrC phosphorylation, and the rate of formation of the open complex [that reflects the rate of communication (Liu et al., 2001)] was measured in a single-round transcription assay. As expected, the rate of enhancer–promoter communication (t1/2 = 1.5 min) is strongly (about 5-fold) decreased in the presence of LacI (t1/2 = 8 min). In fact, the data suggest that even this slow enhancer–promoter communication occurs only on a fraction of the templates; the majority of templates (70%) do not support transcription initiation in the presence of LacI (Figure 2C). In summary, LacI-induced formation of the DNA loop (see below) results in a strong decrease in the rate of enhancer–promoter communication.
Fig. 2. Topological isolation of the enhancer from the promoter reduces the rate of enhancer–promoter communication. (A) Transcription assay for analysis of the effect of LacI on enhancer–promoter communication. The pLY10(1-1)S supercoiled template was pre-incubated with all components of the transcription machinery, and then incubated with or without LacI. Then ATP was added to allow enhancer–promoter communication. The rate of open complex formation was measured in a single-round transcription assay. NtrC activator octamer bound to the enhancer (E) and RNA polymerase bound to the promoter (P) are shown. Other designations are as in Figure 1C. (B) Analysis of labeled specific transcripts by denaturing PAGE. (C) The rate of enhancer– promoter communication is strongly decreased in the presence of LacI. DNA bands containing specific transcripts (B) were quantified and plotted as a function of time.
LacI interacts with two lac operators and forms a stable loop on supercoiled DNA
The data described above are consistent with LacI-induced formation of a DNA loop on supercoiled DNA. The ability of LacI to form loops on linear DNA is well-documented (Hsieh et al., 1987; Kramer et al., 1987, 1988; Oehler et al., 1990). It has been suggested that LacI can also form loops on supercoiled DNA (Eismann and Muller-Hill, 1990; Whitson et al., 1987) but it has not been analyzed directly.
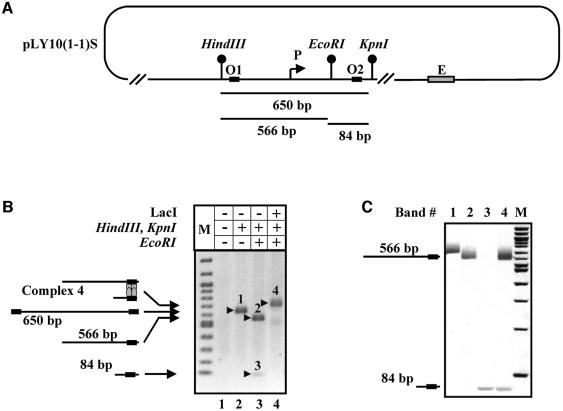
To analyze DNA loop formation, supercoiled pLY10(1-1)S template having two lac operators surrounding the P1 promoter (Figure 3A) was incubated in the presence of LacI and then digested with restriction enzymes producing 566 and 84 bp DNA fragments, each containing one lacO (Figure 3A). If the LacI tetramer interacts with both operators at the same time and forms a DNA loop, both fragments are expected to be bound to LacI and to migrate as one complex in a native gel. This was the case: neither 566 nor 84 bp free DNA fragments were detected after pre-incubation of supercoiled DNA with LacI and subsequent digestion (Figure 3B, compare lanes 3 and 4). Instead, a single complex having slower mobility in a native gel (complex 4) was detected. As expected, analysis of DNA extracted from this complex indicates that it contains both 566 and 84 bp DNA fragments present at a molar ratio of ∼1:1 (Figure 3C).
Fig. 3. LacI interacts with two lac operators and forms a stable loop on supercoiled DNA. (A) Partial restriction map of the pLY10(1-1)S plasmid. If LacI interacts with both lac operators (O1 and O2) on supercoiled DNA, two DNA fragments (84 and 566 bp) generated after digestion with restriction enzymes HindIII, EcoRI and KpnI are expected to be bound to LacI and migrate as a single DNA–protein complex in a native gel. (B) LacI-induced DNA loop formation on supercoiled DNA: analysis of DNA–protein complexes. Supercoiled pLY10(1-1)S template was pre-incubated with or without LacI, then digested by different combinations of restriction enzymes and analyzed in a native agarose gel. The expected products of digestion are shown on the left. Different bands in the gel are arbitrarily numbered. M, 100 bp DNA ladder (NEB). (C) LacI-induced DNA loop formation on supercoiled DNA: analysis of DNA composition of different DNA–protein complexes. DNA was purified from different bands [1–4 in (B)] and analyzed by PAGE. M, 100 bp DNA ladder (NEB).
The data suggest that the loop forms on supercoiled DNA and not after digestion with the restriction enzymes. Indeed, if LacI interacted with the DNA fragments after digestion, various combinations of the 566 and 84 bp DNA fragments (566–566, 566–84 and 84–84) were expected to be found in different complexes with LacI (data not shown). In contrast, only one discrete complex containing both fragments was observed (complex 4, Figure 3B) suggesting that the loop was formed on supercoiled DNA. The efficiency of loop formation was >80%. In summary, LacI binding to the lac operators flanking the promoter results in almost quantitative loop formation on supercoiled DNA.
Lac operators have many properties characteristic for eukaryotic insulators
To further analyze insulator properties of lac operators, a set of plasmids having 2.5 kb enhancer–promoter spacing but containing a different number of ‘ideal’ lac operators positioned differently was constructed (Figure 4). Many of these plasmids were designed similarly to the constructs used for analysis of the mechanism of insulator action in vivo. This allowed comparison of the properties of lac operators in vitro with the properties of natural insulators in vivo. These constructs were transcribed in the presence and in the absence of LacI (Figure 4) and the levels of transcription inhibition by LacI were compared.
