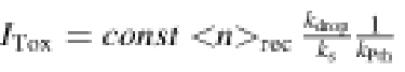Abstract
The expression of very short open reading frames in Escherichia coli can lead to the inhibition of translation and an arrest in cell growth. Inhibition occurs because peptidyl-tRNA hydrolase fails to recycle sufficiently rapidly peptidyl-tRNA released from ribosomes at the stop signal in competition with normal termination, causing starvation for essential species of tRNA. Previous studies have shown that the last sense codon, the strength of the Shine–Dalgarno sequence and the nature and context of the stop codon affect the toxicity associated with mini-gene expression. Here, several important parameters are studied as a function of the length of the mini-gene coding sequence. The rate of peptidyl-tRNA drop-off catalysed by translation factors decreases dramatically for peptides longer than a hexamer. The probability that ribosomes recycle without dissociation of the mini-gene mRNA varies strongly with the length of the coding sequence. The peptidyl-tRNA hydrolase rap mutant, unlike the wild-type enzyme, is highly sensitive to the length and sequence of the peptide. Together, these parameters explain the length dependence of mini-gene toxicity.
Keywords: mini-gene/peptidyl-tRNA/release factor/ribosome recycling/translation termination
Introduction
During normal protein synthesis in bacterial cells, ribosomes attach to mRNA at the ribosome binding site, translate the open reading frame (ORF) until a termination codon is reached, and with the aid of codon-specific termination (or release) factors (RF), release the completed polypeptide chain into the cytoplasm by hydrolysis on the ribosome of the ester linkage between the polypeptide and tRNA (Hershey, 1987; Buckingham et al., 1997). Another reaction can, however, compete with normal termination, whereby peptidyl-tRNA dissociates from the ribosome before hydrolysis of the ester bond (Menninger, 1976; Hernández et al., 1997; Heurgué-Hamard et al., 1998). The peptidyl-tRNA released is then hydrolysed by the enzyme peptidyl-tRNA hydrolase (Pth), allowing the tRNA to be charged by the cognate aminoacyl-tRNA synthetase and reutilized in protein synthesis. Premature dissociation of peptidyl-tRNA from the ribosome (‘drop-off’) is a normal accompaniment of protein synthesis, and Pth is essential to cell growth (Atherly and Menninger, 1972; Schmitt et al., 1997).
Recent work has shown that two sets of proteins catalyse drop-off. One set is composed of three factors involved in the process of ribosome recycling (Karimi et al., 1999): termination factor RF3, ribosome recycling factor RRF and elongation factor EF-G, the last of which primarily catalyses the translocation of peptidyl-tRNA from A- to P-site (Haenni and Lucas-Lenard, 1968). The initiation factors IF1 and IF2 comprise a second set of proteins stimulating drop-off (Karimi et al., 1998). Mutations reducing the level of expression of RRF, and others inactivating RF3, reduce drop-off in vivo (Heurgué-Hamard et al., 1998). In vitro, the three proteins RF3, RRF and EF-G can stimulate the drop-off of dipeptidyl-tRNA at least 30-fold (Dinçbas et al., 1999). Although the experiments in vivo offer convincing evidence for the physiological significance of RRF and RF3 in the drop-off reaction, it remains unknown, however, whether their role in stimulating drop-off is restricted to short peptides or whether a wide range of chain lengths is concerned.
When the frequency of drop-off exceeds the capacity of Pth to recycle the tRNAs sequestered as peptidyl-tRNA, starvation for essential tRNA isoacceptors occurs, leading to an inhibition of protein synthesis and eventually to cell death. This is believed to account for the shutdown in protein synthesis that takes place when a temperature-sensitive pth mutant is shifted to 43°C (Atherly and Menninger, 1972). A related phenomenon appears when certain mutants of Escherichia coli, rap, are infected with bacteriophage λ. The rap locus is identical to pth, and rap mutants exhibit reduced Pth activity for model substrates (Garcia Villegas et al., 1991; Schmitt et al., 1997). These mutants are unable to maintain normal bacteriophage vegetative growth, due to the expression of very short, phage-encoded ORFs, ‘mini-genes’, encoded by the bar loci of phage λ. When the bar regions are carried on plasmids, the induction of their expression can kill rap mutant cells. The transcription of the barI and barII regions yields RNA molecules containing Shine–Dalgarno (SD)-type sequences, followed by the translation initiation codon AUG, a single additional sense codon and a translational stop signal. Further experiments indicate that the transcripts are translated, producing fMet-Ile-tRNAIle, but that dipeptide release brought about by termination factors RF1 and RF2 is partially defective, leading to dipeptidyl-tRNAIle drop-off and starvation for tRNAIle (Hernández-Sánchez et al., 1998).
Recent studies have shown that multiple parameters related to the sequence of the mini-gene transcript contribute to the growth inhibitory effect (Dinçbas et al., 1999). Thus, when the relatively weak ribosome binding sites of the natural bar SD regions are replaced by an SD sequence that interacts strongly with 16S rRNA, mini-gene expression can be lethal even in cells with wild-type Pth, confirming observations by Tenson et al. (1999). In part, this is due to the increased probability that ribosomes retranslate the same mini-mRNA without dissociating from it after peptide release. Three further parameters expected to affect the rate of peptidyl-tRNA accumulation, namely, termination efficiency, rate of drop-off and rate of hydrolysis of the resulting peptidyl-tRNA by Pth, were shown to vary according to the last sense codon in the mini-mRNA, the stop codon present and the nucleotide following the stop codon (Dinçbas et al., 1999).
Here, we study the effect of the length of mini-genes on the parameters affecting peptidyl-tRNA accumulation. The rate of peptidyl-tRNA drop-off increases dramatically for peptidyl-tRNA species <7 amino acids long, due to a greater stimulation of drop-off by RF3, RRF and EF-G. In addition, the rate of translation termination catalysed by RF2 increases when the number of mini-gene codons varies from 2 to 8. The variation in activity of wild-type Pth according to the length of the peptide is weak, but this becomes an important factor in the case of the Pth rap mutant. Finally, the probability that ribosomes recycle without leaving the mini-mRNA varies strongly with the length of the coding sequence. Taken together, these parameters provide an explanation for the fact that toxic mini-genes preferentially encode very short peptides.
Results
The development of an in vitro translation with purified components that is able to translate short synthetic mRNAs has made it possible to isolate ribosomal complexes paused with a stop codon in the A-site and containing a peptidyl-tRNA of known composition in the P-site (Freistroffer et al., 1997). Such termination complexes are obtained by initiating translation on mini-gene transcripts using initiation factors and fMet-tRNAMet, and translating the short ORF in the presence of all other necessary components but in the absence of termination factors. The paused complexes may then be purified from other components by gel filtration and used to study the kinetics of termination or drop-off of peptidyl-tRNA (Heurgué-Hamard et al., 1998; Dinçbas et al., 1999).
