Abstract
Intimin is a bacterial adhesion molecule involved in intimate attachment of enteropathogenic and enterohaemorrhagic Escherichia coli to mammalian host cells. Intimin targets the translocated intimin receptor (Tir), which is exported by the bacteria and integrated into the host cell plasma membrane. In this study we localized the Tir-binding region of intimin to the C-terminal 190 amino acids (Int190). We have also determined the region’s high-resolution solution structure, which comprises an immunoglobulin domain that is intimately coupled to a novel C-type lectin domain. This fragment, which is necessary and sufficient for Tir interaction, defines a new super domain in intimin that exhibits striking structural similarity to the integrin-binding domain of the Yersinia invasin and C-type lectin families. The extracellular portion of intimin comprises an articulated rod of immunoglobulin domains extending from the bacterium surface, conveying a highly accessible ‘adhesive tip’ to the target cell. The interpretation of NMR-titration and mutagenesis data has enabled us to identify, for the first time, the binding site for Tir, which is located at the extremity of the Int190 moiety.
Keywords: Escherichia coli/intimin/NMR/Tir
Introduction
Enteropathogenic (EPEC) and enterohaemorrhagic (EHEC) Escherichia coli constitute a significant risk to human health worldwide. EPEC is the cause of severe infantile diarrhoeal disease in many parts of the developing world, while EHEC are the etiological agents of a food-borne disease that can cause acute gastro-enteritis, bloody diarrhoea, haemorrhagic colitis and haemolytic uraemic syndrome (HUS) (reviewed by Nataro and Kaper, 1998).
EPEC colonize the small intestinal mucosa and, by subverting intestinal epithelial cell function, produce a characteristic histopathological feature known as the ‘attaching and effacing’ (A/E) lesion (Moon et al., 1983). The A/E lesion is characterized by localized destruction (effacement) of brush border microvilli, intimate attachment of the bacillus to the host cell membrane and the formation of an underlying pedestal-like structure in the host cell consisting of polymerized actin, α-actinin, ezrin, talin and myosin (reviewed by Frankel et al., 1998a). A/E lesion formation is essential for pathogenicity and similar lesions have been associated with several other bacterial mucosal pathogens, most notably in EHEC (Donnenberg et al., 1993a,b).
The first gene to be associated with A/E activity was eae encoding the intimate EPEC and EHEC adhesin, intimin (Jerse et al., 1990). Intimin exists as at least five antigenically distinct subtypes that have been named intimin α, β, γ, δ and ε (Adu-Bobie et al., 1998; Oswald et al., 2000 ). EPEC/EHEC intimins exhibit homology at their N-termini to the invasin polypeptides of Yersinia (Isberg et al., 1987) and, like Yersinia invasin (Leong et al., 1990), intimin harbours receptor-binding activity at the C-terminus of the polypeptide (Frankel et al., 1994, 1995). A 76-amino acid motif enclosed by a disulfide bridge between two cysteines lies within the C-terminal domain of intimin. This is absolutely required for intimin binding to the host cell, A/E lesion formation and colonization of mucosal surfaces (Frankel et al., 1995, 1996, 1998b; Hicks et al., 1998; Higgins et al., 1999a,b). The C-terminal domain of invasin also harbours two Cys residues, in similar locations to those found in intimin (Leong et al., 1993).
Recently, we have determined the global fold of the C-terminal 280 amino acids of intimin α (Int280α) by a combination of perdeuteration, site-specific protonation and multidimensional nuclear magnetic resonance (NMR) (Kelly et al., 1998, 1999). The structure shows that Int280α is ∼90 Å in length and is built from three globular domains. The first two domains (residues 1–91 and 93–181) each comprise β-sheet sandwiches that resemble the immunoglobulin super family (IgSF). Despite no significant sequence homology, the topology of the C-terminal domain (residues 183–280) is reminiscent of the C-type lectins, a family of proteins responsible for cell-surface carbohydrate recognition. A domain between residues 558 and 650 within the extracellular portion of intimin aligns with 35% identity (44% conservation) with the first domain of Int280, identifying a further Ig-like domain (Kelly et al., 1999). This produces at least four domains that protrude from the bacterial membrane for interaction with the host cell. For the purposes of future discussion, the domains in Int280 are therefore renamed as D2, D3 and D4 respectively. The presence of a disulfide bridge in the C-type lectin-like module of Int280α is essential for correct folding of this domain and for carbohydrate binding by other C-type lectins (Weis and Drickamer, 1996). Modelling other intimin types (including the EHEC intimin γ) (Yu and Kaper, 1992; Adu-Bobie et al., 1998) would suggest they have similar structures, and define a new family of bacterial adhesion molecule.
