Abstract
Maintenance of telomeres is implicated in chromosome stabilization and cell immortalization. Telomerase, which catalyzes de novo synthesis of telomeres, is activated in germ cells and most cancers. Telomerase activity is regulated by gene expression for its catalytic subunit, TERT, whereas several lines of evidence have suggested a post-translational regulation of telomerase activity. Here we identify the 14-3-3 signaling proteins as human TERT (hTERT)-binding partners. A dominant-negative 14-3-3 redistributed hTERT, which was normally predominant in the nucleus, into the cytoplasm. Consistent with this observation, hTERT-3A, a mutant that could not bind 14-3-3, was localized into the cytoplasm. Leptomycin B, an inhibitor of CRM1/exportin 1-mediated nuclear export, or disruption of a nuclear export signal (NES)-like motif located just upstream of the 14-3-3 binding site in hTERT impaired the cytoplasmic localization of hTERT. Compared with wild-type hTERT, hTERT-3A increased its association with CRM1. 14-3-3 binding was not required for telomerase activity either in vitro or in cell extracts. These observations suggest that 14-3-3 enhances nuclear localization of TERT by inhibiting the CRM1 binding to the TERT NES-like motif.
Keywords: hTERT/nuclear localization/telomerase/14-3-3
Introduction
The telomere is a unique structure found in the termini of most eukaryotic chromosomes. Tandem repeats of the highly conserved telomeric DNA (TTAGGG in vertebrates) (Moyzis et al., 1988) shorten after each cell cycle due to the end replication problem of chromosomal DNA (Harley et al., 1990; Hastie et al., 1990). Since critically shortened telomeres are often associated with the replicative senescence of normal somatic cells, telomeres have been thought to be a mitotic clock that counts the number of times a cell can divide before eliciting the senescence program (Harley et al., 1990; Hastie et al., 1990; Allsopp et al., 1992; Counter et al., 1992). Based on these situations, maintenance of telomeres is implicated in chromosome stabilization and cell immortalization (Zakian, 1995; Greider, 1996).
Most immortal cells, including germ line cells and cancer cells, possess enzymatic activity of telomerase (Kim et al., 1994; Shay and Bacchetti, 1997), which catalyzes de novo synthesis of telomeric repeats at chromosome ends (Greider and Blackburn, 1985; Morin, 1989). Since introduction of the telomerase catalytic subunit gene (TERT) (Meyerson et al., 1997; Nakamura et al., 1997) into normal somatic cells extends the life of the cells (Bodnar et al., 1998), the immortalized phenotype of most cancer cells would involve activation of telomerase. Consistently, genetic disruption of the telomerase RNA component (mTR), which abolishes telomerase activity, causes telomere erosion and chromosomal aberrations, resulting in functional defects in highly proliferative organs (Blasco et al., 1997; Lee et al., 1998). Based on these observations, it is likely that cancer cells bypass telomere crisis by activating telomerase.
Recent studies have demonstrated that c-Myc directly induces TERT transcription and telomerase activity (J.Wang et al., 1998; Wu et al., 1999). In some immortal cells, differentiation induction downregulates telomerase activity with transcriptional repression of the TERT gene (Meyerson et al., 1997). These observations indicate that telomerase activity is mainly regulated by TERT transcription. On the other hand, telomerase activity could also be regulated by post-transcriptional and/or post-translational modifications of the enzyme. For example, decline in telomerase activity in confluent NIH 3T3 fibroblasts is not associated with downregulation of TERT expression (Greenberg et al., 1998). Similarly, telomerase activity in human lymphocytes is not correlated with the levels of human TERT (hTERT) transcript (Liu et al., 1999).
One possible mechanism for the post-translational modification of telomerase is the interaction of hTERT with accessory proteins, including other telomerase subunits, enzymes, adaptors or chaperones. Introduction of hTERT gene into normal cells induces telomerase activity and confers a continuous replicative capacity (Bodnar et al., 1998) whereas alteration of the C-terminus of hTERT disrupts its abilities to maintain telomere length and to immortalize a cell in spite of relevant telomerase activity (Counter et al., 1998). These observations suggest that the C-terminus of hTERT interacts with other factors required for telomere maintenance and cell immortalization.
The 14-3-3 proteins, which constitute a highly conserved isoform of homo- and heterodimeric molecules, associate with a number of different signaling proteins, including Raf-1, Cbl, nitrate reductase, Bad, the epithelial keratins K8/18, Cdc25C and the IGF-1 receptor (Liao and Omary, 1996; Moorhead et al., 1996; Muslin et al., 1996; Zha et al., 1996; Craparo et al., 1997; Liu et al., 1997; Peng et al., 1997). Based on their interaction with various ligands, 14-3-3 proteins have been proposed to be important in controlling intracellular signaling pathways (Aitken, 1996). In addition, 14-3-3 proteins work as molecular chaperones or regulate intracellular localization of their binding partners (Liao and Omary, 1996; Kumagai and Dunphy, 1999; Yang et al., 1999).
Here, we have identified 14-3-3 family proteins as hTERT-binding partners. We found that hTERT and 14-3-3 specifically bound at their respective C-termini, and this interaction was required for efficient accumulation of hTERT in the nucleus. We also demonstrated that 14-3-3 repressed the interaction of hTERT with CRM1/exportin 1, a receptor for the nuclear export machinery. These observations suggest that 14-3-3 is a post-translational modifier of telomerase that functions by controlling the intracellular localization of hTERT.
Results
Identification of 14-3-3 proteins as hTERT-interacting partners with a yeast two-hybrid screening
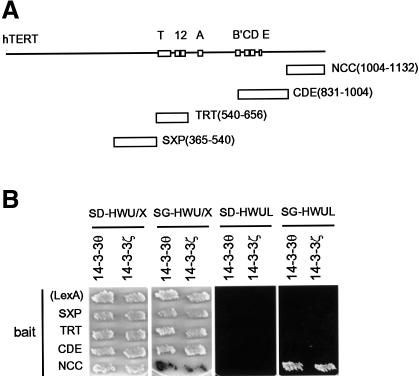
Using the C-terminal 129 amino acids of hTERT (non-characterized C-terminus, NCC; Figure 1A) as bait, we performed a yeast two-hybrid screening either on a HeLa or a human testis library. As a result, we obtained positive clones, two from HeLa and the other two from testis (Figure 1B). Both the HeLa clones contained the same cDNA fragment of 14-3-3θ, an isoform of the 14-3-3 family proteins. These clones lacked the N-terminus (residues 1–69) of the intact protein. The testis clones contained cDNAs of another 14-3-3 isoform, 14-3-3ζ, also lacking the N-terminus (residues 1–56 and 1–53, respectively). These prey clones did not interact with other hTERT domains (Figure 1B) or with other unrelated baits, such as a part of the TEP1 telomerase subunit (data not shown).
