Abstract
Spo11, a type II topoisomerase, is likely to be required universally for initiation of meiotic recombination. However, a dichotomy exists between budding yeast and the animals Caenorhabditis elegans and Drosophila melanogaster with respect to additional roles of Spo11 in meiosis. In Saccharomyces cerevisiae, Spo11 is required for homolog pairing, as well as axial element (AE) and synaptonemal complex (SC) formation. All of these functions are Spo11 independent in C.elegans and D.melanogaster. We examined Spo11 function in a multicellular fungus, Coprinus cinereus. The C.cinereus spo11-1 mutant shows high levels of homolog pairing and occasionally forms full-length AEs, but no SC. In C.cinereus, Spo11 is also required for maintenance of meiotic chromosome condensation and proper spindle formation. Meiotic progression in spo11-1 is aberrant; late in meiosis basidia undergo programmed cell death (PCD). To our knowledge, this is the first example of meiotic PCD outside the animal kingdom. Ionizing radiation can partially rescue spo11-1 for both AE and SC formation and viable spore production, suggesting that the double-strand break function of Spo11 is conserved and is required for these functions.
Keywords: apoptosis/Coprinus/meiotic spindle/spo11/synaptonemal complex
Introduction
In many organisms, recombination events are crucial for the successful completion of meiosis (reviewed in Baker et al., 1976; Roeder, 1997). The functional role of meiotic recombination extends beyond the rearrangement of genetic loci, in that crossovers produced by meiotic recombination are instrumental in maintaining the meiotic bivalent until anaphase I. In Saccharomyces cerevisiae, meiotic recombination is initiated by DNA double-strand breaks (DSBs; Cao et al., 1990; Sun et al., 1991) likely to have been created by the type II topoisomerase Spo11 (Keeney et al., 1997). Although type II topoisomerases transiently cleave double-stranded DNA (reviewed in Wang, 1996), in S.cerevisiae the Mre11–Rad50 complex functions in conjunction with Spo11 to create an irreversible DSB (Bergerat et al., 1997; Keeney et al., 1997).
The absence of functional Spo11 leads to notable differences in meiotic phenotype, depending on the organism studied (Atcheson et al., 1987; Giroux et al., 1989; Dernburg et al., 1998; McKim et al., 1998). In S.cerevisiae, Spo11 is required for homolog pairing (Loidl et al., 1994; Weiner and Kleckner, 1994), as well as for axial element (AE) and synaptonemal complex (SC) formation (Dresser and Giroux, 1988; Giroux, 1988). In contrast, homolog pairing and SC formation are independent of Spo11 function in Caenorhabditis elegans and Drosophila melanogaster (Dernburg et al., 1998; McKim and Hayashi-Hagihara, 1998; McKim et al., 1998). Accordingly, aspects of the mechanism used by C.elegans and D.melanogaster for establishing aligned pachytene chromosomes must differ from those of S.cerevisiae. However, Spo11, and presumably its role in creating DSBs, are required for recombination and chromosome segregation in S.cerevisiae, Schizosaccharo myces pombe, C.elegans and D.melanogaster (Lin and Smith, 1994; Dernburg et al., 1998; McKim and Hayashi-Hagihara, 1998; McKim et al., 1998).
In many organisms, lack of meiotic recombination results in the formation of univalents (Giroux et al., 1989; Dernburg et al., 1998; Woods et al., 1999). In mammals, univalents cannot attach properly to the metaphase I spindle (Woods et al., 1999), and the cells are often eliminated prior to the completion of meiosis I (Edelmann et al., 1996). Programmed cell death (PCD) is a fundamental aspect of normal gametogenesis in both male and female mammals (Coucouvanis et al., 1993; Yin et al., 1997; McGee et al., 1998). However, enhanced meiotic PCD is seen in mouse spermatogonia with a single asynaptic chromosome (XYO and T16/Y; Odorisio et al., 1998). Spermatocytes from Atm-, Mlh1- and Msh5-deficient mice have many univalents during the late stages of prophase I and prometaphase I, and defective cells are subsequently eliminated by PCD (Baker et al., 1996; Edelmann et al., 1996; Xu et al., 1996; Barlow et al., 1998; de Vries et al., 1999). Thus, PCD is likely to be an efficient method of preventing the development of potentially aneuploid gametes.
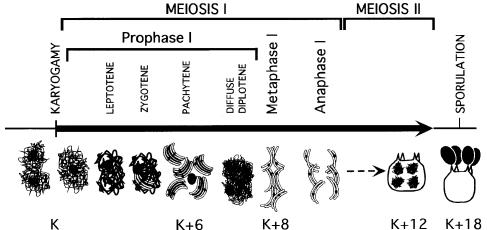
Although meiosis has been studied in numerous organisms, the synchronous meiosis of Coprinus cinereus (Figure 1) makes it particularly amenable for the temporal dissection of meiotic events (see Pukkila et al., 1992; Ramesh and Zolan, 1995; Seitz et al., 1996; Li et al., 1999; Gerecke and Zolan, 2000). In C.cinereus, external cues (Lu, 1974), which can be controlled experimentally, stimulate a filamentous culture to undergo a programmed developmental process that culminates in the formation of a mushroom. Further, the cap is composed primarily of meiotic cells that synchronously give rise to basidiospores (Figure 1). In this paper we describe the characterization of the C.cinereus homolog of SPO11 and follow its roles sequentially through meiosis. The C.cinereus spo11-1 mutant fails to complete meiosis. It exhibits homolog pairing during meiosis and variable amounts of chromosome condensation as well as AE formation, but it fails to form SCs. We demonstrate that unlike C.cinereus wild-type (WT) basidia, which complete meiosis and produce four basidiospores, spo11-1 basidia initiate a sequence of events that ends in cell death. Furthermore, we demonstrate that ionizing radiation can partially suppress the requirement for Spo11 during meiosis, indicating that Spo11 DSB activity is likely to be necessary for meiosis in this organism.
Fig. 1. Schematic of chromosome arrangements during the synchronous meiotic progression of C.cinereus. In C.cinereus, meiosis begins with karyogamy (nuclear fusion), K, as defined in Seitz et al. (1996), the timepoint at which approximately half of all basidia have fused nuclei. Most AmutBmut nuclei (96%) are in pachytene by 6 h post karyogamy (K+6) and many (66%) have completed meiosis II by 12 h post karyogamy (K+12). Sporulation, the production of four haploid, black basidiospores per basidium, occurs by 18 h post karyogamy (K+18).
Results
A spo11 homolog in C.cinereus
We used restriction enzyme-mediated integration (REMI) to create tagged insertional mutants in C.cinereus that are defective in spore formation (Cummings et al., 1999). A pseudohomothallic C.cinereus strain, AmutBmut (Swamy et al., 1984), was used for REMI. Whereas WT and AmutBmut mushroom caps produce copious quantities of black basidiospores, meiotic mutants can be identified because they are defective in basidiospore formation and produce white mushroom caps (reviewed in Pukkila, 1994). We determined that one of the REMI-generated mutants, R126-49, has an insertion in a C.cinereus homolog of SPO11 (Cummings et al., 1999); the translation of a 0.5 kb DNA sequence adjacent to the insertion site matched Spo11 homologs (Cummings et al., 1999). To recover a complete spo11 gene, we used the 0.5 kb DNA fragment as a probe to screen a C.cinereus genomic cosmid library (May et al., 1991). spo11 mapped to a 5.4 kb EcoRI DNA fragment found in two cosmids. This fragment was subcloned and sequenced, and subsequent analysis (described below) showed that it contained the entire genomic sequence of the C.cinereus spo11 gene (DDBJ/EMBL/GenBank accession No. AF214638). To predict the beginning and end of the polypeptide and the location of introns in the transcript, we used RT–PCR (see Materials and methods).
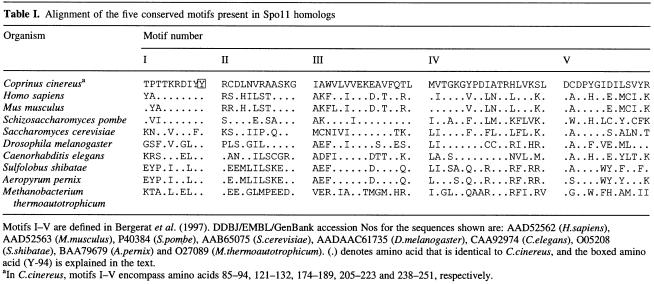
A predicted open reading frame (ORF), encoding a polypeptide of 369 amino acids (40 954 Da), is most similar (33.1% identical) to the Spo11 homolog in humans. Alignment of Spo11 homologs has revealed the presence of five conserved motifs (Bergerat et al., 1997). Alignment of C.cinereus Spo11 to nine Spo11 homologs shows that these conserved motifs are present in C.cinereus Spo11 (Table I). Motif I (Table I) contains a conserved tyrosine (Y-94; Table I, boxed), which we predict is required for the transesterification reaction (Bergerat et al., 1997) in C.cinereus.
