Abstract
Monoubiquitylation is a well-characterized signal for the internalization and sorting of integral membrane proteins to distinct cellular organelles. Recognition and transmission of monoubiquitin signals is mediated by a variety of ubiquitin-binding motifs such as UIM, UBA, UEV, VHS and CUE in endocytic proteins. The yeast Vps27 protein requires two UIMs for efficient interactions with ubiquitin and for sorting cargo into multivesicular bodies. Here we show that the individual UIMs of Vps27 exist as autonomously folded α-helices that bind ubiquitin independently, non-cooperatively and with modest affinity. The Vps27 N-terminal UIM engages the Leu8–Ile44–Val70 hydrophobic patch of ubiquitin through a helical surface conserved in UIMs of diverse proteins, including that of the S5a proteasomal regulatory subunit. The Leu8–Ile44–Val70 ubiquitin surface is also the site of interaction for CUE and UBA domains in endocytic proteins, consistent with the view that ubiquitin- binding endocytic proteins act serially on the same monoubiquitylated cargo during transport from cell surface to the lysosome.
Keywords: endocytosis/monoubiquitin signaling/protein sorting/ubiquitin-binding motif/UIM
Introduction
Intracellular membrane protein transport relies on the presence of specific signals within the cytosolic domains of cargo proteins. Short linear peptide motifs comprise a well-characterized class of signals that are recognized by components of coat protein complexes associated with the cytosolic membrane surface (Bonifacino and Traub, 2003). Monoubiquitylation of membrane proteins is another signal for internalization and endosomal sorting, and constitutes an important mechanism of downregulation of activated cell surface receptors (Di Fiore et al., 2003). Multiple proteins in the endocytic pathway mediate recognition and transmission of the monoubiquitin signal through direct physical association. Efficient and accurate targeting to the destination organelle relies on ubiquitin-binding motifs within these proteins including the UIM (ubiquitin interacting motif), UBA (ubiquitin-associated), UEV (ubiquitin E2 variant), VHS (Vps27/Hrs/STAM) and CUE (similar to a domain in the yeast Cue1 protein).
The UIM is a recurring motif in endocytic proteins (Hofmann and Falquet, 2001). UIMs of several endocytic proteins bind ubiquitin directly in vitro. These include the epsins, eps15 and eps15R proteins that function at the internalization step of endocytosis and also the Vps27/Hrs and Hse1/STAM proteins associated with sorting functions at the early and late endosomes (Bilodeau et al., 2002; Oldham et al., 2002; Polo et al., 2002; Raiborg et al., 2002; Shih et al., 2002). Epsins and Vps27/Hrs are thought to function as molecular adapters that link membrane cargo to vesicle budding machinery because they harbor additional domains with potential or demonstrated phospholipid and clathrin coat protein binding activities. Deletions of UIMs and point mutations of conserved UIM residues not only hinder ubiquitin binding in vitro (Bilodeau et al., 2002; Oldham et al., 2002; Polo et al., 2002; Raiborg et al., 2002; Shih et al., 2002; Fisher et al., 2003), but also compromise transport of ubiquitylated cargo in vivo (Bilodeau et al., 2002; Shih et al., 2002).
The UIM is a short 15-residue sequence motif deduced from similarity searches using the ubiquitin-binding sequences of the S5a regulatory subunit of the proteasome (Young et al., 1998; Hofmann and Falquet, 2001). In addition to several endocytic proteins and the S5a subunit itself, the motif is found in ubiquitin ligases, deubiquitylating enzymes and proteins with other cellular functions (Hofmann and Falquet, 2001). Intriguingly, many UIM-containing proteins harbor multiple copies of the ubiquitin-binding motif, while several proteins with single UIMs are found in non-covalent complexes with other UIM-containing proteins. Mutations within a single UIM in multi-UIM-containing proteins produce larger effects than expected if these UIMs functioned in a redundant fashion (Polo et al., 2002; Shih et al., 2002). Further, the relatively modest affinity of single UIMs for monoubiquitin (Raiborg et al., 2002; Shekhtman and Cowburn, 2002; Fisher et al., 2003) leaves open the possibility that mechanisms of high-affinity binding may involve cooperation between multiple UIMs.
Previous genetic, biochemical and biophysical analyses have provided important clues regarding the molecular mechanism of ubiquitin binding by UIMs, but more detailed models of the interaction have not been described. These studies have implicated conserved residues within the UIM in ubiquitin binding (Polo et al., 2002; Raiborg et al., 2002; Shih et al., 2002; Fisher et al., 2003) and highlighted the importance of a hydrophobic patch on the ubiquitin surface in UIM interactions (Beal et al., 1996; Shekhtman and Cowburn, 2002; Shih et al., 2002; Walters et al., 2002; Fisher et al., 2003). An overlapping hydrophobic patch is involved in binding to the CUE ubiquitin-binding motif and high-resolution structures for two CUE–ubiquitin complexes have recently been described (Kang et al., 2003; Prag et al., 2003). These studies also revealed a structural similarity between the CUE and UBA ubiquitin-binding motifs and implied a conserved mode of interaction with ubiquitin. However, no such relationship between the UIM and CUE/UBA motifs has been recognized, suggesting that the mode of engagement of ubiquitin by UIMs may be distinct from that described for CUE.
Here we present the results of our structural analyses and characterization of the ubiquitin-binding properties of a domain of yeast Vps27 that spans both copies of the UIM in the protein (henceforth designated the tandem Vps27 UIM) (Figure 1A). We also present the solution structure of the complex formed by the N-terminal UIM of Vps27 and ubiquitin, test the predictions of the structure via mutational analysis and propose a structural model for the tandem Vps27 UIM–ubiquitin interaction.
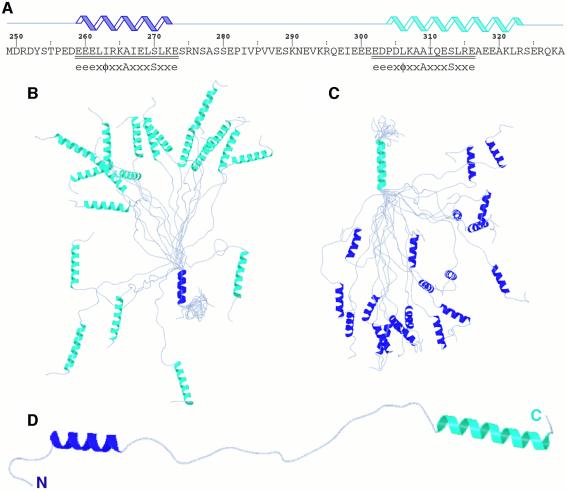
Fig. 1. Solution structure of the unbound tandem Vps27 UIM. (A) Sequence and location of secondary structural elements in the tandem UIM fragment. UIM residues are underlined and aligned with the proposed motif (Hofmann and Falquet, 2001). Ensemble of 20 NMR structures superimposed on the polypeptide backbones of (B) the αA-helix (blue) and (C) the αB-helix (cyan). (D) A representative structure from the NMR ensemble. These views emphasize the high definition of the helical regions, the unstructured nature of the linker region and the lack of tertiary contacts between helices.