Fig. 1. Interaction of hTERT with 14-3-3 in the yeast two hybrid system. (A) Schematic representation of hTERT bait constructs. Bar indicates the bait domain used in the analysis. Values in parentheses indicate the positions of amino acid residues. SXP, a region containing a conventional 14-3-3 binding motif-like sequence; TRT, telomerase reverse transcriptase motif; CDE, reverse transcriptase motifs C, D and E (Nakamura et al., 1997); NCC, non-characterized C-terminus. (B) Interaction of hTERT with 14-3-3 in yeast cells. The prey clones used are 14-3-3θ (residues 70–245) and 14-3-3ζ (residues 57–245). The black (actually blue) signal on the SG-HWU/X plate and the growth on the SG-HWUL plate indicate activation of the reporter genes, lacZ and LEU2, respectively. S, synthetic; G, galactose; X, X-Gal; D, glucose; H, histidine (–); W, tryptophan (–); U, uracil (–); L, leucine (–).
hTERT and 14-3-3 specifically bind via their respective C-termini
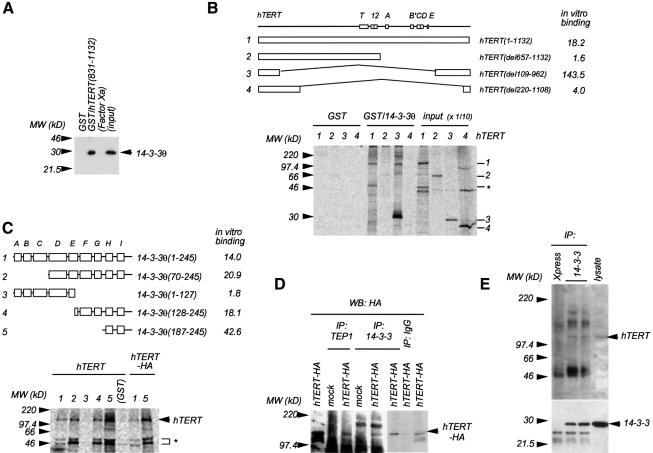
To confirm the direct interaction between hTERT and 14-3-3, we analyzed the binding of recombinant 14-3-3θ and hTERT (residues 831–1132) fused to glutathione S-transferase (GST). As shown in Figure 2A, these purified proteins bound directly in vitro. Furthermore, in vitro-translated full-length hTERT and hTERT(del109–962) bound to GST–14-3-3θ(70–245) but not to the control GST (Figure 2B). In the case of full-length hTERT, another band with lower molecular weight also bound to 14-3-3θ(70–245) (asterisk in Figure 2B). Since this smaller band was similarly detected in hTERT tagged with HA at the C-terminus (hTERT-HA) when analyzed by anti-HA immunoblotting (data not shown), it seemed to be a degraded product of the hTERT C-terminus. Neither hTERT(del220–1108) nor hTERT(del657–1132) bound to 14-3-3θ(70–245). These observations were consistent with the results of the two-hybrid analysis (Figure 1), because only hTERT constructs with an intact NCC region could bind to 14-3-3θ.
Fig. 2. Interaction of hTERT with 14-3-3 via their respective C-termini. Values in parentheses indicate the positions of amino acid residues. (A) Direct interaction between hTERT and 14-3-3. The GST fusion protein was immobilized on glutathione–Sepharose and incubated with recombinant 14-3-3θ. Bound fractions were analyzed by western blotting with anti-14-3-3θ. Factor Xa was used to prepare the recombinant 14-3-3θ and a trace of it (inactivated) remained in the preparation. (B) Binding properties of hTERT deletion mutants with 14-3-3. del, deleted. The GST fusion protein was immobilized on glutathione–Sepharose and incubated with each [35S]hTERT. The radioactivity bound to the beads was detected by phosphoimaging. Control radioactivity (interaction of each hTERT with GST alone) was given a value of 1. The input does not have the Sepharose material. Asterisk indicates an hTERT carboxyl fragment probably due to the degradation of the full-length protein (lane 1). (C) Binding properties of 14-3-3θ deletion mutants with hTERT. Boxes in the upper scheme indicate α-helices. The GST fusion protein was immobilized on glutathione–Sepharose and incubated with 35S-labeled full-length hTERT or hTERT-HA. The specific signal was detected by phosphoimaging. Control radioactivity (interaction of hTERT with GST alone) was given a value of 1. (D) Association of hTERT-HA and 14-3-3 in intact cells. The lysate of hTERT-HA-transfected 293T cells was incubated with either anti-TEP1 (Santa Cruz Biotechnology), anti-14-3-3 or mouse normal IgG fixed on protein G–Sepharose and the bound fractions were detected by anti-HA western blot analysis. IP, immunoprecipitation: WB, western blot. (E) Association of endogenous hTERT and 14-3-3 in intact cells. The A2780 cell lysate was immunoprecipitated with either anti-14-3-3 or anti-Xpress (control, Invitrogen) and the bound fractions were analyzed by anti-hTERT western blotting (upper panel). The same blot was re-probed with anti-14-3-3θ (lower panel).
Next, we examined the binding of the full-length hTERT to various regions of 14-3-3θ. 14-3-3 contains nine α-helices, A–I (Liu et al., 1995; Xiao et al., 1995), and all 14-3-3θ constructs that had the C-terminus (helices H and I) could bind hTERT (Figure 2C). Helix G, required for binding to phosphorylated c-Raf (Gu and Du, 1998), was dispensable for the hTERT binding. The C-terminal fragment of degraded hTERT again bound to these fusion proteins except 14-3-3θ(1–127) and GST itself. Similarly, hTERT with an HA-epitope tag also interacted with the 14-3-3θ C-terminus. We also observed co-immunoprecipitation of endogenous 14-3-3 with ectopically expressed hTERT-HA or with endogenous hTERT (Figure 2D and E). These observations indicate that hTERT binds to 14-3-3 via each C-terminus both in vitro and in intact cells.
14-3-3θ carboxyl fragment inhibits nuclear accumulation of hTERT
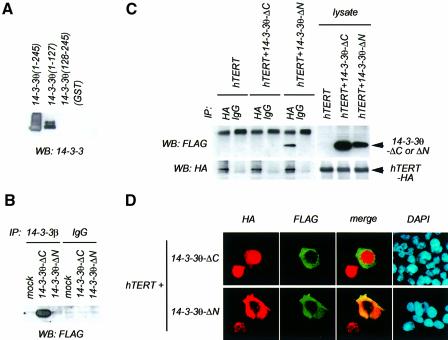
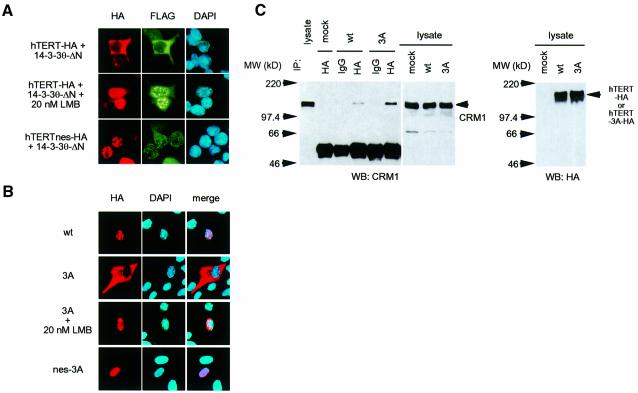
14-3-3 forms homo- and heterodimers (Aitken, 1996) that seemed to be required for successful involvement in signaling events, because mutant monomeric forms of 14-3-3, although able to bind Raf-1, do not enable Raf-1 to be activated in vivo, nor do they restore Raf-1 activity after displacement of 14-3-3 in vitro (Tzivion et al., 1998). Consistent with a previous report (Luo et al., 1995), 14-3-3θ(128–245) (14-3-3θ-ΔN), which lacked the N-terminal dimerization domain, could not dimerize with endogenous 14-3-3 in vitro or in intact cells (Figure 3A and B), whereas it could still bind efficiently to hTERT (Figures 2C and 3C). As a control, 14-3-3θ(1–127) (14-3-3θ-ΔC) could bind to endogenous 14-3-3 but not to hTERT (Figure 3A–C). Interestingly, 14-3-3θ-ΔN inhibited the nuclear accumulation of hTERT-HA expressed in 293T cells (Figure 3D, lower panel). Among the cells transfected with hTERT-HA in combination with an empty vector or 14-3-3θ-ΔC, 82% (55 of 67) or 79% (55 of 70) exhibited punctate accumulation of hTERT-HA in their nuclei, respectively (Figure 3D, upper panel). On the other hand, among the cells co-transfected with hTERT-HA and 14-3-3θ-ΔN, only 41% (15 of 37) did. As shown in Figure 3C, the hTERT–14-3-3θ-ΔN co-localization was observed in 59.5% (22 of 37) of co-transfected cells (either in the cytoplasm or in the nucleus). On the contrary, the hTERT–14-3-3θ-ΔC co-localization was observed in only 2.9% (2 of 70) of co-transfected cells, because hTERT-HA and 14-3-3θ-ΔC existed predominantly in the nucleus and in the cytoplasm, respectively. Furthermore, 14-3-3θ-ΔNV176D, in which the hydrophobic property of the ligand-binding site was altered (H.Wang et al., 1998), lost 80% of its ability to bind hTERT (data not shown). This mutant did not inhibit the nuclear accumulation of hTERT-HA [91% (31 of 34) of co-transfected cells exhibited punctate localization of hTERT-HA in the nuclei; data not shown], indicating that the inhibition of the nuclear accumulation of hTERT-HA required the hTERT–14-3-3θ-ΔN interaction. None of the mutations affected the total amount of ectopically expressed hTERT-HA (Figure 3C, lower panel). Since a similar 14-3-3 carboxyl fragment inhibits the Raf-1 activation by the full-length 14-3-3 in vitro and excludes Raf-1 from its activation process in vivo (Luo et al., 1995), it is likely that 14-3-3θ-ΔN, as a dominant-negative mutant, blocks the association of the wild-type 14-3-3 dimers with hTERT, inhibiting nuclear accumulation of hTERT. In fact, 14-3-3θ-ΔN but not 14-3-3θ-ΔC inhibited the association of hTERT-HA with endogenous 14-3-3 in 293T cells, as determined by immunoprecipitation assay (data not shown).