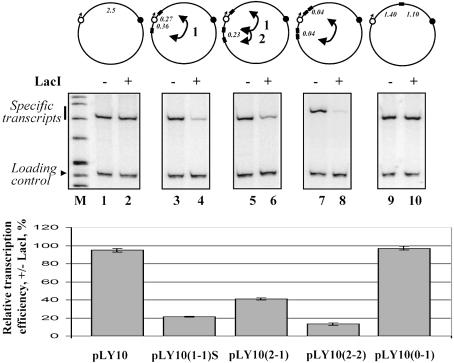
Fig. 4. Topological isolation of the enhancer from the promoter is required for insulation. Various constructs (upper panel), all containing the enhancer and promoter spaced by 2.5 kb but different numbers of ‘ideal’ lac operators positioned differently were transcribed in the presence or in the absence of LacI. The arrows between lac operators indicate the expected LacI-induced DNA loops. Two alternative loops formed on the pLY10(2-1) plasmid are indicated (arrows 1 and 2); other designations are as in Figure 1C. All constructs were transcribed in the same experiment; transcripts were analyzed by denaturing PAGE (middle panel). Equal amounts of a labeled DNA fragment were added to all samples immediately after termination of transcription (loading control). The histogram (lower panel) shows the ratio of the amounts of specific transcripts accumulated in the presence and in the absence of LacI. The data are averages of at least three experiments (standard deviations are indicated).
It has been shown that duplication of insulators can improve their activity (Chung et al., 1993). Duplication of lac operators also increases their activity ∼2-fold (Figure 4, compare lanes 4 and 8), presumably because LacI forms a DNA loop that topologically isolates the enhancer from the promoter more efficiently. In this construct, short spacing (40 bp) between lac operators within each pair is likely to prevent interaction of the same LacI tetramer with two operators positioned next to each other. LacI bends DNA away from the repressor by 40° (Lewis et al., 1996), and once the tetramer binds to one DNA site the second DNA-binding surface is unlikely to reach the second site. This results in binding of two tetramers to each pair of operators and in more efficient formation of the DNA loop.
Duplication of some insulators placed between an enhancer and promoter can also considerably compromise activities of both insulators in vivo (Cai and Shen, 2001; Muravyova et al., 2001, but see also Melnikova et al., 2002; Kuhn et al., 2003). These findings can be recapitulated with lac operators: duplication of one of the two insulator-like elements flanking the promoter results in ∼2-fold decrease in the repression (Figure 4, compare lanes 4 and 6). In this case, two lac operators positioned between the enhancer and promoter were spaced by 228 bp. This spacing is optimal for DNA loop formation by LacI (Law et al., 1993). Therefore, on this construct the DNA loop can be formed in either of two ways: the loop can include the promoter or be formed between the enhancer and promoter. In the former case, the loop (loop 1, Figure 4) inhibits enhancer–promoter communication (lane 4). If the loop is formed between the enhancer and promoter (loop 2), it is not expected to topologically isolate them from each other and would be neutral for promoter activity. In fact, this is the case (data not shown). However, since formation of loops 1 and 2 are competing processes, the overall effect of formation of loop 2 is a decrease in the efficiency of formation of loop 1 and promoter repression by LacI.
It has been proposed that natural insulators can work in pairs (Cai and Shen, 2001; Muravyova et al., 2001) or large clusters (Gerasimova et al., 2000; Gerasimova and Corces, 1998; Ishii et al., 2002). As expected, a single lacO placed between the enhancer and promoter did not inhibit enhancer-dependent transcription (Figure 4, compare lanes 4 and 10).
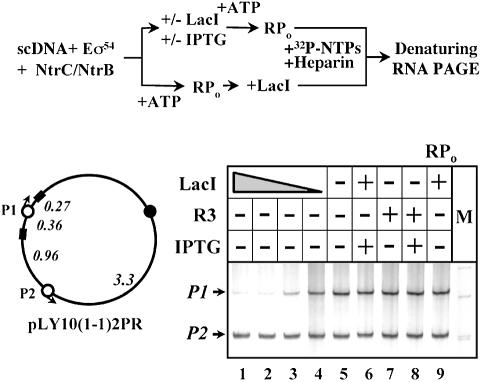
Finally, it has been shown that many insulators are ‘neutral’ DNA elements that do not inactivate promoters but presumably prevent enhancer–promoter communication in vivo (Cai and Levine, 1995; Scott and Geyer, 1995, but see also Wei and Brennan, 2000, 2001). To find out whether this property of insulators can be recapitulated in vitro, the pLY10(1-1)2PR plasmid was constructed (Figure 5). It contains two identical glnAp2 promoters (P1 and P2), both activated by the same enhancer over distances of 2.5 and 3.3 kb, respectively. Only the P1 promoter is flanked by the lac operators. As expected, in the absence of LacI both enhancer-dependent promoters are active (lane 5) suggesting that the enhancer can work efficiently over a distance up to at least 3.3 kb. Adding increasing concentrations of LacI (lanes 4 to 1) results in progressive, selective and strong (up to 10-fold) inhibition of P1; P2 remains fully active. The data suggest that LacI-induced DNA loop formation does not inactivate either promoter P2 or enhancer. At the same time, activity of the promoter P1 localized within the DNA loop that topologically isolates the promoter from the enhancer is strongly inhibited. As expected, addition of IPTG reverses the inhibition (lane 6), inhibition was not observed after formation of the open complex (lane 9), and the R3 mutant repressor was unable to inhibit transcription (lanes 7 and 8).
