Abstract
KLF11 is a biochemical paradigm for a subset of proteins that repress transcription via a Mad1-like mSin3A interacting domain (SID). The biological role of these proteins and the significance of their biochemical activity, however, remain to be established. We report that KLF11 is downregulated in human cancers, inhibits cell growth in vitro and in vivo, and suppresses neoplastic transformation. Transgenic mice for KLF11 display a downregulation of genes encoding the oxidative stress scavengers SOD2 and Catalase1. Chromatin immunoprecipitation assays confirm that, indeed, these genes are bonafide targets of KLF11. KLF11 expression renders cells more sensitive to oxidative drugs, an effect that is rescued by infection with recombinant adenoviruses expressing SOD2 and Catalase1. KLF11-regulated functions require the Mad1-like SID, indicating that these target genes involved in these phenomena are regulated via this corepressor system. These results demonstrate that SID-containing KLF repressor proteins can inhibit cell growth and neoplastic transformation, and outline for the first time cellular and molecular mechanisms by which these functions may be achieved.
Keywords: apoptosis/cell growth/KLF11/mSin3A/SID
Introduction
Krüppel-like transcription factors (KLFs) constitute an expanding group of Sp1-like proteins that are present in organisms ranging from yeast to vertebrates. The structure of these proteins is defined by the presence of three highly homologous, C-terminal Cys2His2 zinc finger domains, which bind DNA, and a variant N-terminal domain that contains transcriptional regulatory motifs (Cook et al., 1999a; Turner and Crossley, 1999; Cook and Urrutia, 2000; Bieker, 2001; Black and Azizkhan-Clifford, 2001). The identification of the entire repertoire of Sp1-like proteins and the characterization of their biochemical properties have been the focus of intensive investigation, primarily during the last 5 years. These studies have revealed that KLF proteins bind to similar, yet distinct GC-rich target sequences, and they function either as activators or repressors (Turner and Crossley, 1999; Bieker, 2001). KLF activator proteins exert their function by interacting with histone acetylases via different types of regulatory domains. For instance, KLF1 and KLF4 activate transcription through the interaction with the coactivator CBP/p300 and p300/CBP-associated factor (PCAF) through a conserved acidic domain in the N-terminal region of these proteins (Geiman et al., 2000; Zhang,W. et al., 2001).
KLF repressor proteins have been the less studied group within the KLF family, although recent studies have revealed the existence of two subfamilies of repressors based on the corepressor system that they utilize. KLF3 and KLF8 repress transcription by forming a complex with CtBP through the use of a canonical PVDLS/T motif, a CtBP-interacting domain (CID) (Turner and Crossley, 1998; van Vliet et al., 2000). KLF10, -11, -13 and -16, on the other hand, repress heterologous promoters via an mSin3A-interacting domain (SID) that is shared with the well characterized tumor suppressor protein Mad1 (Zhang,J.S. et al., 2001). Thus, these data provide an emerging theoretical framework for better understanding the biochemical properties of KLF proteins.
In spite of the intensive biochemical characterization of KLF proteins, the study of their biological role is rather a nascent area of investigation. Initial data, mainly derived from knockout and transgenic animals, suggest that few KLF proteins, in particular KLF transcriptional activators, play a role in morphogenesis. For instance, mice null for Sp1, the founding member of the KLF family, are retarded in development, show a broad range of organ abnormalities, and die around embryonic day 11 (Marin et al., 1997). KLF1, a red blood cell-specific activator, is required for normal erythropoiesis. KLF1-knockout mice develop a fatal anemia during early fetal life when hematopoiesis has switched to the liver (Perkins et al., 1995; Tewari et al., 1998). Unfortunately, the mechanisms behind this function are currently poorly understood. In addition, whether the biochemical properties of these proteins have a clear impact on their cell biological function remains unclear. Similarly, the biological role of KLF repressor proteins needs to be better defined. Thus, studies that focus on the cellular function of these proteins are of paramount importance for a more complete characterization of this important, evolutionarily conserved family of transcription factors.
Our laboratory has been investigating the role of novel members of the KLF family of transcription factors in gene silencing. We have previously identified and defined the domain structure and biochemical properties of mSin3A-mediated KLF repressors (KLF10, -11, -13 and -16) (Cook et al., 1998, 1999b; Kaczynski et al., 2001, 2002; Zhang,J.S. et al., 2001; Ellenrieder et al., 2002). In this study, we have investigated the cellular role of the ubiquitously expressed SP1-like protein, KLF11, a structural and functional paradigm for this subset of KLF proteins. Using a large battery of both in vitro and in vivo experiments, we find that the expression of KLF11 is reduced in epithelial cancers and that this gene inhibits cell growth and suppresses neoplastic transformation. In addition, KLF11 inhibits cell growth in vivo when targeted to epithelial cells in transgenic mice. Furthermore, the function of KLF11 involves the downregulation of genes encoding oxidative stress scavengers, such as superoxide dismutase 2 (SOD2) and Catalase1. More importantly, the presence of an intact SID is required for both the growth suppression properties and regulation of oxidative stress scavengers. Taken together, these results demonstrate that KLF11 is a negative regulator of cell growth and proposes for the first time a mechanism by which KLF11 may achieve its growth regulatory function. Moreover, this is also the first evidence that SID links the repressor activity of a KLF protein with its cell biological functions, and may bear relevance to the understanding of other members of this group, which work using identical biochemical mechanisms.