Fig. 3. Effects of 14-3-3θ deletion mutants on intracellular localization of hTERT. (A) Failure in dimerization of 14-3-3θ(128–245) with endogenous 14-3-3 in vitro. 293T cellular lysate was prepared and was incubated with each GST–14-3-3θ fusion protein immobilized on glutathione–Sepharose. Endogenous 14-3-3 bound to the beads was detected by anti-14-3-3 western blot analysis. (B) Failure in dimerization of 14-3-3θ-ΔN with endogenous 14-3-3 in intact cells. 293T cells were transfected with pFLAG-CMV-2–14-3-3θ(1–127) (ΔC) or pFLAG-CMV-2–14-3-3θ(128–245) (ΔN). Cellular lysates were prepared, and immunoprecipitation followed by western blot analysis was performed. Note that the antibody used for immunoprecipitation directly recognizes the endogenous 14-3-3 but not 14-3-3θ-ΔC or 14-3-3θ-ΔN (data not shown). (C) Interaction of hTERT with 14-3-3θ-ΔN but not with 14-3-3θ-ΔC in intact cells. 293T cells were transfected with pCR3–hTERT-HA, pCR3–hTERT-HA and pFLAG-CMV-2–14-3-3θ-ΔC, or pCR3–hTERT-HA and pFLAG-CMV-2–14-3-3θ-ΔN. Cellular lysates were prepared, and immunoprecipitation followed by western blot analysis was performed. (D) Effects of 14-3-3θ deletion mutants on intracellular localization of hTERT. 293T cells were transfected as in (C) and indirect immunofluorescence staining was performed. Expression of hTERT-HA (colored in red by rhodamine or Texas Red) and FLAG-14-3-3θ-ΔC or -ΔN (colored green by fluorescein) were detected. Note the cytoplasmic localization of hTERT in the hTERT–14-3-3θ-ΔN-coexpressed cell. Also shown are nuclei as colored in blue by DAPI.
hTERT mutant that cannot bind to 14-3-3 fails to localize into the nucleus
At present, conventional 14-3-3 binding motifs, such as RSXpSXP (pS indicates a critical phosphoserine), have been identified in most 14-3-3 binding proteins (Yaffe et al., 1997). However, there is no such consensus motif in the hTERT NCC region. Consistently, alkaline phosphatase treatment of hTERT did not diminish its interaction with 14-3-3θ (data not shown), suggesting that this interaction was independent of phosphorylation, which is usually required for the association of 14-3-3 with its partners. Also, the SXP bait, which contained RQHSSP, resembling the consensus RSXSXP, did not interact with 14-3-3θ or 14-3-3ζ (Figure 1). In addition, two-hybrid screenings using the SXP region as a bait either on a HeLa prey cDNA library or on a normal human testis prey cDNA library did not isolate any 14-3-3-related gene fragments as positive clones (H.Seimiya, M.Shimizu and T.Tsuruo, unpublished observations). These situations suggest that the interaction of hTERT with 14-3-3 is mediated by an alternative mode that is independent of serine phosphorylation.
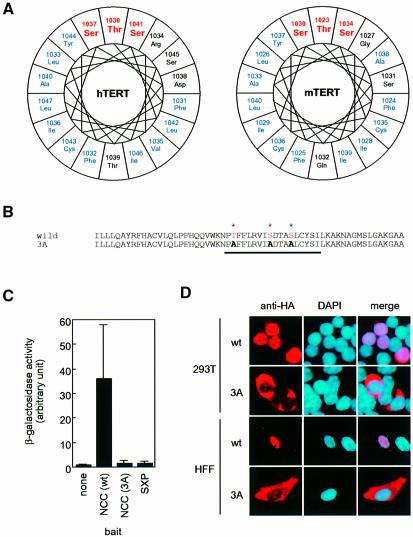
Crystal structure analysis predicts that 14-3-3 forms an amphipathic groove and its binding partners would possess an amphipathic helix to fit into the groove (Liu et al., 1995; Xiao et al., 1995). Since both hTERT and mTERT (mouse homolog) NCC regions have a putative amphipathic helix with a characteristic serine/threonine cluster (Du et al., 1996; Figure 4A), we constructed a mutant hTERT, in which Thr1030, Ser1037 and Ser1041 in the serine/threonine cluster were all mutated to alanine (hTERT-3A; Figure 4B). As expected, this mutant lost the ability to interact with 14-3-3θ both in yeast (Figure 4C) and in 293T cells (data not shown). Finally, we found that hTERT-3A-HA diminished its ability to localize into the nucleus and was accumulated in the cytoplasm both in 293T cells and in normal foreskin fibroblasts (Figure 4D). The percentage of cells with punctate accumulation of hTERT in the nuclei were 87% (87 of 100, 293T/wt), 46% (46 of 100, 293T/3A), 72% (31 of 43, HFF/wt) and 31% (12 of 39, HFF/3A), respectively. In the fibroblasts, the efficiency of ectopic expression of hTERT-HA was much lower than that in 293T cells (as described below; Figure 7B) and it is unlikely that the failure of the mutant in nuclear accumulation is due to artificial overexpression of the protein. These observations suggest that 14-3-3 is involved in nuclear localization of hTERT.
Fig. 4. Intracellular localization of an hTERT mutant that can not bind to 14-3-3. (A) Putative amphipathic helices in the NCC domains of hTERT and mTERT. Wheel structure analysis was performed by DNASIS (Hitachi Software). Serine/threonine cluster (Du et al., 1996) and hydrophobic residues are indicated by red and blue letters, respectively. (B) Partial amino acid sequence of hTERT and hTERT-3A. Underline indicates a putative amphipathic helix. Residues with asterisks form a serine/threonine cluster as in (A). (C) Two hybrid analysis between NCC-3A and 14-3-3θ(70–245). LacZ activation by the baits was monitored by measurement of β-galactosidase levels in liquid cultures grown on galactose. (D) Indirect immunofluorescence staining. Expression of hTERT-HA or hTERT-3A-HA was detected (colored in red by rhodamine). Also shown are nuclei as colored in blue by DAPI. wt, wild-type hTERT; 3A, hTERT-3A; HFF, human foreskin fibroblasts.