Results
Structure of the tandem Vps27 UIM
The NMR spectrum of the tandem Vps27 UIM was characterized by limited chemical shift dispersion in the amide proton region (∼0.8 p.p.m.) that suggested the widespread presence of unstructured and/or helical regions. Analysis of the secondary chemical shifts of backbone carbon nuclei revealed the existence of two discrete helical segments. The polypeptide backbone within these segments appeared to be relatively constrained compared with other regions of the molecule, as the measured {1H}–15N heteronuclear nuclear Overhauser effect (NOE) values were significantly higher than the values measured for the entire domain (0.43 ± 0.11 versus 0.09 ± 0.42, respectively).
Because relatively stable isolated helices are uncommon, and since multiple UIMs are commonly found in UIM-containing proteins or protein complexes, we sought to test whether the individual helices associated with each other to give rise to a higher-order structure. Toward this end, we determined the NMR structure of the tandem Vps27 UIM using a combination of 1H–1H NOE-based distance restraints and chemical shift and scalar coupling constant based torsion angle restraints (Table I). Complete NOE assignments were obtained following an objective iterative automated approach (Linge et al., 2003). Twenty structures in excellent agreement with the input experimental restraints were selected for analysis (Table I).
Table I. NMR structure determination statistics.
| Restraint statistics |
Tandem Vps27 UIM |
Vps27 UIM-1–ubiquitin complex |
| Distance restraints | 858 | 2138 |
| Unambiguous NOE distance restraints | 412 | 1625 |
| Intraresidue | 256 | 846 |
| Sequential (|i – j| = 1) | 124 | 295 |
| Medium range (1 < |i – j| ≤ 4) | 32 | 231 |
| Intramolecular long range (|i – j| > 4) | 0 | 177 |
| Intermolecular | 76 | |
| Hydrogen-bonding distance restraints | 50 | 76 |
| Ambiguous NOE distance restraints | 396 | 437 |
| Torsion angle restraints |
66 (33 φ, 33 ψ) |
112 (51 φ, 51 ψ, 10 χ1) |
| Structure quality of NMR ensemble |
|
|
| Restraint satisfaction | ||
| RMS differences for distance restraints (Å) | 0.007 ± 0.000 | 0.011 ± 0.000 |
| RMS differences for torsion angle restraints (°) | 0.012 ± 0.007 | 0.098 ± 0.012 |
| Deviations from ideal covalent geometry | ||
| Bond lengths (Å) | 0.001 ± 0.000 | 0.001 ± 0.000 |
| Bond angles (°) | 0.305 ± 0.003 | 0.327 ± 0.002 |
| Impropers (°) | 0.149 ± 0.003 | 0.214 ± 0.004 |
| Ramachandran plot statistics | ||
| Residues in most favored regions (%) | 65.4 | 87.3 |
| Residues in allowed regions (%) | 29.4 | 11.8 |
| Residues in disallowed regions (%) |
5.1 |
0.9 |
| Average atomic RMSDs from the average structure |
|
|
| All atoms (Å) | 11.4 | 1.90 |
| All atoms except disordered regionsa (Å) | 10.0 | 1.32 |
| Backbone atoms (N, Cα, C′) | ||
| All residues (Å) | 10.8 | 1.31 |
| All residues excluding disordered regionsa (Å) | 9.1 | 0.57 |
| Secondary structural elements (Å) | 0.27 and 0.36b | 0.53 |
aDisordered regions in the tandem Vps27 UIM include residues 249–258, 273–303 and 324–329, while those in the Vps27 UIM-1–ubiquitin complex include residues 256–257 and 274–278 of Vps27 and 73–76 of ubiquitin.
bReported values are for the αA and αB helices corresponding to residues 259–272 and 304–323, respectively.
The solution structure of the tandem UIM is poorly defined (Table I). Indeed, with the exception of the two helical segments that are defined to high precision at the backbone level (Figure 1B and C), the intervening linker region as well as the N- and C-termini, which constitute more than 50% of the protein, are essentially unstructured and contribute toward the poor definition of the overall structure. Tertiary interactions between the two helices are non-existent, consistent with the absence of long-range NOEs (Table I), implying that the helices specify structurally independent regions (Figure 1D). The αA-helix extends from Glu259 to Lys272, matching almost exactly with the boundaries of the 15-residue UIM near the N-terminus, while the αB-helix is considerably longer, extending from Pro304 to Arg323, and overlaps, albeit imperfectly, with the C-terminal UIM that spans from Glu301 to Glu315 (Figure 1A). Circular dichroism data for the tandem Vps27 UIM indicated 39% helical content (data not shown), which compares well with the predicted helical content of 44% based on the NMR structure and assuming that helices are fully formed.
Vps27 UIM–ubiquitin interactions
To test whether the tandem Vps27 UIM bound ubiquitin cooperatively, we titrated the 15N-labeled protein with unlabeled ubiquitin and monitored changes in the NMR spectrum. The titrations induced significant perturbations in the amide proton and nitrogen chemical shifts of residues within the individual UIMs, implicating these regions in specific ubiquitin binding. Residues outside the UIMs, including the linker region, were largely unaffected. The strongly perturbed amide resonances ‘shifted’ to new positions as a function of added ubiquitin, implying a rapidly dissociating and reassociating complex (Figure 2A). Residues at equivalent positions in the two UIMs exhibited distinctive binding isotherms (Figure 2B). Specifically, a conspicuous initial ‘lag phase’ characterized the isotherms of N-terminal UIM residues that suggested a higher ubiquitin-binding affinity for the C-terminal UIM. However, quantitative interpretation of the isotherms was impeded by the lack of a suitable analytical expression relating chemical shift deviations as a function of added ligand. The binding curves were fit a posteriori using the approach described below.
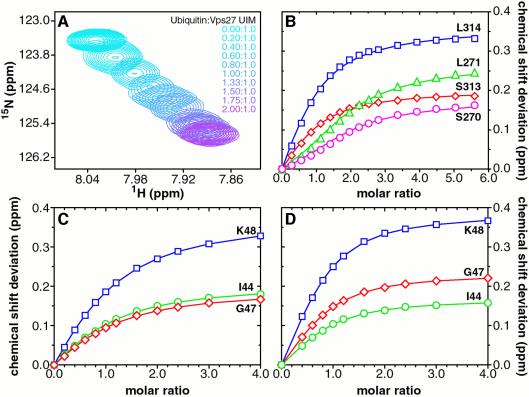
Fig. 2. The UIMs of Vps27 bind ubiquitin with modest affinity, independently and non-cooperatively. (A) Overlays of an expanded region of 1H–15N correlated spectra of the tandem Vps27 UIM showing changes in the amide chemical shifts of Leu314 as a function of added ubiquitin. The peaks are colored from cyan (UIM:ubiquitin molar ratio 1:0) to purple (molar ratio 1:2). (B) Weighted average chemical shift deviations Δav plotted as a function of [ubiquitin]T/[Vps27 UIM]T molar ratio for Ser270 (magenta) and Leu271 (green) in the N-terminal UIM, and for equivalent residues Ser313 (red) and Leu314 (blue) in the C-terminal UIM. The solid lines trace fitted curves as described in the text. Graphs of Δav for ubiquitin residues Ile44 (green), Gly47 (red) and Lys48 (blue) plotted as a function of (C) [Vps27 UIM-1]T/[ubiquitin]T and (D) [Vps27 UIM-2]T/[ubiquitin]T molar ratios. The solid lines connect data points derived from fitting functions. The UIM-1 and UIM-2 peptides bind ubiquitin independently and with similar affinity as in the tandem Vps27 UIM.