Table I. Alignment of the five conserved motifs present in Spoll homologs.
Motifs I–V are defined Bergerat et al. (1997). DDBJ/EMBL/GenBank accession Nos for the sequences shown are: AAD52562 (H.sapiens), AAD52563 (M.musculus), P40384 (S.pombe), AAB65075 (S.cerevisiae), AADAAC61735 (D.Melanogaster), CAA92974 (C.elegans), O05208 (S.shibatae), BAA79679 (A.pernix) and O27089 (M.thermoautotrophicum). (.) denotes amino acid that is identical to C.cinereus, and the boxed amino acid (Y-94) is eaplained in text.
aIn C.cinereus, motifs I–V encompass amino acids 85–94, 121–132, 174–189, 205–223 and 238–251, respectively.
We used Northern blots to analyze the induction of the spo11 transcript during WT meiosis. In C.cinereus, meiosis begins with karyogamy (Figure 1 legend). No spo11 transcript was detected in vegetative tissue or at 1 h prior to karyogamy (K-1; see Figure 1), but an ∼6-fold induction of a 1.3 kb transcript was apparent by 6 h after karyogamy (K+6), a timepoint when essentially all WT and AmutBmut C.cinereus meiotic nuclei are in pachytene (data not shown).
The predicted polypeptide in spo11-1 is severely truncated
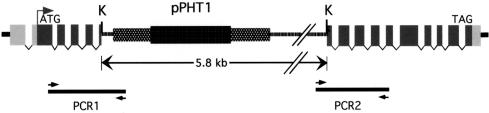
DNA from REMI mutant R126-49 (spo11-1) was analyzed to determine its precise molecular defect in spo11. We had shown previously that this mutant has an insertion of a single copy of the pPHT1 selectable marker, and that this co-segregates with the spore formation defect (Cummings et al., 1999). We analyzed the genomic DNA sequences adjacent to the site of insertion of pPHT1 to determine the location of pPHT1 within spo11. PCR (Cummings et al., 1999; see Materials and methods) was performed on R126-49 to amplify portions of pPHT1 with the genomic DNA adjacent to each side of the site of insertion in R126-49 (Figure 2, PCR1, PCR2). Analysis of the sequences of the products showed that pPHT1 had inserted precisely into the only KpnI site present in the spo11 gene (Figure 2). No genomic DNA was lost in the process. If C.cinereus spo11-1 produces a stable protein, it represents only the first third of the predicted polypeptide and would contain only the first of the five conserved Spo11 motifs (Figure 2; Table I).
Fig. 2. Structure of the meiotic mutant spo11-1. The spo11-1 gene diagram shows predicted exons (dark gray boxes), 14 introns (white bars), and the 5′ and 3′ UTRs (light gray boxes). pPHT1 is inserted into the sole KpnI site (K) in the spo11 gene. PCR products (PCR1 and PCR2; black bars) were sequenced to analyze the insert–genomic DNA junctions, as described in Materials and methods.
To determine whether the meiotic defects (described in detail below) in R126-49 were a consequence of the spo11 mutation alone, we transformed this strain with either a WT or a truncated copy of spo11 (see Materials and methods). Only transformants that contained a WT copy of spo11 were able to rescue the meiotic defects of R126-49 (data not shown).
Spo11 is required for prophase I chromosome condensation but is partially dispensable for homolog pairing
To determine whether the sporulation defect in the C.cinereus spo11-1 mutant (described above) reflects defects in meiotic chromosome behavior, we examined spo11-1 meiotic chromosomes using 4′,6-diamidino-2-phenylindole (DAPI), and compared them with those of its AmutBmut progenitor (Figure 3). At the K+6 timepoint, chromosomes of all meiotic AmutBmut nuclei examined (N = 69) were at pachytene and were resolvable into pairs of homologs (Figure 3A). Similar results have been shown in WT meiotic nuclei (Seitz et al., 1996). We examined 250 meiotic spo11-1 nuclei from three mushroom caps. A portion of the nuclei (16.8%) appeared well condensed (Figure 3B), and many individual chromosomes were discernible in these nuclei (Figure 3B, arrows). Most (63.2%) showed limited amounts of condensation (Figure 3C). The remaining 20% of the nuclei showed little or no detectable condensation (Figure 3D). We then examined the amount of condensation in spo11-1 nuclei at an earlier timepoint, K+3. Of 169 nuclei, 48% appeared well condensed, 46% showed limited amounts of condensation and only 5.9% showed little or no detectable con densation. Our results suggest that unlike in WT strains, in which chromosome condensation increases between K+3 and K+6 (Seitz et al., 1996), in spo11-1, chromosome condensation decreases between K+3 and K+6.
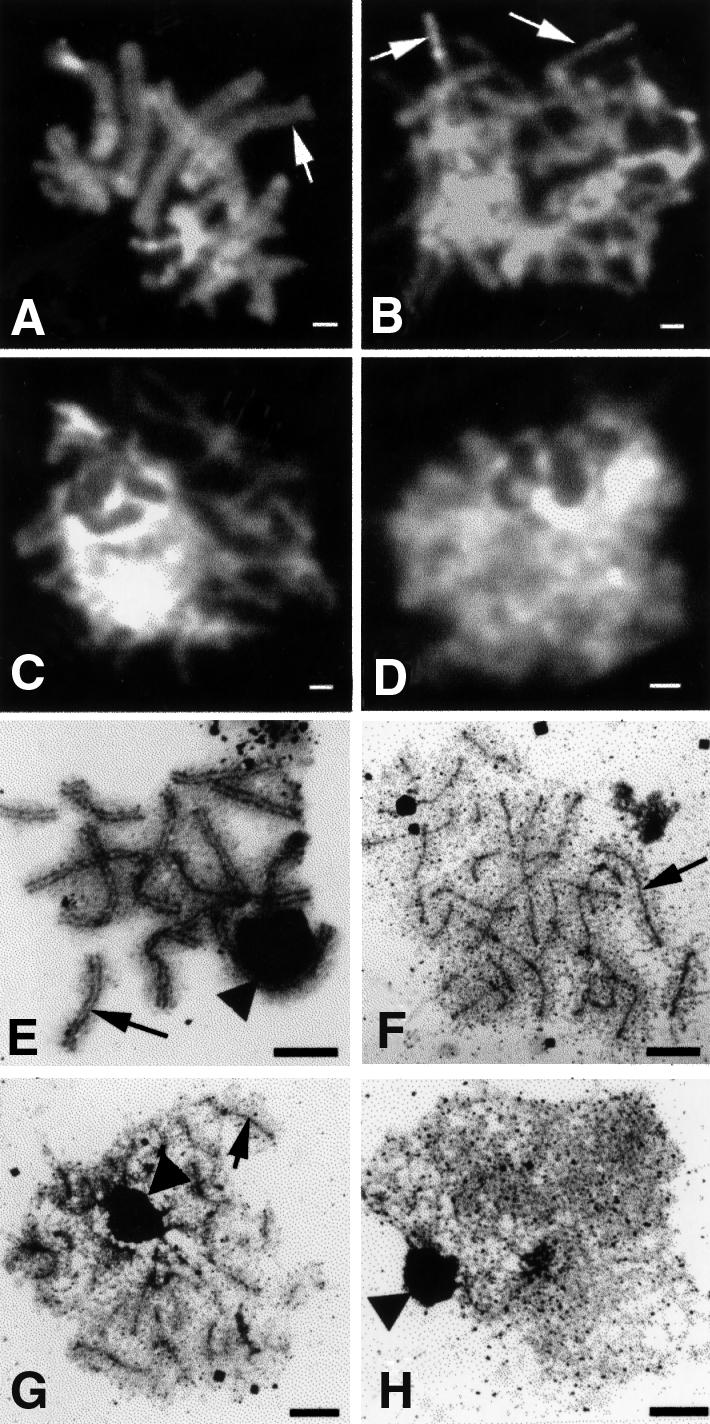
Fig. 3. Meiotic nuclei from C.cinereus strains AmutBmut and spo11-1 isolated at 6 h post karyogamy. (A–D) are fluorescence micrographs of nuclei stained with DAPI; bar, 1 µm. (E–H) are electron micrographs of nuclei stained with silver nitrate; bar, 2 µm. (A) AmutBmut pachytene nucleus; arrow indicates a meiotic bivalent. (B–D) spo11-1 nuclei exhibiting various amounts of chromosome condensation, as described in the text; arrows indicate individual homologs. (E) AmutBmut pachytene nucleus; arrow indicates an example of a full-length SC between homologs. (F–H) spo11-1 nuclei showing variable quantities of AEs: many (F), some (G) and none (H). Arrows indicate examples of unsynapsed AEs. The dark-staining structure (indicated by arrowheads) is the nucleolus.