Pth activity for different substrates
Previous work (Dinçbas et al., 1999), in which the activity of Pth (rap) was measured on peptidyl-tRNA substrates differing by the nature of the tRNA species and the last amino acid, showed that fMet-Lys-tRNALys was the least and fMet-Phe-tRNAPhe the most efficiently cleaved among the substrates studied. The rates of hydrolysis differed by a factor close to four. Two constructs showed furthermore that the cleavage efficiency could be greatly improved merely by increasing the length of the peptidyl-tRNA by two amino acids. We have now studied more systematically the activity of both the rap mutant of Pth and the wild-type enzyme on substrates varying in peptide length from 2 to 8 amino acids with Lys as the last amino acid, the smallest being fMet-Lys-tRNALys. This peptidyl-tRNA arises from the translation of a mini-gene shown to be the most toxic among a substantial set when expressed in the pth (rap) mutant. The Pth substrates were obtained from release complexes consisting of 70S ribosomes bound to mRNA, with the stop codon UAA located at the A-site and peptidyl-tRNA at the P-site. The complexes were incubated with RRF, RF3 and EF-G, known to promote efficient dissociation of peptidyl-tRNA (Heurgué-Hamard et al., 1998; Dinçbas et al., 1999). The kcat/Km values for the Pth (rap) mutant and wild-type Pth are shown in Table I (columns 3 and 4, respectively) and a typical experiment in Figure 1A. The kcat/Km values for wild-type Pth and Pth (rap) cleavage of fMK-tRNALys, to which other values are normalized, were estimated at 1.45 × 106 M–1s–1 and 1.95 × 104 M–1s–1, respectively.
Table I. Parameters related to peptidyl-tRNA accumulation and toxicity index ITox for different peptidyl-tRNAs.
| Plasmid | Peptidyl-tRNA | Pth activity (× 10–5) kcat/Km (M–1s–1) |
Dissociation rate (× 102) s–1 |
Termination by RF1kcat/Km (µM–1s–1) | Termination by RF2kcat/Km (µM–1s–1) | Recycle numbera<n>rec | IToxb Pth (rap) | IToxb Pth wild type | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Pth (rap) | Pth wild type | No factors | RRF, EF-G, RF3 | |||||||
| SD7 | fMK-tRNALys | 0.195 (±0.012) | 14.5 (±0.5) | 0.13 (±0.01) | 1.71 (±0.17) | 70 (±2) | 2.00 (±0.42) | >100 | >250 | >3.5 |
| SD8 | fMFK-tRNALys | 0.590 (±0.031) | 16.1 (±1.1) | 0.11 (±0.01) | 1.17 (±0.05) | 70 (±10) | 2.18 (±47) | 46 (±8) | 27 (±9) | 1.0 |
| SD9 | fMFIK-tRNALys | 0.489 (±0.037) | 15.8 (±1.9) | 0.23 (±0.02) | 2.07 (±0.11) | 97 (±5) | 4.65 (±1.0) | 46 (±15) | 39 (±16) | 1.21 (±0.56) |
| SD10 | fMFITK-tRNALys | 0.594 (±0.027) | 21.4 (±2) | 0.23 (±0.01) | 1.90 (±0.01) | 89 (±6) | 6.8 (±1.6) | 33 (±14) | 21 (±10) | 0.58 (±0.32) |
| SD11 | fMFITQK-tRNALys | 0.100 (±0.011) | 10.2 (±1.6) | 0.13 (±0.01) | 1.46 (±0.19) | 71 (±10) | 7.2 (±1.5) | 14 (±1) | 47 (±15) | 0.45 (±0.18) |
| SD12 | fMFITQLK-tRNALys | 0.755 (±0.025) | 20.0 (±2.0) | 0.08 (±0.02) | 0.27 (±0.03) | 77 (±8) | 10.4 (±2.3) | 11 (±2) | 0.74 (±0.24) | 0.028 (±0.011) |
| SD13 | fMFITQLTK-tRNALys | 1.525 (±0.06) | 23.6 (±1.5) | 0.16 (±0.02) | 0.33 (±0.06) | 86 (±12) | 8.9 (±1.9) | 6.6 (±0.6) | 0.26 (±0.09) | 0.017 (±0.007) |
Values are given with the standard deviation from the mean, calculated from at least three determinations.
aRecycle number is the average number of times a ribosome translates a mRNA without the 30S subunit leaving the mRNA.
bThe definition of relative toxicity index is given in the text; it is calculated using an average of the kcat/Km values for RF1 and RF2, weighted according to their relative concentrations (1:5) in the cell (Adamski et al., 1994). The dissociation rate used is that in the presence of factors RRF, RF3 and EF-G.
Values are normalized to that for SD8 with wild-type Pth.
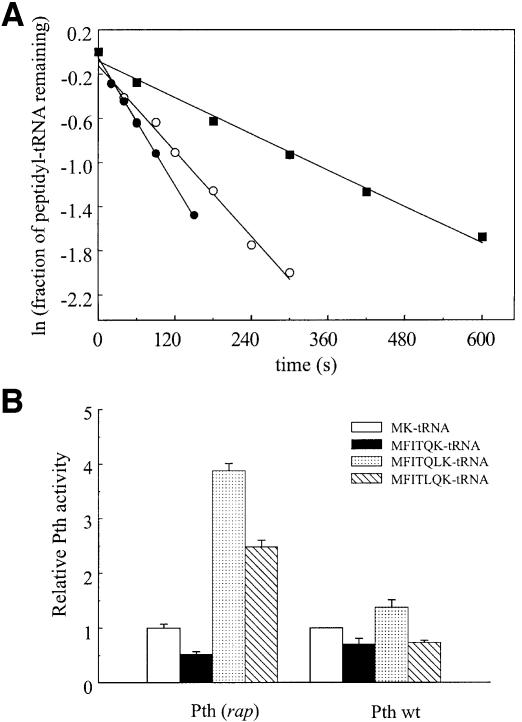
Fig. 1. Pth (rap) activity on different peptidyl-tRNAs. (A) Hydrolysis of fMet-Lys-tRNALys (filled squares), fMet-Phe-Ile-Lys-tRNALys (open circles), fMet-Phe-Ile-Thr-Gln-Leu-Lys-tRNALys (filled circles) in the presence of 0.13 µM Pth (rap). (B) Hydrolysis of different peptidyl-tRNALys in the presence of 0.13 µM Pth (rap) or 4 nM Pth wild type (in competition experiments). Relative activities are given using an activity of 1 for fMet-Lys-tRNALys hydrolysis.
Pth (rap) cleaved the longest peptidyl-tRNA fMFITQLTK-tRNALys with an efficiency almost eight times higher than the dipeptidyl-tRNA fMK-tRNALys. Within this set the cleavage efficiency increased by a factor of three with the transition from two to three amino acids, remained almost constant until the length reached five amino acids, and then increased again. However, anomalous behaviour was observed for fMFITQK-tRNALys (Table I, line 5), which is cleaved even more poorly than fMK-tRNALys. An attempt was made to see whether the C-terminal dipeptide sequence might be responsible by synthesizing a new mRNA (02.MFITLQK.Oa; see Materials and methods for mRNA nomenclature) differing from 02.MFITQLK.Oa by inversion of the two codons next to the last. A significantly reduced Pth (rap) activity was indeed observed towards the new substrate (relative activity of 2.5 versus 3.9, Figure 1B), but not to the extent of fully explaining the anomalous behaviour of fMFITQK-tRNALys. The very inefficient cleavage of this substrate may be due to a combination of peptide sequence (Gln-Lys as the last amino acids) and peptide length.
The activity of wild-type Pth is much less affected by peptide length, but is qualitatively similar (Table I, column 4). The large differences reported previously by Shiloach et al. (1975) in the case of wild-type Pth activity towards (Gly)n-Val-tRNA of different length was not apparent in our experiments. As in the case of the rap mutant of Pth, we observed that fMFITQK-tRNALys was inefficiently cleaved. In this case inverting the two amino acids next to the last of fMFITQLK-tRNALys also decreased the relative activity, by ∼2-fold (relative activity of 0.73 versus 1.37, Figure 1B).