Fig. 5. LacI-induced DNA loop topologically isolates the enhancer from the promoter but does not inactivate them. The pLY10(1-1)2PR plasmid (upper panel) containing two identical glnAp2 promoters (P1 and P2) under the control of one enhancer and two lac operators flanking promoter P1 was transcribed in the presence or in the absence of LacI and IPTG (see the diagram at the top). The regions transcribed from the P1 and P2 promoters are 401 and 309 nt long, respectively. The transcripts were analyzed by denaturing PAGE (lower panel). The template concentration was 3 nM and lac repressor was added to the following concentrations: lane 4, 2 nM; lane 3, 4 nM; lane 2, 6 nM and lanes 1 and 6–9, 8 nM. Other designations are as in Figure 1C.
In summary, many features of eukaryotic insulators observed in vivo can be recapitulated using lac operators in a highly purified enhancer–promoter system in vitro. Lac operators only work in pairs; placement of the enhancer and promoter in different topologically isolated domains is essential for insulator activity. Neither promoter nor enhancer is inactivated by the formation of the loop per se, but enhancer–promoter communication occurs with much lower efficiency.
Lac operators can insulate over a large distance
In all of the experiments described above, lac operators were inhibiting transcription over a relatively short distance: 0.27–0.36 kb from the promoter. Can lac operators, like eukaryotic insulators, work over a much larger distance? In order to address this question and to discriminate between the slithering and branch collision models of enhancer action (Figure 1A and B), pLY10(1-1)L template was constructed (Figure 6). On this plasmid, one lacO is positioned between the enhancer and promoter (1.4 kb downstream of the promoter); the second operator is placed at the maximal distance (3.7 kb) from the first one. The slithering model suggests that placement of the enhancer and promoter in topologically isolated domains is sufficient to inhibit communication between them: the exact positions of lac operators and spacing between them are not essential. At the same time, the pY10(1-1)L template is optimized for branch collision: the enhancer and promoter are localized almost symmetrically relative to the lac operators and over a large distance from the base of the loop. The large distance from the base of the loop maximizes flexibility of the ‘branches’ formed on supercoiled DNA. If the enhancer and promoter communicate by the branch collision mechanism, transcription of the pY10(1-1)L template would be inhibited by LacI less than transcription of the pY10(1-1)S plasmid where the enhancer and promoter are localized asymmetrically relative to the operators, the promoter is positioned much closer to the base of the loop and the DNA loop is much smaller.
Transcription of the pLY10(1-1)L plasmid is strongly (5- to 10-fold) inhibited by LacI (Figure 6A, compare lanes 7 and 8). Addition of IPTG almost completely reverses the inhibition (lane 9). There is no inhibition when the repressor is added after formation of the open complex (lanes 6 and 12). As expected, mutant R3 repressor does not have any effect on transcription (lanes 4, 5, 10 and 11). Neither repressor affects transcription of the control plasmid not containing lac operators (lanes 1–6). The data suggest that LacI can strongly and specifically inhibit transcription initiation when its binding sites are positioned 1.4–2.3 kb from the promoter and 3.7 kb from each other. The inhibition requires the ability of the repressor to form the DNA loop. The data are entirely consistent with the slithering model of enhancer action (Figures 1B and 6B).
Discussion
In summary, a pair of lac operators can work as a strong insulator-like element blocking enhancer action over a large distance in a highly purified in vitro system (Figure 1). Insulator activity requires formation of a stable LacI-induced DNA loop (Figure 3) that topologically isolates the enhancer from the promoter (Figure 4), but does not inactivate either enhancer or promoter (Figure 5). As a result, the rate of enhancer–promoter communication is greatly inhibited (Figure 2). The exact positioning of lac operators is flexible: they can work over distances of 0.3–2.3 kb from the promoter and 0.63–3.7 kb from each other (Figures 1 and 6).
The data have implications for the mechanism of enhancer action over a large distance. Two models were proposed to explain how enhancer–promoter communication over a distance could be facilitated on supercoiled DNA: the slithering and branch collision models (Figure 1A and B). The data are consistent with the slithering model of enhancer action. In particular, the ability of lac operators to prevent enhancer–promoter communication over a large distance and the data on partial inhibition of insulator activity by duplication of lac operators [Figure 4, construct pLY10(2-1)] are difficult to explain by the branch collision model. In contrast, the slithering model provides an immediate explanation for all of the experimental data. After LacI brings two lac operators together and forms a DNA loop, two topologically isolated domains are formed. Slithering is possible within each domain but not between them (Figure 6B). Therefore, the enhancer and promoter efficiently communicate only when they are located within the same domain on supercoiled DNA; communication from one domain to another is almost impossible, even when the domains are large (up to at least 3.7 kb). Communication between the domains by the branch collision mechanism (Figure 1A) could become possible when the size of the loops is >10 kb (Huang et al., 2001).