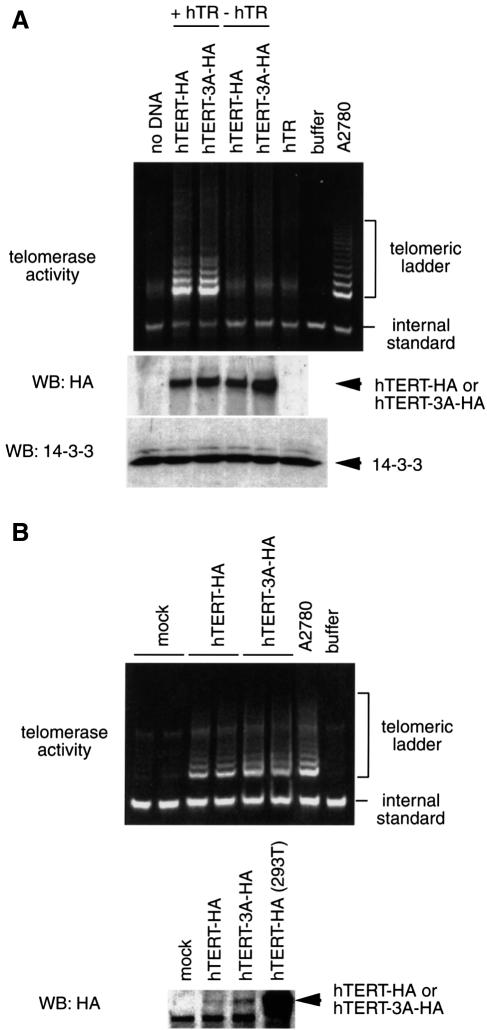
Fig. 7. 14-3-3 binding is not required for telomerase activity. (A) In vitro reconstitution (IVR) of telomerase activity. IVR telomerase was prepared by using the plasmids for each hTERT construct and hTR RNA subunit as described previously (Weinrich et al., 1997). Telomerase activity in each preparation was detected by the TRAP assay (Kim et al., 1994). Also shown are the results of western blot analysis on each IVR preparation with anti-HA or anti-14-3-3. Note that the reticulocyte lysate used for in vitro translation contained its own 14-3-3 proteins. (B) Telomerase activity in the hTERT-transfected fibroblasts. Human foreskin fibroblasts at population doubling 37 were transfected with indicated expression vectors. Portions of A2780 cell lysate (300 cells) or the fibroblast lysate (2 × 104 cells; much larger cell numbers than in case of A2780 were due to low efficiency of transfection and expression of exogenous hTERT) were analyzed by the TRAP assay. Also shown is the result of anti-HA western blot analysis on each total lysate.
Identification of a nuclear export signal-like motif in hTERT
Since hTERT protein is too large to diffuse through a nuclear pore complex channel (Nigg, 1997), the cytoplasmic localization of hTERT observed in Figures 3 and 4 might involve regulated machinery for nuclear export. We examined the hTERT primary amino acid sequence to determine whether it contains a leucine-rich sequence of conserved spacing and hydrophobicity that fits the criteria established for nuclear export signal (NES) (Fischer et al., 1995; Wen et al., 1995). As shown in Figure 5A, we found that residues between amino acids 970 and 981 conform to this motif, as indicated by the similarity to other known NESs such as p53 (Stommel et al., 1999). This putative NES was located just upstream of the 14-3-3 binding amphipathic helix (residues 1030–1047) and was conserved in mTERT. To confirm the interaction between CRM1 and the NES-like motif in hTERT, we performed an in vitro pull-down assay of CRM1 with GST–hTERT(831–1132). As shown in Figure 5B, CRM1 specifically associated with GST–hTERT(831–1132) but not with GST alone. We generated a mutant construct, GST–hTERTnes(831–1132), in which three conserved leucines, Leu974, Leu978 and Leu980, were replaced by alanines. This nes mutant significantly diminished its association with CRM1, indicating that this motif was actually recognized by CRM1.

Fig. 5. Identification of an NES-like motif in hTERT. (A) Comparative alignment of the NES-like motifs identified in TERT C-termini with other known NESs. Bold letters indicate the conserved Leu/Met/Ile residues. hTERTnes, a mutant in which three conserved leucines were replaced by alanines (asterisks). (B) Interaction between hTERT and CRM1 in vitro. The GST fusion protein was immobilized on glutathione–Sepharose and incubated with the 293T cell lysate (3 × 106 cells/sample). After the extensive washing, bound fractions were analyzed by western blotting with anti-human CRM1 antibody. Equal loading of each GST fusion protein was confirmed by Coomassie Blue staining of the SDS–PAGE gel (data not shown).
14-3-3 enhances nuclear localization of hTERT by inhibiting the CRM1 binding to the hTERT NES-like motif
To determine whether the NES/CRM1-mediated nuclear export (Fornerod et al., 1997; Fukuda et al., 1997; Ossareh-Nazari et al., 1997) is involved in cytoplasmic localization of hTERT by 14-3-3θ-ΔN, we first examined the effect of Leptomycin B (LMB), a potent inhibitor of NES/CRM1-mediated nuclear export (Nishi et al., 1994; Kudo et al., 1998, 1999). As shown in Figure 3, 14-3-3θ-ΔN inhibited the nuclear accumulation of hTERT (Figure 6A, top). When the cells were treated with 20 nM LMB for 2 h, this effect of 14-3-3θ-ΔN was completely abolished (Figure 6A, middle). Similarly, cytoplasmic localization of hTERT-3A-HA in foreskin fibroblasts was reversed by treatment with LMB (Figure 6B). LMB did not affect the intracellular localization of the wild-type hTERT-HA (data not shown).
Fig. 6. Involvement of the CRM1-mediated nuclear export in the cytoplasmic localization of hTERT. (A) Effects of LMB and the nes mutation on 14-3-3θ-ΔN-mediated inhibition of nuclear localization of hTERT. 293T cells were transfected with pCR3–hTERT-HA and pFLAG-CMV-2–14-3-3θ-ΔN or pCR3–hTERTnes-HA and pFLAG-CMV-2–14-3-3θ-ΔN. After 24 h of transfection, cells were fixed and indirect immunofluorescence staining was performed. Middle panel, cells were treated with 20 nM LMB for 2 h before the methanol fixation. Expression of hTERT-HA (colored red by rhodamine) and FLAG-14-3-3θ-ΔN (colored green by fluorescein) were detected. (B) Effects of LMB and the nes mutation on the 3A mutation-derived cytoplasmic localization of hTERT. Human foreskin fibroblasts were transfected with pCR3–hTERT-HA (wt), pCR3–hTERT-3A-HA (3A) or pCR3–hTERTnes-3A-HA (nes-3A). After 24 h of transfection, cells were fixed and indirect immunofluorescence staining was performed. Expression of each hTERT-HA construct (colored red by rhodamine) was detected. The percentage of cells with punctate accumulation of hTERT in the nuclei were 72% (31 of 43, wt), 33% (43 of 131, 3A), 60% (62 of 103, 3A + LMB), and 55% (78 of 143, nes-3A), respectively. (C) hTERT–CRM1 interaction in intact cells. 293T Cells were transfected with pCR3–hTERT-HA (wt) or pCR3–hTERT-3A-HA (3A). Cellular lysates were prepared, and immunoprecipitation followed by western blot analysis was performed.
