Abstract
Leptin’s effects are mediated by interactions with a receptor that is alternatively spliced, resulting in at least five different murine forms: Ob-Ra, Ob-Rb, Ob-Rc, Ob-Rd, and Ob-Re. A mutation in one splice form, Ob-Rb, results in obesity in mice. Northern blots, RNase protection assays, and PCR indicate that Ob-Rb is expressed at a relatively high level in hypothalamus and low level in several other tissues. Ob-Ra is expressed ubiquitously, whereas Ob-Rc, -Rd, and -Re RNAs are only detectable using PCR. In hypothalamus, Ob-Rb is present in the arcuate, ventromedial, dorsomedial, and lateral hypothalamic nuclei but is not detectable in other brain regions. These nuclei are known to regulate food intake and body weight. The level of Ob-Rb in hypothalamus is reduced in mice rendered obese by gold thioglucose (GTG), which causes hypothalamic lesions. The obesity in GTG-treated mice is likely to be caused by ablation of Ob-Rb-expressing neurons, which results in leptin resistance.
Leptin, a hormone secreted by adipocytes, regulates the size of the adipose tissue mass while affecting satiety and energy metabolism (1–3). Leptin has pleiotypic effects on a number of organ systems and affects reproduction, metabolism, and glucose homeostasis (4–6). Leptin is more potent when given centrally versus peripherally suggesting that some of its effects are mediated via direct actions on receptors in brain (refs. 3 and 7; unpublished data). The leptin receptor, Ob-R, is alternatively spliced, and five different splice forms of the receptor have been identified (8, 9). One form of the receptor, Ob-Rb, is mutant in C57BL/Ks db/db mice (9, 10). Both reverse transcription–PCR and RNase protection assays indicate that this splice form is expressed at a relatively high level in hypothalamus, suggesting that leptin interacts with neurons in this brain region (9, 11).
The Ob-Rb receptor is different from other forms of the receptor in that it has a long cytoplasmic region with consensus amino acid sequences involved in receptor binding to JAK tyrosine kinases (9). JAK tyrosine kinases are required for signal transduction of receptors in the Ob-R class (12). The cytoplasmic region of Ob-Rb also contains a motif required for binding of the Stat3 transcription factor, a known substrate of JAKs (8, 13). Injections of leptin in vivo result in a dose-dependent activation of Stat3 in hypothalamus within 15 min in wild-type and ob mice but not in db mice (13). The other forms of Ob-R have truncated cytoplasmic regions and presumably cannot by themselves activate JAKs or Stats.
In aggregate, these data suggest that leptin’s effects are mediated, in part, via binding to the Ob-Rb isoform of the receptor in the hypothalamus. Here we define the tissue distribution of the different Ob-R isoforms in mice and use in situ hybridization to localize the different receptor isoforms in mouse brain and other tissues. We also report the exon and intron boundaries of the different splice forms. Finally, the levels of Ob-Rb are quantitated in mutant mice, and mice with hypothalamic lesions induced by gold thioglucose.
MATERIALS AND METHODS
Northern Analysis of Ob-Rs.