DNA supercoiling is essential for enhancer–promoter communication over a distance in vitro (Liu et al., 2001). Can DNA supercoiling facilitate enhancer–promoter interaction in vivo? The bulk of the eukaryotic genome does not contain unconstrained DNA supercoiling (Sinden et al., 1980) and there is no enzyme (gyrase) to generate it. However, localized negative DNA supercoiling could be generated by nucleosome displacement or remodeling (Camerini-Otero and Felsenfeld, 1977; Havas et al., 2000), and by transcript elongation (Liu and Wang, 1987; Wu et al., 1988). Indeed, transcriptionally active genome regions contain considerable levels of unconstrained DNA supercoiling (Kramer et al., 1999). Moreover, action of eukaryotic transcriptional and recombination enhancers over a large distance in vitro requires DNA supercoiling (Bagga and Emerson, 1997; Barton et al., 1997). It may seem unlikely that bulky chromatin structure would allow enhancer–promoter communication by slithering. However efficient enhancer–promoter communication occurs on supercoiled DNA containing bound R3 repressor (Figures 1 and 6) indicating that at least some bulky DNA–protein complexes are transparent for slithering. Many more studies are clearly required to establish to what extent the observations described here can be applied to a much more complicated intranuclear environment. In particular, it would be important to analyze whether DNA supercoiling is required for enhancer action on DNA organized in chromatin.
When the enhancer and promoter are positioned within different DNA loops on supercoiled DNA, changes in the structure of the complex that destabilize or stabilize the loops facilitate or inhibit enhancer–promoter communication, respectively. The loop can be destabilized using mutant R3 repressor or by disruption of LacI–DNA interactions by IPTG. Alternatively, the efficiency of formation of the ‘inhibitory’ loop can be decreased by formation of a competitive loop placing the enhancer and promoter in the same topological domain and thus allowing efficient communication between them [Figure 4, construct pLY10(2-1)]. In all cases a decrease in the efficiency of the ‘inhibitory’ loop formation results in transcription activation. In contrast, an increase in the stability of the loop results in stronger inhibition of transcription [Figure 4, construct pLY10(2-2)]. Thus, the data are entirely consistent with and support the slithering model of enhancer action (Chirico and Langowski, 1996; Vologodskii and Cozzarelli, 1996). The data suggest that two lac operators form a ‘slithering barrier’ at the border of the LacI-induced DNA loop.
LacI-dependent insulation by lac operators is clearly distinct from repression of the lac promoter that also involves two lac operators and DNA loop formation by LacI (Borowiec et al., 1987; Choy and Adhya, 1992; Flashner and Gralla, 1988; Oehler et al., 1990). In the case of repression, one lacO overlaps the promoter and the function of the second operator is solely to increase promoter occupancy by LacI; the second lacO is only required at low concentration of LacI (Choy and Adhya, 1992; Oehler et al., 1990). Once bound at the promoter, LacI interferes with the escape of the polymerase from the promoter (Schmitz and Galas, 1979; Straney and Crothers, 1987). The presence of LacI at the promoter is essential for the repression. In contrast, insulation by lac operators does not require LacI binding at the promoter; in fact, lac operators can be placed 1.4 kb away from the promoter and still strongly inhibit transcription. At the same time, the presence of both lac operators and formation of the loop are essential for the insulator function.
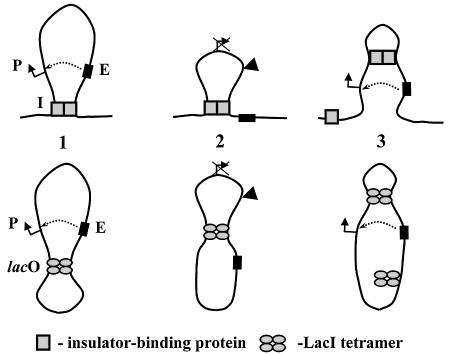
Lac operators recapitulate many important properties of eukaryotic insulators observed in vivo (Figure 7). Many natural eukaryotic insulators are ‘neutral’ elements working in a position-dependent way: transcription is inhibited only when an insulator is positioned between an enhancer and promoter (not upstream of the enhancer or downstream of the promoter) and the promoters remain functional (Cai and Levine, 1995; Scott and Geyer, 1995, but see also Wei and Brennan, 2000, 2001). Most likely, insulators prevent enhancer–promoter communication. Two lac operators behave similarly: transcription is only inhibited when the promoter is topologically isolated from the enhancer by formation of a DNA loop; and neither enhancer nor promoter is inactivated by loop formation per se (Figure 5). Lac operators strongly decrease the rate of enhancer–promoter communication (Figure 2). A natural insulator can inhibit transcription independently of its precise location (as long as it is positioned between an enhancer and promoter) and can work over a large distance (Dorsett, 1993; Golovnin et al., 1999). Likewise, precise positioning of the two lac operators is not essential for their insulator activity, and they can work over a large distance. Finally, in some cases duplication of an insulator positioned between an enhancer and promoter results in loss of insulator activity (Cai and Shen, 2001; Muravyova et al., 2001, but see also Melnikova et al., 2002; Kuhn et al., 2003). Duplication of similarly positioned lac operators also significantly decreases repression of the promoter [Figure 4, construct pLY10(2-1)], presumably because the efficiency of formation of the ‘inhibitory’ loop is decreased. Taken together, the data suggest that topological isolation could be sufficient for insulation in vivo.
Fig. 7. Similarities between action of natural insulators in vivo and lac operators in vitro. An enhancer (E) and promoter (P) can efficiently communicate when flanked by insulators (I) or lac operators (lacO, structure 1). Insertion of an insulator or lacO between an enhancer and promoter may result in formation of a DNA loop and topological isolation of the enhancer from the promoter that prevents enhancer– promoter communication (structure 2). Incorporation of a second insulator between the enhancer and promoter (indicated by arrowhead in structure 2) may result in formation of a competitive DNA loop (structure 3). As a result, the enhancer and promoter would reside in the same topological domain and can efficiently communicate with each other. Many enhancer-blocking properties of insulators and lac operators can be rationalized in the framework of the slithering model of enhancer action (see Discussion). Note that only interactions between two eukaryotic insulators are shown (upper panel) while in vivo insulators form large ‘insulator bodies’ (Gerasimova and Corces, 1998; Gerasimova et al., 2000; Ishii et al., 2002).