Results
Transgenic expression of KLF11 in the mouse exocrine pancreas results in decreased cell proliferation, increased apoptosis and reduced organ size
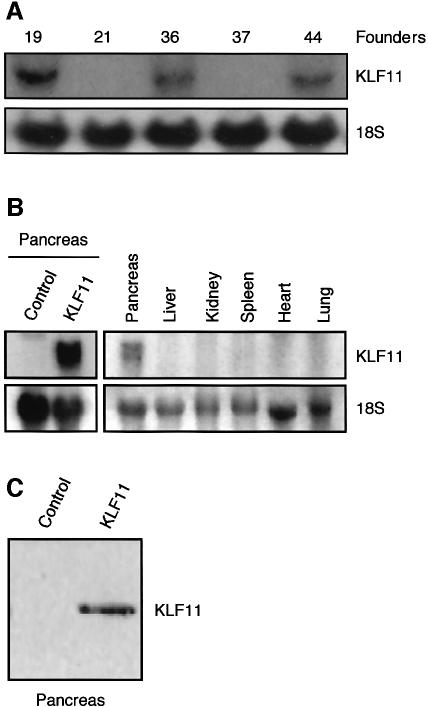
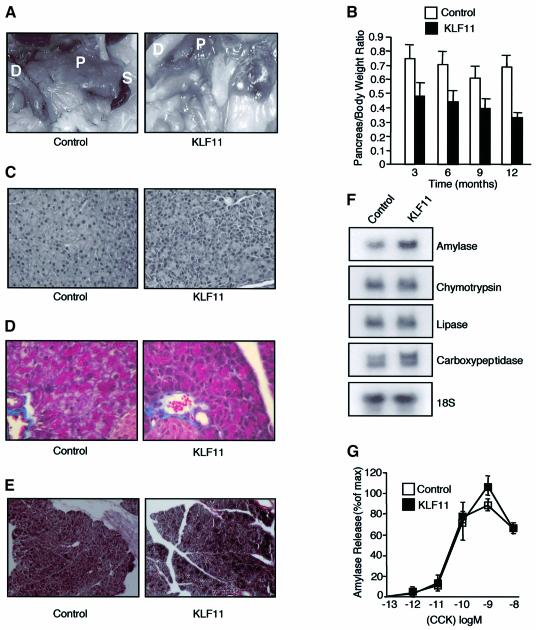
To examine the in vivo role of KLF11 in cell growth, we generated transgenic mice using the elastase1 promoter (Ela1). Ela1 has been used for many years to target the expression of transgene specifically to the acinar cell of the pancreas (Hammer et al., 1987). The major advantage of using this construct in our study is that it would allow us to characterize the in vivo effect of KLF11 without leading to a lethal phenotype. Several transgenic lines, 19, 36 and 44, displaying a similar transgene expression and phenotype, were developed (Figure 1A). Expression of the transgene was detected by northern blot analysis in the pancreata of KLF11 transgenic mice, but not in the liver, kidney, spleen, heart or lung (Figure 1B). Western blot analysis showed a strong signal of KLF11 in the pancreata of KLF11 transgenic animals compared with the control (Figure 1C). For the remainder of this study, we will report data derived from line 44, but similar results were obtained with other lines. The KLF11 mice had significantly smaller pancreata than controls. Grossly, these organs appeared lighter with higher transparency, both features suggestive of pancreatic atrophy (Figure 2A). The weight of the pancreas, expressed as a percentage of total body weight, was also significantly reduced. This reduction in organ size was age dependent, reaching a 35% decrease at 3 months (n = 14) and up to 50% at 12 months (n = 12) (Figure 2B). Histological analysis of the pancreas of KLF11 mice showed mild changes such as a slight degree of cellular pleomorphism (Figure 2C). These changes gave the appearance of an incipient loss of normal acini-like organization that is reminiscent of primary pancreatic acinar atrophy, an alteration in epithelial cells without fibrosis as revealed by Masson and reticulin staining (Figure 2D and E). These morphological alterations were evident at 6 weeks and remained identical throughout the life span of the mouse (data not shown). Expression of acinar markers such as amylase, lipase trypsin, carboxypeptidase, chymotrypsin and elastase revealed no change in KLF11 mice versus controls, suggesting that the differentiation of acinar cells was not altered (Figure 2F). Acinar cells contribute to ∼95% of the total mass of the pancreas, whereas ductular and islet cells together make up the remaining 5%. This remarkable abundance of acinar cells explains why, when these cells are properly matured, a decrease of ∼50% in the mass of the pancreas (Figure 2A and B), without significant expansion of another non-acinar cell population, does not significantly change the relative levels of acinar markers. This normal expression of acinar markers is in agreement with a quasi-normal histology and an appropriate functional maturation of these cells as shown by their unaltered secretory properties, as assayed by measuring the release of amylase in response to cholecystokinin (CCK) (Figure 2G).
Fig. 1. KLF11 expression in transgenic mice. (A) Northern blot analysis was used to determine transgene expression. Similar mRNA levels of transgene were detected in the different transgenic lines. (B) Expression of the transgene could be detected in the pancreata of positive mice, but not in the liver, kidney, spleen, heart or lung. (C) Control western blot analysis shows KLF11 expression in transgenic animals.
Fig. 2. KLF11 transgenic mice show reduced organ size. (A) Macroscopic inspection of the KLF11 transgenic mice revealed that the pancreata appeared smaller, lighter and had a higher transparency when compared with negative controls, suggestive of pancreatic atrophy. D, duodenum; P, pancreas; S, spleen. (B) The relative pancreatic weight expressed as a percentage of total body weight was significantly decreased in the KLF11 transgenic mice for all age groups compared with wild-type mice. (C) The pancreas of KLF11-overexpressing mice showed mild but consistent changes in the pancreas architecture, such as a slight degree of cellular pleomorphism and an incipient loss of normal acini-like organization. The typical spheroidal collections of acinar cells that form an acini unit in the negative control animals were mildly disrupted in KLF11 transgenic mice. (D and E) Mason’s and reticulin staining did not reveal any differences between KLF11 transgenic and control animals. (F) Expression analysis of the acinar enzymes amylase, lipase trypsin, carboxypeptidase, chymotrypsin and elastase revealed no change in KLF11 mice in any age group as compared to controls. (G) The release of amylase induced by CCK in isolated acini of pancreas mice showed no differences between control and KLF11 mice.
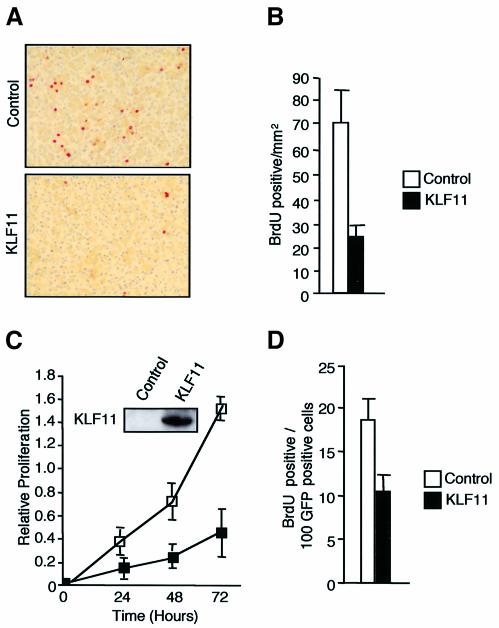
To determine whether the reduction in organ size was due to an alteration in cell proliferation, we performed BrdU incorporation. Since under basal conditions the adult pancreas shows negligible cell proliferation, we examined the proliferative response of the pancreas using a model of regeneration induced by dl-ethionine administration (Walker et al., 1993). The pancreas of KLF11 transgenic mice showed a significant decrease in BrdU-labeling compared with controls (Figure 3A and B). Similar results were obtained in the PANC1 exocrine pancreatic epithelial cell line transfected with KLF11 (Figure 3C and D).