Drop-off of peptidyl-tRNA with varying peptide length
Drop-off rates of peptidyl-tRNA were measured in vitro from a series of release complexes with peptidyl-tRNA in P-site and a UAA stop codon in A-site (see Materials and methods) as previously described (Heurgué-Hamard et al., 1998), in the absence of normal termination. The essential difference between the peptidyl-tRNA species studied was the length of the peptide. The same last sense codon, stop codon and downstream sequence were used throughout the series to avoid effects when a sense codon replaced the stop codon, a different stop codon was used, or the A-site codon was changed (Heurgué-Hamard et al., 1998; Dinçbas et al., 1999). Drop-off for each peptidyl-tRNA was measured in either the presence or the absence of factors RRF, RF3 and EF-G known to stimulate drop-off (Heurgué-Hamard et al., 1998). The results are shown in Figure 2A and Table I (columns 5 and 6). In the absence of translation factors, the drop-off rate does not depend regularly on peptide length. Thus, the dissociation rate of fMFITQLTK-tRNALys is almost the same as that for fMK-tRNALys. In contrast, the dissociation rate of peptidyl-tRNA in the presence of RRF, RF3 and EF-G depends strongly on peptide length, decreasing dramatically when it reaches 7 amino acids (Figure 2B). The low value for tripeptidyl-tRNA in relation to dipeptidyl- and tetrapeptidyl-tRNA indicates a dependence on sequence of both factor-catalysed and non-catalysed dissociation (Table I, lines 1–3).
Fig. 2. (A) Dissociation of peptidyl-tRNA from ribosomal complexes in the presence of 1 µM RRF, RF3 and EF-G: fMet-Phe-Ile-Thr-Gln-Leu-Lys-tRNALys (filled circles), fMet-Phe-Ile-Thr-Gln-Lys-tRNALys (filled triangles), fMet-Phe-Ile-Lys-tRNALys (open circles), fMet-Lys-tRNALys (filled squares). (B) Dissociation of peptidyl-tRNA stimulated by RRF, RF3 and EF-G at 1 µM concentration as a function of peptide length of peptidyl-tRNALys.
Termination of translation of mini-genes
Termination rates catalysed by RF1 and RF2 were measured from the same release complexes used to measure drop-off rates of peptidyl-tRNA. When two release complexes compete for limited amounts of RF, it is possible to estimate relative kcat/Km values for the termination reaction (Pavlov et al., 1998). Absolute values for the dipeptidyl-tRNA substrate were determined as described previously (Dinçbas et al., 1999), and used to calculate kcat/Km values for the remaining substrates (Table I, columns 7 and 8). Significant variations were observed for RF2, but not for RF1, termination becoming more efficient as peptide length increases. As in the case of the previous parameters, the peptide sequence may play an active role, in line with reports that the amino acids present at the C-termini of proteins affect peptide release (Björnsson et al., 1996).
Recycling of ribosomes on mini-mRNAs
Previous work showed that minigene toxicity could not be fully understood by consideration of only the parameters described above: Pth activity, drop-off rate and termination efficiency (Dinçbas et al., 1999). Efficiency of translation was clearly a further critical point, especially when the strength of the SD sequence was different. Although minigene toxicity was first described in the case of bar mRNA expression in the pth (rap) mutant, it is not restricted to Pth-deficient strains (Dinçbas et al., 1999; Tenson et al., 1999). Toxicity depends to a considerable extent on the nature of the SD sequence, which in the λ bar sequences is poorly complementary to 16S RNA. Replacement of the bar SD sequence by a more efficient one, like SD 002 (Calogero et al., 1988) makes the barI minigene toxic in wild-type Pth cells, pointing to the efficiency of translation as an important parameter.
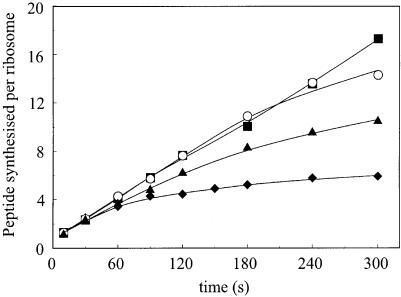
In the in vitro system it is possible to measure the ribosomal recycling time (Pavlov et al., 1997a,b), which represents the average time for a ribosome between two initiations on mRNA, and includes the time required for initiation, elongation, termination and recycling. Initiation may take place either on the same mRNA molecule, or on another molecule following dissociation of the 30S subunit from the mRNA. However, if a second messenger with a different sequence is added in excess after initiation on the first mRNA, the probabilities of these alternatives may be measured. Previously, we showed that the probability of reinitiating translation on the same mRNA molecule was almost 100% for a messenger with SD 002, and 40% for a weaker SD from the λ bar1 gene, a large difference that could explain the longer recycling time for messengers with weak SD sequence. We have now measured this probability for the series of different mRNAs, all possessing the same strong SD 002 but containing from two to eight sense codons. These results are shown in Table I, column 8 and in Figure 3. They show that the mRNA encoding fMK will on average be translated >100 times before leaving the ribosome, and reveal a strong inverse relationship between this recyling number and length of the mini-gene coding sequence.
Fig. 3. Recycling of ribosomes on mRNA 02.MK.Oa (filled squares), 02.MFIK.Oa (open circles), 02.MFITQK.Oa (filled triangles) and 02.MFITQLTK.Oa (filled diamonds) in the presence of chase mRNA. The curves were analysed (see Materials and methods) to determine the average number of times <n>rec that an mRNA is translated before it leaves the ribosome and is replaced with another mRNA.
Growth inhibition due to expression of mini-genes of varying length: relation with a toxicity index (ITox) defined from biochemical parameters
Mini-genes were cloned in the pTrc99c vector (Amann et al., 1988) downstream of the trc promoter, which allows expression to be induced by the addition of isopropyl-β-d-thiogalactopyranoside (IPTG). The vector carries the lacIq gene to maintain repression in the absence of IPTG. The effect of inducing mini-gene expression was studied both in the pth (rap) mutant and in wild-type bacteria by measurement of growth rates (generations by hour) in rich medium (Table II). As expected, mini-gene expression is more inhibitory to growth in the pth (rap) strain than the wild-type strain, and a lower level of expression, obtained at a sub-saturating level of IPTG (0.1 mM) was required in this case to allow a more progressive change in growth rate over the series of mini-genes.
Table II. Effect of mini-gene expression on cell growth: growth rates (generations/h) of Xac rap and Xac strains at 37°C following IPTG induction.
| Plasmid | Mini-gene overexpressed | Xac rap 0.1 mM IPTG | Xac 1 mM IPTG |
|---|---|---|---|
| SD7 | 02.MK.Oa | no transformant | 1.24 |
| SD8 | 02.MFK.Oa | 0.50 | 1.91 |
| SD9 | 02.MFIK.Oa | 0.63 | 1.92 |
| SD10 | 02.MFITK.Oa | 1.49 | 2.06 |
| SD11 | 02.MFITQK.Oa | 1.02 | 2.05 |
| SD12 | 02.MFITQLK.Oa | 2.29 | 2.12 |
| SD13 | 02.MFITQLTK.Oa | 2.32 | 2.11 |
| pTrc99c | none | 2.33 | 2.12 |
pTrc99c is the control vector without insert.
Among the mini-genes studied, that encoding fMet-Lys is the most toxic. It was impossible to transform the Xac rap strain with plasmid SD7, as previously reported in the case of the minigene with the same SD sequence but encoding fMet-Ile (Dinçbas et al., 1999). In both wild-type and rap strains, the toxicity decreases (growth rate increases) with mini-gene length. In the rap strain, plasmid SD11, encoding fMFITQK, is again an exception, and appears more toxic than SD10 encoding fMFITK.