The intimin-encoding eae gene is part of the large pathogenicity islands found in EPEC and EHEC, termed the LEE (locus of enterocyte effacement) (McDaniel et al., 1995; Perna et al., 1998). In addition to intimin, the LEE also encodes a type III secretion system (Jarvis et al., 1995; Jarvis and Kaper, 1996), an intimin receptor [translocated intimin recepor (Tir)/EspE] (Kenny et al., 1997; Deibel et al., 1998) and three secreted proteins, EspA, EspB and EspD, which are required for signal transduction in host cells and A/E lesion formation (Donnenberg et al., 1993c; Kenny et al., 1996; Lai et al., 1997). EspA is a structural protein and a major component of a large filamentous organelle that is transiently expressed on the bacterial surface and interacts with the host cell during the early stage of A/E lesion formation (Ebel et al., 1998; Knutton et al., 1998). EspA filaments may contribute to bacterial adhesion but, of greater significance, they appear to be a component of a translocation apparatus and as such are essential for the translocation of EspB (Knutton et al., 1998; Wolff et al., 1998) and Tir (Kenny et al., 1997) into host cells.
Int280 can bind directly to uninfected host cells (Frankel et al., 1994, 1996). Moreover, a recent study has demonstrated that intimin can induce a T helper cell-type 1 immune response in the colonic mucosa and colonic hyperplasia in mice (Higgins et al., 1999a). In addition, the bacterial protein Tir (EspE) can also act as an EPEC/EHEC intimin receptor (Kenny et al., 1997; Deibel et al., 1998). Recently, the intimin-binding region of Tir has been localized to a stretch of amino acids residues that resides between the two putative membrane-spanning domains of the polypeptide (termed Tir-M) (de Grado et al., 1999; Hartland et al., 1999; Kenny, 1999). Immunofluorescence staining of infected cells using polyclonal antisera raised against the N- and C-terminal peptides of Tir (Tir-N and Tir-C, respectively) demonstrate that these two regions are located within the host cell where they can induce the polymerization of actin and other cytoskeletal proteins to produce the characteristic pedestal-like structure (Kenny et al., 1997; Hartland et al., 1999).
We report that an active Tir-binding fragment of intimin spans the IgSF-like domain, D3, and the lectin-like domain, D4. We have also determined the high-resolution structure of the portion, comprising the C-terminus 190 amino acids, which form a super domain capable of interaction with Tir. Using chemical shift titration experiments we have also further localized, for the first time, the binding site of Tir to a concerted patch of residues at the tip of the structure (D4), and propose a plausible model for the intimin–Tir interaction.
Results and discussion
Localization of the minimal Tir-binding region of intimin
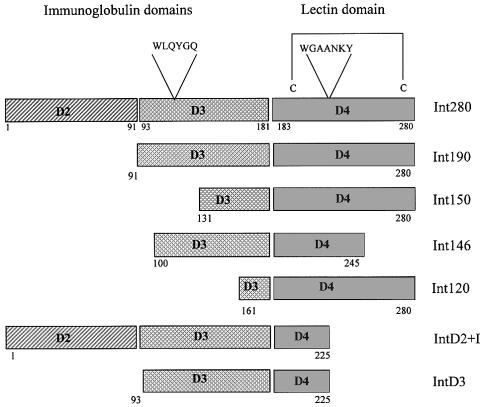
Based on low resolution structural information reported previously (Kelly et al., 1998, 1999), a number of truncated Int280α derivatives, expressed as maltose-binding protein (MBP) fusion proteins, were constructed and used together with purified Tir-M (Hartland et al., 1999) in a gel overlay protein-binding assay. The MBP–Int280α derivatives are shown schematically in Figure 1 and include Int280, Int190, Int150 and Int120. We have shown before that the D3 domain forms numerous hydrophobic contacts with D4 (Kelly et al., 1999). Accordingly, we included the first 42 amino acids of D4 in the derivatives designed to assess the potential Tir-binding activity of the IgSF-like domains (IntD2 and D3; Figure 1). MBP–Int146, which harbours D3 and 62 amino acids of D4, encompasses both conserved WLQYGQ and WGAANKY motifs (Adu-Bobie et al., 1998) (Figure 1). The gel overlay protein-binding assays revealed that Tir-M bound to the immobilized Int280α and Int190α fusion proteins (Figure 2A). However, no binding was observed with any other MBP–Int fusion proteins or MBP alone (Figure 2A; data not shown). Similar results were obtained when the binding assay was performed with immobilized Tir-M and soluble MBP–Int fusion proteins (data not shown). These results are consistent with those reported recently for intimin γ from EHEC (Liu et al., 1999).