Mouse multiple tissue northern blots were purchased from CLONTECH and hybridized to Ob-R cDNA probes following the manufacturer’s instructions. All probes are derived from PCR products using primers that are specific for each Ob-R isoform. Hybridization was carried out at 42°C for 18 hr. Blots were washed with 2× SSC and 0.05 percent SDS at room temperature for 30 min, then with 0.1× SSC and 0.1 percent SDS at 50°C for 30 min. Exposure was performed using Kodak X-Omat AR film with intensifying screen at −70°C. Primer sequences for each Ob-R isoform are as follows: common region, forward primer 5′-CAATGTGGATCAGGATCAACC3-′; reverse primer 5′-GCCAGAACTGTAACAGTGTG-3′; product size, 0.6 kb; Ob-Ra, forward primer 5′-AGAACGGACACTCTTTGAAGTC-3′; reverse primer 5′-ACCANAGCGACAATGTGNCCAC-3′; product size, 0.25 kb; Ob-Rb, forward primer 5′-TGGATAAACCCTTGCTCTTCA-3′; reverse primer 5′-GGTCTCAGAGCACCCAGGTA-3′; product size, 0.20 kb; Ob-Rc, forward primer 5′-AGGTCTTAATAAGTCTCAGCCA-3′; reverse primer 5′-TGTACTAACTCCCTTGCCTCG-3′; product size, 0.20 kb; Ob-Rd, forward primer 5′-TGGCAGATTCTCACTGTGG-3′; reverse primer 5′-TTGTCTCTGACACTCATCC-3′; product size, 0.36 kb; Ob-Re, forward primer 5′-GTATGTGTACTGTACTTTTCA-3′; reverse primer 5′-GAGGGTGTCAGTAAATGATTT-3′; product size, 0.07 kb.
In Situ Hybridization.
In situ hybridization using 33P-labeled ribonucleotide probe was done according to previously published methods (14, 15). Tissue sections (10 μm) were fixed (4% paraformaldehyde in PBS), washed (0.1 M PBS), treated with triethanolamine (0.1 M, pH 8.0), washed (2× SSC), and dehydrated. Sense and antisense RNA probes from the common region (624 nucleotides, amino acids 123–331) and the cytoplasmic region of Ob-R (492 nucleotides, amino acids 899-1063) were transcribed in vitro with T7 RNA polymerase and 33P-labeled UTP. Hybridization (50% formamide, 0.3 M NaCl, 20 mM Tris, 0.5 mM EDTA, 10 mM NaH2PO4, 10% dextran sulfate, 1× Denhardt’s, and 0.5 mg/ml tRNA) was carried out at 55°C for 30 hr in a moist chamber with 1 × 106 cpm of labeled probe per 50 μl of hybridization solution. After wash (2× SSC, 50% formamide, and 10 mM DTT at 55°C for 30 min; 2× SSC at 55°C for 30 min), RNase treatment (37°C for 30 min), and final wash (0.5× SSC at 55°C for 30 min; 0.2× SSC at 55°C for 30 min twice), sections were dehydrated and dipped in Kodak NTB2 emulsion. Exposure time was 14 days.
Sequencing and Genomic Mapping.
A 129/SVJ mouse genomic library (Stratagene) was screened with probes from different regions of Ob-R. Sequencing was carried out using dye-termination method in the Sequencing Facility of Rockefeller University (New York). Genomic mapping was performed by restriction digestion and PCR by using genomic λ clones.
RNase Protection Assay.
The antisense RNA probe used for detecting specific leptin receptors was synthesized by in vitro run-off transcription from T7 promoters using linearized templates. The cytoplasmic-region probe, used in Fig. 4, is the one that corresponds to mouse Ob-Rb cytoplasmic (C-terminal) region from amino acid 907 to amino acid 1064. The probe will protect a 417-bp region of the leptin receptor coding sequence. The Ob-Rb probe used in Fig. 1B corresponded to amino acids 857-1064. This probe protects a 621-nt-long RNA. Twenty micrograms of total RNA was used for all tissues in the RNase protection assays. Total RNAs were used at a level of 20 μg per assay in all experiments. Specific transcripts were determined by their size and the relatively faster migrating rates than the probe. Hybridization was carried out in 80% formamide, 40 mM Pipes (pH 6.7), 0.4 M NaCl, and 1 mM EDTA at 47°C overnight. The reaction mixture was treated with 30 units per ml of RNase T2 at 30°C for 60 min. The reaction was treated with 0.5% of SDS and 0.5 mg/ml of proteinase K at 37°C for 15 min. After phenol-chloroform extraction and ethanol precipitation, the RNA fragments were resolved on denaturing 6% acrylamide gel.