Eukaryotic insulators are likely to have other activities in addition to the enhancer-blocking activity. Thus, it is clear that boundary activity of insulators is mechanistically different from the enhancer-blocking activity (Mutskov et al., 2002; Recillas-Targa et al., 2002); only the enhancer-blocking activity was recapitulated in this study. Moreover, some insulators can functionally interact with promoters (Wei and Brennan, 2000, 2001) suggesting that this activity is part of the mechanism of insulator action. Finally, the ability of some insulators to block enhancer action in trans (Krebs and Dunaway, 1998) cannot be immediately explained by the slithering barrier model of insulator action. However, the model could provide a useful framework for further analysis of the mechanism of insulator action.
Materials and methods
Plasmid construction
All templates are derivatives of the pLY10 plasmid containing the enhancer and the glnAp2 promoter (Liu et al., 2001). The plasmids were constructed using PCR amplification of desired fragments, followed by digestion of the fragment and the target plasmids with restriction enzymes and ligation. The pTH8 plasmid was described by Hunt and Magasanik (1985). The cloning approaches outlined in Table I were used.
Table I. Cloning approaches used in this study.
| Plasmids | Oligonucleotides | Amplified from | Cloned into | Restriction enzymes |
|---|---|---|---|---|
| pLY10(1-1)S | 5′-CTCTAAGCTTAAATTGTGAGCGCTCACAATTCCACGAAGACCTTTATTCAGAAGGG | pLY10 | pLY10 | HindIII |
| 5′-CCAGGTACCAATTGTGAGCGCTCACAATTAATGCCTTTCCAGCCGCCAATCGAGG | KpnI | |||
| pLY10(1-1)2PR | 5′-CGCCTTTTCGGCCGAATTTAAAAGTTGGCACAGATTTCGC | pTH8 | pLY10 | EagI |
| 5′-TTTTCCCAGTCCGGACGTTGTAAAACGACGGCCAGTGCC | (1-1)S | BspEI | ||
| pLY10(2-1)S | 5′-CGGGTCGCCGGTGAAATTGTGAGCGCTCACAATTCGCTAGCAGCACGCCATAGTGACTGG | pLY10 | pLY10 | SgrAI |
| 5′-CCAGGTACCAATTGTGAGCGCTCACAATTAATGCCTTTCCAGCCGCCAATCGAGG | (1-1)S | KpnI | ||
| pLY10(2-2)S | 5′-GGTGCAAGCTTGCAATTGTGAGCGCTCACAATTAAACTACCGCATTAACGCTTAAATTGTGAGC | pLY10 | pLY10 | HindIII |
| 5′-ATTAGGTACCAAATTGTGAGCGCTCACAATTCAGGTGTTTCCTCCAGCTACCAATTGTGAGC | (1-1)S | KpnI | ||
| pLY10(0-1) | 5′-CTCTCAGTAAGATCTACAAATATGTTGTGC | pLY10 | pLY10 | BglII |
| 5′-GCATCAATTGCAATTGTGAGCGCTCACAATTCGGCTTTCAGGAACTCGCGGTCCAGATC | MfeI | |||
| pLY10(1-1)L | 5′-AGATCCGGAAAATTGTGAGCGCTCACAATTACTTCCGCGTTTCCAGACTTTACGAAAC | pLY10 | pLY10 | BspEI |
| 5′-CATGGCGGCCGACGCGCTGGGCTAC | (0-1) | EagI |
In vitro transcription assay
All protein components of transcription machinery were purified as described previously (Bondarenko et al., 2002; Liu et al., 2001). LacI and the R3 mutant repressor were purified as described by Barry and Matthews (1997). Their purity was >95% according to silver staining. In vitro transcription reactions were optimized for maximal utilization of promoter on supercoiled templates. Single round transcription assay was carried out in 50 µl aliquots containing 3 nM DNA template, 130 nM core RNA polymerase, 200 nM σ54, 150 nM NtrC, 50 nM NtrB in buffer [50 mM Tris–HCl (pH 7.5), 50 mM KCl, 10 mM MgCl2, 0.1 mM EDTA (pH 8.0)]. The reaction mixture was incubated at 37°C for 10 min. LacI or its mutant R3 were added to the reaction (final concentration 30 nM) and incubated at 37°C for an additional 10 min. To release LacI from DNA, IPTG was added to 1 mM final concentration. ATP was added to the reaction to 0.5 mM final concentration at 37°C for 15 min or for variable time. To start transcription, NTP (final concentration 80 µM), 2.5 µCi [α-32P]GTP, RNase inhibitor (final concentration 0.2 U/µl) and heparin (final concentration 80 µg/ml) were added to the reaction. After incubation at 37°C for 15 min transcription was terminated by adding 100 µl of phenol/chloroform (1:1). End-labeled 227 bp DNA fragment (loading control) was added to the mixture in 50 µl of H2O. The samples were extracted with phenol/chloroform, precipitated with ethanol, washed with 70% ethanol and dissolved in formamide. The samples were separated on 8% denaturing PAGE, dried and analyzed on a phosphoimager.