Fig. 1. Schematic representation of the overlapping Int280-derived polypeptides. The two IgSF-like domains (D2 and D3), the C-type lectin-like domain (D4) and the conserved motifs in Int280 are shown at the top. The position of W150 within Int190 is indicated. Numbers on both sides of the fragments mark the first and last amino acids of each fragment within the Int280 domain.
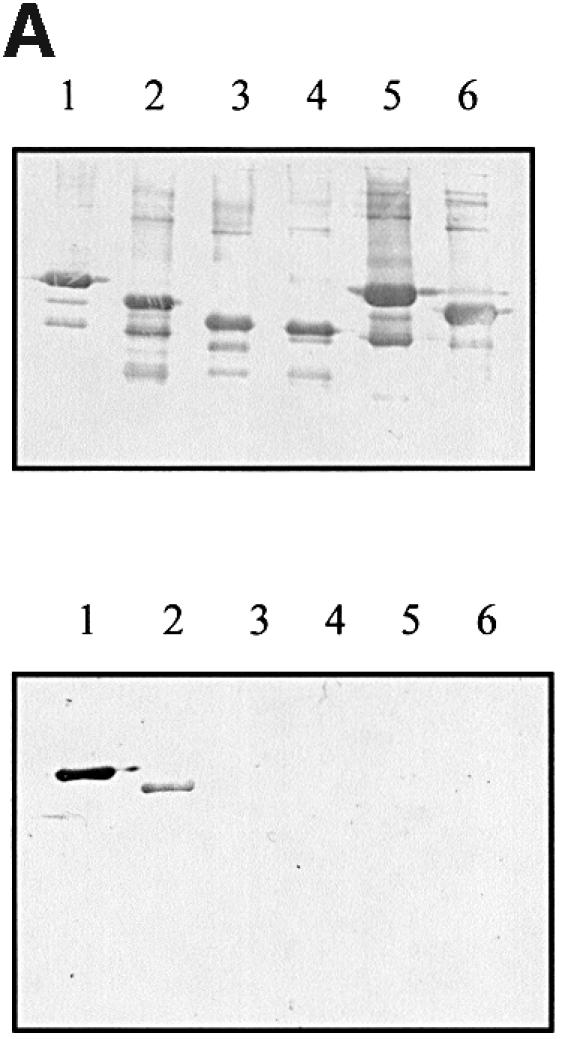
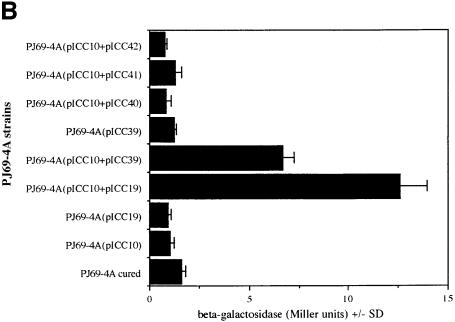
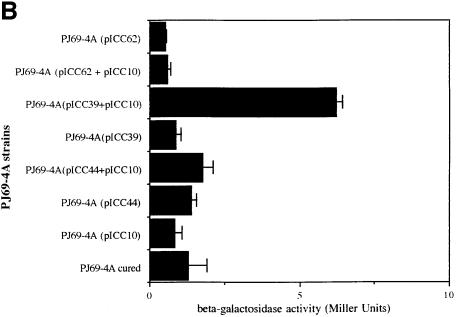
Fig. 2. (A) Detection of Int–Tir interactions using gel overlays. Western blots of MBP–Int derivatives were reacted with a rabbit MBP antiserum (top) or overlayed with Tir-M (bottom). Similar levels of MBP–Int280 (lane 1), MBP–Int190 (lane 2), MBP–Int150 (lane 3), MBP–Int146 (lane 4), MBP–IntD3&D4 (lane 5) and MBP–IntD3 (lane 6); fusion proteins were detected with polyclonal antiserum (top), while Tir-M only bound to MBP–Int280 (bottom, lane 1) and MBP–Int190 (bottom, lane 2). (B) Detection of Int–Tir interactions using the yeast two-hybrid system. β-galactosidase assays showing a 14-fold increase in enzymatic activity in strains co-expressing the whole Tir polypeptide and Int280 and a 7.5-fold increase when Int190 was co-expressed with Tir compared with the parent, cured and the other single and double transformants.