Synthetic peptides bearing individual Vps27 UIMs and encompassing the αA- and αB-helices (residues 256–278 and 299–329, henceforth designated UIM-1 and UIM-2, respectively) were generated and titrated with 15N-labeled ubiquitin. Significant but selective perturbations in the ubiquitin spectrum were observed for both UIM peptides, indicative of specific binding. Non-linear least-squares fits of the binding isotherms of three strongly perturbed ubiquitin residues, including Ile44, Gly47 and Lys48, yielded equilibrium dissociation constants Kd of 277 ± 8 and 117 ± 17 µM for the UIM-1–ubiquitin and UIM-2–ubiquitin complexes, respectively (Figure 2C and D). The extent of saturation of the N-terminal UIM in the context of the tandem UIM fragment was evaluated numerically (see Materials and methods) based on knowledge of the saturation of the C-terminal UIM and the Kd of the UIM-1–ubiquitin complex. Satisfactory fits to the binding isotherms for residues in the N-terminal UIM could be achieved without invoking more complicated models (Figure 2B), implying that the individual UIMs of Vps27 bind ubiquitin independently and not cooperatively.
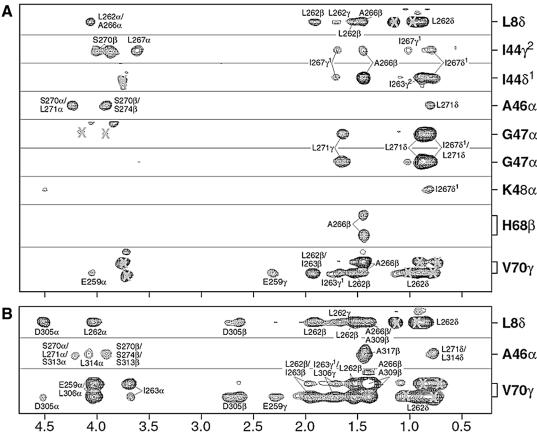
The individual UIMs of Vps27 targeted the same ubiquitin surface and interacted with ubiquitin in an analogous manner. This conclusion was confirmed through comparative chemical shift perturbation mapping analyses of the respective UIMs as well as ubiquitin in the tandem Vps27 UIM–ubiquitin, UIM-1–ubiquitin and UIM-2–ubiquitin complexes (data not shown). Conclusive evidence is provided by the pattern of intermolecular NOEs in the tandem Vps27 UIM–ubiquitin and UIM-1–ubiquitin complexes (Figure 3A and B). Specifically, the side-chain protons of Leu8 and Val70 exhibited intermolecular NOEs with the side-chain protons of Leu262 and Asp305—residues located at equivalent positions in the two UIMs (Figures 1A and 3). Similarly, the Ala46 alpha proton showed NOEs to the side-chain protons of Ser274 and Ala317 that immediately follow the UIMs. The UIMs of Vps27 share a high degree of sequence similarity, especially at positions mediating direct contacts with ubiquitin. This similarity generated a high level of degeneracy in chemical shifts and ambiguity in the interpretation of intermolecular NOEs for equivalent UIM residues in the tandem Vps27 UIM–ubiquitin complex. Therefore we determined the solution structure of the UIM-1–ubiquitin complex instead.
Fig. 3. Intermolecular NOEs establishing a comparable mode of ubiquitin interaction for the two UIMs of Vps27. Selected strips from 3D 13C-filtered 13C-edited NOESY spectra highlighting intermolecular NOEs in (A) the Vps27 UIM-1–ubiquitin complex and (B) the tandem Vps27 UIM–ubiquitin complex. Labels inside and outside the box identify NOEs involving UIM and ubiquitin protons, respectively. Incompletely suppressed signals from 13C-bound protons or other spectral artifacts are denoted by crosses. NOE patterns are conserved for equivalent residues in the UIMs, implying a similar mode of interaction.
Structure of the Vps27 UIM-1–ubiquitin complex
The solution structure of the UIM-1–ubiquitin complex was determined to high precision using approaches similar to those described for the unbound tandem Vps27 UIM (Figure 4A and Table I). Vps27 UIM-1 interacts with ubiquitin through an α-helical structure (Figure 4B). The length and location of the UIM-1 αA-helix in the complex increases by one residue in both directions of the polypeptide chain relative to that found in the unbound tandem Vps27 UIM. The backbone conformation within the helix is well defined compared with the regions flanking the helix. The structure of ubiquitin in the complex consists of a five-stranded β-sheet and two α-helices corresponding to the well-characterized α+β ubiquitin fold. The structure is well defined overall, except for the four C-terminal residues. The 72-residue ‘core’ region closely resembles the high-resolution crystal structure of human ubiquitin (Vijay-Kumar et al., 1987), as indicated by the atomic root mean square deviation (RMSD) of 0.9 Å for polypeptide backbone atoms. Thus binding to UIMs did not cause a significant structural change in ubiquitin.
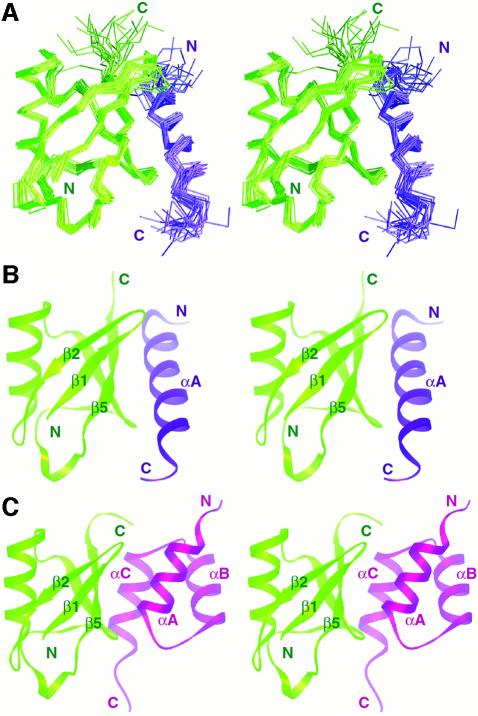
Fig. 4. Solution structure of the Vps27 UIM-1–ubiquitin complex and comparison with the Cue2 CUE–ubiquitin complex. (A) A stereo view of the Cα trace of the ensemble of 20 NMR structures of the Vps27 UIM-1–ubiquitin complex following a best-fit superposition of backbone atoms in well-ordered regions. Stereo views of a representative structure from the NMR ensembles of (B) the Vps27 UIM-1– ubiquitin complex and (C) the Cue2 CUE–ubiquitin complex (Kang et al., 2003). Ubiquitin is colored green, while UIM-1 and CUE are colored blue and purple, respectively. The UIM and CUE ubiquitin-binding motifs bind to overlapping ubiquitin surfaces.