Fig. 3. KLF11 is a negative regulator of cell proliferation in pancreatic epithelial cells both in vitro and in vivo. (A) BrdU immunohistochemistry showing a representative staining of pancreas from wild-type and KLF11 transgenic mice. (B) The pancreas of the KLF11 transgenic mice showed increases in BrdU label compared with wild-type control in the DL-ethionine-induced atrophy model. (C) KLF11-transfected cells display a significant reduction in cell proliferation measured by MTS as compared with cells transfected with the control plasmid. Control western blot shows expression of the KLF11 full-length protein. (D) A similar effect is also observed in BrdU incorporation in PANC1 cells transfected with human full-length KLF11 and control vector.
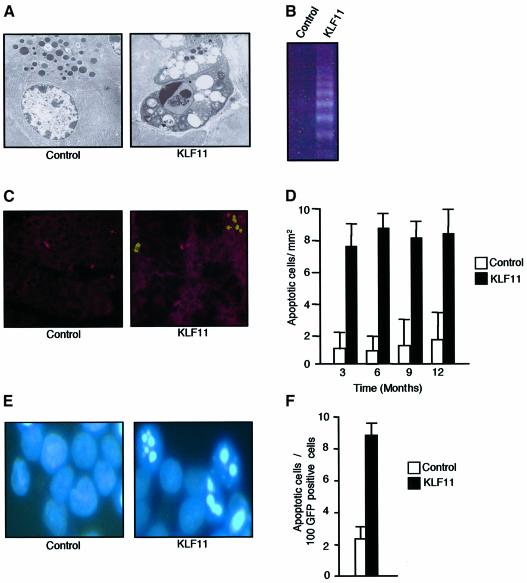
In addition, the pancreas of the KLF11 transgenic mice showed an increase in the number of apoptotic cells, but not necrosis. Apoptosis was evidenced by appearance of the typical signs of apoptosis such as compaction and segregation of chromatin, condensation of the nuclei, the formation of cytoplasmic blebs and apoptotic bodies as seen using electron microscopy, and an increase in DNA fragmentation as revealed by the formation of the typical DNA ladder (Figure 4B). Subsequently we measured the number of TUNEL-positive cells per mm2. KLF11 transgenic mice showed a 5-fold increase in the number of apoptotic cells at all ages examined (Figure 4C and D). Similar results were also observed in PANC1 cells transfected with KLF11 (Figure 4E and F). These changes in cell growth explain the reduction in organ size since the apoptosis observed in the pancreas throughout the life of the animal (Figure 4C and D) cannot be compensated by new cell proliferation due to a significant decrease in the regeneration capabilities of acinar cells (Figure 3A and B). Thus, together these results demonstrate that KLF11 is a negative regulator of epithelial cell growth in vitro and in vivo, through a decrease in proliferation and an increase in apoptosis.
Fig. 4. KLF11 overexpression shows an increase in the rate of apoptosis both in vivo and in vitro. (A) Electron microscopic examination revealed typical signs of apoptosis in transgenic mice, such as the formation of cytoplasmic blebs, and apoptotic bodies were also found in transgenic mice in addition to chromatin condensation. (B) KLF11 transgenic mice showed the formation of a typical DNA ladder as a result of internucleosomal fragmentation. (C) TUNEL staining showing a representative pancreas section of 3-month-old KLF11 transgenic and wild-type mice. (D) To quantify the degree of apoptosis, we counted the number of TUNEL-positive cells per mm2 and calculated the change of apoptosis. As demonstrated, a 5-fold increase in the number of apoptotic cells was observed in KLF11 transgenic mice compared with wild-type mice. This increase in apoptosis was consistent in KLF11-overexpressing animals of all ages. (E) Nuclear Hoechst staining showed the typical morphological changes of the apoptosis KLF11-transfected PANC1 cells. (F) KLF11 overexpression induces 3- to 4-fold increases in the rate of apoptosis in PANC1 cells compared with cells transfected with the empty vector.
KLF11 suppresses oncogene-induced neoplastic transformation
A subset of proteins that negatively regulates cell growth also suppresses neoplastic transformation induced by potent oncogenic pathways. These data suggest that proteins of this type participate in a cellular response that is designed to be dominant over both the proliferative and survival function of constitutively active oncogenic mutations. Constitutively active mutations such as K-ras are found in preneoplastic lesions, which are thought to transform only after the inactivation of suppressive genes (Urrutia et al., 2002). Thus, the identification of these proteins, which have a dominant effect over oncogenic pathways, is important for understanding better the molecular machinery that is sufficient to inhibit cell growth in a wide variety of biological and pathobiological contexts. Therefore, here we assessed whether the negative regulation of cell growth by KLF11 was sufficient to suppress neoplastic transformation, using foci formation assays in NIH 3T3 cells. KLF11 was found to significantly reduce the number of foci induced by oncogenic H-Ras compared with the cells transfected with the control plasmid. This inhibitory effect on ras-mediated transformation was not due to a decrease in H-Ras expression as controlled by northern blot (Figure 5A and C). In addition to inhibiting H-Ras, KLF11 suppressed transformation by K-RasA, K-RasB, mutant Gα12 and middleT (Figure 5D and E), suggesting that this protein can inhibit the oncogenic cascades previously described to induce cancer in a variety of tissues. Taken together, these results indicate that KLF11 can function as a potent suppressor of neoplastic cell growth.
Fig. 5. KLF11 suppresses oncogene-mediated transformation in an mSin3A-dependent manner. (A) The overexpression of KLF11 in NIH 3T3 cells significantly reduces the number of foci induced by a mutant H-ras when compared with cells transfected with H-ras alone. (B) This inhibition of the ras-mediated anchorage-dependent growth was not due to decreased H-ras expression caused by co-transfection with KLF11, as shown by northern blot analysis. (C) Cells transfected with H-ras alone had ∼8-fold differences compared with those cotransfected with H-ras and KLF11 in the number of foci. (D and E) KLF11 inhibits not only ras-mediated, anchorage-dependent growth, but also anchorage-dependent growth induced by other well characterized oncogenes such as Gα12, K-ras A, K-ras B and middleT, compared with cells transfected with oncogene alone. Taken together, these results indicate that KLF11 can function as a potent suppressor of anchorage-dependent cell growth induced by several well characterized oncogenes in vitro. (F) KLF11 deletion mutants that lack SID or contain only the zinc finger domain are not able to inhibit proliferation. Control western blot shows that the expression of the mutants is similar to the full length KLF11. (G) The KLF11 full-length protein can inhibit anchorage-dependent growth induced by H-ras, whereas the deletion mutants that lack SID or contains only the zinc finger can not. (H) KLF11 is strongly suppressed in tumors derived from pancreas, breast, kidney, lung, ovary, colon and stomach when compared with corresponding normal cDNA. The results shown are representative examples of the tumors examined.