These results prompted us to examine further the functional significance of the NES-like motif in hTERT. We introduced the nes mutation (Figure 5A) into hTERT-HA and examined the intracellular localization of the resulting mutant, hTERTnes-HA. In contrast to the wild-type hTERT-HA, 14-3-3θ-ΔN failed to redistribute hTERTnes-HA into the cytoplasm of 293T cells. Nuclear localization of hTERTnes-HA was observed in 87% (26 of 30) or 90% (27 of 30) of the transfected cells in the presence (Figure 6A, bottom) or the absence (data not shown) of 14-3-3θ-ΔN, respectively. Furthermore, when the nes mutation was introduced into hTERT-3A-HA (14-3-3-unbound), the resulting mutant, hTERTnes- 3A-HA (14-3-3 and CRM1-unbound), resumed the ability to accumulate into the nucleus (Figure 6B, bottom). Similarly, subcellular fractionation and western blot analysis revealed that hTERT-3A-HA was predominantly detected in the cytoplasmic fraction but either hTERT-HA or hTERTnes-3A-HA were not (data not shown). These results indicate that cytoplasmic localization of hTERT by 14-3-3θ-ΔN or of hTERT-3A are dependent on the NES-like motif near the 14-3-3-binding site in hTERT. Finally, we observed the association of hTERT with CRM1 in 293T cells (Figure 6C). Strikingly, the 3A mutation in hTERT enhanced the hTERT–CRM1 binding (3-fold increase, as determined by a densitometric analysis), suggesting a functional masking of the NES-like motif in hTERT by the 14-3-3 binding. Taken together, these observations suggest that 14-3-3 enhances nuclear localization of hTERT by inhibiting the hTERT–CRM1 interaction.
14-3-3 binding is not required for telomerase activity either in vitro or in intact cells
We examined telomerase activity of hTERT-3A. First, in vitro-translated hTERT-HA exhibited telomerase activity in a telomerase RNA template (hTR)-dependent manner (Figure 7A). In the reaction mixture, there were enormous amounts of 14-3-3 proteins derived from the rabbit reticulocyte lysate used for in vitro translation. Similar to the wild-type hTERT-HA, hTERT-3A-HA showed telomerase activity. We did not observe any difference in the signal intensity between hTERT-HA and hTERT-3A-HA. Next, telomerase-negative human foreskin fibroblasts were transfected with these constructs. Consistent with previous reports (Bodnar et al., 1998; Nakayama et al., 1998), the wild-type hTERT-HA conferred telomerase activity in the fibroblasts. Again, hTERT-3A-HA exhibited comparable telomerase activity in the cells (Figure 7B). We did not observe any effects of recombinant full-length or C-terminal 14-3-3θ proteins on telomerase activity, at least as determined by the in vitro TRAP assay (Kim et al., 1994; H.Seimiya, H.Sawada and T.Tsuruo, unpublished observations). These observations indicate that the hTERT–14-3-3 interaction is not required for telomerase activity either in vitro or in intact cells.
Discussion
One possible mechanism for the post-translational regulation of telomerase is modulation of the relative telomerase activity per holoenzyme by signaling factors (Li et al., 1997, 1998). Another possible mechanism, in which the relative telomerase activity is not necessarily affected, is the regulation of telomerase access to native telomeres (van Steensel and de Lange, 1997). Here, we demonstrated an involvement of sophisticated nuclear export machinery in the hTERT localization, suggesting a novel mode for regulation of telomerase functions. Our observation that the wild-type hTERT and the mutant hTERT-3A exhibited comparable telomerase activity in the whole-cell lysates (Figure 7B) indicates that the alteration of telomerase functions which depends on the intracellular redistribution of TERT would not be detected by the conventional TRAP assay. Our findings also suggest that the exclusion of telomerase from the nucleus would have some physiological significance in telomere dynamics of telomerase-positive cells. Actually, our preliminary experiments have shown the cytoplasmic localization of hTERT especially in pre-mitotic cells, indicating that hTERT is physiologically shuttled between the nucleus and the cytoplasm (H.Seimiya and T.Tsuruo, unpublished observations). The phenotype of the cells expressing an hTERT mutant without capability of cytoplasmic localization remains to be elucidated.
14-3-3 proteins often regulate intracellular localization of their binding partners. For example, phosphorylation of Cdc25 by Chk1 kinase creates a binding site in Cdc25 for 14-3-3 (Peng et al., 1997; Sanchez et al., 1997). The subsequent 14-3-3 binding to Cdc25 markedly reduces the nuclear import rate of Cdc25, allowing nuclear export mediated by an NES present in the N-terminus of Cdc25 to predominate (Yang et al., 1999). Since Cdc25 has a functional nuclear localization signal (NLS) that lies adjacent to the site of 14-3-3 binding, 14-3-3 binding might sterically block the access of Cdc25 to the nuclear import machinery (Kumagai and Dunphy, 1999; Yang et al., 1999). In fact, a Cdc25 mutant that cannot associate with 14-3-3 binds significantly better than the wild-type Cdc25 to the importin-α/β heterodimer, a nuclear import receptor for NLS (Kumagai and Dunphy, 1999; Yang et al., 1999). Although 14-3-3 has an NES-like motif in the C-terminus, it is not necessarily the case that the 14-3-3 binding to the ligands results in nuclear export of the ligands via direct interaction of CRM1 and the 14-3-3 NES. First, the Cdc25 mutant incapable of binding 14-3-3 is fully competent for nuclear export mediated by the Cdc25-intrinsic NES (Yang et al., 1999). Secondly, a mutant Cdc25 lacking functional NESs is not exported from the nucleus despite its continued ability to bind 14-3-3 (Kumagai and Dunphy, 1999; Yang et al., 1999). In fact, the ligand binding site and the NES in 14-3-3 are partially overlapped and the 14-3-3–CRM1 interaction is significantly inhibited by the phosphoserine peptide derived from a 14-3-3 binding protein (Rittinger et al., 1999). Based on these observations, masking and exposure of NLS/NES sequences that lie adjacent to the sites of 14-3-3 binding could determine the cytoplasmic or nuclear localization of such types of 14-3-3 binding proteins.
Our study suggests that 14-3-3 enhances the nuclear localization of hTERT by interfering the access of CRM1 to the hTERT NES-like motif near the 14-3-3 binding site. According to this model, it is likely that endogenous 14-3-3 dimers cannot interact with the monomeric 14-3-3θ-ΔN-bound hTERT, in which the NES-like motif is exposed without masking by a 14-3-3 dimer. Similarly, the NES masking of p53 tumor suppressor protein regulates intracellular localization and functions of p53 (Stommel et al., 1999). Alternatively, even if 14-3-3 binding does not actually ‘mask’ the NES-like motif in hTERT, 14-3-3 binding might result in a conformational change of hTERT that disrupts the CRM1 binding. We observed that some hTERT mutants in which portions of amino acid residues encompassing the 14-3-3 binding site were deleted completely lost the telomerase activity, suggesting an artificial misfolding of the proteins (H.Seimiya and T.Tsuruo, unpublished observations). This suggests that the hTERT domain around the 14-3-3 binding site might be an important platform for switching the conformation of hTERT.
Since the telomerase complex is too large to pass through a nuclear pore complex by simple diffusion (Nigg, 1997), sophisticated nuclear import machinery would be involved in re-entry of the cytoplasmic telomerase to the nucleus. Among the 293T cells in which hTERT–14-3-3θ-ΔN colocalization was observed (Figure 3D), 32% (7 of 22) still exhibited nuclear accumulation of hTERT (data not shown). Similarly, ∼30% of the hTERT-3A-transfected fibroblasts still had an ability to accumulate the mutant protein in the nuclei (Figure 6B), suggesting a continued ability of this mutant to recruit the nuclear import machinery. In fact, according to the PSORT II analysis (http://psort.ims.u-tokyo.ac.jp/; Human Genome Center, University of Tokyo), hTERT has several NLS-like motifs (e.g. residues 233–239) although whether these motifs can work as functional NLSs is still to be determined. Also, another hTERT-binding protein that has an NLS may enhance nuclear accumulation of hTERT. Taken together, these observations suggest that the cytoplasmic or nuclear localization of hTERT is determined by the balance between two machineries: an unidentified NLS-mediated nuclear import and the NES-mediated nuclear export that is modulated by the 14-3-3 binding. Regulation of the hTERT–14-3-3 binding is of great interest to give further insights into the physiological significance of the cytoplasmic localization of telomerase.