In the pth wild-type strain the inverse relationship between peptide length and growth inhibition is still observed, but is less striking: a higher toxicity of the gene encoding the dipeptide fMK (plasmid SD7) is evident, but it is barely possible to distinguish any of the remaining plasmids by their effect on growth in liquid medium. Strains transformed with plasmid SD7 cease growing after 2 h (2.5 generations), whereas the remaining transformed strains continue growing for ∼5 h (10 generations). On solid medium (LB agar), the relationship between peptide length and growth is also very clear in both Xac and Xac rap strains (results not shown). For example, in the presence of 1 mM IPTG no overnight growth at 37°C is observed in the case of transformants of strain Xac by plasmids SD7, SD8 or SD9. The detailed biochemical and growth rate analysis we report here has been obtained with a set of peptidyl-tRNAs with varying length but fixed sequences. Some of these in vivo experiments have, however, been repeated with peptidyl-tRNAs of a different sequence, a subset of fMIEKLTK-tRNA. The dependence of toxicity on length was very similar to the series based on fMFITQLK (L.Mora, unpublished results).
It was shown previously (Dinçbas et al., 1999) that in vivo toxicity can be predicted from in vitro results with the aid of a toxicity index that estimates the total rate of cellular drop-off normalized to Pth activity in terms of biochemically measurable parameters. An extended version of the index (Materials and methods) contains the rate of initiation of transcription of mini-genes (kT), the average number <m>TR of times that a particular mRNA is bound to ribosomes and translated rather than degraded, the average (or ‘recycle’) number <n>rec of times that a transcript is recycled without leaving its 30S subunit, the rate of drop-off of peptidyl-tRNA (kdrop), kcat/KM for the interaction between release factors RF1 or RF2 and the ribosome (kS), kcat/KM for the interaction between peptidyl-tRNA and Pth (kPth) according to
ITox = kT <m>TR <n>rec (kdrop/kS⋅1/kPth)
Defined this way, ITox becomes approximately proportional to the fraction of tRNA sequestered in the peptidyl-tRNA form.
The in vitro experiments described above yield data for <n>rec, kS, kdrop and kPth for the different mRNA and peptidyl-tRNA, and for the comparisons of different in vivo situations it is assumed that the other parameters in ITox do not vary from case to case. Relative ITox values (with fMFK-tRNALys and wild-type Pth as a reference) are indicated in Table I and may be compared with the growth rate data presented in Table II. In the Xac rap strain, the smallest mini-gene is highly toxic, whereas mini-genes encoding hepta- and octa-peptides exhibit no toxicity. The higher toxicity of plasmid SD11 compared with SD10 in Xac rap is clearly reflected in the toxicity index, and is due to low Pth cleavage efficiency. To compare growth inhibition with ITox, we have defined an index of in vivo toxicity as:
I = (µ0/µ) – 1
where µ0 is the growth rate in the presence of the parent plasmid pTrc99c, and µ the growth rate with the mini-gene under study. When growth is not impaired, µ will tend to µ0 and I to 0. Figure 4 shows that there is an approximately linear relation between in vivo and in vitro toxicity in the case of the pth wild-type strain. This is much less true for the pth (rap) strain, in which the levels of inhibition are much higher, even at 0.1 mM IPTG. It was clear from experiments at lower IPTG concentrations (results not shown) that the order of inhibition by different plasmids varied according to the level of mini-gene expression. Thus, SD9 was less inhibitory than SD8 at 0.1 mM IPTG (Table II) but more inhibitory than SD8 at 10 µM IPTG, whereas the parameters measured in vitro predict that they should be similar (Table I).
Fig. 4. Comparison of in vivo toxicity with ITox determined by the different biochemical parameters in strain Xac and Xac(rap); for the latter, see insert to figure. The index
and the in vivo toxicity index I are described further in the text.
Discussion
The drop-off of short peptidyl-tRNA molecules from the ribosome is of interest for at least two reasons. The first, of more general importance, concerns peptidyl-tRNA drop-off near the beginning of normal genes in the absence of in-frame termination codons. It should be recalled that the strong stimulation of short peptidyl-tRNA drop-off by RRF, EF-G and RF3 occurs irrespective of whether the codon in the ribosomal A-site is a sense or a stop codon (Heurgué-Hamard et al., 1998). The possible physiological interest of this phenomenon will be discussed below. The second reason is to understand the mechanism of mini-gene toxicity.
The clearest example of mini-gene expression known to impair bacterial physiology concerns the λ mini-genes barI and barII (Ontiveros et al., 1997; Hernández-Sánchez et al., 1998), both of which encode the dipeptide fMet-Ile. Recent studies by Tenson et al. (1999) showed that mini-gene toxicity was preferentially associated with very short ORFs. These authors selected in vivo toxic mini-genes from a library encoding peptides of up to 5 amino acids in length and obtained preferentially those with in-frame stop codons (i.e. shorter than pentapeptide).
The accepted explanation for mini-gene toxicity depends on an accumulation of peptidyl-tRNA corresponding to the last codon of the mini-gene, causing subsequent starvation for this tRNA. Specific accumulation of this particular peptidyl-tRNA and suppression of toxicity by overexpression of the corresponding tRNA isoacceptor has been shown experimentally for the natural or synthetic mini-genes encoding fMet-Ile, fMet-Lys and fMet-Arg (Ontiveros et al., 1997; Tenson et al., 1999).
Our primary purpose in the experiments described here was to understand why toxic mini-genes are small. To do so, we measured on a series of constructs varying in length the biochemical parameters likely to modify the pool of peptidyl-tRNA in the cell. Four important quantities that determine the cellular pool of peptidyl-tRNA that were directly measured in vitro as a function of peptide length are: (i) the average number of times an mRNA is translated before it is degraded, here scored as the average number of times, <n>rec, an mRNA is translated without leaving the 30S subunit; (ii) the rate of peptidyl-tRNA dissociation from the ribosome at each translation round; (iii) normal termination catalysed by release factors, the principal reaction competing with drop-off; and (iv) Pth activity, which allows recycling of peptidyl-tRNA thereby avoiding tRNA sequestration. The results we report here confirm that mini-gene toxicity is inversely correlated with the length of the coding sequence, and identify four reasons for this. For the series of mini-genes studied, those exceeding four sense codons in length did not affect the growth of pth wild-type cells, and those exceeding six sense codons (with one exception) did not affect the growth of pth (rap) cells. The use of a toxicity index, scoring the total flux towards the free peptidyl-tRNA pool, normalized to peptidyl-tRNA hydrolase activity, helped to identify the particular parameters associated with the growth inhibitory behaviour of individual mini-genes. Thus, the high toxicity due to fMet-Lys synthesis is caused essentially by a combination of three parameters: an inefficient Pth activity towards fMK-tRNALys; a high rate of dissociation catalysed by RRF, EF-G and RF3 of this dipeptidyl-tRNA from the ribosome; and a high efficiency of mini-gene translation (large <n>rec). As the peptide encoded increases in length beyond five amino acids, the rate of factor-catalysed drop-off decreases dramatically, to <20% of the value for short peptidyl-tRNAs. The phenomenon may be amplified by the fact that RF2-dependent peptide release shows the opposite dependence on length, increasing in efficiency as the peptide chain lengthens. This variation is not, however, evident for RF1, which is much more active than RF2 though present at a 5-fold lower concentration in E.coli K12 strains (Adamski et al., 1994). In contrast, over the range of peptide lengths studied, uncatalysed peptidyl-tRNA drop-off shows no general dependence on length.