Suppression of cell proliferation and neoplastic transformation by KLF11 requires the Mad1-like SID
The interaction of KLF11 with the corepressor mSin3A via the SID is required for transcriptional repression (Zhang,J.S. et al., 2001), although the cell biological importance of this mechanism remains poorly understood. To determine whether the interaction with mSin3A is important for KLF11-mediated inhibition of cell proliferation, we performed an MTS cell proliferation assay in Chinese hamster ovary (CHO) cells transfected wild-type KLF11 or with a SID-deleted KLF11 mutant. As predicted from the results described previously (Figure 3C and D), 72 h after transfection the wild-type KLF11 decreased proliferation by 60% (Figure 5F). In contrast, cells that were transfected with SID-deleted KLF11 proliferated similar to control (Figure 5F). Similar results were observed using a mutant lacking the entire N-terminal domain of the protein, indicating that other sequences present in the SID-deleted KLF11 mutant are unlikely contribute to regulation of cell proliferation (Figure 5F). Mutants lacking the SID domain also lose the ability to inhibit neoplastic transformation (Figure 5G). Thus, these results suggest that mSin3A is required for both KLF11-mediated cell proliferation inhibition and suppression of neoplastic transformation.
The expression of KLF11 mRNA is downregulated in human cancers
We gathered initial evidence on the downregulation of KLF11 in human tumors through virtual SAGE and Digital Differential Display PCR analysis of human genome data, available from public databases. This approach suggested that the levels of KLF11 are reduced in a variety of tumors, in particular breast, pancreas, kidney, lung, ovary and stomach cancers. To confirm this observation experimentally, we used a multi-tissue array containing matched normal and tumor samples combined with Taqman PCR analysis. KLF11 expression was significantly reduced in 50% of tumors derived from pancreas (n = 20), breast (n = 53) and kidney (n = 20), and 10–20% of the tumors from lung (n = 21), ovary (n = 14), colon (n = 38) and stomach (n = 28). Downregulation of KLF11 mRNA levels in this assay was taken as any reduction in the tumor greater than 40% of that found in the matched normal cDNA used as control (Figure 5H). These results are also in agreement with multitumor array data deposited in the Source database (http://source.stanford.edu/cgi-bin/sourceSearch). Therefore, together, bioinformatic evidence and experimental data demonstrate that KLF11 mRNA expression is reduced in human tumors.
KLF11 dowregulates key genes oxidative stress scavenger genes in vitro and in vivo, requiring the SID
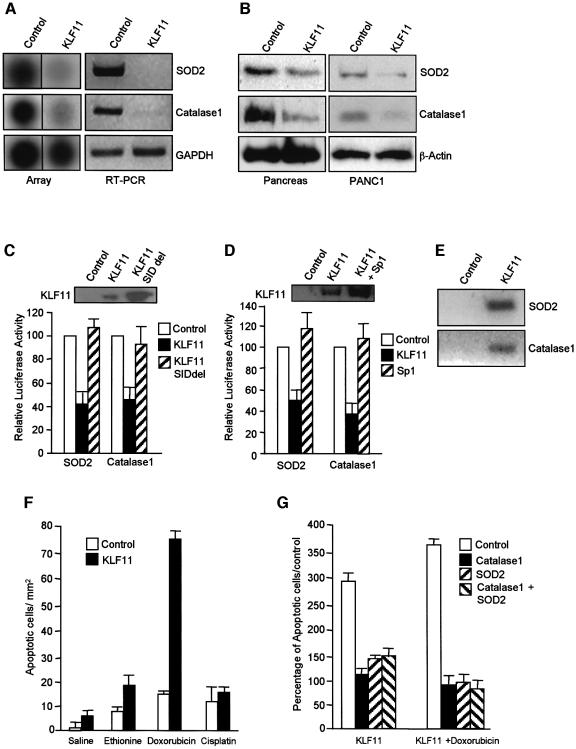
Based on the phenotype of the transgenic mice, we hypothesized that KLF11 may be involved in the regulation of genes that participate in apoptosis, cell cycle pathways and/or a general stress response pathway traditionally associated with growth regulation. The level of expression of several cell cycle regulators, apoptotic, and DNA repair genes does not change in the transgenic compared to wild-type animals as well as in cells transfected with KLF11 (see Supplementary data, available at The EMBO Journal Online). However, a cDNA array analysis of genes from functional classes, including oxidative stress, DNA repair, general stress response and DNA replication, revealed that KLF11 transgenic mice displayed a decrease in the expression of genes encoding key scavengers of oxidative stress such as SOD2 and Catalase1 (Figure 6A). The decreases in the levels of SOD2 and Catalase1 detected using array experiments was confirmed by RT–PCR and western blot analysis (Figure 6A and B). Interestingly, the promoters of these genes contain GC-rich sequences of the type previously shown to be downregulated by KLF11 (Nenoi et al., 2001; Xu et al., 2002). Therefore, we tested whether KLF11 can regulate these promoters using a luciferase reporter assay. Indeed, transfection of PANC1 cells with KLF11 results in repression of the promoter activity of these genes by >50% of controls levels (Figure 6C). Furthermore, deletion of the SID domain from KLF11 leads to a significant reduction in KLF11-mediated repression of the SOD2 and Catalase1 promoters (Figure 6C), indicating that this repression involves interactions with mSin3A. In addition, the expression of exogenous Sp1 relieved the repression mediated by KLF11 in a dose-dependent manner (Figure 6D), supporting the model of competition between Sp1 and SP1-like repressors previously proposed by our laboratory (Kaczynski et al., 2001, 2002, 2003). Moreover, chromatin immunoprecipitation assay demonstrated that SOD2 and Catalase1 are direct targets of KLF11 (Figure 6E).
Fig. 6. KLF11 downregulates oxidative stress scavengers. (A) KLF11 transgenic mice displayed a decrease in the expression of genes encoding key scavengers of oxidative stress such as SOD2 and Catalase1, as shown by array analysis and RT–PCR. (B) Western blot analysis shows a decrease in the protein levels of SOD2 and Catalase1 in KLF11 transgenic mice and PANC1 cells transfected with KLF11 (C) KLF11 overexpression in PANC1 results in the reduction of the promoter activity of both SOD2 and Catalase1. The mutant that lacks the Mad1-like SID motif loses the repressor activity on the promoters of oxidative stress scavengers. Control western blot shows expression of the KLF11 wild-type and mutant proteins. (D) Competition with Sp1 relieves the repression mediated by KLF11 compared with KLF11alone. Control western blot shows expression of the KLF11 wild-type protein. (E) Chromatin immunoprecipitation assay shows that KLF11 can bind the SOD2 and Catalase1 promoters in PANC1 cells. (F) TUNEL assay shows that the KLF11 animals were highly sensitive to the oxidative agent doxorubicin. (G) SOD2 and Catalase1 recombinant adenovirus are able to rescue the apoptotic phenotype induced by KLF11 and doxorubicin.