While 14-3-3 also works as a molecular chaperone (Liao and Omary, 1996), its binding to hTERT was not required for telomerase activity, at least as determined by the conventional TRAP assay (Figure 7). However, we still cannot exclude the possibility that 14-3-3 binding might modulate telomerase functions especially toward the native telomeres in intact cells. According to Counter and colleagues, the telomerase activity detectable with the in vitro TRAP assay is not sufficient for maintenance of telomere length or cell immortalization (Counter et al., 1998). It would be of interest to examine the ability of hTERT-3A mutant to immortalize normal cells.
Since 14-3-3 family proteins can dimerize via their N-terminal domains, they sometimes work as adaptors for two different binding partners (Braselmann and McCormick, 1995). Since 14-3-3-binding proteins include various signaling factors, such as kinases and phosphatases (Aitken, 1996), and some phosphorylation/dephosphorylation events affect telomerase activity (Li et al., 1997, 1998), there might be an intracellular mechanism in which telomerase functions are regulated by another 14-3-3 binding protein via a 14-3-3 dimer as an adaptor. Also, since 14-3-3 is involved in various signaling events, including proliferation, apoptosis, and check point control, interaction of TERT and 14-3-3 suggests a functional coupling of telomerase and signal transduction machinery in such cellular events.
Materials and methods
Expression vectors for hTERT constructs
Full-length hTERT cDNA was amplified from thymus cDNA (Clontech) and was cloned into a pCR3 vector (Invitrogen). hTERT C-terminal fragment tagged with the HA epitope was amplified with 5′-GGCTGTGCCACCAAGCATTCC-3′ and 5′-CGCTCTAGACTAAG CGTAGTCTGGGACGTCGTATGGGTAGTCCAGGATGGTCTTGAA GTC-3′ by using pCR3–hTERT as a template. The fragment was trimmed with BsmI and XbaI and replaced with the BsmI–XbaI fragment in pCR3–hTERT. hTERT-3A-HA, hTERTnes-HA and hTERTnes-3A-HA were constructed, essentially as described previously (Imai et al., 1991). To diminish concern about artificial overexpression of exogenous hTERT, we did not modify the constructs for increased translational efficiency as described previously (Bodnar et al., 1998). The expression vectors for deletion mutants of hTERT, hTERT(del109–962), hTERT(del220–1108) and hTERT(del657–1132), were constructed by digesting pCR3–hTERT with SacII, SmaI and XhoI, respectively, and were self-ligated. The sequences were confirmed using an Applied Biosystems PRISM 310 automated DNA sequencer.
Yeast two-hybrid screening
Yeast two-hybrid screening was performed essentially as described previously (Gyuris et al., 1993).
Expression vectors for GST fusion proteins
Full-length human 14-3-3θ cDNA was amplified from thymus cDNA. Fusion vectors for GST–14-3-3θ, GST–hTERT(831–1132) or GST–hTERTnes(831–1132) were constructed by cloning each restriction fragment of 14-3-3θ, hTERT or hTERTnes into appropriate sites of pGEX-5X vectors (Pharmacia). The sequence was confirmed using the DNA sequencer. Recombinant 14-3-3θ was prepared by cleaving GST–14-3-3θ with Factor Xa, followed by removal of the cleaved GST and uncleaved GST–14-3-3θ with glutathione–Sepharose.
In vitro hTERT–14-3-3 binding assay
Each hTERT protein labeled with [35S]methionine was prepared by using the TNT T7 transcription-translation system (Promega), according to the instruction manual. Portions of each [35S]hTERT protein were incubated with the GST–14-3-3θ-bound glutathione–Sepharose in the 1× IP buffer consisting of 25 mM HEPES pH 7.4, 100 mM NaCl, 5 mM MgCl2, 100 mM EDTA, 0.2 mg/ml bovine serum albumin, 0.1% Tween-20 and 1 mM phenylmethylsulfonyl fluoride (PMSF). In other experiments, recombinant 14-3-3θ was incubated with the GST–hTERT(831–1132) beads in TNE lysis buffer, consisting of 10 mM Tris–HCl pH 7.8, 1% Nonidet P-40, 150 mM NaCl, 1 mM EDTA, 10 µg/ml aprotinin and 1 mM PMSF. The beads were extensively washed with the buffer and subjected to SDS–PAGE. The specific signals were detected with a BAS 2500 phosphoimager (Fuji film) or by western blot analysis (see below).
Immunoprecipitation and western blot analysis
293T cells were transfected with appropriate vectors using a standard calcium phosphate method. For the hTERT–14-3-3 binding, total lysates from the 293T or A2780 cells were prepared with TNE lysis buffer. For the hTERT–CRM1 binding, cell lysates were prepared as described previously (Rittinger et al., 1999). Portions of the lysates were preincubated with protein G–Sepharose and the resulting supernatants were incubated with appropriate antibodies and protein G–Sepharose. The beads were extensively washed with each lysis buffer and subjected to SDS–PAGE. The specific signals were detected by western blot analysis, as described previously (Seimiya and Tsuruo, 1998), with anti-HA, anti-hTERT, anti-14-3-3θ (Santa Cruz Biotechnology), anti-FLAG (Kodak), anti-14-3-3 (H-8; this antibody cross-reacts with all isoforms of 14-3-3 proteins at their N-termini; Santa Cruz Biotechnology) and anti-CRM1 (kindly provided by Dr Minoru Yoshida, University of Tokyo) as primary antibodies.
Immunofluorescence microscopy
Cells were transfected with appropriate expression vectors on a chamber slide that was pre-coated with 0.2% gelatin. After culturing for 24–48 h, cells were fixed with cold methanol, and soaked in phosphate-buffered saline (PBS) containing 10% fetal bovine serum (FBS). In some cases, cells were treated with 20 nM LMB (kindly provided by Dr Minoru Yoshida) for 2 h before the methanol fixation. The slide was washed with PBS and incubated with rabbit anti-HA (and mouse anti-FLAG). Then, the slide was washed with PBS and incubated with rhodamine- or Texas Red-conjugated anti-rabbit Ig (and fluorescein-conjugated anti-mouse Ig) (Cappel, Amersham). The nuclei were counterstained with 4′,6- diamidine-2-phenylindole (DAPI). The slides were analyzed by fluorescence microscopy in the presence of Anti-Fade reagent (Molecular Probes).
In vitro hTERT–CRM1 binding assay
The 293T cell lysate was prepared as above. Portions of the lysate were incubated with the GST, GST–hTERT(831–1132) or GST–hTERTnes(831–1132)-bound glutathione–Sepharose for 3 h. The beads were extensively washed with 0.1% Nonidet P-40/PBS and subjected to SDS–PAGE. The specific signals were detected by anti-CRM1 western blot analysis.
In vitro reconstitution of telomerase and TRAP assay
The RNA expression vector for hTR was constructed and IVR telomerase was prepared as described previously (Weinrich et al., 1997). TRAP assay was performed essentially as described previously (Kim et al., 1994), with a set of primers (TS, 5′-AATCCGTCGAGCAGAGTT-3′; ACX, 5′-GCGCGGCTTACCCTTACCCTTACCCTAACC-3′; NT, 5′-ATCGCTTCTCGGCCTTTT-3′) and an internal standard, TSNT (5′-AATCCGTCGAGCAGAGTTAAAAGGCCGAGAAGCGAT-3′) (Kim and Wu, 1997). The telomeric products were separated by PAGE and visualized by staining with SYBR Green™ (Takara, Kyoto, Japan). For in vivo experiments, normal foreskin fibroblasts were transfected with each hTERT-HA construct as described above. After incubation for 24 h, the TRAP lysates were prepared and the TRAP assay was performed (Kim et al., 1994).
Acknowledgments
Acknowledgements
We are grateful to Ms Naomi Wada for skillful assistance with the experiments and Dr Roger Brent for providing us the plasmids and the yeast EGY48 strain for two hybrid analysis. We thank Dr Minoru Yoshida for providing us Leptomycin B and anti-human CRM1 antibody. This work was supported by a Grant-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Science, Sports and Culture, Japan, and Foundation for Promotion of Cancer Research in Japan.