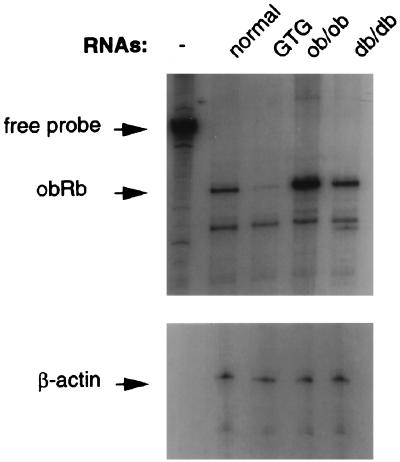
Figure 4.
RNase protection assay of mRNA levels of Ob-R in obese mice. The animals used for the preparation of hypothalamic RNAs are indicated across the top. The positions of the free probe and protected bands are indicated on the left. In addition to Ob-Rb probe, β-actin was also probed to the same RNAs in a parallel experiment as a control.
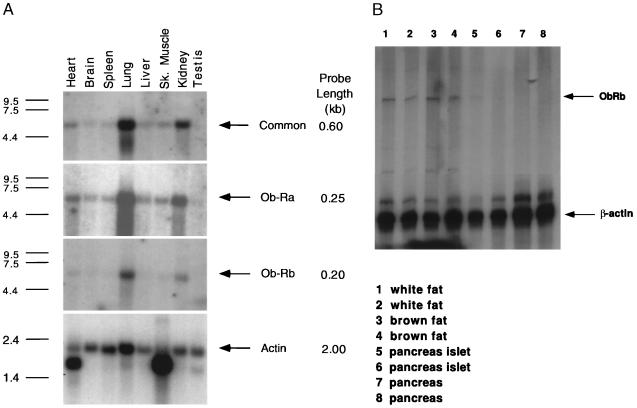
Figure 1.
(A) Northern blot analysis of the expression of Ob-R isoforms in mouse tissues. Multiple tissue northern blots (CLONTECH) were hybridized to cDNA probes specific for Ob-Ra, Ob-Rb, Ob-Rc, Ob-Rd, Ob-Re, as well as one that recognizes all Ob-R isoforms. Tissue origins are marked on top (Sk. muscle, skeleton muscle). All blots were also probed with human β-actin cDNA to control for RNA loading. The size of the cDNA probes used for hybridization to each Ob-R isoform is indicated on the right. Exposure times for each probe are as follows: common region, −70°C for 3 days; Ob-Ra, −70°C, overnight; Ob-Rb, −70°C for 2 weeks; actin, room temperature for 30 min. (B) RNase protection assay of mRNA levels of Ob-Rb in fat and pancreas. RNase protection assays were performed for white fat, brown fat, pancreatic islets, and whole pancreas. Each sample was assayed in duplicate, and β-actin was used as an internal control in every sample. The protected RNA bands for Ob-Rb and β-actin are indicated by arrows.
RESULTS
A series of Northern blots were performed using several Ob-R cDNA probes (Fig. 1). These included a probe common to all splice forms of the receptor and probes specific for each of the alternatively spliced mRNAs, designated Ob-Ra through Ob-Re (see Fig. 1). β-Actin was used as a positive control. As previously reported, a probe common to all the splice forms detects a high level of Ob-R expression in lung and kidney with lower levels in all other tissues tested (Fig. 1 Top) (8). A near-identical tissue distribution is seen when Northern blots are hybridized to a probe specific for Ob-Ra, a short form of the receptor that lacks the Stat3 binding site (9, 10, 13). The intensity of hybridization is similar for both probes, suggesting that the majority of Ob-R in these tissues is Ob-Ra. Hybridizations with Ob-Rb reveal a similar tissue distribution (Fig. 1 Bottom). However, the signal intensity was much lower as evidenced by the 2-week exposure of autoradiographs for the Ob-Rb probe as compared with the 3-day exposures for Ob-R and Ob-Ra. [The length of the Ob-Ra (250-nt) and Ob-Rb (200-nt) probes was similar.) Signals were not detected with probes specific for Ob-Rc, Ob-Rd, and Ob-Re, suggesting that the abundance of these RNAs is low among the tissues tested. Although these blots did not include hypothalamic RNA, previous data have indicated that there is a high level of Ob-Rb in this brain region (9, 11). To further refine the tissue distribution of Ob-Rb, its RNA levels were assayed in white fat, brown fat, and pancreas and were measured using an RNase protection assay. (Ob-Rb was not detected in these tissues using Northern blots.) Low levels of Ob-Rb RNA were observed in white and brown fat but none was detected in mouse pancreatic islets or whole pancreas.