The interaction of the different Int280α derivatives with Tir was also investigated using the yeast two-hybrid system, designed to identify protein–protein interactions through the functional restoration of the yeast GAL4 transcriptional activator in vivo (James et al., 1996). For this, selected DNA fragments encoding truncated Int280α polypeptides were sub-cloned into pGAD424 to generate plasmids pICC39, pICC40, pICC41 and pICC42 (Table I). We previously reported that in the yeast two-hybrid system, the interaction of Int280 (pICC19) with the whole Tir polypeptide is more efficient than the interaction of Int280 with Tir-M (Hartland et al., 1999). Accordingly, the DNA fragment encoding the whole Tir polypeptide, expressed from the second yeast two-hybrid system vector, pGBT9 (pICC10; Hartland et al., 1999), was used in this part of the study.
Table I. List of plasmids.
| Plasmid | Description | Source/reference |
|---|---|---|
| pGBT9 | A yeast GAL4 DNA-BD cloning vector | Clontech |
| pGAD424 | A yeast GAL4 DNA-AD cloning vector | Clontech |
| pMal-C2 | Vector for expression of MBP-tagged proteins | NEB |
| pET3d | Vector for expression of proteins for purification without a tag | Novagen |
| pICC10 | pGBT9 expressing Tir | Hartland et al. (1999) |
| pICC19 | pGAD424 expressing Int280 | Hartland et al. (1999) |
| pICC39 | pGAD424 expressing Int190 | this study |
| pICC40 | pGAD424 expressing Int150 | this study |
| pICC41 | pGAD424 expressing Int146 | this study |
| pICC42 | pGAD424 expressing Int120 | this study |
| pICC44 | pGAD424 expressing Int190W150A | this study |
| pICC45 | pMal expressing MBP–Int280 | Frankel et al. (1994) |
| pICC46 | pMal expressing MBP–Int190 | this study |
| pICC47 | pMal expressing MBP–Int150 | Frankel et al. (1994) |
| pICC48 | pMal expressing MBP–Int146 | this study |
| pICC49 | pMal expressing MBP–Int120 | Frankel et al. (1994) |
| pICC50 | pMal expressing MBP–IntD2+D3 | this study |
| pICC51 | pMal expressing MBP–IntD3 | this study |
| pICC52 | pMal expressing Int190W150A | this study |
| pCVD438 | pACYC184 containing the eae gene of E2348/69 | Donnenberg and Kaper (1991) |
| pICC53 | pCVD438W899A | this study |
| pICC54 | pET3a expressing Int190 | this study |
| pICC59 | pET3a expressing Int188 | this study |
| pICC62 | pGAD424 expressing Int280A240 | this study |
| pICC63 | pMal expressing Int280A240 | this study |
Plasmid pICC10 was co-transformed with each of the different pGAD424-based plasmids into a derivative of the yeast stain PJ69-4A selected previously as a reporter for intimin–Tir interaction (Hartland et al., 1999). Replica plating these colonies onto selective media yielded vigorously growing colonies, and hence a positive two-hybrid phenotype, in yeast strains expressing both Tir and Int280 (pICC10 and pICC19) and Tir and Int190 (pICC10 and pICC39). No yeast colonies were observed using Tir with any of the other Int280 truncations or single plasmid transformants (data not shown).