The five-stranded β-sheet of ubiquitin constitutes the principal interaction surface for the Vps27 UIM-1 αA-helix. The helix binds in an antiparallel orientation relative to the C-terminal β-strand, which allows it to interact with the β4- and β5-strands and the β1–β2 and β4–β5 loops (Figure 4B). Thus the interaction surface of ubiquitin resembles that of the recently described structure of the CUE–ubiquitin complex (Figure 4C) (Kang et al., 2003; Prag et al., 2003). Unlike the CUE domain, which engages the ubiquitin surface through a pair of helices, the UIM binds to ubiquitin via a single helix in a comparable-affinity interaction. The intermolecular interface in the UIM-1–ubiquitin complex covers ∼400 Å2 in each protein, consistent with a modest affinity interaction.
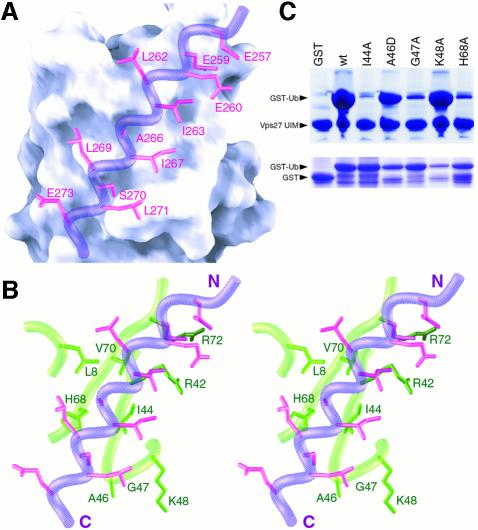
Like other UIMs, Vps27 UIM-1 is dominated by hydrophilic residues. A contiguous hydrophobic patch comprising Leu262, Ile263, Ala266, Ile267, Leu269 and Leu271 on the helical surface constitutes the principal binding site for ubiquitin. Leu262, Ile263, Ala266 and Ile267 directly engage the complementary Leu8–Ile44–Val70 hydrophobic patch on the ubiquitin surface, virtually burying the side chains of Ile44 and Val70 in the process (Figure 5A and B). Similarly, the side chain of Ala266, an extremely highly conserved residue in the UIM (Figure 6A), is buried in a shallow pocket formed by the Leu8, Ile44, His68 and Val70 side chains. The backbone carbonyl of Ala266 is also positioned to form a hydrogen bond with the protonated His68 side chain. Leu269 interacts peripherally with His68, while Ile267 and Leu271 interact more extensively with Gly47.
Fig. 5. Non-covalent interactions at the Vps27 UIM-1–ubiquitin interface and effects of mutations on the overall stability of the complex. (A) A view of the molecular surface of ubiquitin shown along with the interacting side chains of Vps27 UIM-1 (magenta). The backbone of UIM-1 is shown in worm representation (blue). (B) A similar view as in (A) shown in stereo except that the molecular surface is replaced by a worm-and-stick representation for interacting ubiquitin residues (green). (C) Binding of GST, GST–ubiquitin or GST–ubiquitin mutants to His6-Vps27 UIM. Bound proteins were resolved via SDS–PAGE and analyzed by Coomassie staining (top panel). One percent of the amount of GST or GST–ubiquitin mutants used in these assays is shown in the bottom panel.
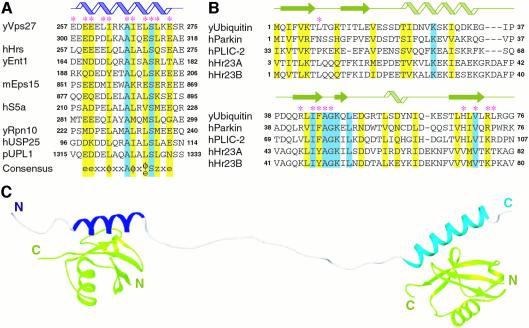
Fig. 6. Sequence conservation in UIMs and ubiquitin-like domains and a structural model for the tandem Vps27 UIM–ubiquitin complex. Multiple- sequence alignments of (A) UIMs and (B) ubiquitin and ubiquitin-like domains from a variety of cellular proteins known to interact with UIMs. The asterisks identify residues in Vps27 UIM–1 and ubiquitin that make direct intermolecular contacts. Invariant and conserved residues are shaded blue and yellow, respectively. The cartoons at the top identify the location of secondary structural elements in the NMR structure of the Vps27 UIM-1– ubiquitin complex. Abbreviated species prefixes are as follows: h, Homo sapiens; m, Mus musculus; p, Arabidopsis thaliana; y, Saccharomyces cerevisiae. The notation for the consensus sequence is as follows: e, negatively charged residue; x, helix-favoring residue; φ, hydrophobic; z, bulky hydrophobic or polar residue with considerable aliphatic content. (C) An NMR-based structural model of the tandem Vps27 UIM–ubiquitin complex. The αA- and αB-helices of the tandem Vps27 UIM are colored blue and cyan, respectively, and ubiquitin is in green.
UIMs are enriched in negatively charged residues at the N-terminus and contain an invariant serine residue near the C-terminus (Figure 6A). The side chains of Glu257, Glu259 and Glu260 at the N-terminus of the motif interact with a positively charged patch comprising Arg42 and Arg72 on the surface of ubiquitin (Figure 5A and B). A favorable intermolecular electrostatic interaction between Glu273, a moderately conserved glutamate in the motif, and His68 is also detected in the NMR ensemble, albeit in only 50% of the structures because of the relatively poor definition of the Glu273 side chain. The side chain of Ser270, the invariant serine residue in the UIM (Figure 6A), is partially buried at the interface and the hydroxyl group forms intermolecular hydrogen bonds with the amide hydrogen of Ala46 and/or Gly47 in the NMR structures.
To confirm whether the UIMs interacted similarly with ubiquitin in the context of the tandem UIM fragment, we determined the solution structure of the protein in complex with ubiquitin using distance and torsion angle restraints derived from NMR data via automated approaches described above. However, the high level of ambiguity associated with the interpretation of intermolecular NOEs (cf. above) yielded structures of low precision. Therefore we consider these structures as ‘models’ of the tandem Vps27 UIM–ubiquitin complex. The UIM–ubiquitin interactions in these models closely resemble those in the UIM-1–ubiquitin complex (Figures 4B and 6C). Moreover, long-range contacts between the αA- and αB-helices are absent, as in the case of the unbound tandem UIM.