Thus, the expression of KLF11 inhibits the expression of key oxidative stress scavengers in cultured cells and in vivo. Because of the in vitro and in vivo effects of KLF11 on scavenger genes, we tested whether the pancreata of the transgenic mice were more sensitive to oxidative stress. Indeed, pharmacological experiments showed that KLF11 animals treated with doxorubicin showed a 6-fold increase in the number of apoptotic cells compared with untreated KLF11 animals (Figure 6F). This chemotherapeutic agent has been shown previously to induce apoptosis via oxidative stress in mice, and these results indicate a synergistic effect between both the presence of the transgene and the oxidative chemical. Conversely, mice that overexpressed SOD2 or Catalase1 have increased resistance for oxidative injury induced by doxorubicin (Kang et al., 1996; Yen et al., 1996). Control KLF11 animals treated with agents that act via other mechanisms such as ethionine (inhibition of protein synthesis) (Walker et al., 1993) or cisplatin (DNA damage) (Gonzalez et al., 2001) only showed additive effects when KLF11 transgenic mice were compared with untreated mice. In addition, restoration of normal expression of SOD2 and Catalase1 using recombinant adenovirus in PANC1 cells rescue the apoptotic phenotype induced by doxorubicin and KLF11 (Figure 6G). Together, these data support a role for KLF11 in the negative regulation of oxidative stress genes.
In summary, the results of this study show that KLF11 is downregulated in a subset of epithelial cell tumors and can inhibit cell growth and neoplastic transformation. The cellular mechanisms through which KLF11 probably mediates these effects are decreasing cell proliferation and increasing apoptosis, as observed both in cultured cells and in vivo. Consistent with these results, the expression of KLF11 in the exocrine pancreas of transgenic animals results in a significant reduction in the size and weight of this organ. We have also demonstrated that doxorubicin, an agent that primarily acts by the generation of oxidative stress, synergizes with KLF11 in the induction of apoptosis. Expression profiling experiments and reporter assays demonstrate that the expression of KLF11 decreases the levels of the oxidative stress scavenger genes superoxide dismutase2 and Catalase1. In addition, chromatin immunoprecipitation assay has been used to demonstrate that these genes are direct targets of KLF11. Furthermore, infection with SOD and Catalase1 recombinant adenoviruses rescue cells from KLF11-induced apoptosis, both in the presence and in the absence of doxorubicin. Together, these results demonstrate that KLF11 displays a suppressor effect on cell growth and neoplastic transformation. These data also suggest that, similar to other genes (Drane et al., 2001; Sharpless and Depinho, 2002; Trinei et al., 2002), the effects on growth regulation of KLF11 probably involve the downregulation of genes that function as scavengers of oxidative stress.
Discussion
The motivation of the current study has been to gain a better insight into the growth-suppressive functions of KLF/Sp1-like genes with potential tumor suppressor activity and to test whether these biological functions correlate with their biochemical activities. Indeed, the data described here suggest a highly mechanistic model for understanding the function of KLF11, the best characterized member of a distinct group of Sp1-like transcriptional repressors. According to this model, KLF11 functions as a negative regulator of neoplastic transformation (pathophysiological event), by decreasing cell growth and increasing apoptosis (cellular mechanisms) via a mechanism that involves downregulation of the oxidative stress genes SOD2 and Catalase1, and an increased susceptibility to oxidative insults (biochemical mechanism). These genes are the first targets of KLF11 to be identified. Interestingly, p53 function has also been associated with the production of oxidative stress, suggesting that the modulation of genes involved in this pathway participate in tumor suppression mechanisms (Drane et al., 2001; Sharpless and Depinho, 2002; Trinei et al., 2002). Furthermore, gradual decreases in the expression of antioxidant enzyme expression have been reported in pancreatic cells from the normal pancreas to pancreatic cancer (Cullen et al., 2003). In our studies, these effects require interaction with the corepressor mSin3A (molecular mechanism). The interaction is mediated by a SID, which was originally described in the Mad family of basic helix–loop–helix transcription factors, and was also shown to be necessary for both their repressor and growth-suppressive activities (Roussel et al., 1996). Yochum et al. have recently reported that the mammalian SID-containing transcription factor Pf1 requires this domain to regulate genes that play a role in cellular senescence (Yochum and Ayer, 2002). Together, these two studies, in conjunction with the results reported here, provide evidence to suggest a wider participation of SID-like domains in transcriptional repression and cell growth regulation.
It is also important to discuss inferences derived from this study that can be used to expand our knowledge on the potential role that the entire family of KLF proteins may play in neoplastic transformation. Although these proteins can be structurally divided into several subfamilies, they share a certain degree of redundancy in the recognition of target DNA sequences and, in certain cases, they regulate the same gene promoters. The function of different KLF proteins at these shared promoter sites can be either synergistic or antagonistic, which supports the prediction that by regulating similar genes but in using opposing mechanisms, other KLF proteins may modulate neoplastic transformation. For instance, KLF6 belongs to a different subfamily of KLF proteins than KLF11. Members of this subfamily do not contain an mSin3A-interacting domain, and thus KLF6 likely works via a slightly different mechanism than the one proposed here (Narla et al., 2001). Finally, this information makes it tempting to speculate that a defining feature of KLF transcription factors is their ability to modulate oncogenesis and tumor suppression, a premise that should serve as the theoretical framework for future functional studies.
In conclusion, the characterization of KLF11 reported here links the biochemical properties of this protein with its cell biological role in growth regulation. These data have potential broader applications since KLF11 currently serves as the paradigm of a subfamily of KLF/Sp1-like repressors that function through an identical biochemical mechanism, namely via and SID. Furthermore, this study, together with emerging data from other laboratories, thus far support the more general prediction that members of the KLF family of transcriptional regulators may have evolved to participate in the modulation of normal morphogenesis and neoplastic transformation.
Materials and methods
Cell lines, culture conditions and transfection procedures
The human pancreatic epithelial cell line PANC1, the CHO cell line and the mouse fibroblasts (NIH 3T3) were obtained from th American Type Culture Collection (ATCC, Rockville, MD). The cells were cultured and transiently transfected as described previously (Cook et al., 1998; Gebelein et al., 1998; Kaczynski et al., 2001; Zhang,J.S. et al., 2001; Ellenrieder et al., 2002).