Like factor-catalysed drop-off, <n>rec decreases strikingly with peptide length. Previous experiments with dipeptide synthesis had shown that the degree of complementarity of the SD sequence to the 16S rRNA was of primary importance to the magnitude of <n>rec (Dinçbas et al., 1999). Here we show how <n>rec varies as a function of the number of amino acids encoded by a mini-gene, from >100 for dipeptide down to ∼6 for octapeptide synthesis. This parameter is clearly of great importance, and the results are relevant not only to mini-gene translation, but also to the more general problem of translational coupling between genes cotranscribed from multicistronic operons, where short distances separate the termination codon of one gene and the ribosome binding site of another (Adhin and van Duin, 1990; Draper, 1996). In our calculation of the toxicity index we assumed that mRNAs are protected from degradation when they are on the 30S subunit, in accordance with previous results that frequently translated mRNAs are protected from degradation (Iost and Dreyfus, 1995). Our data suggest an inverse relationship between the length of the coding sequence and mRNA lifetime, as indeed found experimentally for a set of mini-genes with varying lengths and with a strong SD sequence (L.Mora, unpublished results).
In the case of pth (rap) cells, a third factor becomes crucial, as the hydrolase activity of this mutant, unlike that of the wild-type enzyme, shows a strong positive correlation with peptide length. An exception was nevertheless observed in the case of the hexapeptidyl-tRNA studied, which was even less rapidly hydrolysed than fMet-Lys-tRNALys by both wild type and the rap mutant of Pth. Accordingly, peptide sequence can modulate the overall dependence on peptide length, a finding that is worthy of further study. A recent report suggests that residue Arg133 in Pth, which is mutated to His in Pth (rap), is one of two positively charged residues that interact with the 5′-phosphate of elongator peptidyl-tRNA (Schmitt et al., 1997; Fromant et al., 1999). We suggest therefore that a weakened interaction with this 5′-phosphate in tRNA may require compensation by a longer peptide chain. If all Pth molecules in our preparations are active, wild-type Pth is ∼75-fold more efficient than the rap mutant in hydrolysing fMet-Lys-tRNALys, and remains 10-fold more efficient in the case of octapeptidyl-tRNA.
Previous work from Shiloach et al. (1975) showed a higher dependence on length of the activity of wild-type Pth on other peptidyl-tRNAs. Thus, the kcat/Km value for Gly-Val-tRNA was 20 times lower than for (Gly)2-Val-tRNA and 55 times lower than for (Gly)n-Val-tRNA where n is three or larger. These differences between their results and ours could be due to the nature of the peptide chain, less hydrophobic in case of the peptidyl-tRNAs we have studied. However, additional experiments with substrates such as fMet-Gly-tRNA yielded values of kcat/Km close to those we report here for other peptidyl-tRNAs (V.Heurgué-Hamard, unpublished results).
The importance of the length of the coding sequence in mini-genes highlights the role of the termination signal. Compared with the translation of sense codons by ternary complex (aminoacyl-tRNA·EF-Tu·GTP), RF-dependent release of the polypeptide chain is quite slow (Freistroffer et al., 1997, 2000), which can explain why the tRNA translating the last sense codon is the species that accumulates preferentially as peptidyl-tRNA. In support of this, changes in the choice of stop codon and its downstream context (factors affecting the kinetics of termination) have significant effects on mini-gene toxicity (Dinçbas et al., 1999).
Two further effects of a stop codon are specific to mini-genes. When present early in the coding sequence (before the eighth position), the stop codon combines the effect of introducing a pause in elongation with the catalytic effect on drop-off of the translation factors. Furthermore, the position of the stop codon in a mini-gene with a strong SD sequence has an overriding influence on the ribosome recycling number <n>rec, which itself has a proportional effect on the overall flux of peptidyl-tRNA dissociating from the ribosome (see Equation 1, Materials and methods). Thus we suggest that these combined effects of a stop codon placed early in the coding sequence, which constitute the peculiarity of a mini-gene, explain in general how even low levels of mini-gene expression can impair growth. Nevertheless, it seems likely that in the normal bacterial cell peptidyl-tRNA drop-off occurs with significant frequency also at sense codons followed by another sense codon. The following facts suggest that peptidyl-tRNAs with small peptide chains arise every time an mRNA is translated (Heurgué-Hamard et al., 1998). First, drop-off of peptidyl-tRNAs from ribosomes early in mRNA decoding is efficiently catalysed by the factors RRF, EF-G and RF3 also with sense rather than stop codons in the A-site. Secondly, mutations affecting RRF and RF3 suppress the thermosensitivity of mutant pth strains in the supposed absence of mini-gene expression, indicating that the stimulation of drop-off by the factors is of general physiological significance.
Although the role of factors in stimulating drop-off has been well demonstrated both in vitro and in vivo (Heurgué-Hamard et al., 1998; Karimi et al., 1998), progress is still needed to explain the mechanism of this phenomenon. By analogy to the termination and recycling process (Karimi et al., 1999), RRF and EF-G could dissociate ribosomal subunits, a step that in the drop-off event would probably need a preliminary disruption of peptide interaction with the 50S subunit. RF3 could eventually play this role by changing ribosome conformation. Previous studies showed that the nature of the nascent peptide interaction with 23S rRNA changed strikingly when the peptide size increased from 4 to 6 amino acids (Stade et al., 1995). When the peptide is small, crosslinks can be made to nucleotides located near the peptidyl-transferase center, and then as longer peptides are examined, the targets of crosslinking move out of this center to other helices of 23S rRNA. It is conceivable that the presence of longer peptides on tRNA in the P-site induces a change in the conformation of the factor binding site on the ribosome that reduces binding of RF3 or RRF, thereby inhibiting factor-stimulated drop-off.
What might be the advantage to the cell of peptidyl-tRNA drop-off early in mRNA translation? One possibility is that factor-induced drop-off early in mRNA translation has evolved, in concert with the stringent response, to help bacteria recover quickly after a nutrient shift-down (Gallant, 1979). After such an event, cells depend on de novo synthesis of amino acid synthesizing enzymes, which must occur when the supply of amino acids may be severely rate limiting. Under such conditions the protein elongation rate is very slow and inversely proportional to the concentration of active ribosomes, which leads to a long delay between initiation of transcription of genes encoding these enzymes and their appearance as active proteins. Quick recovery would therefore require a rapid reduction in the fraction of ribosomes that consume amino acids, since this would increase protein elongation rate and thereby reduce the delay before emergence of vital enzymes.
It is likely that the probability of translation-dependent drop-off early in mRNA translation increases dramatically after nutrient down-shifts, due to slow protein elongation. In those cases the 30S subunit may also reinitiate several times on the same mRNA so that a substantial fraction of ribosomes in the cell may idle and produce short peptides. Since these peptides return rapidly to the amino acid pools due to the action of Pth and peptidases, this idling reaction effectively reduces the fraction of ribosomes that consume the amino acid pools, and this will accelerate protein elongation on those ribosomes that successfully pass the early region of mRNA. In this way, the waiting time for new amino acid-synthesizing enzymes could be reduced dramatically. When the rate of supply of amino acids has recovered and protein elongation is fast again, the proposed drop-off-driven idling mechanism will automatically be turned off.