To evaluate the contributions of ubiquitin residues in stabilizing the tandem Vps27 UIM–ubiquitin complex, we mutated selected residues and assayed these mutants for binding to the Vps27 UIM fragment. The Ile44Ala mutation was previously shown to weaken Vps27 UIM binding considerably (Shih et al., 2002), consistent with its key role in contacting Vps27 UIM-1. Ile44Ala ubiquitin was included in our studies as a control. Ubiquitin carrying the single site mutation Ile44Ala, Gly47Ala or His68Ala failed to bind Vps27 UIM efficiently, whereas Ala46Asp and Lys48Ala mutants bound comparable amounts as wild-type ubiquitin (Figure 5C). The deleterious effects of the alanine mutations can be attributed to the introduction of steric bulk in the case of Gly47 and to the loss of favorable intermolecular hydrogen bonding and electrostatic interactions in the case of His68. The neutral effect of the Ala46Asp and Lys48Ala mutations is also consistent with the predictions of the NMR structure, since both Ala46 and Lys48 side chains are well exposed to solvent and are not involved in engaging Vps27 UIM directly.
Discussion
The presence of multiple ligand-binding sites within a protein or multiprotein complex affords opportunities for multiple simultaneous interactions (Klotz, 1997). Cooperative interactions can significantly enhance (positive cooperativity) or reduce (negative cooperativity) the efficiency of ligand binding compared with binding to a single site. The presence of multiple copies of the UIM in endocytic proteins, as well as the severe endocytic defects associated with single-site UIM mutations in Vps27, suggested the presence of positive cooperativity in UIM–ubiquitin interactions. Indeed, such a mechanism for ubiquitin binding has recently been described for the CUE ubiquitin-binding motif of an endocytic protein (Prag et al., 2003).
Our studies of the Vps27 UIMs described herein explore possible cooperativity in UIM–monoubiquitin interactions. NMR titration experiments reveal that the individual UIMs of Vps27 bind ubiquitin independently, with modest affinity, and with comparable affinity in isolation as well as when tethered to each other in the context of the tandem UIM fragment (Figure 2). Based on chemical shift perturbation mapping and intermolecular NOE data, the UIMs appear to bind ubiquitin similarly both in isolation and in tandem (Figure 3). These results thus argue against a cooperative binding mechanism, at least at the level of the tandem UIM binding to monoubiquitin.
Additional evidence for the lack of cooperativity in ubiquitin binding is provided by the structure of the tandem Vps27 UIM. In both the ubiquitin-free and the ubiquitin-bound states, the tandem UIM consists of two autonomously folded and structurally independent α-helices that overlap with the individual UIMs (Figures 1 and 6C). Interestingly, a recent structure of the Vps27 C-terminal UIM suggests that the motif can form a tetramer in the crystalline state (Fisher et al., 2003). However, we find no evidence for homotypic association involving individual UIMs in solution up to millimolar concentrations from analytical ultracentrifugation and NMR analyses (data not shown). Similar results were obtained in the aforementioned study (Fisher et al., 2003).
The UIMs, like the CUE ubiquitin-binding motif, bind ubiquitin through preformed surfaces. Since isolated helices are uncommon in proteins, we analyzed the tandem Vps27 UIM sequence using AGADIR, a widely used algorithm to predict helical content of peptides (Lacroix et al., 1998). AGADIR predicted an unusually high helical content for the protein (13% helical content overall and average values of 48% and 16% for regions corresponding to the αA- and αB-helices, respectively), suggesting that it may be an intrinsic property of UIM sequences. In the NMR structure of the unbound protein, the individual helices differ in length as well as in location relative to the UIMs. In particular, the αB-helix is longer by six residues relative to αA in the ubiquitin-free state and extends seven residues beyond the final residue in the motif, implying that the helix start- and endpoints may not be coincident with the location of the UIM in other proteins. Indeed, a peptide encompassing the C-terminal Vps27 UIM motif but otherwise lacking more than one turn of the αB-helix binds ubiquitin with 10-fold reduced affinity (Fisher et al., 2003) compared with the value we measured for the Vps27 UIM-2 peptide (cf. above), underscoring an important role for secondary structure in determining binding affinity.
The 15-residue UIM motif e–e–e–x–φ–x–x–A–x–x–x–S–x–x–e (where e denotes the negatively charged residue and φ the hydrophobic residue) proposed by Hofmann and Falquet contains conserved residues at seven positions (Hofmann and Falquet, 2001), six of which are located at the intermolecular interface in the Vps27 UIM-1–ubiquitin complex (Figure 6A). A wealth of mutational data reported previously for the UIMs of a variety of cellular proteins suggests that residues at these positions play an important role in stabilizing the interaction with ubiquitin. For example, non-conservative substitution of the hydrophobic residue at the fifth position, which is in direct contact with the ubiquitin hydrophobic patch, strongly diminishes ubiquitin binding (Young et al., 1998; Polo et al., 2002). Consistent with their role in mediating favorable electrostatic interactions, single- and multisite alanine mutations of negatively charged residues at the N- and C-termini of the motif also reduce ubiquitin binding (Fisher et al., 2003). Mutation of the invariant serine to alanine has a comparably modest effect on the stability of the complex (Fisher et al., 2003). In contrast, serine to aspartate or glutamate mutation strongly diminishes binding, consistent with the partial burial of the side chain at the intermolecular interface (Raiborg et al., 2002; Shih et al., 2002). The invariant alanine residue, by virtue of its short side chain, likely plays a crucial role in facilitating close approach of the interacting chains.
Sequence conservation data and our structural analyses reveal that a bulky hydrophobic residue or a polar residue with a substantial aliphatic content at the 13th position of the motif is likely to be an important affinity determinant as well. Similar considerations as well as mutational data suggest that a bulky hydrophobic residue at the ninth position of the motif may have an analogous role (Young et al., 1998; Polo et al., 2002). A negatively charged residue at the 11th position of the motif may also serve as an affinity determinant by interacting favorably with His68 of ubiquitin. In those instances where the UIM is involved in binding ubiquitin-like (Ubl) domains that contain a hydrophobic residue at the equivalent location (Figure 6B), a bulky hydrophobic residue may play a similar role. Therefore we propose a rather more elaborate motif for UIMs: e–e–x–x–φ–x–x–A–φ–x–e/φ–S–z–x–e, where x is a helix-favoring residue and z is a bulky hydrophobic or polar residue with considerable aliphatic content (Figure 6A).
UIMs were first described in the S5a component of the proteasome and were shown to be required for efficient binding of Lys48-linked polyubiquitin chains (Young et al., 1998). At the sequence level, the S5a UIMs appear to have the necessary affinity determinants to engage monoubiquitin (Figure 6A). Further, since the UIM helix is oriented such that the side chain of Lys48 is fully exposed to solvent, Lys48-linked polyubiquitin chains should be capable of binding UIMs in a similar manner to monoubiquitin. Such interactions may provide a platform for the assembly of a higher-order complex with enhanced stability, since S5a has been shown to bind polyubiquitin more efficiently than monoubiquitin. S5a also interacts with Ubl domains in a variety of cellular proteins including parkin, hPLIC-2 and hHr23A/B (Hiyama et al., 1999; Walters et al., 2002; Sakata et al., 2003). Ubiquitin residues involved in making contacts with Vps27 UIM-1 are largely conserved in these Ubl domains (Figure 6B), suggesting that these domains most likely associate with S5a UIMs in a similar manner as the Vps27 UIM-1 associates with ubiquitin.