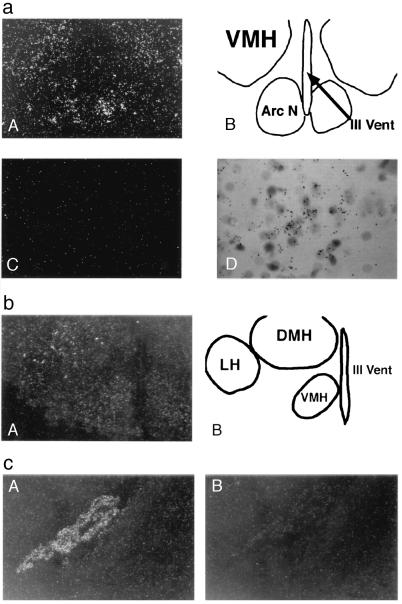
The precise anatomic localization of Ob-R in mouse brain was determined using in situ hybridization with 33P-labeled riboprobes specific for each splice form. In all cases, sense probes were used as negative controls. In situ hybridization of a probe from the extracellular region revealed intense hybridization to the choroid plexus (Fig. 2). Hybridization to choroid plexus was not seen with the Ob-Rb-specific probe. When using the extracellular region probe, a lower intensity of hybridization was seen in several hypothalamic nuclei including the arcuate, DMH, VMH, and LH nuclei. A probe specific for Ob-Rb hybridized strongly to the aforementioned hypothalamic nuclei but did not hybridize to the choroid plexus. In contrast to data in other reports, Ob-Rb was also detected in lateral hypothalamus but was not detected elsewhere in brain (16). Quantitation of the signal intensity indicated that the majority of hypothalamic Ob-R RNA is accounted for by Ob-Rb (i.e., similar intensity in this brain region is noted using both the common and Ob-Rb-specific probes; data not shown). The percentage of Ob-Rb-positive cells in the hypothalamic nuclei was estimated to be 15% in arcuate, 20% in VMH, and 15% in the LH.
Figure 2.
(a) In situ hybridization of Ob-Rb in an adult mouse brain. Dark-field photomicrograph showing Ob-Rb antisense (A) and sense strand (C) riboprobe hybridization to continuous coronal sections through hypothalamus. (B) Camera lucida drawing of the region shown in A, which is showing the location of the ventromedial hypothalamic (VMH) nuclei, the arcuate nuclei (Arc N), and the third ventricle (III Vent). (D) High-magnification bright-field photomicrograph of Ob-Rb mRNA hybridization in VMH. (b) In situ hybridization of Ob-Rb in an adult mouse brain coronal section. (A) Dark-field photomicrograph showing Ob-Rb reactivity in the VMH, dorsomedial hypothalamic (DMH) nuclei, and lateral hypothalamic (LH) nuclei. (B) Camera lucida drawing of A showing the location of the VMH, DMH, LH, and III Vent. (c) Dark-field photomicrograph of Ob-R in situ hybridization in an adult mouse brain adjacent to coronal sections through lateral ventricle. A specific antisense riboprobe corresponding to the extracellular region (common probe) (A) and the cytoplasmic region (Ob-Rb-specific probe) (B) of Ob-R cDNA sequence was used for hybridization. Reactivity observed is in the choroid plexus.