Gel-shift assay for DNA loop detection
Supercoiled pLY10(1-1)S template at concentration 15 nM was incubated with or without 50 nM LacI in the binding buffer [50 mM Tris–HCl (pH 7.5), 50 mM KCl, 10 mM MgCl2, 0.1 mM EDTA (pH 8.0)] at 37°C for 20 min in 20 µl aliquots. Then 10 U of HindIII and KpnI only or together with EcoRI were added and incubated at 37°C for 30 min. The reaction was terminated by adding 4 µl of 5× loading buffer containing 50 mM EDTA, 5× TAE buffer and 50% glycerol. The samples were immediately analyzed in 1% agarose gel in TAE buffer. The gel was stained with EtBr and DNA fragments of interest were extracted from the gel by using QIAGEN gel-extraction kit followed by phenol/chloroform extraction and ethanol precipitation. The purified DNA fragments were separated in 6% PAG in TAE buffer and detected by EtBr staining.
Acknowledgments
Acknowledgements
We are grateful to Drs G.Felsenfeld, P.Georgiev, T.Gerasimova, P.Geyer, M.Kashlev, L.Lutter, A.Ninfa, W.Walter and H.-Y.Wu for critical reading of the manuscript and/or helpful comments. We would like to thank Dr K.S.Matthews for providing wt and R3 lac repressor-producing strains and purified lac repressor.
References
- Bagga R. and Emerson,B.M. (1997) An HMG I/Y-containing repressor complex and supercoiled DNA topology are critical for long-range enhancer-dependent transcription in vitro. Genes Dev., 11, 629–639. [DOI] [PubMed] [Google Scholar]
- Barkley M.D., Riggs,A.D., Jobe,A. and Burgeois,S. (1975) Interaction of effecting ligands with lac repressor and repressor-operator complex. Biochemistry, 14, 1700–1712. [DOI] [PubMed] [Google Scholar]
- Barry J.K. and Matthews,K.S. (1997) Ligand-induced conformational changes in lactose repressor: a fluorescence study of single tryptophan mutants. Biochemistry, 36, 15632–15642. [DOI] [PubMed] [Google Scholar]
- Barton M.C., Madani,N. and Emerson,B.M. (1997) Distal enhancer regulation by promoter derepression in topologically constrained DNA in vitro. Proc. Natl Acad. Sci. USA, 94, 7257–7262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondarenko V., Liu,Y., Ninfa,A. and Studitsky,V.M. (2002) Action of prokaryotic enhancer over a distance does not require continued presence of promoter-bound sigma54 subunit. Nucleic Acids Res., 30, 636–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifer C., Vidal,M., Grosveld,F. and Sippel,A.E. (1990) Tissue specific and position independent expression of the complete gene domain for chicken lysozyme in transgenic mice. EMBO J., 9, 2843–2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borowiec J.A., Zhang,L., Sasse-Dwight,S. and Gralla,J.D. (1987) DNA supercoiling promotes formation of a bent repression loop in lac DNA. J. Mol. Biol., 196, 101–111. [DOI] [PubMed] [Google Scholar]
- Buck M. and Cannon,W. (1992) Activator-independent formation of a closed complex between sigma 54-holoenzyme and nifH and nifU promoters of Klebsiella pneumoniae. Mol. Microbiol., 6, 1625–1630. [DOI] [PubMed] [Google Scholar]
- Buck M., Gallegos,M.T., Studholme,D.J., Guo,Y. and Gralla,J.D. (2000) The bacterial enhancer-dependent sigma(54) (sigma(N)) transcription factor. J. Bacteriol., 182, 4129–4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai H. and Levine,M. (1995) Modulation of enhancer-promoter interactions by insulators in the Drosophila embryo. Nature, 376, 533–536. [DOI] [PubMed] [Google Scholar]
- Cai H.N. and Shen,P. (2001) Effects of cis arrangement of chromatin insulators on enhancer-blocking activity. Science, 291, 493–495. [DOI] [PubMed] [Google Scholar]
- Camerini-Otero R.D. and Felsenfeld,G. (1977) Supercoiling energy and nucleosome formation: the role of the arginine-rich histone kernel. Nucleic Acids Res., 4, 1159–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Alberti,S. and Matthews,K.S. (1994) Wild-type operator binding and altered cooperativity for inducer binding of lac repressor dimer mutant R3. J. Biol. Chem., 269, 12482–12487. [PubMed] [Google Scholar]
- Chirico G. and Langowski,J. (1996) Brownian dynamics simulations of supercoiled DNA with bent sequences. Biophys. J., 71, 955–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy H.E. and Adhya,S. (1992) Control of gal transcription through DNA looping: inhibition of the initial transcribing complex. Proc. Natl Acad. Sci. USA, 89, 11264–11268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung J.H., Whiteley,M. and Felsenfeld,G. (1993) A 5′ element of the chicken β-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell, 74, 505–514. [DOI] [PubMed] [Google Scholar]
- Dorsett D. (1993) Distance-independent inactivation of an enhancer by the suppressor of Hairy-wing DNA-binding protein of Drosophila. Genetics, 134, 1135–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eismann E.R. and Muller-Hill,B. (1990) lac repressor forms stable loops in vitro with supercoiled wild-type lac DNA containing all three natural lac operators. J. Mol. Biol., 213, 763–775. [DOI] [PubMed] [Google Scholar]
- Flashner Y. and Gralla,J.D. (1988) Dual mechanism of repression at a distance in the lac operon. Proc. Natl Acad. Sci. USA, 85, 8968–8972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerasimova T.I. and Corces,V.G. (1998) Polycomb and trithorax group proteins mediate the function of a chromatin insulator. Cell, 92, 511–521. [DOI] [PubMed] [Google Scholar]