Immediately following karyogamy, homologous chromosomes of WT C.cinereus begin to pair, as demonstrated by fluorescence in situ hybridization (FISH; Li et al., 1999). Allelic foci are defined as paired if the distance between homologous probe signals is 1.1 µm or less (Li et al., 1999). Both chromosomes 8 and 13 are paired in 91% of WT nuclei at K+1, and in 94% of the nuclei by K+6 (Li et al., 1999). We examined homolog pairing in AmutBmut and spo11-1 using FISH analysis with interstitial probes specific for chromosomes 8 and 13. For AmutBmut, homolog pairing occurs as efficiently as in WT; by K+6 both loci examined were paired in 92.7% of nuclei (Table II). In spo11-1 nuclei, only 36% had both chromosome 8 and 13 loci paired at K+3, but by K+6, 66.6% had both loci paired and 91% had either one or both of the loci paired (Table II).
Table II. FISH data for spo11-1 meiotic nuclei.
| Strain | Timepoint | Na | Percentage of meiotic nuclei that exhibit pairing |
|||
|---|---|---|---|---|---|---|
| Chrom.8 | Chrom.13 | Both 8 and 13 | Neither 8 nor 13 | |||
|
AmutBmut |
K+6 |
69 |
93 |
98 |
93 |
0 |
| spo11-1 | K+3 | 169 | 58 | 58 | 36 | 19 |
| K+6 | 75 | 77 | 80 | 67 | 9.0 | |
| K+6 | 55 | 73 | NT | NA | NA | |
Homolog pairing was examined using chromosome-specific FISH probes, as described in Materials and methods. Nuclei were examined at the timepoints K+3 and K+6, 3 and 6 h after karyogamy (as defined in the text), respectively. NT, not tested. NA, not applicable.
aNumber of nuclei examined.
We tested to see whether chromosome condensation correlated with homolog pairing. The spo11-1 nuclei examined by FISH at K+3 and K+6 were scored as either well condensed, variable or uncondensed. At K+3, 28 of the 81 well condensed nuclei had both loci paired, and two of the 10 uncondensed nuclei had both loci paired. At K+6, nine of the 18 well condensed nuclei had both loci paired, and three of the seven uncondensed nuclei had both loci paired.
Coprinus cinereus spo11-1 makes AEs but not SCs
Critical to the progression of meiosis is the formation of the SC, a proteinaceous structure that assembles between aligned homologs along their lengths. Single AEs are observed in WT nuclei soon after karyogamy; by K+1, 92% of nuclei contain long AEs (Seitz et al., 1996). AEs become the lateral elements (LEs) of the SC. By K+6, 90% of WT nuclei have full-length SCs (Seitz et al., 1996).
We examined single AEs and the LEs of SCs in AmutBmut and spo11-1 nuclei at K+6. In most AmutBmut nuclei (96%), the SCs resembled those of WT (N = 45; Figure 3E). The remaining 4.0% of the nuclei appeared uncondensed, and SC structures were not visible (data not shown). Strikingly, of the 60 spo11-1 nuclei examined, none exhibited any mature SC structures (Figure 3F–H). In 18% of the spo11-1 nuclei, at least 25 of the predicted 26 AEs were observed (Figure 3F). In 36% of the nuclei, AEs were present in variable number and length (Figure 3G). In the remaining 46% of the nuclei, no structures of any kind were visible (Figure 3H). Similar results have been obtained in a spo11-1 strain containing WT A and B mating-type loci (S.T.Merino, W.J.Cummings, S.N.Acharya and M.E.Zolan, submitted).
We were curious to know whether the AEs present in spo11-1 were similar in length to the LEs of the SC of AmutBmut. Measurements of structures were made on silver-stained chromosome spreads from AmutBmut and spo11-1 nuclei at K+6. For AmutBmut, the average total length of LEs was 135.3 ± 39 µm (N = 10 nuclei). Because there are two LEs per SC, this number should be twice the total length of SC, and our results agree with published WT SC lengths of 70 ± 4.7 µm (Pukkila and Lu, 1985) and 76 ± 9.5 µm (Seitz et al., 1996). For spo11-1, only nuclei in which at least 25 AEs were present were measured. Four of the 10 spo11-1 nuclei that were analyzed had >26 AEs. We suspect that these represent either discontinuous AEs or fragmented chromosomes; nonetheless, all were included in the calculations. The average total AE length of spo11-1 was 98.6 ± 7.2 µm (N = 10 nuclei), a value that overlaps within one SD of the LEs of AmutBmut. Thus, the AE values for spo11-1 can approach those of a WT cell.
Coprinus cinereus spo11-1 makes aberrant meiotic spindles and fails to complete meiosis
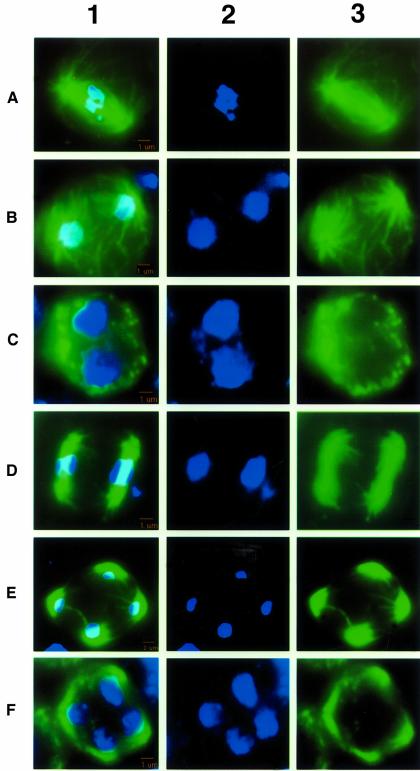
We used monoclonal antibodies to α-tubulin to analyze spindle formation and meiotic progression in AmutBmut and spo11-1 by fluorescence microscopy (Figures 4 and 5). Normal mid-to-late meiotic progression of AmutBmut is shown in Figure 4; images of DNA (column 2) and α-tubulin (column 3) were merged to create the images shown in column 1. spo11-1 basidia at K+8 are shown in Figure 5, rows A–D; the corresponding stage in AmutBmut, metaphase I, is shown in Figure 4A. In spo11-1, the degree of chromatin condensation was variable (Figure 5, rows A and B). Some of the nuclei (Figure 5, row A) appeared to be less condensed, and although polymerized α-tubulin was present in these basidia, the spindle, when present, was aberrant. When only the DNA was examined (Figure 5, column 2), the chromosomes sometimes appeared to be arranged on a single plane (Figure 5, rows C and D). However, analysis of the same nuclei in the context of the meiotic spindle (Figure 5, column 1) revealed that the chromosomes were actually located randomly along the spindle or along the length of the spindle, but not perpendicular to it as in AmutBmut (Figure 4A). No spindles containing α-tubulin were distinguishable in spo11-1 basidia at K+10 (Figure 5, rows E and F). In some of these basidia, the tubulin is amorphic; in others, the basidia lacked any detectable α-tubulin (not shown). In comparison, at K+10 most WT nuclei have completed meiosis I and are in the process of meiosis II (Figure 4B–D).