Materials and methods
Chemicals and buffers
GTP and ATP were from Pharmacia, Sweden. Putrescine, spermidine, phosphoenolpyruvate (PEP) and myokinase (MK) were from Sigma (St Louis, MO). Pyruvate kinase (PK) was from Boehringer Mannheim. All radioactive components were from Amersham (Buckinghamshire, UK). A standard factor mix used in the in vitro experiments contained (in 50 µl): 0.1 µmol ATP, 1 µmol PEP, 0.1 µmol GTP, 5 µg of PK, 0.3 µg of MK in polymix buffer (Jelenc and Kurland, 1979), unless specified otherwise.
Preparation of DNAs and mRNAs
Template DNAs for in vitro transcription were prepared by annealing the following oligonucleotides at the complementary sequences (underlined) and filling the gaps by PCR. Each mRNA contains the same ribosomal binding site as 002 mRNA (Calogero et al., 1988). The mRNA nomenclature was described previously (Dinçbas et al., 1999) and indicates the type of (SD) sequence (02 is from 002 mRNA), the encoded amino acid sequence, the stop codon (O indicates the Opal codon UAA) and the next nucleotide. Lower case nucleotides are mini-gene coding sequences.
The same forward oligonucleotide was used for all constructs: ′-ctctctGGTACCGAAATTAATACGACTCACTATAgggAATTCGG GCCCTTGTTAACAATTAAGGAGG. Specific reverse oligonucleotides were used for each construct. 02.MK.Oa: 5′-T21CTGCAGatttatttcatAGTATACCTCCTTAATTGTTAACAAGGGCCCG; 02.MFK.Oa: 5′-T21 CTGCAGatttatttgaacatAGTATACCTCCTTAATTGTTAACAAGGGCCCG; 02.MFIK.Oa: 5′-T21CTGCAGatttatttgatgaacatAGTATACCTCCTTAATTGTTAACAAGGGCCCG; 02.MFITK.Oa: 5′-T21CTGCAGatt tatttggtgatgaacatAGTATACCTCCTTAATTGTTAACAAGGGCCCG; 02.MFITQK.Oa: 5′-t21CTGCAGatttatttctgggtgatgaacatAGTATACCTC CTTAATTGTTAACAAGGGCCCG; 02.MFITQLK.Oa: 5′-T21CTGCA GatttatttcagctgggtgatgaacatAGTATACCTCCTTAATTGTTAACAAG GGCCCG; 02.MFITLQK.Oa: 5′-T21CTGCAGATttatttctgcagggtgatgaa catAGTATACCTCCTTAATTGTTAACAAGGGCCCG; 02.MFITQL TK.Oa: 5′-T21CTGCAGatttatttggtcagctgggtgatgaacatAGTATACCTCC TTAATTGTTAACAAGGGCCCG. The sequence of 02.MK.Oa is 5′-GGGAAUUCGGGCCCUUGUUAACAAUUAAGGAGG UACUAUGAAAUAAAUCUGCAGA21.
The ORF and stop codons are in bold type, and the SD region is in italic.
The sequence of mRNAs 02.MFK.Oa, 02.MFIK.Oa, 02.MFITK.Oa, 02.MFITQK.Oa, 02.MFITQLK.Oa, 02.MFITLQK.Oa and 02.MFITQLTK.Oa are similar, but the ORFs are, respectively: AUGUUCAAA, AUGUUCAUCAAA, AUGUUCAUCACCAAA, AUGUUCAUCACCCAGAAA, AUGUUCAUCACCCAGCUGAAA, AUGUUCAUCACCCUGCAGAAA, AUGUUCAUCACCCAGCUGACCAAA, and the names indicate the sequences encoded using the single letter amino acid abbreviations.
Protein purifications
Bacterial elongation factors, initiation factors, and other components needed for the preparation of ribosomal complexes paused at the stop codon of mini-gene mRNA were prepared as described previously (Ehrenberg et al., 1990; Freistroffer et al., 1997; Dinçbas et al., 1999; Karimi et al., 1999). RF2 was prepared from W3110 cells according to Tate and Caskey (1990), RF1 was purified from MRE600 cells as described by Dinçbas et al. (1999), and wild-type and rap mutant Pth was as described by Dinçbas et al. (1999).
In vitro assays
Determination of kcat/Km values for RF1 and RF2. Relative kcat/Km values for hydrolysis of the different peptidyl-tRNAs in the ribosomal P-site by RF2 were measured in competition experiments (Pavlov et al., 1998). Ribosomal complexes paused at a stop codon were prepared as described previously (Freistroffer et al., 1997; Pavlov et al., 1998). Two differently labelled ribosomal release complexes in standard factor mix (typically 1 pmol of each), were set to compete for a limiting amount of release factor (typically 0.2 pmol). The extents of hydrolysis of the peptidyl-tRNAs were analysed as described previously (Dinçbas et al., 1999) and data were evaluated according to Pavlov et al. (1998). Absolute values of kcat/Km for RF1 and RF2 were determined as described by Dinçbas et al. (1999), using ribosome-release complexes programmed with the messenger encoding the dipeptide MK. kcat/Km values for RF1 and RF2-dependent termination for the other constructs were calculated using relative values obtained in competition experiments.
Determination of absolute values of kcat/Km for wild-type Pth and Pth (rap). Ribosome release complexes were prepared as above with ribosomes programmed with 02.MK.Oa mRNA. Ribosomal release complexes were incubated for 30 min at 37°C with RRF, RF3 and EF-G; 75 pmol of each; in 70 µl of standard factor mix for dissociation of peptidyl-tRNAs from the ribosomal P-site. Following incubation, 5 µl of Pth (rap) (10 pmol) or Pth (wild type) (0.3 pmol) were added to 70 µl of the release complex mix. Samples were withdrawn at different times and the reaction was stopped by cold TCA precipitation. The kcat/Km was determined from the linear relationship with time of the remaining fraction r(t) of peptidyl-tRNA:
ln r(t) = – (kcat/Km)⋅[Pth]⋅t
The concentrations of active Pth were approximated to the total Pth concentrations measured as described by Bradford (1976).
Determination of Pth (rap) activity. Reactions were done as indicated above, but stopped by adding HCOOH at a final concentration of 10%. Samples were spun in an Eppendorf centrifuge at 14 000 r.p.m. for 15 min. The pellet was dissolved in 200 µl of 0.5 M KOH and incubated for 15 min at room temperature for hydrolysis of the oligopeptide from peptidyl-tRNA, whether on or off the ribosome. A volume of 10 µl of concentrated HCOOH (98–100%) was added to the dissolved sample for precipitation of large RNA and large proteins. Samples were centrifuged at 14 000 r.p.m. for 15 min and the soluble peptides were separated on an RP18 (Waters) column equilibrated with various concentrations of methanol (MK: 18%, MFK: 34%; MFIK and MFITK: 42%; MFITQK: 36%; MFITQLK, MFITLQK and MFITQLTK: 50%) and 0.1% trifluoroacetic acid. Radioactivity was monitored by on-line radiometry (Ramona). Due to high Km values for Pth (rap) interaction with peptidyl-tRNAs, kcat/Km for the hydrolysis of free peptidyl-tRNAs could be measured under these conditions.