The engagement of an overlapping ubiquitin surface by UIMs and the related CUE/UBA ubiquitin-binding motifs (Kang et al., 2003; Prag et al., 2003) has important implications in endocytosis. Since these motifs are found in multiple endocytic proteins along the pathway, it suggests that these proteins act serially on the same monoubiquitylated cargo during transport from the cell surface to the lysosome. The modest affinity of the interactions, at least at the level of monoubiquitin interactions, suggests that the endocytic pathway is tuned for rapid cargo transport. However, the profound effect caused by a modest reduction in ubiquitin-binding sites at a particular step in the pathway is intriguing (Shih et al., 2002). Given that these ubiquitin-binding motifs appear to bind monoubiquitin in a non-cooperative manner, studies directed at unraveling other mechanisms of cooperativity will be important.
Materials and methods
Production of Vps27 UIM and ubiquitin
The coding sequence of the yeast tandem Vps27 UIM, corresponding to residues 250–329, was amplified by PCR and inserted into a pET expression vector (Novagen). Escherichia coli BL21(DE3) cells containing the vector were grown at 37°C in Luria–Bertani media. Protein expression was induced using 1 mM ispropyl-β-d-thiogalactopyranoside (IPTG) when A600 nm was ∼0.7 and cells were harvested 5 h thereafter. Cell pellets were suspended in 50 mM imidazole buffer (pH 7) containing 1 mM phenyl-methylsulfonyl fluoride (PMSF), 1 µM leupeptin, 1 mM pepstatin, 0.1% Triton X-100 and 1000 units DNase I (Takara Biomedicals), lysed via sonication and centrifuged, and the supernatant was loaded onto a Hi-Trap Q column (Amersham Pharmacia). Bound proteins were eluted using 50 mM imidazole buffer (pH 5) containing 0.1 M NaCl. Vps27 UIM-containing fractions were purified further via reversed-phase high-pressure liquid chromatography (HPLC) using a C18 column and a mobile phase comprising 80% acetonitrile and 0.1% trifluoroacetic acid. The identity of the protein was confirmed by electrospray ionization mass spectrometry (ESI-MS). Vps27 UIM samples uniformly labeled with 15N and/or 13C isotopes were produced as described above, except that cells were grown in M9 minimal media containing [15N]ammonium sulfate (Martek) and/or [13C6]d-glucose (Isotec), respectively. Unlabeled and uniformly 15N- and 15N,13C-labeled samples of yeast ubiquitin were produced as described previously (Beal et al., 1996; Kang et al., 2003). The extent of isotope incorporation in uniformly labeled samples determined by ESI-MS was >97% for 15N and >96% for 13C.
Vps27 UIM-1 and UIM-2 peptides, corresponding to Vps27 residues 256–278 and 299–329, respectively, were synthesized using automated procedures and purified to homogeneity via reversed-phase HPLC. The peptides contained a non-native tyrosine residue at the N-terminus to facilitate concentration determination.
Generation of UIM–ubiquitin complexes and NMR sample preparation
Tandem Vps27 UIM–ubiquitin complexes of 1:2 molar ratio were generated for NMR studies by titrating unlabeled Vps27 UIM with 15N- or 15N,13C-ubiquitin and vice versa. An equimolar complex of unlabeled Vps27 UIM-1 and 15N,13C-ubiquitin was generated in a similar manner. The progress of the titrations was monitored by recording one-dimensional (1D) 1H and two-dimensional (2D) 1H–15N correlated spectra. Protein concentrations for NMR structural analyses ranged from 0.75 to 1 mM for the tandem Vps27 UIM and Vps27 UIM-1 and from 1 to 2 mM for ubiquitin. All NMR samples contained 20 mM sodium phosphate buffer (pH 6) and 0.2% NaN3.
NMR spectroscopy
NMR data were acquired on a Varian Inova 600 MHz spectrometer at 25°C. NMR data processing and analysis were performed using Felix 98 software (Accelrys) incorporating tools developed in-house for accelerated resonance and NOE assignment (Radhakrishnan et al., 1999). Backbone and side-chain 1H, 15N and 13C resonances for Vps27 UIM and ubiquitin were assigned by analyzing three-dimensional (3D) HNCACB, CBCA(CO)NH, C(CO)NH-TOCSY, HNCO, H(CCO)NH-TOCSY, 15N-edited TOCSY, HCACO, HCCH-COSY and HCCH-TOCSY spectra (Bax and Grzesiek, 1993; Ferentz and Wagner, 2000). Aromatic side-chain resonances were assigned from 2D [1H–13C]HSQC and 13C-double-half-filtered TOCSY spectra (Otting and Wüthrich, 1990). Backbone and side-chain 1H resonances of Vps27 UIM-1 were assigned from an analysis of 15N,13C-double-half-filtered TOCSY spectra recorded in H2O and D2O. {1H}–15N heteronuclear NOEs for backbone amides of the tandem Vps27 UIM were measured in quadruplicate using 2D sensitivity-enhanced water-flipback methods (Farrow et al., 1994).
NMR restraint generation and structure determination
Structures were calculated using ARIA (version 1.2) in conjunction with CNS (Brünger et al., 1998; Linge et al., 2003). NOE restraints for the unbound tandem Vps27 UIM were obtained from 3D 15N-edited NOESY (mixing time τm of 100 ms) and 3D aliphatic 13C-edited NOESY (τm = 60 ms) spectra. NOE restraints for the Vps27 UIM-1–ubiquitin complex were acquired from 3D 15N-edited NOESY (τm = 80 ms), 3D aliphatic and aromatic 13C-edited NOESY (τm of 50 ms and 60 ms, respectively), 3D 13C-filtered 13C-edited NOESY (τm = 120 ms) (Zwahlen et al., 1997) and 2D 15N,13C-double-half-filtered NOESY spectra recorded in H2O and D2O (τm of 80 ms and 100 ms, respectively). Intermolecular NOEs were assigned manually and were calibrated indirectly by calculating a scaling factor for the intensities of well-resolved peaks in 13C-edited NOESY and 13C-filtered 13C-edited NOESY spectra. The NOEs were assigned upper bounds of 3, 4, 5 and 6 Å. All other NOEs were calibrated and assigned iteratively and automatically by ARIA. NOE assignments were checked manually during the refinement.
Backbone φ and ψ torsion angle restraints were derived from an analysis of Hα, Cα, Cβ, C′ and backbone 15N chemical shifts using TALOS (Cornilescu et al., 1999). Restraints were imposed only for those residues that exhibited TALOS reliability scores ≥ 9. Side-chain χ1 restraints for isoleucine and valine residues were derived from measured 3JNCγ and 3JC′Cγ values (Bax et al., 1994).