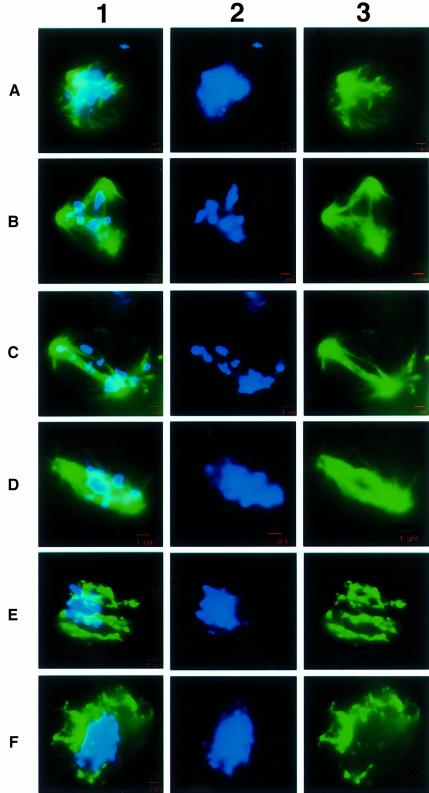
Fig. 4. Normal mid-to-late meiotic progression of C.cinereus as seen in AmutBmut. Fluorescence micrographs of AmutBmut basidia in which chromosomes are stained with DAPI (column 2) and spindles are detected using antibodies to α-tubulin (column 3). Images in columns 1 and 2 are merged to create the images shown in column 1. Bars, 1 µm. (A) metaphase I; (B) anaphase I; (C) telophase I; (D) metaphase II; (E) anaphase II; (F) telophase II. We have posted a higher resolution electronic version of this image at http://sunflower.bio.indiana.edu/~mzolan/zolanlab/Spo11%20Microscopy.html
Fig. 5. Mid-to-late meiotic progression in C.cinereus spo11-1. Fluorescence micrographs of spo11-1 basidia, in which chromosomes are stained with DAPI (column 2) and spindles are detected using antibodies to α-tubulin (column 3). Images in columns 2 and 3 are merged to create the images shown in column 1. Bars, 1 µm. (A–D) Karyogamy plus 8 h. (E and F) Karyogamy plus 10 h. We have posted a higher resolution electronic version of this image at http://sunflower.bio.indiana.edu/~mzolan/zolanlab/Spo11%20Microscopy.html
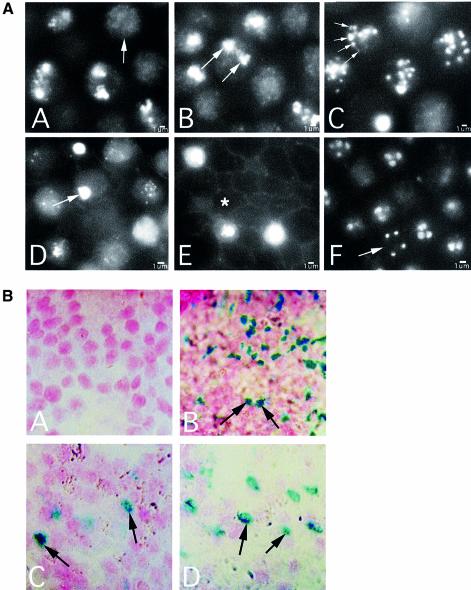
To determine the fate of spo11-1 nuclei over time, we examined spo11-1 chromosomes using DAPI at later meiotic timepoints (Figure 6A, panels A–E). At K+8, 42% of the spo11-1 basidia (N = 125) had a single nucleus in which the chromosomes appeared somewhat diffuse (Figure 6A, panel A, arrow), and 53% of the cells had a single, highly compacted nucleus (similar to Figure 6A, panel D, arrow). The remaining 5% of the basidia contained two DNA bodies. This may indicate that although spo11-1 nuclei do not progress through a normal metaphase I, some of the nuclei undergo a separation of chromosomes. By K+10, ∼40% of the spo11-1 basidia contained two small regions of intense staining (Figure 6A, panel B, arrows); the remainder contained a single, apparently diffuse nucleus. Surprisingly, a dramatic change was observed in the spo11-1 basidia at K+11 (Figure 6A, panel C). At this timepoint, the nuclear material appeared to be condensing and was found as distinct, highly compacted fragments arranged as a ring at the cell periphery (Figure 6A, panel C, arrows). The number of fragments and their size were variable, and not all nuclei exhibited this phenomenon. By K+12 (Figure 6A, panel D), the nuclear material appeared dispersed throughout the cell in the majority of the basidia (85%). A small proportion (5%) contained a single highly compacted nucleus (Figure 6A, panel D, arrow), and 10% of the basidia appeared empty. By K+14, most cells (85%) were devoid of DAPI-stained material (Figure 6A, panel E, asterisk); the remaining 15% contained a single DNA body of varying intensity. This nuclear fragmentation followed by DNA loss was restricted to basidia; paraphyses, vegetative cells that act as spacers, did not appear to be affected (not shown). For comparison, Figure 6A, panel F is a section of gill tissue from AmutBmut at K+12. By this timepoint, many of the basidia (66%) had completed meiosis, and four nuclei were present (Figure 6A, panel F, arrow).
Fig. 6. Late meiotic progression in C.cinereus spo11-1 and AmutBmut. (A) Fluorescence micrographs of basidia in which nuclei are stained with DAPI. Bars, 1 µm. (Panel A) spo11-1 at karyogamy plus 8 h; arrow indicates a diffuse nucleus. (Panel B) spo11-1 at karyogamy plus 10 h; arrows indicate two compact nuclei within a basidium. (Panel C) spo11-1 at karyogamy plus 11 h; arrows indicate ring of compact DNA at cell periphery. (Panel D) spo11-1 at karyogamy plus 12 h; arrow indicates a single, highly compacted nucleus. (Panel E) spo11-1 at karyogamy plus 14 h; asterisk indicates a basidium devoid of DNA. (Panel F) AmutBmut at karyogamy plus 12 h; arrow indicates four haploid nuclei within a basidium. (B) TUNEL assay for nuclear fragmentation in C.cinereus AmutBmut and spo11-1. Sections of gill tissue from caps of AmutBmut and spo11-1 were examined in TUNEL assays. Fragmented DNA ends were detected using TACS Blue, and nuclei were counter-stained using Nuclear Fast Red. (Panel A) AmutBmut gill tissue at karyogamy plus 10 h. (Panel B) Nuclease-pretreated AmutBmut gill tissue at karyogamy plus 10 h; arrows indicate TUNEL-positive nuclei. (Panels C and D) spo11-1 gill tissue at karyogamy plus 10 h; arrows indicate TUNEL-positive nuclei.
In mammalian cells, condensation of nuclear chromatin followed by nuclear fragmentation and marginalization is consistent with apoptosis (Kerr et al., 1972; Clifford et al., 1996). A standard procedure used to identify apoptosis in animal cells is the TUNEL assay (Gavrieli et al., 1992, 1993), which involves labeling DNA ends using terminal deoxynucleotidyl transferase (TdT), detecting with TACS Blue, and counter-staining nuclei with Fast Red. Figure 6B, panel A shows a section of AmutBmut gill at K+10; few nuclei (0.5%; N = 200) stained blue in a TUNEL assay, indicating almost no fragmentation of DNA in these basidia. Figure 6B, panel B is a nuclease-treated control: AmutBmut gill tissue at K+10 was treated with TACS-nuclease, an enzyme that generates DNA breaks, prior to detection with TACS Blue. The blue staining of nuclei, indicative of DNA fragmentation, is present in 52% of the basidia (N = 200; Figure 6B, panel B, arrows). Figure 6B, panels C and D show the results of the TUNEL assay using gill tissue of spo11-1 at K+10. Of the 300 meiotic spo11-1 nuclei examined, 17.3% were positive for the TUNEL assay (Figure 6B, panels C and D, arrows), indicating that the DNA in the spo11-1 nuclei is fragmented.
Coprinus cinereus spo11-1 has decreased spore production and viability
In WT and AmutBmut C.cinereus, meiosis culminates in the production of four haploid nuclei. Each nucleus migrates into one of four basidiospores developing on the apical surface of the basidium. We observed a considerable amount of variability in normal spore production, indicated by large values of standard deviation. Spore production is somewhat lower in AmutBmut than in WT: 5.5 × 105 ± 2 × 105 and 6.8 × 106 ± 5 × 106 basidiospores were produced, respectively, per milligram of cap tissue. However, spore viability is high in both strains: 96% in AmutBmut (n = 727 spores) and 95% in WT (Ramesh and Zolan, 1995). In contrast, spo11-1 produced a sparse number of basidiospores (3.2 × 103 ± 3 × 103/mg cap tissue; 0.58% of the amount produced by AmutBmut), and of those, few (1.6 ± 1%) were viable.
The meiotic defects in spo11-1 can be partially suppressed by ionizing radiation
Based on sequence similarity, we predicted that C.cinereus Spo11 is responsible for catalyzing DNA DSBs during meiosis. We reasoned that if one aspect of Spo11 function is to act as a molecular DNA ‘scissor’, then this function could be supplied by treating mushrooms with ionizing radiation. Because spo11-1 makes no detectable SC, we reasoned that Spo11 activity is required before synapsis, as in S.cerevisiae (Alani et al., 1990; Cao et al., 1990; Padmore et al., 1991; Sun et al., 1991; Keeney et al., 1997), and therefore we irradiated fruiting bodies of spo11-1 and AmutBmut at K+1.5, a timepoint at which karyogamy should be complete, but most nuclei should not have full-length SCs. Following irradiation, AmutBmut strains still produced black caps with copious quantities of basidiospores. However, these basidiospores showed a decrease in viability; only 52.4 ± 20% were able to germinate. Unlike unirradiated spo11-1, which produced white caps with few viable spores (1.6 ± 1%), irradiated spo11-1 caps were gray, indicating that more spores were produced (1.8 × 103 ± 8 × 102 basidiospores/mg cap tissue). Also, spore viability was enhanced considerably, to 14.4 ± 2%. These results indicate that ionizing radiation can partially suppress the meiotic requirement for Spo11 in C.cinereus.