References
- Aitken A. (1996) 14-3-3 and its possible role in co-ordinating multiple signaling pathways. Trends Cell Biol., 6, 341–347. [DOI] [PubMed] [Google Scholar]
- Allsopp R.C., Vaziri,H., Patterson,C., Goldstein,S., Younglai,E.V., Futcher,A.B., Greider,C.W. and Harley,C.B. (1992) Telomere length predicts replicative capacity of human fibroblasts. Proc. Natl Acad. Sci. USA, 89, 10114–10118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasco M.A., Lee,H.-W., Hande,M.P., Samper,E., Lansdorp,P.M., DePinho,R.A. and Greider,C.W. (1997) Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell, 91, 25–34. [DOI] [PubMed] [Google Scholar]
- Bodnar A.G. et al. (1998) Extension of life-span by introduction of telomerase into normal human cells. Science, 279, 349–352. [DOI] [PubMed] [Google Scholar]
- Braselmann S. and McCormick,F. (1995) BCR and RAF form a complex in vivo via 14-3-3 proteins. EMBO J., 14, 4839–4848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Counter C.M., Avilion,A.A., LeFeuvre,C.E., Stewart,N.G., Greider,C.W., Harley,C.B. and Bacchetti,S. (1992) Telomere shortening associated with chromosome instability is arrested in immortal cells which express telomerase activity. EMBO J., 11, 1921–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Counter C.M., Hahn,W.C., Wei,W., Caddle,S.D., Beijersbergen,R.L., Lansdorp,P.M., Sedivy,J.N. and Weinberg,R.A. (1998) Dissociation among in vitro telomerase activity, telomere maintenance and cellular immortalization. Proc. Natl Acad. Sci. USA, 95, 14723–14728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craparo A., Freund,R. and Gustafson,T.A. (1997) 14-3-3 (ε) interacts with the insulin-like growth factor I receptor and insulin receptor substrate I in a phosphoserine-dependent manner. J. Biol. Chem., 272, 11663–11669. [DOI] [PubMed] [Google Scholar]
- Du X., Fox,J.E. and Pei,S. (1996) Identification of a binding sequence for the 14-3-3 protein within the cytoplasmic domain of the adhesion receptor, platelet glycoprotein Ibα. J. Biol. Chem., 271, 7362–7367. [DOI] [PubMed] [Google Scholar]
- Fischer U., Huber,J., Boelens,W.C., Mattaj,I.W. and Luhrmann,R. (1995) The HIV Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell, 82, 475–483. [DOI] [PubMed] [Google Scholar]
- Fornerod M., Ohno,M., Yoshida,M. and Mattaj,I.W. (1997) CRM1 is an export receptor for leucine-rich nuclear export signals. Cell, 90, 1051–1060. [DOI] [PubMed] [Google Scholar]
- Fukuda M., Asano,S., Nakamura,T., Adachi,M., Yoshida,M., Yanagida,M. and Nishida,E. (1997) CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature, 390, 308–311. [DOI] [PubMed] [Google Scholar]
- Greenberg R.A., Allsopp,R.C., Chin,L., Morin,G.B. and DePinho,R.A. (1998) Expression of mouse telomerase reverse transcriptase during development, differentiation and proliferation. Oncogene, 16, 1723–1730. [DOI] [PubMed] [Google Scholar]
- Greider C.W. (1996) Telomere length regulation. Annu. Rev. Biochem., 65, 337–365. [DOI] [PubMed] [Google Scholar]
- Greider C.W. and Blackburn,E.H. (1985) Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell, 43, 405–413. [DOI] [PubMed] [Google Scholar]
- Gu M. and Du,X. (1998) A novel ligand-binding site in the ζ-form 14-3-3 protein recognizing the platelet glycoprotein Ibα and distinct from the c-Raf-binding site. J. Biol. Chem., 273, 33465–33471. [DOI] [PubMed] [Google Scholar]
- Gyuris J., Golemis,E., Chertkov,H. and Brent,R. (1993) Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell, 75, 791–803. [DOI] [PubMed] [Google Scholar]
- Harley C.B., Futcher,A.B. and Greider,C.W. (1990) Telomeres shorten during ageing of human fibroblasts. Nature, 345, 458–460. [DOI] [PubMed] [Google Scholar]
- Hastie N.D., Dempster,M., Dunlop,M.G., Thompson,A.M., Green,D.K. and Allshire,R.C. (1990) Telomere reduction in human colorectal carcinoma and with ageing. Nature, 346, 866–868. [DOI] [PubMed] [Google Scholar]
- Imai Y., Matsushima,Y., Sugimura,T. and Terada,M. (1991) A simple and rapid method for generating a deletion by PCR. Nucleic Acids Res., 19, 2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N.W. and Wu,F. (1997) Advances in quantitation and characterization of telomerase activity by the telomeric repeat amplification protocol (TRAP). Nucleic Acids Res., 25, 2595–2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N.W. et al. (1994) Specific association of human telomerase activity with immortal cells and cancer. Science, 266, 2011–2015. [DOI] [PubMed] [Google Scholar]
- Kudo N., Wolff,B., Sekimoto,T., Schreiner,E.P., Yoneda,Y., Yanagida,M., Horinouchi,S. and Yoshida,M. (1998) Leptomycin B inhibition of signal-mediated nuclear export by direct binding to CRM1. Exp. Cell Res., 242, 540–547. [DOI] [PubMed] [Google Scholar]
- Kudo N., Matsumori,N., Taoka,H., Fujiwara,D., Schreiner,E.P., Wolff,B., Yoshida,M. and Horinouchi,S. (1999) Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. Proc. Natl Acad. Sci. USA, 96, 9112–9117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai A. and Dunphy,W.G. (1999) Binding of 14-3-3 proteins and nuclear export control the intracellular localization of the mitotic inducer Cdc25. Genes Dev., 13, 1067–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.-W., Blasco,M.A., Gottlieb,G.J., Horner,J.W.,II, Greider,C.W. and DePinho,R.A. (1998) Essential role of mouse telomerase in highly proliferative organs. Nature, 392, 569–574. [DOI] [PubMed] [Google Scholar]
- Li H., Zhao,L.-L., Funder,J.W. and Liu,J.-P. (1997) Protein phosphatase 2A inhibits nuclear telomerase activity in human breast cancer cells. J. Biol. Chem., 272, 16729–16732. [DOI] [PubMed] [Google Scholar]
- Li H., Zhao,L., Yang,Z., Funder,J.W. and Liu,J.P. (1998) Telomerase is controlled by protein kinase Cα in human breast cancer cells. J. Biol. Chem., 273, 33436–33442. [DOI] [PubMed] [Google Scholar]
- Liao J. and Omary,M.B. (1996) 14-3-3 proteins associate with phosphorylated simple epithelial keratins during cell cycle progression and act as a solubility cofactor. J. Cell Biol., 133, 345–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Bienkowska,J., Petosa,C., Collier,R.J., Fu,H. and Liddington,R. (1995) Crystal structure of the ζ isoform of the 14-3-3 protein. Nature, 376, 191–194. [DOI] [PubMed] [Google Scholar]