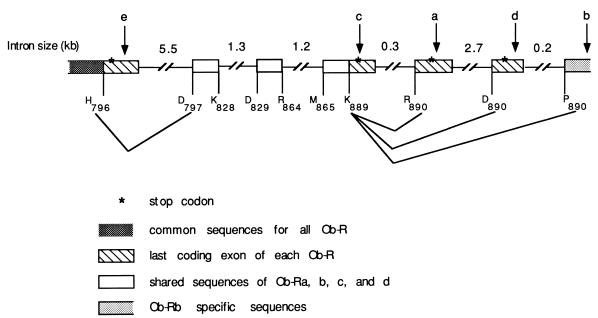
The level of expression and tissue distribution of the Ob-Ra and Ob-Rb splice variants of Ob-R suggest that these forms may play important physiological roles. The low level of expression of the other forms could indicate that one or more of these RNAs are by-products of RNA splicing. To further explore the biogenesis of these RNAs, the genomic organization of the 3′ end of the Ob-R gene was further characterized (Fig. 3). Genomic clones were isolated and mapped using several restriction enzymes. Ob-Ra and Ob-Rb are the result of alternative splicing and polyadenylylation. As predicted, the Ob-Ra terminal exon is 5′ to the Ob-Rb terminal exon and the alternative 3′ ends are approximately 2.9 kb apart (9, 11). Ob-Rc and Ob-Rd encode short forms of the receptor, while Ob-Re predicts a secreted form (9). Further analysis of the genomic structure revealed that the Ob-Rc nucleotide sequence downstream of the splice donor at Lys-889 is identical to that of genomic DNA. Thus, this splice variant is synthesized when splicing does not occur after the splice donor at Lys-889. Ob-Re RNA is found when the splice donor at His-796 is not used, because the sequence downstream of His-796 is also identical to genomic DNA (GenBank accession no. U97032). Ob-Rd results when a short exon is spliced between the Ob-Rb splice donor at Lys-889 and its splice acceptor at Pro-890 (GenBank accession no. U97034). The abundance of the Ob-Rd splice form is low and is not different among any of the db mutants (data not shown).
Figure 3.
Genomic DNA organization of the 3′ end of the Ob-R gene. The genomic structure of the exons that encode amino acids H796–P890 of Ob-Rb is shown (see ref. 9). All other splicing variants of the receptor, Ob-Ra, Rc, Rd, and Re, end within the 13-kb region shown. Each exon is identified at the top with an arrow. Introns are represented by solid lines, with their approximate sizes labeled above (not drawn to scale).
The levels of Ob-Rb RNA were quantitated in several models of rodent obesity (Fig. 4). The levels of Ob-Rb were elevated 5-fold in ob and 2-fold in db mice. In mice with hypothalamic lesions induced by GTG, the level of Ob-Rb RNA was decreased 4-fold. GTG administered in a single intraperitoneal dose causes obesity as a result of specific lesions in the VMH and arcuate nuclei (17).
DISCUSSION
The biologic effects of leptin are likely to be mediated, in part, by interactions with the Ob-Rb form of the leptin receptor in hypothalamic neurons (13, 16, 17). This conclusion is supported by the observed expression of Ob-Rb (the receptor that is mutant in C57BL/Ks db/db mice) in hypothalamus, the increased potency of leptin-administered intracerebroventricular injection, and the activation of Stat3 in hypothalamus after peripheral administration of leptin (unpublished data; refs. 3, 7, 13, and 16). The distribution of Ob-Rb RNA in the VMH, LH, DMH, and arcuate nuclei, all of which are known to regulate feeding behavior, adds further evidence in support of a hypothalamic site of leptin action. There are, however, several notable differences between the neuronatomic data presented here and those published previously (16). In our in situ hybridization, Ob-Rb was not detected in any brain regions outside the hypothalamus but was detected in the lateral hypothalamus. This distinction is important, as our data suggest that the hypothalamus is the principle central nervous system target of leptin action.