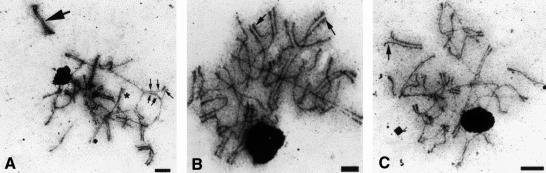
We next compared spo11-1 nuclei at K+6 from unirradiated and irradiated (at K+1.5) mushrooms to establish whether the ionizing radiation caused any apparent cytological changes that could explain the partial suppression of the spo11-1 meiotic defect. In contrast to unirradiated spo11-1, where 46% of the nuclei had no structures of any kind (Figure 3F and see above), all (100%; N = 66) irradiated spo11-1 had AEs. Also, in contrast to unirradiated spo11-1, in which no SC was present (Figure 3F–H), 74% of the irradiated spo11-1 meiotic nuclei exhibited some SC (Figure 7; N = 66). The remaining 26% of the nuclei examined had only AEs and no SC (data not shown). In the nuclei in which SC was observed, the quantity of SC ranged from short regions of synapsis (Figure 7A, asterisk) to apparently full-length SC (Figure 7A, large arrow). Occasionally, SC terminated in knobs, consistent with formation at chromosome ends (Figure 7B, arrows; Seitz et al., 1996), but in some cases only the central length of the chromosomes was synapsed, and apparent end associations specifically were excluded (Figure 7C, arrow). Much of the SC appeared to be promiscuous. Periodically, complex synapses involving three or more chromosomes were present; in one such example (Figure 7A, small arrows), a single chromosome was synapsed with two different chromosomes, one at either end, and the central region of the third chromosome remained as a stretch of unpaired AE. These and other more complicated aggregations involving several chromosomes indicate that not all synaptic events reflected interactions between homologs. Alternatively, translocations caused by the irradiation could result in aberrant synaptic figures.
Fig. 7. Electron micrographs of meiotic nuclei from C.cinereus strain spo11-1 irradiated at karyogamy plus 1.5 h and isolated at karyogamy plus 6 h. Nuclei were stained with silver nitrate. The dark-staining structure in each micrograph is the nucleolus (arrowhead). Three different nuclei from the same irradiated cap are shown in (A–C). Bar, 2 µm. (A) Asterisk denotes short region of synapsis, small arrows indicate promiscuous synapsis and large arrows indicate apparent full synapsis. (B) Arrows indicate SC terminated in knobs. (C) Arrow indicates interstitial SC where ends are specifically excluded.
Discussion
We have shown that Spo11 is required for meiosis in C.cinereus. The predicted amino acid sequence has similarity to all Spo11 homologs analyzed to date and includes all five motifs characteristic of these proteins. Analysis of the DNA sequence in the spo11-1 mutant indicates that the insertion into spo11 occurred within the first third of the gene. If a Spo11 polypeptide exists as a stable protein in spo11-1, it would contain only motif I. The spo11-1 mutant produces a sparse number of basidiospores, and these have low viability (1.6% versus 96% in AmutBmut). spo11 mutants from other organisms have correspondingly similar reduced viabilities of meiotic progeny. In S.cerevisiae diploids, spo11Δ mutants exhibit <1% spore viability, compared with 94% spore viability in heterozygous strains (Atcheson et al., 1987). In C.elegans, spo11 hermaphrodites produce a normal number of fertilized eggs, but few (<0.6%) survive past the embryo stage (Dernburg et al., 1998). Thus, there is an apparently universal requirement for Spo11 protein in meiotic DNA metabolism.
Chromosome condensation and AE formation
In C.cinereus WT nuclei, prophase I condensation begins prior to karyogamy and continues progressively until pachytene (Seitz et al., 1996). Although some chromosome condensation also occurs in the spo11 mutants, it is not progressive. On the contrary, fewer condensed nuclei (16.8%) are apparent at K+6 than at K+3 (48%), suggesting that Spo11 function is required for the maintenance of the condensed state.
Is chromosome condensation a prerequisite for AE formation? In spo11-1, the percentage of uncondensed nuclei (14%) is less than the percentage of nuclei with no detectable AE (46%). This result suggests that at least some chromosome condensation is required for AEs to form, and thus at least some chromosome condensation must be AE independent. The Spo11-dependent condensation is likely to be necessary, but not sufficient, for AE formation. Meiotic mutants in other organisms also demonstrate condensation in the absence of AE formation. For example, in S.cerevisiae, RED1 is likely to encode a component of the LE (Smith and Roeder, 1997), and red1 meiotic nuclei fail to form AE but undergo some condensation (61% of WT; Nag et al., 1995).
Meiotic chromosomes in spo11-1 pair remarkably well
Compared with S.cerevisiae, C.cinereus spo11-1 is able to undergo a strikingly high amount of pairing; 67% of the nuclei have both loci on two chromosomes paired, and 91% of the nuclei have at least one of the two loci paired. Saccharomyces cerevisiae spo11Δ shows drastic reductions in the amount of pairing; in one study, only 10% of spo11Δ nuclei demonstrated pairing for a given locus, compared with 61% in WT (Weiner and Kleckner, 1994). Furthermore, Weiner and Kleckner (1994) suggest that the 10% may represent essentially an absence of interstitial homolog pairing, since non-homologous (random background) pairing was observed in 6% of nuclei in these experiments. In contrast, complete homologous pairing was observed in spo11 mutants of C.elegans (Dernburg et al., 1998) and D.melanogaster (McKim and Hayashi-Hagihara, 1998; McKim et al., 1998). Clearly, in these latter two organisms homolog pairing does not rely on Spo11.
Recently, Cha et al. (2000) have demonstrated that unlike spo11Δ, the S.cerevisiae spo11-Y135F DSB catalysis-defective mutant shows WT levels of homolog pairing. This separation-of-function mutant demonstrates that Spo11 has at least two meiotic roles, catalyzing DSBs and pairing homologs, and that these roles can be independent. By extension, either C.cinereus spo11-1 is a null, and homolog pairing is entirely independent of Spo11 function in this organism, or the N-terminal third of the Spo11 protein remaining in spo11-1 is sufficient for homolog pairing.
Does chromosome condensation correlate with homolog pairing? Unlike WT nuclei, in which there is a continuous progression of both chromosome condensation and homolog pairing, in C.cinereus spo11-1 these two phenomena are separated. Progressively more nuclei are paired but fewer are condensed. Pairing interactions between less condensed homologs could be less susceptible to physical disruption, such as that caused by our spreading technique. Alternatively, the process of condensation may disrupt transient or weak homolog pairing interactions that have not been progressively stabilized (Weiner and Kleckner, 1994). Although our sample sizes are small (N = 121 for K+3 and N = 37 for K+6), our data suggest that our measurement of homolog pairing in spo11-1 is independent of the degree of chromosome condensation; at both timepoints, the percentage of nuclei with paired homologs is similar in both well condensed and uncondensed nuclei.
Basidial cell death
Meiotic progression in C.cinereus spo11-1 is aberrant. Instead of normal mid-to-late meiotic events, morphological changes are triggered that appear similar to those of cells undergoing PCD. By K+10, spo11-1 DNA is fragmented, as indicated by a positive TUNEL assay (Figure 6B). By K+11, spo11-1 basidia have undergone pyknosis as well as nuclear fragmentation with condensation and marginalization (Figure 6A, panel C). It is unclear what is precipitating this cascade of events. It is unlikely that PCD is triggered directly by the absence of a meiotic DSB; other C.cinereus mutants that are presumably defective in meiotic DSB formation (based on sequence similarity to known DSB repair proteins) do not appear to undergo this progression to cell death. mre11-1 basidia at K+15 contain a single nucleus (Gerecke and Zolan, 2000), unlike spo11-1 basidia, which are devoid of DNA by the same timepoint. Intriguingly, the spo11-1 late meiotic phenotype is somewhat reminiscent of C.cinereus rad9-1 nuclei observed at late timepoints (Valentine et al., 1995). It is unclear whether rad9-1 also undergoes PCD, but we are investigating this possibility (S.T.Merino and M.E.Zolan, in preparation).
Although PCD has been described in fungi (Lu, 1991; Leslie and Zeller, 1996; Madeo et al., 1997; Roze and Linz, 1998), meiotic PCD has been described previously only in animal cells. Meiotic PCD occurs in normal mammalian gametogeneses (Coucouvanis et al., 1993; Yin et al., 1997; McGee et al., 1998), and relatively more PCD can occur in meiotic mutants (Edelmann et al., 1996; Xu et al., 1996; Barlow et al., 1998; Odorisio et al., 1998; de Vries et al., 1999). The phenotype of C.cinereus spo11-1 meiotic nuclei is strikingly similar to those of Mlh1-deficient mice. The chromosomes from Mlh1- deficient mice spermatogonia fail to form chiasmata (Baker et al., 1996), and the resultant univalents are likely to be unable to make the necessary bipolar attachments to the meiotic spindle (Woods et al., 1999). Also, the meiotic spindle in Mlh1-defective females is aberrant; unequal poles, collapse of the spindle, elongated spindle and tripolar spindles were observed. Woods et al. (1999) and McKim and Hawley (1995) suggest that bivalent chromosomes have a role in meiotic spindle formation, in that they establish bipolar attachments to the antiparallel arrays of microtubules to tether the spindle. In C.cinereus spo11-1, homologs fail to undergo synapsis, thus univalents are likely to be present. Chromosomes fail to attach properly to the meiotic spindle and spindles are grossly aberrant. Similar to the situation in Mlh1-deficient mice, the PCD in C.cinereus spo11-1 meiotic nuclei could be triggered as a consequence of unattached univalent chromosomes.