- Liu K., Schoonmaker,M.M., Levine,B.L., June,C.H., Hodes,R.J. and Weng,N.-P. (1999) Constitutive and regulated expression of telomerase reverse transcriptase (hTERT) in human lymphocytes. Proc. Natl Acad. Sci. USA, 96, 5147–5152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.C., Liu,Y., Elly,C., Yoshida,H., Lipkowitz,S. and Altman,A. (1997) Serine phosphorylation of Cbl induced by phorbol ester enhances its association with 14-3-3 proteins in T cells via a novel serine-rich 14-3-3-binding motif. J. Biol. Chem., 272, 9979–9985. [DOI] [PubMed] [Google Scholar]
- Luo Z.J., Zhang,X.F., Rapp,U. and Avruch,J. (1995) Identification of the 14.3.3 ζ domains important for self-association and Raf binding. J. Biol. Chem., 270, 23681–23687. [DOI] [PubMed] [Google Scholar]
- Meyerson M. et al. (1997) hEST2, the putative human telomerase catalytic subunit gene, is up-regulated in tumor cells and during immortalization. Cell, 90, 785–795. [DOI] [PubMed] [Google Scholar]
- Moorhead G., Douglas,P., Morrice,N., Scarabel,M., Aitken,A. and MacKintosh,C. (1996) Phosphorylated nitrate reductase from spinach leaves is inhibited by 14-3-3 proteins and activated by fusicoccin. Curr. Biol., 6, 1104–1113. [DOI] [PubMed] [Google Scholar]
- Morin G.B. (1989) The human telomere terminal transferase enzyme is a ribonucleoprotein that synthesizes TTAGGG repeats. Cell, 59, 521–529. [DOI] [PubMed] [Google Scholar]
- Moyzis R.K., Buckingham,J.M., Cram,L.S., Dani,M., Deaven,L.L., Jones,M.D., Meyne,J., Ratliff,R.L. and Wu,J.R. (1988) A highly conserved repetitive DNA sequence, (TTAGGG)n, present at the telomeres of human chromosomes. Proc. Natl Acad. Sci. USA, 85, 6622–6626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muslin A.J., Tanner,J.W., Allen,P.M. and Shaw,A.S. (1996) Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine. Cell, 84, 889–897. [DOI] [PubMed] [Google Scholar]
- Nakamura T.M., Morin,G.B., Chapman,K.B., Weinrich,S.L., Andrews,W.H., Lingner,J., Harley,C.B. and Cech,T.R. (1997) Telomerase catalytic subunit homologs from fission yeast and human. Science, 277, 955–959. [DOI] [PubMed] [Google Scholar]
- Nakayama J. et al. (1998) Telomerase activation by hTRT in human normal fibroblasts and hepatocellular carcinomas. Nature Genet., 18, 65–68. [DOI] [PubMed] [Google Scholar]
- Nigg E.A. (1997) Nucleocytoplasmic transport: signals, mechanisms and regulation. Nature, 386, 779–787. [DOI] [PubMed] [Google Scholar]
- Nishi K., Yoshida,M., Fujiwara,D., Nishikawa,M., Horinouchi,S. and Beppu,T. (1994) Leptomycin B targets a regulatory cascade of crm1, a fission yeast nuclear protein, involved in control of higher order chromosome structure and gene expression. J. Biol. Chem., 269, 6320–6324. [PubMed] [Google Scholar]
- Ossareh-Nazari B., Bachelerie,F. and Dargemont,C. (1997) Evidence for a role of CRM1 in signal-mediated nuclear protein export. Science, 278, 141–144. [DOI] [PubMed] [Google Scholar]
- Peng C.-Y., Graves,P.R., Thoma,R.S., Wu,Z., Shaw,A.S. and Piwnica-Worms,H. (1997) Mitotic and G2 checkpoint control: regulation of 14-3-3 protein binding by phosphorylation of Cdc25C on serine-216. Science, 277, 1501–1505. [DOI] [PubMed] [Google Scholar]
- Rittinger K., Budman,J., Xu,J., Volinia,S., Cantley,L.C., Smerdon,S.J., Gamblin,S.J. and Yaffe,M.B. (1999) Structural analysis of 14-3-3 phosphopeptide complexes identifies a dual role for the nuclear export signal of 14-3-3 in ligand binding. Mol. Cell, 4, 153–166. [DOI] [PubMed] [Google Scholar]
- Sanchez Y., Wong,C., Thoma,R.S., Richman,R., Wu,Z., Piwnica-Worms,H. and Elledge,S.J. (1997) Conservation of the Chk1 checkpoint pathway in mammals: linkage of DNA damage to Cdk regulation through Cdc25. Science, 277, 1497–1501. [DOI] [PubMed] [Google Scholar]
- Seimiya H. and Tsuruo,T. (1998) Functional involvement of PTP-U2L in apoptosis subsequent to terminal differentiation of monoblastoid leukemia cells. J. Biol. Chem., 273, 21187–21193. [DOI] [PubMed] [Google Scholar]
- Shay J.W. and Bacchetti,S. (1997) A survey of telomerase activity in human cancer. Eur. J. Cancer, 33, 787–791. [DOI] [PubMed] [Google Scholar]
- Stommel J.M., Marchenko,N.D., Jimenez,G.S., Moll,U.M., Hope,T.J. and Wahl,G.M. (1999) A leucine-rich nuclear export signal in the p53 tetramerization domain: regulation of subcellular localization and p53 activity by NES masking. EMBO J., 18, 1660–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzivion G., Luo,Z. and Avruch,J. (1998) A dimeric 14-3-3 protein is an essential cofactor for Raf kinase activity. Nature, 394, 88–92. [DOI] [PubMed] [Google Scholar]
- van Steensel B. and de Lange,T. (1997) Control of telomere length by the human telomeric protein TRF1. Nature, 385, 740–743. [DOI] [PubMed] [Google Scholar]
- Wang H., Zhang,L., Liddington,R. and Fu,H. (1998) Mutation in the hydrophobic surface of an amphipathic groove of 14-3-3ζ disrupt its interaction with Raf-1 kinase. J. Biol. Chem., 273, 16297–16304. [DOI] [PubMed] [Google Scholar]
- Wang J., Xie,L.Y., Allan,S., Beach,D. and Hannon,G.J. (1998) Myc activates telomerase. Genes Dev., 12, 1769–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinrich S.L. et al. (1997) Reconstitution of human telomerase with the template RNA component hTR and the catalytic protein subunit hTRT. Nature Genet., 17, 498–502. [DOI] [PubMed] [Google Scholar]
- Wen W., Meinkoth,J.L., Tsien,R.Y. and Taylor,S.S. (1995) Identification of a signal for rapid export of proteins from the nucleus. Cell, 82, 463–473. [DOI] [PubMed] [Google Scholar]
- Wu K.-J., Grandori,C., Amacker,M., Simon-Vermot,N., Polack,A., Lingner,J. and Dalla-Favera,R. (1999) Direct activation of TERT transcription by c-MYC. Nature Genet., 21, 220–224. [DOI] [PubMed] [Google Scholar]
- Xiao B., Smerdon,S.J., Jones,D.H., Dodson,G.G., Soneji,Y., Aitken,A. and Gamblin,S.J. (1995) Structure of a 14-3-3 protein and implications for coordination of multiple signaling pathways. Nature, 376, 188–191. [DOI] [PubMed] [Google Scholar]
- Yaffe M.B., Rittinger,K., Volinia,S., Caron,P.R., Aitken,A., Leffers,H., Gamblin,S.J., Smerdon,S.J. and Cantley,L. (1997) The structural basis for 14-3-3: phosphopeptide binding specificity. Cell, 91, 961–971. [DOI] [PubMed] [Google Scholar]
- Yang J., Winkler,K., Yoshida,M. and Kornbluth,S. (1999) Maintenance of G2 arrest in the Xenopus oocyte: a role for 14-3-3-mediated inhibition of Cdc25 nuclear import. EMBO J., 18, 2174–2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakian V.A. (1995) Telomeres: beginning to understand the end. Science, 270, 1601–1607. [DOI] [PubMed] [Google Scholar]
- Zha J., Harada,H., Yang,E., Jockel,J. and Korsmeyer,S.J. (1996) Serine phosphorylation of death agonist BAD in response to survival factor results in binding to 14-3-3 not BCL-X. Cell, 87, 619–628. [DOI] [PubMed] [Google Scholar]