The expression of Ob-Rb in the arcuate, ventromedial, and lateral hypothalamic nuclei is consistent with leptin’s effects on food intake and body weight. Bilateral lesions of the VMH and, to a lesser extent, the arcuate nucleus have been reported to cause obesity in rodents and humans, whereas lesions of the LH decrease body weight and food intake (18). In addition, stimulation of the VMH results in reduced food intake and stimulation of the LH increases food intake (18, 19). Because the VMH and LH nuclei have opposite effects on weight, the anatomic localization of Ob-Rb raises the possibility that leptin has different effects on neurons localized to these nuclei. The mechanism by which leptin affects neuronal activity is not known.
The localization of Ob-Rb to the arcuate nucleus suggests the possibility that some of leptin’s effects are mediated via projections from this hypothalamic nucleus. Neurons in the arcuate nucleus express high levels of NPY, a neurotransmitter known to increase food intake and body weight (20). The levels of NPY RNA are increased in ob mice, and injections of leptin result in a 4-fold decrease in its level of RNA (7). αMSH, another neuropeptide that modulates food intake, is also expressed in this nucleus (21, 22). Efferent neurons from the arcuate nucleus project to a number of brain regions including the infundibulum, where release of growth factor-releasing hormone (GRH) directly into the portal venous system of the pituitary stimulates secretion of growth hormone (19). Further studies are required to establish whether leptin affects secretion of GRH or other hypothalamic-releasing hormones.
The increased level of Ob-Rb RNA in ob and db mice suggests that the level of leptin receptor is secondarily increased in animals with defective leptin signaling. The decreased level of Ob-Rb in GTG-treated mice suggests that the obesity in these mice results from the ablation of some Ob-Rb-positive neurons, which results in leptin resistance (17). This possibility is supported by the increase in leptin levels in GTG-treated mice and the resistance of VMH-lesioned animals to recombinant leptin (23, 24). Although the levels of Ob-Rb in GTG-treated mice are decreased, Ob-Rb is still detectable. These data suggest that Ob-Rb is also expressed in neurons that are not affected by GTG. It is possible that, in GTG-treated mice, leptin can still act on a subset of neurons that are unaffected by this drug. This may explain why VMH-lesioned mice are not as obese as C57BL/Ks db/db mice.
GTG has been suggested to induce VMH lesions as a result of glucose uptake by glucose-sensitive neurons found in this nucleus (25). The GTG sensitivity of some Ob-Rb neurons in the VMH, as suggested by the reduced levels of Ob-Rb RNA in hypothalamus of GTG-treated mice, could suggest that both the size of fat stores as well as glucose levels are sensed by a subset of neurons. Further studies are required to explore this possibility. The function of Ob-Rb in other mouse tissues is unclear. Expression of Ob-Ra in choroid plexus may be important in leptin uptake and/or efflux from the cerebrospinal fluid (8, 26, 27).
The importance of the Ob-Rc, Ob-Rd, and Ob-Re splice variants is unknown. Although each of the short forms of Ob-R (a, c, and d) could transduce a signal by forming a heterodimer with a cell surface protein such as gp130 (as has been reported for the 80-kDa interleukin 6 receptor; ref. 28), the abundance of these molecular forms is extremely low among the tissues tested. Further studies are required to evaluate the functional role of these receptors.
These data suggest that leptin interacts directly with receptors on neurons in several hypothalamic nuclei (16). Further characterization of this potentially heterogeneous group of neurons and their connections will have important implications for our understanding of leptin action and the molecular mechanisms regulating body weight.