In C.cinereus spo11-1, ionizing radiation induces AE and SC formation, and enhances spore viability
Based on the similarities of C.cinereus Spo11 to other Spo11 homologs, we reasoned that WT C.cinereus may also be making DSBs that lead to meiotic recombination. We found that ionizing radiation markedly increased C.cinereus basidiospore production and viability. The 9-fold increase in viability (from 1.6 to 14%) is even more striking when compared with the lethality of the treatment (spore viability in unirradiated AmutBmut was 96%, and that in irradiated AmutBmut was 52%). When spo11 mutants in both S.cerevisiae (Thorne and Byers, 1993) and C.elegans (Dernburg et al., 1998) were exposed to radiation during meiosis, gamete viability also increased. Thus DNA breaks, promoted by Spo11, are probably a common mechanism for initiating meiotic recombination events that lead to chiasmata and accurate homolog segregation.
When C.cinereus spo11-1 is treated with ionizing radiation, two key phenotypes are observed: an increase in AE formation and the establishment of SC. Compared with unirradiated spo11-1, in which only 54% of meiotic nuclei have AE, after irradiation all spo11-1 meiotic nuclei had AE. We have demonstrated that chromosome condensation is necessary for AE formation (see above), and that Spo11 is required for maintaining chromosome condensation (see above). Because ionizing radiation leads to an increase in AE formation, we propose that the DSB function of Spo11 is required for chromosome condensation, AE formation or both. Since γ irradiation also leads to SC formation in spo11-1 (Figure 7), meiotic DNA breaks normally precede and are required for SC formation in C.cinereus as they are in S.cerevisiae (Alani et al., 1990; Cao et al., 1990; Padmore et al., 1991; Sun et al., 1991). This differs from D.melanogaster and C.elegans, which may make DNA breaks after SC formation (Dernburg et al., 1998; McKim and Hayashi-Hagihara, 1998; McKim et al., 1998).
In C.cinereus, as in S.cerevisiae (Loidl et al., 1994) and Zea mays (McClintock, 1933; Maguire and Riess, 1994), SC formation does not depend on homolog pairing (Seitz et al., 1996; Gerecke and Zolan, 2000). Coprinus cinereus spo11-1 shows a substantial amount of homolog pairing (66%) and no SC formation. After irradiation, spo11-1 makes promiscuous SC, as synapsis among multiple AEs was observed (Figure 7), but the increase in spore viability is likely to be a result of SC between homologs, and this may be facilitated by the high level of intrinsic homolog pairing in C.cinereus spo11-1.
Materials and methods
Strains and culture conditions
The dikaryons of C.cinereus used in this study (WT=J6;5-4×J6;5-5; Seitz et al., 1996) and AmutBmut (Swamy et al., 1984) have been described previously. The WT monokaryons Java-6 and Okayama-7 are described in Binninger et al. (1987) and Wu et al. (1983), respectively. The spo11-1 strain (R126-49) was created by insertional mutagenesis (Cummings et al., 1999) by transforming a dominant selectable marker (pPHT1) that confers resistance to hygromycin B into AmutBmut. Culture conditions, matings and fruiting conditions for C.cinereus were as described by Zolan et al. (1988).
Screening the cosmid library
Forty-three pools of the C.cinereus genomic cosmid library (May et al., 1991) were screened using standard procedures (Sambrook et al., 1989), with a PCR-generated 0.5 kb DNA fragment of the C.cinereus spo11 gene (Cummings et al., 1999).
DNA sequencing and analysis
Subcloning and DNA sequencing were as described in Gerecke and Zolan (2000). DNA sequences obtained were translated in all six reading frames and compared with all non-redundant polypeptides in the translated NCBI database (GenBank, Bethesda, MD), using the program BLAST (Altschul et al., 1990). Polypeptide alignments were performed using CLUSTAL W (Thompson et al., 1994).
PCR amplification of the cDNA
Poly(A) RNA (440 ng), purified from WT cap tissue at K+6 (Yeager Stassen et al., 1997), was used to generate cDNAs (Gerecke and Zolan, 2000). A 643 bp mid-region of the spo11 cDNA was amplified using the primers OL-162 (5′-AATGGGCGAGGCACAATT) and OL-163 (5′-GCTCATGCTTTGGCTCCC) and standard conditions. The 5′ and 3′ ends of the spo11 cDNA were amplified and cloned separately. Primers (5′-TTGAGCTGTTGAAGTCAGCG and 5′-GATCCATAC GGAATCGACAT) were designed based on the genomic DNA that encodes the predicted ORF of C.cinereus spo11. These were used in amplifications independently with primers designed to the T3 and T7 promoters, present within the λZAP vector. A λZAP cDNA library of C.cinereus early meiotic transcripts (Yeager Stassen et al., 1997) was used as the template for both the 5′ and 3′ end amplifications.
Alignment of the cDNA (1305 bases) and genomic sequences allowed us to identify 13 introns in 1.8 kb of coding sequence, as well as one intron in the 5′ UTR. The full-length cDNA sequence was translated and compared with all non-redundant polypeptides in the translated NCBI database to identify similar sequences. Additionally, the spo11 cDNA was aligned to the genomic DNA in order to determine the UTRs and the intron boundaries.
Transformation of C.cinereus
A fragment of genomic DNA that contains the ORF of the spo11 gene plus ∼2 kb both upstream and downstream of the predicted start and stop codons was released from one of the cosmids (207A7) using EcoRI (New England Biolabs). The 5.4 kb EcoRI DNA fragment was ligated into the linearized (EcoRI) plasmid pPAB1-2 (Granado et al., 1997). The resulting construct, pPS1, was amplified and purified from Escherichia coli using a plasmid DNA extraction kit (BIGGERprep; 5 PRIME→3 PRIME). The insert orientation was confirmed by restriction digests, and the DNA sequence of the 5.4 kb insert was obtained. pPS1 was then used to create a second construct (pPS2), which contains a truncated spo11: pPS1 was digested with XhoI and then re-circularized (T4 ligase; New England Biolabs; 16°C, overnight). pPS2 was amplified in E.coli, purified, and the truncation of spo11 was confirmed with restriction digests.
Protoplasts of C.cinereus were transformed using the method described in Binninger et al. (1987). For transformation into the C.cinereus strain R126-49, 28 µg of either pPS1 or pPS2 were used. Transformants were selected by restoration of p-aminobenzoic acid (paba) prototrophy in the presence of hygromycin B (Calbiochem). Transformants were induced to fruit, and cap color was noted.
Spore production and viability
The number of basidiospores produced per milligram of cap tissue was determined using the procedure described in Ramesh and Zolan (1995). Viability of basidiospores was established using the spotted drops method described in Ramesh and Zolan (1995). For each strain tested, basidiospores from 3–5 caps were analyzed; the average and standard deviation are reported.
Electron microscopy
Spreads of C.cinereus chromosomes were prepared and stained with silver nitrate as described previously (Pukkila and Lu, 1985), and grids were viewed with a JEOL-1010 electron microscope. The sums of the lengths of AEs or LEs were obtained by first scanning EM prints of silver-stained chromosomes into Adobe Photoshop v. 4.0 and subsequently measuring the lengths using NIH Image v. 1.56 (Wayne Rasband, public domain software).
γ irradiations
AmutBmut and spo11-1 strains were induced to form fruiting bodies using standard light and temperature conditions (Zolan et al., 1988). At 1.5 h post-karyogamy, fruiting bodies were irradiated in a 137Cs irradiator (J.L.Shephard and Associates; model Mark I-68A), using 20 krad at a dose rate of 191.25 rad/min. After irradiations, fruiting bodies were returned to standard light and temperature conditions.
Fluorescence microscopy
The progression of nuclei through meiosis was monitored in gill tissue by staining DNA with DAPI (Vector Laboratories).
FISH procedures were as described in Li et al. (1999), and the same chromosome 8- and 13-specific probes were used as described in Gerecke and Zolan (2000). Meiotic nuclei from spo11-1 fruiting bodies were examined at K+3 and K+6.