Determination of relative kcat/Km values of Pth wild type. Relative kcat/Km values for hydrolysis of the different free peptidyl-tRNAs by Pth were measured in competition experiments. Two differently labelled ribosomal release complexes (typically 1 pmol of each) (Freistroffer et al., 1997; Pavlov et al., 1998) in standard factor mix were incubated for 30 min at 37°C with RRF, RF3 and EF-G at a final concentration of 1 µM each to promote peptidyl-tRNA dissociation. Peptidyl-tRNAs were then set to compete for a limiting amount of Pth (typically 0.6 pmol). The extents of hydrolysis of the peptidyl-tRNAs were analysed by withdrawing aliquots at different time points for quenching in equal volumes of 20% ice-cold formic acid. After centrifugation at 14 000 r.p.m. in an Eppendorf centrifuge for 12 min at 4°C, the radioactivity in the supernatant was counted in Aquasafe 300 plus scintillation fluid (Zinsser). Data were evaluated by plotting the natural logarithm of the peptidyl-tRNA fraction remaining in function of time.
Drop-off rate constants. Peptidyl-tRNA drop-off experiments were as described (Heurgué-Hamard et al., 1998). Dissociation rate constants were measured in the presence of EF-G (1 µM), RRF (1 µM), RF3 (1 µM) or in the absence of translation factors. Translation factors were pre-warmed together with Pth (wild type) (120 pmol) for 2 min at 37°C in standard factor mix buffer. An equal volume of ribosomal release complex, pre-warmed for 1 min at 37°C, was added to the factor mix. The reaction was stopped at specified times by addition of HCOOH to a final concentration of 10% and the peptides released by KOH treatment were analysed by HPLC as described above [see determination of Pth (rap) activity].
Probabilities of dissociation of mRNA from ribosomes after termination. Recycling experiments on seven different mRNAs coding for fMK, fMFK, fMFIK, fMFITK, fMFITQK, fMFITQLK and fMFITQLTK were done as described previously (Pavlov et al., 1997a,b; Dinçbas et al., 1999) with the following modifications: factor mix 1 contained per 60 µl modified quantities of these components: 0.3 mg of tRNAbulk (prepared as described by Ehrenberg et al., 1990), 2 nmol of EF-Tu, 200 pmol of EF-Ts, 750 pmol of chase mRNA, which contained SD 002 and encoded fMFITR but contained no stop codon. Data were analysed as described by Dinçbas et al. (1999).
Bacterial strains, plasmids and bacteriophage
The E.coli K12 strains (Xac, Xac rap) and B strain (VH998) used for overexpression of Pth (rap) are described in Dinçbas et al. (1999). Minigenes were cloned from the constructions used in the in vitro experiments (see Table II for plasmid names). PCRs were cut by KpnI and PstI and cloned into pTrc99c digested by the same enzymes. All plasmid constructions were sequenced by the method of Sanger et al. (1977).
Growth conditions
Luria Broth (LB) medium was supplemented according to the requirements. Antibiotics were added at the following final concentration: 50 µg/ml kanamycin; 200 µg/ml ampicillin. When induction was necessary to express mini-genes, 1 M IPTG was added to LB plates or liquid medium to the indicated final concentration. Growth was monitored either by streaking transformed strains on plates or in most cases in liquid medium. An overnight culture was diluted to an OD600 of 0.05 and grown until 0.5 in LB–ampicillin medium without IPTG. This culture was again diluted to 0.05 grown in LB–ampicillin medium containing the IPTG concentration indicated. Absorbance at 600 nm was monitored as a function of time for at least 6 h. All growth measurements were done at 37°C.
Recombinant DNA manipulations and genetic manipulations
General procedures for DNA recombinant techniques, plasmid extraction, etc. were performed as described by Sambrook et al. (1989). Purification of fragments on agarose gel was by Jetsorb gel extraction (Bioprobe). Phage P1 lysates, transductions and transformations were performed as described by Miller (1992).
Modification of toxicity index to include recycling of ribosomes
In the following treatment, kT is the rate of initiation of transcription of a mini-gene, kTR is the rate of initiation of translation of an mRNA, kd is the rate constant of degradation of a free mRNA (it will be assumed that ribosome-bound mRNA is not degraded) and it follows immediately that <m>TR = kTR/kd is the average number of times that a chosen mRNA binds from its free state to any ribosome and becomes translated one or several times. P is the probability that an mRNA leaves the 30S subunit after termination with release factor, [RF]kS is the total rate of termination by release factor and kdrop is the rate of drop-off of peptidyl-tRNA. The probability of drop-off per ribosome cycle is Pdrop = kdrop / (kdrop + [RF]kS).
When kdrop<<kS[RF], Pdrop can be replaced by kdrop/[RF]kS and for relative Itox values, the release factor concentration can often be neglected. Defining the toxicity index as the total flux to the peptidyl-tRNA pool of a certain tRNA isoacceptor normalized to the peptidyl-tRNA hydrolase activity and assuming that P is the same when peptide synthesis is terminated by release factors and drop-off, gives
ITox = kT <m>TR <n>rec (kdrop/kS⋅1/kPth) (Eqn 1)
since then 1/P= <n>rec, the average number of times that an mRNA is translated before it leaves the ribosome and is replaced with another molecule.
With the same general definition of Itox but assuming instead that an mRNA leaves the 30S subunit with certainty after a drop-off event then the toxicity index would instead be
ITox = kT <m>TR⋅1/[1 + (1/Pdrop – 1)⋅P]⋅1/kPth (Eqn 2)
When Pdrop<<P equations (1) and (2) converge except that the latter also contains the concentration of release factor 1 or 2 in the denominator. Preliminary experiments indicate that P is the same after termination with release factor and by drop-off and we have therefore chosen Equation (1) for main text and data evaluation.
The global in vivo life time (1/kD) of an mRNA can be expressed in biochemically measurable parameters as
1/kD = <m>TR⋅<n>rec/k + 1/kd (Eqn 3)
where k is a rate constant for ribosome recyling, i.e. the inverse of the time per ribosome cycle when the 30S and mRNA remain associated. The first term in Equation 3 describes the average time that an mRNA is protected by ribosomes assuming that the protection time in the first ribosome cycle is the same as in subsequent ones. The second term is the average time that an mRNA spends in free state.
Acknowledgments
Acknowledgements
This work was supported by the Swedish Research Council for Engineering Sciences, the Swedish Natural Science Research Council and the Centre National pour la Recherche Scientifique (UPR9073). V.H.-H. thanks the Human Frontier Science Programme for support.