Plasmid construct and generation of transgenic mice
For cell growth and transformation studies, the full-length KLF11, KLF11 deletion mutants and Sp1 were cloned into the pcDNA3.1/HisA vector (InVitrogen, Carlsbad, CA) and sequenced. The H-ras, K-rasA and K-rasB plasmids were a gift from Dr E.Santos [National Institutes of Health (NIH), Bethesda, MD], the G12 and G13 plasmids were kindly provided by Dr L.Vitale-Cross, the middle T plasmid was a gift from Dr D.J.McKean (Mayo Clinic, Rochester, MN) and the elastase I promoter was a gift from Dr R.McDonald (University of Pennsylvania, PA). The transgene cassette was made using a pBluescript II SK+ vector (Strategene, La Jolla, CA) as a backbone. The vector was opened up with HindIII/BamHI, and the acinar-specific control region (–500 to +8) from the rat elastase I gene was ligated (20). This elastase/pBluescript vector was digested with BamHI/NotI and the human growth hormone (hGH) 3′ untranslated region (UTR) (+500 to +2657). This construct was digested with BamHI, filled in, dephosphorylated and ligated with human KLF11. Full-length cDNA was released from pcDNA 3.1 His A (Invitrogen) using HindIII/XbaI, filled in and ligated into the pBEG vector, which had been digested with BamHI, filled in and dephosphorylated. A 4 kb HindIII/NotI fragment was isolated and used for microinjections into in-bred FVB zygotes. After two injections, 19 (founders) out of 120 pups tested positive. Genomic DNA was prepared from mice tails and tested either by Southern blot (founders) or PCR (F1 and beyond). Seventeen founders were capable of propagating, and each line was expanded. Founders were sacrificed and the organs were isolated for light and electron microscopy, RNA and protein. Pancreata sections were stained with hematoxylin and eosin (H&E). Lines 19, 36, 37, 44 and 72 showed similar phenotypes, and line 44 was chosen for full analysis because of its high copy number.
Morphological examination and CCK-induced release of amylase
KLF11-transgenic and wild-type control mice were sacrificed at varying ages and a body weight measurement was taken. The whole pancreas was removed, fixed and weighed. The pancreas weight:body weight ratio was calculated. Pancreata were fixed in 10% formaldehyde, embedded in paraffin, sectioned and stained with H&E for the overall examination of the organ. In addition, Trichromic Mason’s and reticulin staining were performed to examine fibrotic marker levels (Histoserv Inc., Gaithersburg, MD). The CCK-induced release of amylase was performed as described previously (Lutz et al., 1993).
Apoptosis assays
PANC1 cells were co-transfected with the KLF11 cDNA (20 µg) and green fluorescent protein (5 µg), which allowed us to separate transfected from untransfected cells by FACS sorting. Cells undergoing apoptosis were identified by the morphological criteria of nuclear fragmentation, nuclear margination and organelle disorganization using the DNA staining Hoechst 33860. Apoptosis was accessed using a TUNEL labeling method (Roche Molecular Biochemicals, Indianapolis, IN) on paraffin-embedded pancreas sections. TUNEL-positive cells were counted in 75 fields, expressed per mm2, and the means calculated for each timepoint. Pancreata from control and KLF11 transgenic mice were fixed in 2.5% glutaraldehyde pH 7.4, and embedded in Epon-Araldite. Samples fixed in aldehyde solutions only were embedded in the hydrophilic London White Resin (London Resin, UK) according to standard procedures. Areas of interest were trimmed, sectioned with a Sorvall Porter-Bloom MT2 ultramicrotome (60–100 nm), collected on uncoated nickel 200–300 mesh grids, and observed in a Zeiss 10CR transmission electron microscope (Carl Zeiss Inc., Oberkochen Germany). For the detection of the nucleosomal ladder we performed a PCR-based DNA Ladder (Clontech Laboratories, Palo Alto, CA). DNA was extracted from pancreata of control and KLF11 transgenic mice, ligated using LM adaptors, and amplified by PCR for 25 cycles. The resulting ladder was visualized on a 1.2% agarose gel. For rescue experiments, KLF11-transfected PANC1 cells were infected with SOD2 (Zwacka et al., 1998) and Catalase1 (Lam et al., 1999) recombinant adenovirus (Gene Transfer Vector Core, University of Iowa). At 24 h post-infection the cells were treated with doxorubicin at concentration of 1 µM for 12 h. Nuclear Hoechst staining was used to quantify the number of apoptotic cells.
Proliferation and transformation assay
MTS proliferation assay with PANC1 and CHO cells were performed as described previously (Cook et al., 1998). BrdU incorporation was used to measure proliferation in KLF11-positive and -negative mice of all ages by performing immunohistochemistry on paraffin-embedded pancreas sections using the anti-BrdU antibody (Amersham Pharmacia Biotech, Freiburg, Germany). Mice were injected with 50 mg/kg body weight BrdU (Amersham Pharmacia Biotech) 12 h before they were sacrificed. Pancreata were extracted and fixed, embedded and processed for immunohistochemistry. After deparaffination the slides were rinsed in phosphate-buffered saline and processed with the Histo-mouse kit (Zymed, South San Francisco, CA). Positively stained cells were counted in 75 fields. Averages and standard deviations were calculated from this data and are shown in Figure 2. The transformation assays were performed as described previously (Gebelein et al., 1998).
Expression studies
We used the stress mouse cDNA expression array with 145 mouse cDNAs spotted on a nylon membrane (Clontech). Six identical membranes were used in parallel, in order to receive the expression profiles of three KLF11 transgenic compared with three control mice. The hybridization pattern was quantified by phosphorimaging followed by Atlas Image™ 1.0 program (Clontech) analysis. For the tumor screening, we used a matched tumor/normal array (Clontech). A KLF11-specific probe was hybridized onto an array containing 241 paired cDNA samples derived from multiple human tumors and corresponding normal tissue from individual patients. The relative expression was assessed by comparing the signal obtained in one membrane (after normalizing to the housekeeping genes) relative to a second membrane. Single-stranded cDNA was generated from 2 µg of total RNA using the first stranded synthesis kit (Stratagene). cDNA (40–80 ng) was amplified in a 50 µl PCR reaction mixture with specific primers for SOD2, Catalase1 and GAPDH. The amplified products were visualized on an ethidium bromide-stained agarose gel. Northern and western blots were performed as described previously (Cook et al., 1998; Gebelein et al., 1998).
Reporter assay
SOD2-luciferase and Catalase1-luciferase constructs were generated as described previously (Kang et al., 1996; Yen et al., 1996). PANC1 cells were cultured in six-well tissue culture plates and co-transfected with KLF11 full-length SINdel mutant, Sp1 and reporter plasmids. The parental pcDNA 3.1 His A vector alone was used as a control for basal transcriptional activity. As a control for transfection efficiency, all conditions included co-transfection with the RSV-Renilla luciferase control plasmid. At 24 h after transfection, the dual luciferase assays were performed with a Turner 20/20 luminometer (Promega, Madison, WI). For SOD2 and Catalase1 reporter assays, the relative luciferase activity ± standard deviation (SD) is the ratio of Renilla-normalized SOD2 and Catalase1 values. All reporter studies were performed in triplicate in at least three independent experiments with similar results.