Meiotic spindles were detected based on the procedures described in Tanabe and Kamada (1994), with modifications. Single gill sections were fixed in 4% (v/v) formaldehyde, 50 mM NaH2PO4·HCl pH 6.5, 5 mM MgCl2, 5% (w/v) PEG 8000 and 5 mM EGTA at room temperature (RT) for 2 h. Sections were washed three times in 50 mM NaH2PO4·HCl pH 6.5, 5 mM MgCl2 for 10 min each, and cell walls were permeabilized [0.4% (w/v) Novozyme 234 (Interspex Products, Inc.) in 50 mM NaH2PO4·HCl pH 6.5, 5 mM MgCl2] for 7–10 min at 37°C, and then washed twice in phosphate-buffered saline (PBS) pH 7.3 for 10 min each. Subsequently, sections were soaked in a detergent solution [100 µl of Triton X-100, 19 mg of EGTA, 9.5 ml of PBS, 0.5 ml of phenylmethylsulfonyl fluoride (PMSF)] at RT for 20 min. Then, sections were washed five times in PBS for 10 min each. DM1A mouse anti-α-tubulin antibody (Accurate Chemical and Scientific Corp.) was used at a 1:250 dilution in PBS containing 1% (w/v) bovine serum albumin (BSA). Excess antibody was removed by three consecutive 10 min washes in PBS containing 1% (w/v) BSA. The primary antibody was detected with the secondary antibody, fluorescein isothiocyanate (FITC)-conjugated anti-mouse IgG F(ab′)2-fragment (Cappel Research Products), at 1:500 for 4 h at 37°C, and excess antibody was removed by three consecutive 10 min washes in PBS pH 8.4. Single sections were mounted in an anti-fade mounting medium with DAPI (Vectashield; Vector Laboratories). A cover slip was then applied and sealed with nail lacquer.
Nuclei in both FISH and immunofluorescence analyses were examined as in Li et al. (1999).
TUNEL assay
Single layers of gill tissue were fixed for 1.5 h in 3.7% formaldehyde in PBS. Gills were rinsed twice for 10 min each in PBS. The tissue was immersed in 80% (v/v) ethanol, and single gill layers were placed onto positively charged Probe-On Plus slides (Fisher Scientific). Slides were dried for 2 h at 45°C, immersed in 70% ethanol for 10 min, and air-dried overnight. Gill tissue was rehydrated by immersing slides in a descending series of aqueous ethanol concentrations (100, 95 and 70%) for 5 min each. Slides were rinsed in PBS for 10 min and used in subsequent analyses. The TACS™ 2 TdT-Blue Label in situ apoptosis detection kit (Trevigen, Inc.) was used for in situ detection of DNA fragmentation within gill tissue nuclei. Detection was performed as per the manufacturer’s instructions. Cells were permeabilized using proteinase K (5 µg/ml), and the final dehydration was performed using p-xylene. Gills were mounted using Permount (Fisher Scientific) and examined under visible light using a Nikon Microphot-FXA microscope.
Acknowledgments
Acknowledgements
We thank E.Larson for preliminary analysis of homolog pairing in spo11-1, S.Gasior for suggesting the dose and dose rate for the irradiation experiments, and C.Walczack for helpful discussions about spindle structure. We also thank E.Gerecke for help with the Northern blot analysis, and S.Keeney, F.Baudat, M.Jasin, P.Romanienko and D.Camerini-Otero for making the mammalian Spo11 sequences available to us prior to publication. Finally we thank E.Gerecke, J.Drummond, S.Acharya and J.Cummings for critical reading of the manuscript. This work was supported by NIH grant GM43930 (to M.E.Z.). M.C. was partially supported by a grant from the Walther Cancer Institute through the Indiana Institute for Molecular and Cellular Biology. S.T.M. was supported by NSF Minority Postdoctoral Research Fellowship DBI-9628899. J.E.S. was a Medic-B scholar (NIH), and A.M.M. was supported by an LS McClung Undergraduate Summer Research Award and a URCAP Award, Indiana University.
References
- Alani E., Padmore,R. and Kleckner,N. (1990) Analysis of wild-type and rad50 mutants of yeast suggests an intimate relationship between meiotic chromosome synapsis and recombination. Cell, 61, 419–436. [DOI] [PubMed] [Google Scholar]
- Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Atcheson C.L., DiDomenico,B., Frackman,S., Esposito,R.E. and Elder,R.T. (1987) Isolation, DNA sequence and regulation of a meiosis-specific eukaryotic recombination gene. Proc. Natl Acad. Sci. USA, 84, 8035–8039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker B.S., Carpenter,A.T., Esposito,M.S., Esposito,R.E. and Sandler,L. (1976) The genetic control of meiosis. Annu. Rev. Genet., 10, 53–134. [DOI] [PubMed] [Google Scholar]
- Baker S.M. et al. (1996) Involvement of mouse Mlh1 in DNA mismatch repair and meiotic crossing over. Nature Genet., 13, 336–342. [DOI] [PubMed] [Google Scholar]
- Barlow C. et al. (1998) Atm deficiency results in severe meiotic disruption as early as leptonema of prophase I. Development, 125, 4007–4017. [DOI] [PubMed] [Google Scholar]
- Bergerat A., de Massy,B., Gadelle,D., Varoutas,P.C., Nicolas,A. and Forterre,P. (1997) An atypical topoisomerase II from Archaea with implications for meiotic recombination. Nature, 386, 414–417. [DOI] [PubMed] [Google Scholar]
- Binninger D.M., Skrzynia,C., Pukkila,P.J. and Casselton,L.A. (1987) DNA-mediated transformation of the basidiomycete Coprinus cinereus.EMBO J., 6, 835–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L., Alani,E. and Kleckner,N. (1990) A pathway for generation and processing of double-strand breaks during meiotic recombination in S.cerevisiae.Cell, 61, 1089–1101. [DOI] [PubMed] [Google Scholar]
- Cha R.S., Weiner,B.M., Keeney,S., Dekker,J. and Kleckner,N. (2000) Progression of meiotic DNA replication is regulated by interchromosomal interaction proteins, negatively by Spo11p and positively by Rec8p. Genes Dev., 14, 493–503. [PMC free article] [PubMed] [Google Scholar]
- Clifford J., Chiba,H., Sobieszczuk,D., Metzger,D. and Chambon,P. (1996) RXRα-null F9 embryonal carcinoma cells are resistant to the differentiation, anti-proliferative and apoptotic effects of retinoids. EMBO J., 15, 4142–4155. [PMC free article] [PubMed] [Google Scholar]
- Coucouvanis E.C., Sherwood,S.W., Carswell-Crumpton,C., Spack,E.G. and Jones,P.P. (1993) Evidence that the mechanism of prenatal germ cell death in the mouse is apoptosis. Exp. Cell Res., 209, 238–247. [DOI] [PubMed] [Google Scholar]
- Cummings W.J., Celerin,M., Crodian,J., Brunick,L.K. and Zolan,M.E. (1999) Insertional mutagenesis in Coprinus cinereus: use of a dominant selectable marker to generate tagged, sporulation-defective mutants. Curr. Genet., 36, 371–382. [DOI] [PubMed] [Google Scholar]
- Dernburg A.F., McDonald,K., Moulder,G., Barstead,R., Dresser,M. and Villeneuve,A.M. (1998) Meiotic recombination in C.elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell, 94, 387–398. [DOI] [PubMed] [Google Scholar]
- de Vries S.S., Baart,E.B., Dekker,M., Siezen,A., de Rooij,D.G., de Boer,P. and de Riele,H. (1999) Mouse MutS-like protein Msh5 is required for proper chromosome synapsis in male and female meiosis. Genes Dev., 13, 523–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dresser M.E. and Giroux,C.N. (1988) Meiotic chromosome behavior in spread preparations of yeast. J. Cell Biol., 106, 567–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelmann W. et al. (1996) Meiotic pachytene arrest in MLH1-deficient mice. Cell, 85, 1125–1134. [DOI] [PubMed] [Google Scholar]
- Gavrieli Y., Sherman,Y. and Ben-Sasson,S.A. (1992) Identification of programmed cell death in situ via specific labeling of nuclear DNA fragmentation. J. Cell Biol., 119, 493–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerecke E.E. and Zolan,M.E. (2000) An mre11 mutant of Coprinus cinereus has defects in meiotic chromosome pairing, condensation and synapsis. Genetics, 154, 1125–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giroux C.N. (1988) Chromosome synapsis and meiotic recombination. In Kucherlapati,R. and Smith,G.R. (eds), Genetic Recombination. American Society for Microbiology, Washington, DC, pp. 465–496. [Google Scholar]
- Giroux C.N., Dresser,M.E. and Tiano,H.F. (1989) Genetic control of chromosome synapsis in yeast meiosis. Genome, 31, 88–94. [DOI] [PubMed] [Google Scholar]
- Gorczyca W., Gong,J. and Darzynkiewicz,Z. (1993) Detection of DNA strand breaks in individual apoptotic cells by the in situ terminal deoxynucleotidyl transferase and nick translation assays. Cancer Res., 53, 1945–1951. [PubMed] [Google Scholar]