The function of the non-selective reporter, lacZ, was also assessed in these strains by measuring β-galactosidase activity (Figure 2B). The host or single plasmid-bearing strains exhibited low levels of β-galactosidase activity, whereas the strains expressing Int280 or Int190 and Tir showed a 14- or 7.5-fold induction of β-galactosidase units, respectively. Based on the β-galactosidase levels of the strains bearing the plasmids pICC10 and pICC19, and pICC10 and pICC39, Int280 appears to interact with Tir with greater affinity than Int190. This implies that regions outside Int190 may be involved in interaction with the whole Tir polypeptide. Nevertheless, taken together, these results show that the region of intimin that spans the C-terminal 190 amino acids is capable of interacting with Tir. In the low-resolution structure of Int280 it can be seen that the D3–D4 junction comprises a small surface area. Therefore, it is unlikely that a truncation at this point will severely affect stability, whereas truncations elsewhere will probably compromise its stability. Localizing the Tir-binding region of intimin to the C-terminal 190 amino acids using gel overlay and yeast two-hybrid system assays defines the minimal functional fragment.
Solution structure of Int190
Using a combined perdeuteration/site-specific protonation and multi-dimensional NMR approach we were able to define the fold of the original 280-amino acid fragment, which is 30.1 kDa (Kelly et al., 1999). The shorter Int190 fragment is ∼20 kDa and its size falls within the applicability of standard multi-resonance NMR methods for determining highly defined structures. The DNA fragment encoding Int190 was sub-cloned into pET3a and overexpressed in BL21. However, Int190 was expressed at levels significantly lower than Int280. We hypothesized that this might be due to the presence of two hydrophobic amino acids (Phe1 and Phe2) at the N-terminus of the Int190 polypeptide. In order to try to improve expression and solubility we removed these two amino acids and cloned into pET3a a DNA fragment encoding Int188. The level of expression was vastly improved but Int188 concentrated into insoluble inclusion bodies. Accordingly, following centrifugation, the inclusion body material was precipitated, solubilized and refolded as described in Materials and methods.
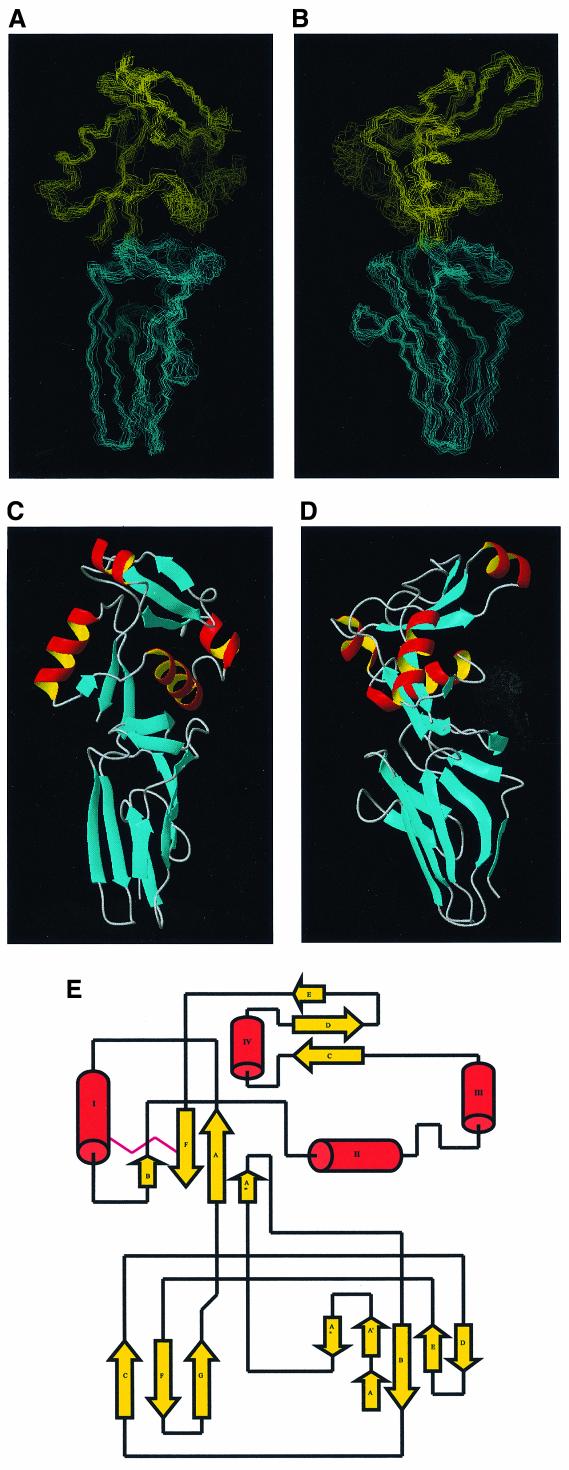
Comparison of the 1H-15N heteronuclear single quantum coherence (HSQC) NMR spectra for Int188 and Int280 indicates that Int188 is fully structured. Peaks corresponding to amides within the N-terminal 92 amino acids of Int280 are absent in the Int188 spectrum but the rest remain largely unchanged. 