Acknowledgments
We thank Susan Korres for assistance in preparing the manuscript. This work was supported by grants from the National Institutes of Health/National Institute of Diabetes and Digestive Kidney Diseases (DK41096) and the National Institute of Neurological Disorders and Stroke (RO1NS34389). H.J.O. and H.F. are recipients of National Research Service Award Postdoctoral Training Grants CA09673-18 and PK09604-01.
ABBREVIATIONS
- VMH
ventromedial hypothalamic
- LH
lateral hypothalamic
- DMH
dorsomedial hypothalamic
Footnotes
References
- 1.Halaas J L, Gajiwala K S, Maffei M, Cohen S L, Chait B T, Rabinowitz D, Lallone R L, Burley S K, Friedman J M. Science. 1995;269:543–546. doi: 10.1126/science.7624777. [DOI] [PubMed] [Google Scholar]
- 2.Pelleymounter M A, Cullen M J, Baker M B, Hecht R, Winters D, Boone T, Collins F. Science. 1995;269:540–543. doi: 10.1126/science.7624776. [DOI] [PubMed] [Google Scholar]
- 3.Campfield L A, Smith F J, Guisez Y, Devos R, Burn P. Science. 1995;269:546–549. doi: 10.1126/science.7624778. [DOI] [PubMed] [Google Scholar]
- 4.Levin N, Nelson C, Gurney A, Vandlen R, de Sauvage F. Proc Natl Acad Sci USA. 1996;93:1726–1730. doi: 10.1073/pnas.93.4.1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chehab F F, Lim M E, Lu R. Nat Genet. 1996;12:318–320. doi: 10.1038/ng0396-318. [DOI] [PubMed] [Google Scholar]
- 6.Ahima R S, Prabakaran D, Mantzoros C, Qu D, Lowell B, Maratos-Flier E, Flier J S. Nature (London) 1996;382:250–252. doi: 10.1038/382250a0. [DOI] [PubMed] [Google Scholar]
- 7.Stephens T W, Bashinski M, Bristow P K, Bue-Valleskey J M, Burgett S G, Hale H, Hoffmann J, Hsiung H M, Krauciunas A, Mackellar W, Rosteck P R, Schoner B, Smith D, Tinsley F C, Zhang X Y, Helman M. Nature (London) 1995;377:530–534. doi: 10.1038/377530a0. [DOI] [PubMed] [Google Scholar]
- 8.Tartaglia L A, Dembski M, Weng X, Deng N, Culpepper J, Devos R, Richards G J, Campfield L A, Clark F T, Deeds J, Muir C, Sanker S, Moriarty A, Moore K J, Smutko J S, Mays G G, Woolf E A, Monroe C A, Tepper R I. Cell. 1995;83:1263–1271. doi: 10.1016/0092-8674(95)90151-5. [DOI] [PubMed] [Google Scholar]
- 9.Lee G H, Proenca R, Montez J M, Carroll K M, Darvishzadeh J G, Lee J I, Friedman J M. Nature (London) 1996;379:632–635. doi: 10.1038/379632a0. [DOI] [PubMed] [Google Scholar]
- 10.Chen H, Charlat O, Tartaglia L A, Woolf E A, Weng X, Ellis S J, Lakey N D, Culpepper J, Moore K J, Breitbart R E, Duyk G M, Tepper R I, Morgenstern J P. Cell. 1996;84:491–495. doi: 10.1016/s0092-8674(00)81294-5. [DOI] [PubMed] [Google Scholar]
- 11.Ghilardi N, Ziegler S, Wiestner A, Stoffel R, Heim M H, Skoda R. Proc Natl Acad Sci USA. 1996;93:6231–6235. doi: 10.1073/pnas.93.13.6231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kishimoto T, Taga T, Akira S. Cell. 1994;76:253–262. doi: 10.1016/0092-8674(94)90333-6. [DOI] [PubMed] [Google Scholar]