Structures were calculated from extended conformations as the starting model. Default settings were used for most parameters in the ARIA run.cns task file. The number of steps in the simulated annealing protocol was doubled for improved convergence. The final force constants for the distance and torsion angle restraints were set to 50 kcal/mol/Å2 and 200 kcal/mol/rad2, respectively. Twenty structures were computed at each iteration and the seven best were used for automated NOE assignment. Eighty structures were computed in the final iteration and the 20 structures with the lowest restraint energies, restraint violations and RMS deviations from ideal covalent geometry were selected for structural analysis.
The final structures were analyzed using PROCHECK (Laskowski et al., 1996), HBPLUS (McDonald and Thornton, 1994), CNS (Brünger et al., 1998) and MSMS (Sanner et al., 1996). Molecular images were generated using Molscript (Kraulis, 1991), RIBBONS (Carson, 1997) and GRASP (Nicholls et al., 1991).
UIM–ubiquitin affinity measurements
The equilibrium dissociation constants for the Vps27 UIM-1–ubiquitin and Vps27 UIM-2–ubiquitin complexes were measured by titrating 0.5 mM [15N]ubiquitin samples with concentrated stocks of the appropriate unlabeled UIM peptide. Protein concentrations were determined spectrophotometrically (Gill and von Hippel, 1989). Weighted average chemical shift deviations Δav were computed using the formula √{[(ΔHN)2 + (ΔN/5)2]/2} and plotted as a function of molar ratio M = [UIM-1 or UIM-2]T/[ubiquitin]T. Non-linear regression assuming a single binding site was performed using the program XMGR employing the fitting function:
y(x) = b({(x + 1 + a) – √[(x + 1 + a)2 – 4x]}/2)
where y(x) is the observed deviation at molar ratio x, and a and b are the fitted parameters related to the dissociation constant and the chemical shift deviation at x = ∞, respectively (Kang et al., 2003).
To evaluate cooperativity in ubiquitin binding, a 0.5 mM sample of 15N-labeled tandem Vps27 UIM was titrated with increasing amounts of unlabeled ubiquitin. Δav for UIM residues were plotted as a function of molar ratio M = [ubiquitin]T/[UIM]T. Because of the lack of a suitable analytical expression for direct non-linear least-squares fitting of the binding isotherms, as in this case of a receptor containing two ligand-binding sites with different, but invariant, affinities (Klotz, 1997), the following strategy was used. Residues in the C-terminal UIM exhibit hyperbolic binding curves reminiscent of single-site binding. ‘Forced’ fitting of these curves to the single-site binding model yielded chemical shift deviations at infinite molar ratio (Δavmax). The extent of saturation of this binding site (B2 = Δav/Δavmax) could thus be estimated for various values of M. The equation
B1 = (M – B1 – B2)/(C1 + M – B1 – B2)
where B1 is the extent of saturation of the N-terminal UIM and C1 = KdUIM-1/[UIM]T, was solved numerically for B1 from a knowledge of C1 and B2 for various values of M. Binding isotherms for the N-terminal UIM residues were simulated by multiplying the resulting B1 curves with Δavmax, optimized to yield the best fit.
Ubiquitin-binding assays
Wild-type and mutant ubiquitin and the tandem Vps27 UIM fragment were expressed in E.coli as GST- and His6-fusion proteins, respectively, and the binding assays were performed as described previously (Shih et al., 2002; 2003). Expression constructs for GST–ubiquitin mutants were generated from GST–ubiquitin constructs using the QuickChange site-directed mutagenesis kit (Stratagene) and confirmed by DNA sequencing. The bound proteins were resolved on an SDS–PAGE gel and analyzed by Coomassie staining.
RCSB PDB accession codes
Accession codes for the unbound tandem Vps27 UIM and Vps27 UIM-1–ubiquitin complex are 1Q0V and 1Q0W, respectively.
Acknowledgments
Acknowledgements
We thank Susan Shih for generating the tandem Vps27 UIM expression construct used in the study and Ben Ramirez for assistance with NMR data collection. The pET-3a vector was a generous gift from Cecile Pickart (Johns Hopkins University). This work was supported by funds from the NIH to I.R. and L.H. K.A.S. was supported by an NIH Molecular Biophysics training grant. We gratefully acknowledge the Lurie Comprehensive Cancer Center for supporting the Center for Structural Biology, Northwestern University, and the Keck Biophysics Facility, Northwestern University, for access to instrumentation (www.biochem.northwestern.edu/Keck/keckmain.html).
References
- Bax A. and Grzesiek,S. (1993) Methodological advances in protein NMR. Acc. Chem. Res., 26, 131–138. [Google Scholar]
- Bax A., Vuister,G.W., Grzesiek,S., Delaglio,F., Wang,A.C., Tschudin,R. and Zhu,G. (1994) Measurement of homonuclear and heteronuclear J-couplings from quantitative J-correlation. Methods Enzymol., 239, 79–105. [DOI] [PubMed] [Google Scholar]
- Beal R., Deveraux,Q., Xia,G., Rechsteiner,M. and Pickart,C. (1996) Surface hydrophobic residues of multiubiquitin chains essential for proteolytic targeting. Proc. Natl Acad. Sci. USA, 93, 861–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilodeau P.S., Urbanowski,J.L., Winistorfer,S.C. and Piper,R.C. (2002) The Vps27p Hse1p complex binds ubiquitin and mediates endosomal protein sorting. Nat. Cell. Biol., 4, 534–539. [DOI] [PubMed] [Google Scholar]
- Bonifacino J.S. and Traub,L.M. (2003) Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu. Rev. Biochem., 72, 395–447. [DOI] [PubMed] [Google Scholar]
- Brünger A.T. et al. (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Carson M. (1997) Ribbons. Macromol. Crystallogr. B, 277, 493–505. [PubMed] [Google Scholar]
- Cornilescu G., Delaglio,F. and Bax,A. (1999) Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR, 13, 289–302. [DOI] [PubMed] [Google Scholar]
- Di Fiore P.P., Polo,S. and Hofmann,K. (2003) When ubiquitin meets ubiquitin receptors: a signalling connection. Nat. Rev. Mol. Cell. Biol., 4, 491–497. [DOI] [PubMed] [Google Scholar]
- Farrow N.A. et al. (1994) Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry, 33, 5984–6003. [DOI] [PubMed] [Google Scholar]