Chromatin immunoprecipitation
PANC1 cells were transfected with KLF11 full-length FLAG-tagged plasmids. At 24 h post-transfection, cells were cross-linked with formaldehyde for 20 min at 25°C, harvested in SDS–lysis buffer (Upstate Biotechnology, Lake Placid, NY), and sheared to fragment DNA (∼500 bp). Samples were then immunoprecipitated using an agarose-conjugated anti-FLAG antibody (Sigma-Aldrich) or agarose beads alone at 4°C overnight. Following immunoprecipitation, samples were washed and eluted using the Chromatin Immunoprecipitation Kit (Upstate Biotechnology) according to the manufacturer’s instructions. Cross-links were removed at 65°C for 4 h and immunoprecipitated DNA was purified using phenol/chloroform extraction (500 µl) and ethanol precipitation. A 200 bp region of the SOD2 promoter and 195 bp of the Catalase1 promoter were detected in immunoprecipitated samples by PCR. PCR products were visualized on a 2% agarose gel.
DL-ethionine-induced atrophy protocol and pharmacological experiments
Mice were randomly separated into four groups. Animals in groups 1 and 2 were fed standard commercial diet, while groups 3 and 4 received protein depletion diet (Harlan, Indianapolis, IN) and were injected daily (i.p.) with DL-ethionine (Sigma Chemical Co., St Louis, MO) in saline solution at a dose of 0.2 g/kg body weight. After 10 and 20 days the animals were killed to study pancreatic weight, histology, proliferation and apoptosis. In the pharmacological experiments, mice were separated into eight groups. Animals in groups 1 and 2 received a daily injection of saline solution, groups 3 and 4 received a daily injection of 0.04 g of DL-ethionine per kilogram of body weight, groups 5 and 6 were injected daily with 20 mg/kg body weight of doxorubicin (Sigma Chemical Co.), and groups 7 and 8 received an injection of 15 mg/kg body weight cisplatin (American Phamaceutical Partners Inc., Los Angeles, CA). After 2 days the mice were sacrificed and the pancreata were extracted and evaluated for apoptosis.
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank Tiffany Cook and Brian Gebelein for their participation in experiments performed in the early phases of this project. We also thank these investigators and Joanna Kaczynski for their invaluable contributions to our writing and helpful discussions. This work was supported by the Lustgarten Foundation for Pancreatic Cancer Research and the National Institutes of Health grant Nos DK52913 and DK56620 to R.U.
References
- Bieker J.J. (2001) Kruppel-like factors: three fingers in many pies. J. Biol. Chem., 276, 34355–34358. [DOI] [PubMed] [Google Scholar]
- Black A.R., Black,J.D. and Azizkhan-Clifford,J. (2001) Sp1 and kruppel-like factor family of transcription factors in cell growth regulation and cancer. J. Cell. Physiol., 188, 143–160. [DOI] [PubMed] [Google Scholar]
- Cook T. and Urrutia,R. (2000) TIEG proteins join the Smads as TGF-β-regulated transcription factors that control pancreatic cell growth. Am. J. Physiol. Gastrointest. Liver Physiol., 278, G513–G521. [DOI] [PubMed] [Google Scholar]
- Cook T., Gebelein,B., Mesa,K., Mladek,A. and Urrutia,R. (1998) Molecular cloning and characterization of TIEG2 reveals a new subfamily of transforming growth factor-β-inducible Sp1-like zinc finger-encoding genes involved in the regulation of cell growth. J. Biol. Chem., 273, 25929–25936. [DOI] [PubMed] [Google Scholar]
- Cook T., Gebelein,B. and Urrutia,R. (1999a) Sp1 and its likes: biochemical and functional predictions for a growing family of zinc finger transcription factors. Ann. N. Y. Acad. Sci., 880, 94–102. [DOI] [PubMed] [Google Scholar]
- Cook T., Gebelein,B., Belal,M., Mesa,K. and Urrutia,R. (1999b) Three conserved transcriptional repressor domains are a defining feature of the TIEG subfamily of Sp1-like zinc finger proteins. J. Biol. Chem., 274, 29500–29504. [DOI] [PubMed] [Google Scholar]
- Cullen J.J., Mitros,F.A. and Oberley,L.W. (2003) Expression of antioxidant enzymes in diseases of the human pancreas: another link between chronic pancreatitis and pancreatic cancer. Pancreas, 26, 23–27. [DOI] [PubMed] [Google Scholar]
- Drane P., Bravard,A., Bouvard,V. and May,E. (2001) Reciprocal down-regulation of p53 and SOD2 gene expression—implication in p53 mediated apoptosis. Oncogene, 20, 430–439. [DOI] [PubMed] [Google Scholar]
- Ellenrieder V., Zhang,J.S., Kaczynski,J. and Urrutia,R. (2002) Signaling disrupts mSin3A binding to the Mad1-like Sin3-interacting domain of TIEG2, an Sp1-like repressor. EMBO J., 21, 2451–2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebelein B., Fernandez-Zapico,M., Imoto,M. and Urrutia,R. (1998) KRAB-independent suppression of neoplastic cell growth by the novel zinc finger transcription factor KS1. J. Clin. Invest., 102, 1911–1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiman D.E., Ton-That,H., Johnson,J.M. and Yang,V.W. (2000) Transactivation and growth suppression by the gut-enriched Kruppel-like factor (Kruppel-like factor 4) are dependent on acidic amino acid residues and protein-protein interaction. Nucleic Acids Res., 28, 1106–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez V.M., Fuertes,M.A., Alonso,C. and Perez,J.M (2001) Is cisplatin-induced cell death always produced by apoptosis? Mol. Pharmacol., 59, 657–663. [DOI] [PubMed] [Google Scholar]
- Hammer R.E., Swift,G.H., Ornitz,D.M., Quaife,C.J., Palmiter,R.D., Brinster,R.L. and MacDonald,R.J. (1987) The rat elastase I regulatory element is an enhancer that directs correct cell specificity and developmental onset of expression in transgenic mice. Mol. Cell. Biol., 7, 2956–2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaczynski J.A., Zhang,J.S., Ellenrieder,V., Conley,A., Duenes,T., Kester,H., van Der Burg,B. and Urrutia,R. (2001) The Sp1-like protein BTEB3 inhibits transcription via the basic transcription element box by interacting with mSin3A and HDAC-1 co-repressors and competing with Sp1. J. Biol. Chem., 276, 36749–36756. [DOI] [PubMed] [Google Scholar]