- 13.Vaisse C, Halaas J L, Horvath C M, Darnell J E, Jr, Stoffel M, Friedman J M. Nat Genet. 1996;14:95–97. doi: 10.1038/ng0996-95. [DOI] [PubMed] [Google Scholar]
- 14.Gibbs R B, Wu D, Hersh L B, Pfaff D W. Exp Neurol. 1994;129:70–80. doi: 10.1006/exnr.1994.1148. [DOI] [PubMed] [Google Scholar]
- 15.Gibbs R B, Pfaff D W. J Comp Neurol. 1994;341:324–339. doi: 10.1002/cne.903410304. [DOI] [PubMed] [Google Scholar]
- 16.Mercer J G, Hoggard N, Williams L M, Lawrence C B, Hannah L T, Trayhurn P. FEBS Lett. 1996;387:113–116. doi: 10.1016/0014-5793(96)00473-5. [DOI] [PubMed] [Google Scholar]
- 17.Maffei M, Fei H, Lee G W, Dani C, Leroy P, Zhang Y, Proenca R, Negrel R, Ailhaud G, Friedman J M. Proc Natl Acad Sci USA. 1995;92:6957–6960. doi: 10.1073/pnas.92.15.6957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bray G A, York D A. Physiol Rev. 1979;59:719–809. doi: 10.1152/physrev.1979.59.3.719. [DOI] [PubMed] [Google Scholar]
- 19.Luiten P G M, Horst G J, Steffens A B. Prog Neurobiol. 1987;28:1–54. doi: 10.1016/0301-0082(87)90004-9. [DOI] [PubMed] [Google Scholar]
- 20.Wilding J P H, Gilbey S, Bailey C, Batt R, Williams G, Chatei M, Bloom S. Endocrinology. 1993;132:1939–1944. doi: 10.1210/endo.132.5.7682936. [DOI] [PubMed] [Google Scholar]
- 21.Mountjoy K G, Mortrud M T, Low M J, Simerly R B, Cone R D. Mol Endocrinol. 1994;8:1298–1308. doi: 10.1210/mend.8.10.7854347. [DOI] [PubMed] [Google Scholar]
- 22.Fan W, Boston B A, Kesterson R A, Hruby V J, Cone R D. Nature (London) 1997;385:165–168. doi: 10.1038/385165a0. [DOI] [PubMed] [Google Scholar]
- 23.Funahashi T, Shimomura I, Hiraoka H, Arai T, Takahashi M, Nakamura T, Nozaki S, Yamashita S, Takemura K, Tokunaga K, Matsuzawa Y. Biochem Biophys Res Commun. 1995;211:469–475. doi: 10.1006/bbrc.1995.1837. [DOI] [PubMed] [Google Scholar]
- 24.Frederich R C, Hamn A, Anderson S, Lollmann B, Lowell B B, Flier B. Nat Med. 1995;1:1311–1314. doi: 10.1038/nm1295-1311. [DOI] [PubMed] [Google Scholar]
- 25.Edelman P M, Schwartz I L, Cronkite E P, Brecher G, Liningston L. J Exp Med. 1965;121:403–413. doi: 10.1084/jem.121.3.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Banks W A, Kastin A J, Huang W, Jaspan J B, Maness L M. Peptides. 1996;17:305–311. doi: 10.1016/0196-9781(96)00025-3. [DOI] [PubMed] [Google Scholar]
- 27.Schwartz M W, Peskind E, Raskind M, Boyko E J, Porte D., Jr Nat Med. 1996;2:589–593. doi: 10.1038/nm0596-589. [DOI] [PubMed] [Google Scholar]
- 28.Taga T, Hibi M, Hirata Y, Yamasaki K, Yasukawa K, Matsuda T, Hirano T, Kishimoto T. Cell. 1989;58:573–581. doi: 10.1016/0092-8674(89)90438-8. [DOI] [PubMed] [Google Scholar]