- Ferentz A.E. and Wagner,G. (2000) NMR spectroscopy: a multifaceted approach to macromolecular structure. Q. Rev. Biophys., 33, 29–65. [DOI] [PubMed] [Google Scholar]
- Fisher R.D., Wang,B., Alam,S.L., Higginson,D.S., Robinson,H., Sundquist,W.I. and Hill,C.P. (2003) Structure and ubiquitin binding of the ubiquitin interacting motif. J. Biol. Chem., 278, 28976–28984. [DOI] [PubMed] [Google Scholar]
- Gill S.C. and von Hippel,P.H. (1989) Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem., 182, 319–326. [DOI] [PubMed] [Google Scholar]
- Hiyama H., Yokoi,M., Masutani,C., Sugasawa,K., Maekawa,T., Tanaka,K., Hoeijmakers,J.H. and Hanaoka,F. (1999) Interaction of hHR23 with S5a. The ubiquitin-like domain of hHR23 mediates interaction with S5a subunit of 26 S proteasome. J. Biol. Chem., 274, 28019–28025. [DOI] [PubMed] [Google Scholar]
- Hofmann K. and Falquet,L. (2001) A ubiquitin-interacting motif conserved in components of the proteasomal and lysosomal protein degradation systems. Trends Biochem. Sci., 26, 347–350. [DOI] [PubMed] [Google Scholar]
- Kang R.S., Daniels,C.M., Francis,S.A., Shih,S.C., Salerno,W.J., Hicke,L. and Radhakrishnan,I. (2003) Solution structure of a CUE–ubiquitin complex reveals a conserved mode of ubiquitin binding. Cell, 113, 621–630. [DOI] [PubMed] [Google Scholar]
- Klotz I.M. (1997) Ligand-Receptor Energetics: A Guide for the Perplexed. Wiley, New York, NY. [Google Scholar]
- Kraulis P.J. (1991) Molscript—a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr., 24, 946–950. [Google Scholar]
- Lacroix E., Viguera,A.R. and Serrano,L. (1998) Elucidating the folding problem of alpha-helices: local motifs, long- range electrostatics, ionic-strength dependence and prediction of NMR parameters. J. Mol. Biol., 284, 173–191. [DOI] [PubMed] [Google Scholar]
- Laskowski R.A., Rullmannn,J.A., MacArthur,M.W., Kaptein,R. and Thornton,J.M. (1996) AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR, 8, 477–486. [DOI] [PubMed] [Google Scholar]
- Linge J.P., Habeck,M., Rieping,W. and Nilges,M. (2003) ARIA: automated NOE assignment and NMR structure calculation. Bioinformatics, 19, 315–316. [DOI] [PubMed] [Google Scholar]
- McDonald I.K. and Thornton,J.M. (1994) Satisfying hydrogen bonding potential in proteins. J. Mol. Biol., 238, 777–793. [DOI] [PubMed] [Google Scholar]
- Nicholls A., Sharp,K.A. and Honig,B. (1991) Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins, 11, 281–296. [DOI] [PubMed] [Google Scholar]
- Oldham C.E., Mohney,R.P., Miller,S.L., Hanes,R.N. and O’Bryan,J.P. (2002) The ubiquitin-interacting motifs target the endocytic adaptor protein epsin for ubiquitination. Curr. Biol., 12, 1112–1116. [DOI] [PubMed] [Google Scholar]
- Otting G. and Wüthrich,K. (1990) Heteronuclear filters in two-dimensional [1H,1H]-NMR spectroscopy: combined use with isotope labelling for studies of macromolecular conformation and intermolecular interactions. Q. Rev. Biophys., 23, 39–96. [DOI] [PubMed] [Google Scholar]
- Polo S., Sigismund,S., Faretta,M., Guidi,M., Capua,M.R., Bossi,G., Chen,H., De Camilli,P. and Di Fiore,P.P. (2002) A single motif responsible for ubiquitin recognition and monoubiquitination in endocytic proteins. Nature, 416, 451–455. [DOI] [PubMed] [Google Scholar]
- Prag G., Misra,S., Jones,E.A., Ghirlando,R., Davies,B.A., Horazdovsky,B.F. and Hurley,J.H. (2003) Mechanism of ubiquitin recognition by the CUE domain of Vps9p. Cell, 113, 609–620. [DOI] [PubMed] [Google Scholar]
- Radhakrishnan I., Perez-Alvarado,G.C., Parker,D., Dyson,H.J., Montminy,M.R. and Wright,P.E. (1999) Structural analyses of CREB-CBP transcriptional activator-coactivator complexes by NMR spectroscopy: implications for mapping the boundaries of structural domains. J. Mol. Biol., 287, 859–865. [DOI] [PubMed] [Google Scholar]
- Raiborg C., Bache,K.G., Gillooly,D.J., Madshus,I.H., Stang,E. and Stenmark,H. (2002) Hrs sorts ubiquitinated proteins into clathrin-coated microdomains of early endosomes. Nat. Cell. Biol., 4, 394–398. [DOI] [PubMed] [Google Scholar]
- Sakata E. et al. (2003) Parkin binds the Rpn10 subunit of 26S proteasomes through its ubiquitin-like domain. EMBO Rep., 4, 301–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanner M.F., Olson,A.J. and Spehner,J.C. (1996) Reduced surface: an efficient way to compute molecular surfaces. Biopolymers, 38, 305–320. [DOI] [PubMed] [Google Scholar]
- Shekhtman A. and Cowburn,D. (2002) A ubiquitin-interacting motif from Hrs binds to and occludes the ubiquitin surface necessary for polyubiquitination in monoubiquitinated proteins. Biochem. Biophys. Res. Commun., 296, 1222–1227. [DOI] [PubMed] [Google Scholar]
- Shih S.C., Katzmann,D.J., Schnell,J.D., Sutanto,M., Emr,S.D. and Hicke,L. (2002) Epsins and Vps27p/Hrs contain ubiquitin-binding domains that function in receptor endocytosis. Nat. Cell. Biol., 4, 389–393. [DOI] [PubMed] [Google Scholar]
- Shih S.C., Prag,G., Francis,S.A., Sutanto,M.A., Hurley,J.H. and Hicke,L. (2003) A ubiquitin-binding motif required for intramolecular monoubiquitylation, the CUE domain. EMBO J., 22, 1273–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijay-Kumar S., Bugg,C.E. and Cook,W.J. (1987) Structure of ubiquitin refined at 1.8 Å resolution. J. Mol. Biol., 194, 531–544. [DOI] [PubMed] [Google Scholar]
- Walters K.J., Kleijnen,M.F., Goh,A.M., Wagner,G. and Howley,P.M. (2002) Structural studies of the interaction between ubiquitin family proteins and proteasome subunit S5a. Biochemistry, 41, 1767–1777. [DOI] [PubMed] [Google Scholar]
- Young P., Deveraux,Q., Beal,R.E., Pickart,C.M. and Rechsteiner,M. (1998) Characterization of two polyubiquitin binding sites in the 26 S protease subunit 5a. J. Biol. Chem., 273, 5461–5467. [DOI] [PubMed] [Google Scholar]
- Zwahlen C., Legault,P., Vincent,S.J.F., Greenblatt,J., Konrat,R. and Kay,L.E. (1997) Methods for measurement of intermolecular NOEs by multinuclear NMR spectroscopy: application to a bacteriophage lambda N-peptide/boxB RNA complex. J. Am. Chem. Soc., 119, 6711–6721. [Google Scholar]