- Kaczynski J.A., Conley,A.A., Fernandez-Zapico,M., Delgado,S.M., Zhang,J.S. and Urrutia,R. (2002) Functional analysis of basic transcription element (BTE)-binding protein (BTEB) 3 and BTEB4, a novel Sp1-like protein, reveals a subfamily of transcriptional repressors for the BTE site of the cytochrome P4501A1 gene promoter. Biochem. J., 366, 873–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaczynski J., Cook,T. and Urrutia,R. (2003) Sp1- and Kruppel-like transcription factors. Genome Biol., 4, 206–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y.J., Chen,Y. and Epstein,P.N. (1996) Suppression of doxorubicin cardiotoxicity by overexpression of catalase in the heart of transgenic mice. J. Biol. Chem., 271, 12610–12616. [DOI] [PubMed] [Google Scholar]
- Lam E.W., Zwacka R., Seftor,E.A., Nieva,D.R., Davidson,B.L., Engelhardt J.F., Hendrix,M.J., Oberley,L.W. (1999) Effects of antioxidant enzyme overexpression on the invasive phenotype of hamster cheek pouch carcinoma cells. Free Radic. Biol. Med., 27, 572–579. [DOI] [PubMed] [Google Scholar]
- Lutz M.P., Sutor,S.L., Abraham,R.T. and Miller,L.J. (1993) A role for cholecystokinin-stimulated protein tyrosine phosphorylation in regulated secretion by the pancreatic acinar cell. J. Biol. Chem., 268, 11119–11124. [PubMed] [Google Scholar]
- Marin M., Karis,A., Visser,P., Grosveld,F. and Philipsen,S. (1997) Transcription factor Sp1 is essential for early embryonic development but dispensable for cell growth and differentiation. Cell, 89, 619–628. [DOI] [PubMed] [Google Scholar]
- Narla G. et al. (2001) KLF6, a candidate tumor suppressor gene mutated in prostate cancer. Science, 294, 2563–2566. [DOI] [PubMed] [Google Scholar]
- Nenoi M., Ichimura,S., Mita,K., Yukawa,O. and Cartwright,I.L. (2001) Regulation of the catalase gene promoter by Sp1, CCAAT-recognizing factors and a WT1/Egr-related factor in hydrogen peroxide-resistant HP100 cells. Cancer Res., 61, 5885–5894. [PubMed] [Google Scholar]
- Perkins A.C., Sharpe,A.H. and Orkin,S.H. (1995) Lethal β-thalassaemia in mice lacking the erythroid CACCC-transcription factor EKLF. Nature, 375, 318–322. [DOI] [PubMed] [Google Scholar]
- Roussel M.F., Ashmun,R.A., Sherr,C.J., Eisenman,R.N. and Ayer,D.E. (1996) Inhibition of cell proliferation by the Mad1 transcriptional repressor. Mol. Cell. Biol., 16, 2796–2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpless N.E. and DePinho,R.A. (2002) p53: good cop/bad cop. Cell, 110, 9–12. [DOI] [PubMed] [Google Scholar]
- Tewari R., Gillemans,N., Wijgerde,M., Nuez,B., von Lindern,M., Grosveld,F. and Philipsen,S. (1998) Erythroid Kruppel-like factor (EKLF) is active in primitive and definitive erythroid cells and is required for the function of 5′HS3 of the β-globin locus control region. EMBO J., 17, 2334–2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinei M. et al. (2002) A p53-p66Shc signalling pathway controls intracellular redox status, levels of oxidation-damaged DNA and oxidative stress-induced apoptosis. Oncogene, 21, 3872–3878. [DOI] [PubMed] [Google Scholar]
- Turner J. and Crossley,M. (1998) Cloning and characterization of mCtBP2, a co-repressor that associates with basic Kruppel-like factor and other mammalian transcriptional regulators. EMBO J., 17, 5129–5140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner J. and Crossley,M. (1999) Mammalian Kruppel-like transcription factors: more than just a pretty finger. Trends Biochem. Sci., 24, 236–240. [DOI] [PubMed] [Google Scholar]
- Urrutia R., Conley,A.A., Ellenrieder,V., Kaczynski,J. and DiMagno,E. (2002) Pancreatic cancer: cellular and molecular mechanisms underlying its development. In Encyclopedia of cancer. Elsevier Science, New York, NY, pp. 250–258. [Google Scholar]
- van Vliet J., Turner,J. and Crossley,M. (2000) Human Kruppel-like factor 8: a CACCC-box binding protein that associates with CtBP and represses transcription. Nucleic Acids Res., 28, 1955–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker N.I., Winterford,C.M., Williamson,R.M. and Kerr,J.F. (1993) Ethionine-induced atrophy of rat pancreas involves apoptosis of acinar cells. Pancreas, 8, 443–449. [DOI] [PubMed] [Google Scholar]
- Xu Y., Porntadavity,S. and St Clair,D.K. (2002) Transcriptional regulation of the human manganese superoxide dismutase gene: the role of specificity protein 1 (Sp1) and activating protein-2 (AP-2). Biochem. J., 362, 401–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen H.C., Oberley,T.D., Vichitbandha,S., Ho,Y.S. and St Clair,D.K. (1996) The protective role of manganese superoxide dismutase against adriamycin-induced acute cardiac toxicity in transgenic mice. J. Clin. Invest., 98, 1253–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yochum G.S. and Ayer,D.E. (2002) Role for the mortality factors MORF4, MRGX and MRG15 in transcriptional repression via associations with Pf1, mSin3A and Transducin-Like Enhancer of Split. Mol. Cell. Biol., 22, 7868–7876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.S., Moncrieffe,M.C., Kaczynski,J., Ellenrieder,V., Prendergast,F.G. and Urrutia,R. (2001) A conserved alpha-helical motif mediates the interaction of Sp1-like transcriptional repressors with the corepressor mSin3A. Mol. Cell. Biol., 21, 5041–5049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Kadam,S., Emerson,B.M. and Bieker,J.J. (2001) Site-specific acetylation by p300 or CREB binding protein regulates erythroid Kruppel-like factor transcriptional activity via its interaction with the SWI-SNF complex. Mol. Cell. Biol., 21, 2413–2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwacka R.M., Zhou,W., Zhang,Y., Darby,C.J., Dudus,L., Halldorson,J., Oberley,L. and Engelhardt,J.F. (1998) Redox gene therapy for ischemia/reperfusion injury of the liver reduces AP1 and NF-κB activation. Nat. Med., 4, 698–704. [DOI] [PubMed] [Google Scholar]