1H, 15N, 13C sequence-specific backbone and side-chain assignments were completed using the standard methodology (Bax et al., 1990; Grzesiek and Bax, 1992a,b; Kay et al., 1994; Muhandiram and Kay, 1994). Structures were calculated on the basis of 1226 nuclear Overhauser effect (NOE) distance, 115 H-bond distance and 307 dihedral angle restraints. The calculations used a hybrid torsion angle and Cartesian co-ordinate dynamics protocol, executed within the program X_PLOR (Nilges et al., 1988; Brünger, 1993). A final family of 15 structures (shown in Figure 3) were produced which contained no distance violations >0.2 Å. Int188 is ∼60 Å in length and comprises two intimate globular domains: D3 (residues 3–90) and D4 (residues 91–190, numbered according to Int190). The overall root-mean square deviations (r.m.s.d.) between the family and mean co-ordinate position are 0.7 ± 0.1 and 0.9 ± 0.1 Å for the backbone atoms of secondary structure element in D3 and D4, respectively. For all heavy atoms of the same region the r.m.s.d. values rise to 1.2 ± 0.1 and 1.4 ± 0.1 Å, respectively. Numerous contacts between D3 and D4 are observed that define the relative orientation of the two domains. The r.m.s.d. for all the backbone atoms of D3–D4 (residues 1–188) from the average structure is 1.5 ± 0.2 Å, indicating the relative orientation of these domains is well defined. The complete set of structural statistics is shown in Table II.
Fig. 3. (A) Cα traces representing the superimposition of the 15 refined Int188 structures. (B) Cα traces representing the superimposition of the 15 refined Int188 structures. The orientations of (A) and (B) are related by a 90° rotation. (C) Schematic representation of Int188 for the orientation displayed in (A). (D) Schematic representation of Int188 domains for the orientation displayed in (B). (E) A ‘flattened’ illustration highlighting the topology of Int188. Helices are represented as open tubes and β-strands as arrows.
Table II. Results from structure calculations for Int188.
| Statistic | <SA>a | |
|---|---|---|
| Restraints | ||
| medium and short range (1 < |i – j| < 5) | 771 | 0.030 ± 0.003 |
| long range (|i – j| > 4) | 455 | 0.025 ± 0.002 |
| hydrogen bonds | 151 | 0.027 ± 0.003 |
| dihedral angles | 307 | 0.94 ± 0.03 |
| Idealized geometry | ||
| bonds (Å) | 0.0024 ± 0.0003 | |
| angles (°) | 0.58 ± 0.009 | |
| improper angles (°) | 0.47 ± 0.02 | |
| Coordinate positionsb | ||
| backbone atoms for secondary structure residues in D3: | ||
| (5–8, 8–14, 20–23, 32–38, 43–48, 54–57, 61–65, 70–77, 82–87) | 0.7 ± 0.1c | |
| heavy atoms for secondary structure residues in D3: | ||
| (5–8, 8–14, 20–23, 32–38, 43–48, 54–57, 61–65, 70–77, 82–87) | 1.2 ± 0.1 | |
| backbone atoms for secondary structure residues in D4: | ||
| (92–96, 104–113, 122–132, 146–151, 162–167, 173–175, 186–190) | 0.9 ± 0.1 | |
| heavy atoms for secondary structure residues in D4: | ||
| (92–96, 104–113, 122–132, 146–151, 162–167, 173–175, 186–190) | 1.4 ± 0.1 | |
| backbone atoms for all residues 3–190 | 1.5 ± 0.2 | |
| all heavy atoms for all residues 3–190 | 1.9 ± 0.2 |
aThe average r.m.s.ds for the final 15 structures.