- Granado J.D., Kertesz-Chaloupkova,K., Aebi,M. and Kues,U. (1997) Restriction enzyme-mediated DNA integration in Coprinus cinereus.Mol. Gen. Genet., 256, 28–36. [DOI] [PubMed] [Google Scholar]
- Keeney S., Giroux,C.N. and Kleckner,N. (1997) Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell, 88, 375–384. [DOI] [PubMed] [Google Scholar]
- Kerr J.F., Wyllie,A.H. and Currie,A.R. (1972) Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. Br. J. Cancer, 26, 239–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie J.F. and Zeller,K.A. (1996) Heterokaryon incompatibility in fungi—more than just another way to die. J. Genet., 75, 415–424. [Google Scholar]
- Li L., Gerecke,E.E. and Zolan,M.E. (1999) Homolog pairing and meiotic progression in Coprinus cinereus.Chromosoma, 108, 384–392. [DOI] [PubMed] [Google Scholar]
- Lin Y. and Smith,G.R. (1994) Transient, meiosis-induced expression of the rec6 and rec12 genes of Schizosaccharomyces pombe.Genetics, 136, 769–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loidl J., Klein,F. and Scherthan,H. (1994) Homologous pairing is reduced but not abolished in asynaptic mutants of yeast. J. Cell Biol., 125, 1191–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu B.C. (1991) Cell degeneration and gill remodelling during basidiocarp development in the fungus Coprinus cinereus.Can. J. Bot., 69, 1161–1169. [Google Scholar]
- Lu B.C. (1974) Meiosis in Coprinus: VI. The control of the initiation of meiosis. Can. J. Genet. Cytol., 16, 115–164. [PubMed] [Google Scholar]
- Madeo F., Frohlich,E. and Frohlich,K.U. (1997) A yeast mutant showing diagnostic markers of early and late apoptosis. J. Cell Biol., 139, 729–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguire M.P. and Riess,R.W. (1994) The relationship of homologous synapsis and crossing over in a maize inversion. Genetics, 137, 281–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May G., Le Chevanton,L. and Pukkila,P.J. (1991) Molecular analysis of the Coprinus cinereus mating type A factor demonstrates an unexpectedly complex structure. Genetics, 128, 529–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B. (1933) The association of non-homologous parts of chromosomes in mid-prophase of Zea mays. In Goldschmidt,R. and Von Mollendroff,W. (eds), Zeitschrift fur Zellforschung und Mikroskopische Anatomie. Verlag von Julius Springer, Berlin, Germany, pp. 191–237. [Google Scholar]
- McGee E.A., Hsu,S.Y., Kaipia,A. and Hsueh,A.J. (1998) Cell death and survival during ovarian follicle development. Mol. Cell. Endocrinol., 140, 15–18. [DOI] [PubMed] [Google Scholar]
- McKim K.S. and Hawley,R.S. (1995) Chromosomal control of meiotic cell division. Science, 270, 1595–1601. [DOI] [PubMed] [Google Scholar]
- McKim K.S. and Hayashi-Hagihara,A. (1998) mei-W68 in Drosophila melanogaster encodes a Spo11 homolog: evidence that the mechanism for initiating meiotic recombination is conserved. Genes Dev., 12, 2932–2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKim K.S., Green-Marroquin,B.L., Sekelsky,J.J., Chin,G., Steinberg,C., Khodosh,R. and Hawley,R.S. (1998) Meiotic synapsis in the absence of recombination. Science, 279, 876–878. [DOI] [PubMed] [Google Scholar]
- Nag D.K., Scherthan,H., Rockmill,B., Bhargava,J. and Roeder,G.S. (1995) Heteroduplex DNA formation and homolog pairing in yeast meiotic mutants. Genetics, 141, 75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odorisio T., Rodriguez,T.A., Evans,E.P., Clarke,A.R. and Burgoyne,P.S. (1998) The meiotic checkpoint monitoring synapsis eliminates spermatocytes via p53-independent apoptosis. Nature Genet., 18, 257–261. [DOI] [PubMed] [Google Scholar]
- Padmore R., Cao,L. and Kleckner,N. (1991) Temporal comparison of recombination and synaptonemal complex formation during meiosis in S.cerevisiae.Cell, 66, 1239–1256. [DOI] [PubMed] [Google Scholar]
- Pukkila P.J. (1994) Meiosis in mycelial fungi. In Wessels,J.G.H. and Meinhardt,F. (eds), The Mycota. Growth, Differentiation and Sexuality. Springer-Verlag, Berlin, Germany, pp. 267–228. [Google Scholar]
- Pukkila P.J. and Lu,B.C. (1985) Silver staining of meiotic chromosomes in the fungus, Coprinus cinereus.Chromosoma, 91, 108–112. [DOI] [PubMed] [Google Scholar]
- Pukkila P.J., Skrzynia,C. and Lu,B.C. (1992) The rad3-1 mutant is defective in axial core assembly and homologous chromosome pairing during meiosis in the basidiomycete Coprinus cinereus.Dev. Genet., 13, 403–410. [Google Scholar]
- Ramesh M.A. and Zolan,M.E. (1995) Chromosome dynamics in rad12 mutants of Coprinus cinereus.Chromosoma, 104, 189–202. [DOI] [PubMed] [Google Scholar]
- Roeder G.S. (1997) Meiotic chromosomes: it takes two to tango. Genes Dev., 11, 2600–2621. [DOI] [PubMed] [Google Scholar]
- Roze L.V. and Linz,J.E. (1998) Lovastatin triggers an apoptosis-like cell death process in the fungus Mucor racemosus.Fungal Genet. Biol., 25, 119–133. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Seitz L.C., Tang,K., Cummings,W.J. and Zolan,M.E. (1996) The rad9 gene of Coprinus cinereus encodes a proline-rich protein required for meiotic chromosome condensation and synapsis. Genetics, 142, 1105–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A.V. and Roeder,G.S. (1997) The yeast Red1 protein localizes to the cores of meiotic chromosomes. J. Cell Biol., 136, 957–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H., Treco,D. and Szostak,J.W. (1991) Extensive 3′-overhanging, single-stranded DNA associated with the meiosis specific double-strand breaks at the ARG4 recombination initiation site. Cell, 64, 1155–1161. [DOI] [PubMed] [Google Scholar]
- Swamy S., Uno,I. and Ishikawa,T. (1984) Morphogenic effects of mutations at the A and B incompatability factors in Coprinus cinereus.J. Gen. Microbiol., 130, 3219–3224. [Google Scholar]
- Tanabe S. and Kamada,T. (1994) The role of astral micotubules in conjugate division in the dikaryon of Coprinus cinereus.Exp. Mycol., 18, 338–348. [Google Scholar]
- Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorne L.W. and Byers,B. (1993) Stage-specific effects of X-irradiation on yeast meiosis. Genetics, 134, 29–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentine G., Wallace,Y.J., Turner,F.R. and Zolan,M.E. (1995) Pathway analysis of radiation-sensitive meiotic mutants of Coprinus cinereus.Mol. Gen. Genet., 247, 169–179. [DOI] [PubMed] [Google Scholar]
- Wang J.C. (1996) DNA topoisomerases. Annu. Rev. Biochem., 65, 635–692. [DOI] [PubMed] [Google Scholar]
- Weiner B.M. and Kleckner,N. (1994) Chromosome pairing via multiple interstitial interactions before and during meiosis in yeast. Cell, 77, 977–991. [DOI] [PubMed] [Google Scholar]
- Woods L.M., Hodges,C.A., Baart,E., Baker,S.M., Liskay,M. and Hunt,P.A. (1999) Chromosomal influence on meiotic spindle assembly: abnormal meiosis I in female Mlh1 mutant mice. J. Cell Biol., 145, 1395–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M.M.J., Cassidy,J.R. and Pukkila,P.J. (1983) Polymorphisms in DNA of Coprinus cinereus.Curr. Genet., 7, 385–392. [DOI] [PubMed] [Google Scholar]
- Xu Y., Ashley,T., Brainerd,E.E., Bronson,R.T., Meyn,M.S. and Baltimore,D. (1996) Targeted disruption of ATM leads to growth retardation, chromosomal fragmentation during meiosis, immune defects and thymic lymphoma. Genes Dev., 10, 2411–2422. [DOI] [PubMed] [Google Scholar]
- Yeager Stassen N.Y., Logsdon,J.M.,Jr, Vora,G.J., Offenberg,H.H., Palmer,J.D. and Zolan,M.E. (1997) Isolation and characterization of rad51 orthologs from Coprinus cinereus and Lycopersicon esculentum and phylogenetic analysis of eukaryotic recA homologs. Curr. Genet., 31, 144–157. [DOI] [PubMed] [Google Scholar]
- Yin Y., DeWolf,W.C. and Morgentaler,A. (1997) p53 is associated with the nuclear envelope in mouse testis. Biochem. Biophys. Res. Commun., 235, 689–694. [DOI] [PubMed] [Google Scholar]
- Zolan M.E., Tremel,C.J. and Pukkila,P.J. (1988) Production and characterization of radiation-sensitive meiotic mutants of Coprinus cinereus.Genetics, 120, 379–387. [DOI] [PMC free article] [PubMed] [Google Scholar]