References
- Adamski F.M., McCaughan,K.K., Jørgensen,F., Kurland,C.G. and Tate,W.P. (1994) The concentration of polypeptide chain release factors 1 and 2 at different growth rates of Escherichia coli. J. Mol. Biol., 238, 302–308. [DOI] [PubMed] [Google Scholar]
- Adhin M.R. and van Duin,J. (1990) Scanning model for translational reinitiation in eubacteria. J. Mol. Biol., 213, 811–818. [DOI] [PubMed] [Google Scholar]
- Amann E., Ochs,B. and Abel,K.J. (1988) Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in Escherichia coli. Gene, 69, 301–315. [DOI] [PubMed] [Google Scholar]
- Atherly A.G. and Menninger,J.R. (1972) Mutant Escherichia coli strain with temperature sensitive peptidyl-transfer RNA hydrolase. Nature, 240, 245–246. [DOI] [PubMed] [Google Scholar]
- Björnsson A., Mottagui-Tabar,S. and Isaksson,L.A. (1996) Structure of the C-terminal end of the nascent peptide influences translation termination. EMBO J., 15, 1696–1704. [PMC free article] [PubMed] [Google Scholar]
- Bradford M.M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem., 72, 248–254. [DOI] [PubMed] [Google Scholar]
- Buckingham R.H., Grentzmann,G. and Kisselev,L. (1997) Polypeptide chain release factors. Mol. Microbiol., 24, 449–456. [DOI] [PubMed] [Google Scholar]
- Calogero R.A., Pon,C.L., Canonaco,M.A. and Gualerzi,C.O. (1988) Selection of the mRNA translation initiation region by Escherichia coli ribosomes. Proc. Natl Acad. Sci. USA, 85, 6427–6431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinçbas V., Heurgué-Hamard,V., Buckingham,R.H., Karimi,R. and Ehrenberg,M. (1999) Shutdown in protein synthesis due to the expression of mini-genes in bacteria. J. Mol. Biol., 291, 745–759. [DOI] [PubMed] [Google Scholar]
- Draper D.E. (1996) Translational initiation. In Neidhart,F.C., Curtis,R.,III, Ingraham,J.L., Lin,E.C.C., Brookslow,K., Magasanik,B., Reznikoff,W.S., Riley,M., Schaechter,M. and Umbarger,H.G. (eds), Escherichia coli and Salmonella: Cellular and Molecular Biology. ASM Press, Washington, DC, pp. 902–908. [Google Scholar]
- Ehrenberg M., Bilgin,N. and Kurland,C.G. (1990) Design and use of a fast and accurate in vitro translation system. In Spedding,G. (ed.), Ribosomes and Protein Synthesis. IRL Press, Oxford University Press, Oxford, UK, pp. 101–129. [Google Scholar]
- Freistroffer D.V., Pavlov,M.Y., MacDougall,J., Buckingham,R.H. and Ehrenberg,M. (1997) Release factor RF3 in E.coli accelerates the dissociation of release factors RF1 and RF2 from the ribosome in a GTP dependent manner. EMBO J., 16, 4126–4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freistroffer D.V., Kwiatkowski,M., Buckingham,R.H. and Ehrenberg,M. (2000) The accuracy of codon recognition by ribosome release factors. Proc. Natl Acad. Sci. USA, 97, 2046–2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromant M., Plateau,P., Schmitt,E., Mechulam,Y. and Blanquet,S. (1999) Receptor site for the 5′-phosphate of elongator tRNAs governs substrate selection by peptidyl-tRNA hydrolase. Biochemistry, 38, 4982–4987. [DOI] [PubMed] [Google Scholar]
- Gallant J. (1979) Stringent control in E.coli. Annu. Rev. Genet., 13, 393–415. [DOI] [PubMed] [Google Scholar]
- Garcia Villegas M.R., De La Vega,F.M., Galindo,J.M., Segura,M., Buckingham,R.H. and Guarneros,G. (1991) Peptidyl-tRNA hydrolase is involved in λ inhibition of host protein synthesis. EMBO J., 10, 3549–3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haenni A.L. and Lucas-Lenard,J. (1968) Stepwise synthesis of a tripeptide. Proc. Natl Acad. Sci. USA, 61, 1363–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández J., Ontiveros,C., Valadez,C., Buckingham,R.H. and Guarneros,G. (1997) Regulation of protein synthesis by minigene expression. Biochimie, 79, 527–531. [DOI] [PubMed] [Google Scholar]
- Hernández-Sánchez J., Valadez,J.G., Herrera,J.V., Ontiveros,C. and Guarneros,G. (1998) λ bar minigene-mediated inhibition of protein synthesis involves accumulation of peptidyl-tRNA and starvation for tRNA. EMBO J., 17, 3758–3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershey J.W.B. (1987) Protein synthesis. In Neidthardt,F.G. (ed.), Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. ASM Press, Washington, DC, pp. 613–647. [Google Scholar]
- Heurgué-Hamard V., Karimi,R., Mora,L., MacDougall,J., Leboeuf,C., Grentzmann,G., Ehrenberg,M. and Buckingham,R.H. (1998) Ribosome release factor RF4 and termination factor RF3 are involved in dissociation of peptidyl-tRNA from the ribosome. EMBO J., 17, 808–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iost I. and Dreyfus,M. (1995) The stability of Escherichia coli lacZ mRNA depends upon the simultaneity of its synthesis and translation. EMBO J., 14, 3252–3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelenc P.C. and Kurland,C.G. (1979) Nucleotide triphosphate regeneration decreases the frequency of translation errors. Proc. Natl Acad. Sci. USA, 76, 3174–3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi R., Pavlov,M.Y., Heurgué-Hamard,V., Buckingham,R.H. and Ehrenberg,M. (1998) Initiation factors IF1 and IF2 synergistically remove small peptidyl-tRNAs from the P-site of E.coli ribosomes. J. Mol. Biol., 281, 241–252. [DOI] [PubMed] [Google Scholar]
- Karimi R., Pavlov,M., Buckingham,R.H. and Ehrenberg,M. (1999) Novel roles for classical factors at the interface between translation termination and initiation. Mol. Cell, 3, 601–609. [DOI] [PubMed] [Google Scholar]
- Menninger J.R. (1976) Peptidyl-transfer RNA dissociates during protein synthesis from ribosomes of E.coli. J. Biol. Chem., 251, 3392–3398. [PubMed] [Google Scholar]
- Miller J.H. (1992) A Short Course in Bacterial Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Ontiveros C., Valadez,G., Hernandez,J. and Guarneros,G. (1997) Inhibition of Escherichia coli protein synthesis by abortive translation of phage λ minigenes. J. Mol. Biol., 269, 167–175. [DOI] [PubMed] [Google Scholar]
- Pavlov M.Y., Freistroffer,D., MacDougall,J., Buckingham,R.H. and Ehrenberg,M. (1997a) Fast recycling of E.coli ribosomes requires both ribosome recycling factor (RRF) and release factor RF3. EMBO J., 16, 4134–4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlov M.Y., Freistroffer,D.V., Heurgué-Hamard,V., Buckingham,R.H. and Ehrenberg,M. (1997b) Release factor RF3 abolishes competition between release factor RF1 and ribosome recycling factor (RRF) for a ribosome binding site. J. Mol. Biol., 273, 389–401. [DOI] [PubMed] [Google Scholar]
- Pavlov M.Y., Freistroffer,D.V., Dinçbas,V., MacDougall,J., Buckingham,R.H. and Ehrenberg,M. (1998) A direct estimation of the context effect on the efficiency of termination. J. Mol. Biol., 284, 579–590. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Sanger F., Nicklen,S. and Coulson,A.R. (1977) DNA sequencing with chain-terminating inhibitors. Proc. Natl Acad. Sci. USA, 74, 5463–5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt E., Mechulam,Y., Fromant,M., Plateau,P. and Blanquet,S. (1997) Crystal structure at 1.2 Å resolution and active site mapping of Escherichia coli peptidyl-tRNA hydrolase. EMBO J., 16, 4760–4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiloach J., Bauer,S., de Groot,N. and Lapidot,Y. (1975) The influence of the peptide chain length on the activity of peptidyl-tRNA hydrolase from E.coli. Nucleic Acids Res., 2, 1941–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stade K., Junke,N. and Brimacombe,R. (1995) Mapping the path of the nascent peptide chain through the 23S RNA in the 50S ribosomal subunit. Nucleic Acids Res., 23, 2371–2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate W.P. and Caskey,C.T. (1990) Termination of protein synthesis. In Spedding,G. (ed.), Ribosomes and Protein Synthesis—A Practical Approach. IRL Press, Oxford, UK, pp. 81–100. [Google Scholar]
- Tenson T., Herrera,J.V., Kloss,P., Guarneros,G. and Mankin,A.S. (1999) Inhibition of translation and cell growth by minigene expression. J. Bacteriol., 181, 1617–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]