- Gerasimova T.I. and Corces,V.G. (2001) Chromatin insulators and boundaries: effects on transcription and nuclear organization. Annu. Rev. Genet., 35, 193–208. [DOI] [PubMed] [Google Scholar]
- Gerasimova T.I., Byrd,K. and Corces,V.G. (2000) A chromatin insulator determines the nuclear localization of DNA. Mol. Cell, 6, 1025–1035. [DOI] [PubMed] [Google Scholar]
- Geyer P.K. (1997) The role of insulator elements in defining domains of gene expression. Curr. Opin. Genet. Dev., 7, 242–248. [DOI] [PubMed] [Google Scholar]
- Geyer P.K. and Corces,V.G. (1992) DNA position-specific repression of transcription by a Drosophila zinc finger protein. Genes Dev., 6, 1865–1873. [DOI] [PubMed] [Google Scholar]
- Geyer P.K. and Clark,I. (2002) Protecting against promiscuity: the regulatory role of insulators. Cell Mol. Life Sci., 59, 2112–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golovnin A., Gause,M., Georgieva,S., Gracheva,E. and Georgiev,P. (1999) The su(Hw) insulator can disrupt enhancer-promoter interactions when located more than 20 kilobases away from the Drosophila achaete-scute complex. Mol. Cell. Biol., 19, 3443–3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havas K., Flaus,A., Phelan,M., Kingston,R., Wade,P.A., Lilley,D.M. and Owen-Hughes,T. (2000) Generation of superhelical torsion by ATP-dependent chromatin remodeling activities. Cell, 103, 1133–1142. [DOI] [PubMed] [Google Scholar]
- Hsieh W.T., Whitson,P.A., Matthews,K.S. and Wells,R.D. (1987) Influence of sequence and distance between two operators on interaction with the lac repressor. J. Biol. Chem., 262, 14583–14591. [PubMed] [Google Scholar]
- Huang J., Schlick,T. and Vologodskii,A. (2001) Dynamics of site juxtaposition in supercoiled DNA. Proc. Natl Acad. Sci. USA, 98, 968–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt T.P. and Magasanik,B. (1985) Transcription of glnA by purified Escherichia coli components: core RNA polymerase and the products of glnF, glnG and glnL. Proc. Natl Acad. Sci. USA, 82, 8453–8457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii K., Arib,G., Lin,C., Van Houwe,G. and Laemmli,U.K. (2002) Chromatin boundaries in budding yeast: the nuclear pore connection. Cell, 109, 551–562. [DOI] [PubMed] [Google Scholar]
- Kellum R. and Schedl,P. (1991) A position-effect assay for boundaries of higher order chromosomal domains. Cell, 64, 941–950. [DOI] [PubMed] [Google Scholar]
- Kellum R. and Schedl,P. (1992) A group of scs elements function as domain boundaries in an enhancer-blocking assay. Mol. Cell. Biol., 12, 2424–2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer H., Niemoller,M., Amouyal,M., Revet,B., von Wilcken-Bergmann,B. and Muller-Hill,B. (1987) lac repressor forms loops with linear DNA carrying two suitably spaced lac operators. EMBO J., 6, 1481–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer H., Amouyal,M., Nordheim,A. and Muller-Hill,B. (1988) DNA supercoiling changes the spacing requirement of two lac operators for DNA loop formation with lac repressor. EMBO J., 7, 547–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer P.R., Bat,O. and Sinden,R.R. (1999) Measurement of localized DNA supercoiling and topological domain size in eukaryotic cells. Methods Enzymol., 304, 639–650. [DOI] [PubMed] [Google Scholar]
- Krebs J.E. and Dunaway,M. (1998) The scs and scs′ insulator elements impart a cis requirement on enhancer-promoter interactions. Mol. Cell, 1, 301–308. [DOI] [PubMed] [Google Scholar]
- Kuhn E.J., Viering,M.M., Rhodes,K.M. and Geyer,P.K. (2003) A test of insulator interactions in Drosophila. EMBO J., 22, 2463–2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law S.M., Bellomy,G.R., Schlax,P.J. and Record,M.T.,Jr (1993) In vivo thermodynamic analysis of repression with and without looping in lac constructs. Estimates of free and local lac repressor concentrations and of physical properties of a region of supercoiled plasmid DNA in vivo. J. Mol. Biol., 230, 161–173. [DOI] [PubMed] [Google Scholar]
- Lewis M., Chang,G., Horton,N.C., Kercher,M.A., Pace,H.C., Schumacher,M.A., Brennan,R.G. and Lu,P. (1996) Crystal structure of the lactose operon repressor and its complexes with DNA and inducer. Science, 271, 1247–1254. [DOI] [PubMed] [Google Scholar]
- Liu L.F. and Wang,J.C. (1987) Supercoiling of the DNA template during transcription. Proc. Natl Acad. Sci. USA, 84, 7024–7027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Bondarenko,V., Ninfa,A. and Studitsky,V.M. (2001) DNA supercoiling allows enhancer action over a large distance. Proc. Natl Acad. Sci. USA, 98, 14883–14888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnikova L., Gause,M. and Georgiev,P. (2002) The gypsy insulators flanking yellow enhancers do not form a separate transcriptional domain in Drosophila melanogaster: the enhancers can activate an isolated yellow promoter. Genetics, 160, 1549–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muravyova E., Golovnin,A., Gracheva,E., Parshikov,A., Belenkaya,T., Pirrotta,V. and Georgiev,P. (2001) Loss of insulator activity by paired Su(Hw) chromatin insulators. Science, 291, 495–498. [DOI] [PubMed] [Google Scholar]
- Mutskov V.J., Farrell,C.M., Wade,P.A., Wolffe,A.P. and Felsenfeld,G. (2002) The barrier function of an insulator couples high histone acetylation levels with specific protection of promoter DNA from methylation. Genes Dev., 16, 1540–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oehler S., Eismann,E.R., Kramer,H. and Muller-Hill,B. (1990) The three operators of the lac operon cooperate in repression. EMBO J., 9, 973–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Gorman R.B., Rosenberg,J.M., Kallai,O.B., Dickerson,R.E., Itakura,K., Riggs,A.D. and Matthews,K.S. (1980) Equilibrium binding of inducer to lac repressor.operator DNA complex. J. Biol. Chem., 255, 10107–10114. [PubMed] [Google Scholar]
- Popham D.L., Szeto,D., Keener,J. and Kustu,S. (1989) Function of a bacterial activator protein that binds to transcriptional enhancers. Science, 243, 629–635. [DOI] [PubMed] [Google Scholar]
- Porter S.C., North,A.K., Wedel,A.B. and Kustu,S. (1993) Oligomerization of NTRC at the glnA enhancer is required for transcriptional activation. Genes Dev., 7, 2258–2273. [DOI] [PubMed] [Google Scholar]
- Recillas-Targa F., Pikaart,M.J., Burgess-Beusse,B., Bell,A.C., Litt,M.D., West,A.G., Gaszner,M. and Felsenfeld,G. (2002) Position-effect protection and enhancer blocking by the chicken β-globin insulator are separable activities. Proc. Natl Acad. Sci. USA, 99, 6883–6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rippe K., Guthold,M., von Hippel,P.H. and Bustamante,C. (1997) Transcriptional activation via DNA-looping: visualization of intermediates in the activation pathway of E. coli RNA polymerase x sigma 54 holoenzyme by scanning force microscopy. J. Mol. Biol., 270, 125–138. [DOI] [PubMed] [Google Scholar]
- Sasse-Dwight S. and Gralla,J.D. (1988) Probing the Escherichia coli glnALG upstream activation mechanism in vivo. Proc. Natl Acad. Sci. USA, 85, 8934–8938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schedl P. and Grosveld,F. (1995) Domains and boundaries. In Elgin,S.C. (ed.), Chromatin Structure and Gene Expression. Oxford University Press, Oxford, UK, pp. 172–196. [Google Scholar]
- Schmitz A. and Galas,D.J. (1979) The interaction of RNA polymerase and lac repressor with the lac control region. Nucleic Acids Res., 6, 111–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott K.S. and Geyer,P.K. (1995) Effects of the su(Hw) insulator protein on the expression of the divergently transcribed Drosophila yolk protein genes. EMBO J., 14, 6258–6267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons A., Tils,D., von Wilcken-Bergmann,B. and Muller-Hill,B. (1984) Possible ideal lac operator: Escherichia coli lac operator-like sequences from eukaryotic genomes lack the central G X C pair. Proc. Natl Acad. Sci. USA, 81, 1624–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinden R.R., Carlson,J.O. and Pettijohn,D.E. (1980) Torsional tension in the DNA double helix measured with trimethylpsoralen in living E. coli cells: analogous measurements in insect and human cells. Cell, 21, 773–783. [DOI] [PubMed] [Google Scholar]
- Straney S.B. and Crothers,D.M. (1987) Lac repressor is a transient gene-activating protein. Cell, 51, 699–707. [DOI] [PubMed] [Google Scholar]
- Su W., Porter,S., Kustu,S. and Echols,H. (1990) DNA-looping and enhancer activity: association between DNA-bound NtrC activator and RNA polymerase at the bacterial glnA promoter. Proc. Natl Acad. Sci. USA, 87, 5504–5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vologodskii A. and Cozzarelli,N.R. (1996) Effect of supercoiling on the juxtaposition and relative orientation of DNA sites. Biophys. J., 70, 2548–2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedel A. and Kustu,S. (1995) The bacterial enhancer-binding protein NTRC is a molecular machine: ATP hydrolysis is coupled to transcriptional activation. Genes Dev., 9, 2042–2052. [DOI] [PubMed] [Google Scholar]
- Wei W. and Brennan,M.D. (2000) Polarity of transcriptional enhancement revealed by an insulator element. Proc. Natl Acad. Sci. USA, 97, 14518–14523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W. and Brennan,M.D. (2001) The gypsy insulator can act as a promoter-specific transcriptional stimulator. Mol. Cell. Biol., 21, 7714–7720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West A.G., Gaszner,M. and Felsenfeld,G. (2002) Insulators: many functions, many mechanisms. Genes Dev., 16, 271–288. [DOI] [PubMed] [Google Scholar]
- Whitson P.A., Hsieh,W.T., Wells,R.D. and Matthews,K.S. (1987) Influence of supercoiling and sequence context on operator DNA binding with lac repressor. J. Biol. Chem., 262, 14592–14599. [PubMed] [Google Scholar]
- Wu H.Y., Shyy,S.H., Wang,J.C. and Liu,L.F. (1988) Transcription generates positively and negatively supercoiled domains in the template. Cell, 53, 433–440. [DOI] [PubMed] [Google Scholar]
- Wyman C., Rombel,I., North,A.K., Bustamante,C. and Kustu,S. (1997) Unusual oligomerization required for activity of NtrC, a bacterial enhancer-binding protein. Science, 275, 1658–1661. [DOI] [PubMed] [Google Scholar]