bNumbered according to Int190, see Figure 4C.
cThe average r.m.s.ds from the average structure.
The topology of Int188 is shown in Figure 3. D3 displays little sequence homology with known IgSF members, but belongs to the Type C set of the IgSF. Interestingly, D4 contains a unique feature that is not seen in mammalian IgSF domains. A prominent β-insertion (A′, A′′) between strands A and A′′′ extends a platform on top of D3 that contacts D4 and helps to define the relative orientation of the two domains. This feature was poorly defined in our earlier determination of the fold of Int280, but in the current study is now characterized by numerous representative β-sheet NOEs and is therefore well defined.
D4 comprises four helices that surround two anti-parallel β-sheets. The C-terminal strand is disulfide-bonded to helix I, and together with the N-terminus of D4 forms the two principal strands of the first sheet. Helix III protrudes from the main structure into the solvent and therefore was not observed in our low-resolution structure of Int280 (Kelly et al., 1999). Interestingly, this helix contains an unusual kink at residue 139, which is replaced by proline in other intimin types. Despite no significant sequence homology (i.e. <10%), the α/β topology of D4 is reminiscent of the C-type lectins domains (CTLDs). Seventy-seven structurally equivalent Cα atoms from D4 of intimin superimpose with a r.m.s.d. of 3.0 Å on the lectin domain from E-selectin (Graves et al., 1994). The CTLDs form a family of calcium-binding proteins responsible for cell-surface carbohydrate recognition (CRD) that includes animal cell-receptors and bacterial toxins. However, several variants on the CTLD theme have recently been characterized. One example of a CTLD that lacks calcium co-ordination but recognizes carbohydrate is the TSG-6 link module that binds haluronan (Kohda et al., 1996). Conversely, the type II antifreeze protein from sea raven, which does co-ordinate calcium, inhibits the formation of ice crystals, hypothesized to bind ice surfaces via the second β-sheet and calcium loop (Gronwald et al., 1998). Intriguingly, the CTLDs are also structurally related to proteins that are involved in protein recognition. These include CD94/NKG2 heterodimer, in which two CTL-like domains form an extensive flat surface involving the second β-sheet that is postulated to interact with the type 1b human leukocyte antigen (Boyington et al., 1999).
In CTLDs, the carbohydrate recognition site lies on the exposed face of the second β-sheet and an extensive loop between strands C and D (30 residues) in which co-ordinated calcium is directly involved in binding. No evidence for calcium binding is available for intimin and, moreover, intimin D4 lacks the extensive calcium binding loop, which is replaced by a six-residue helix (helix IV). Further differences between the CTL-like domain from intimin and archetypal CTLDs also exist. D4 from intimin contains a single disulfide bond whereas CTLDs retain several that are conserved. Intimin also contains a larger proportion of helical structure than known CTLDs and the relative orientations of these regions are subtly different. However, the similarity with CTLDs does raise the question of an intimin function involving carbohydrate recognition, but evidence has yet to be provided. Therefore, this remains highly speculative.
Structural comparison with invasin from Yersinia pseudotuberculosis
Intimin from EPEC and invasin from enteropatho genic Yersinia pseudotuberculosis are probably the best-characterized bacterial cell-adhesion paradigms. Research has now culminated in the high-resolution three- dimensional structure information on both proteins.
Intimin and invasin belong to a family of outer membrane proteins that mediate bacterial adherence. Members of this family of proteins are ∼900 amino acids in length and possess highly similar N-termini: >36% identity exists within the first 500 amino acids, part of which is hypothesized to be sequestered within the bacterial outer membrane. Despite being highly related, these proteins perform remarkably divergent tasks, and this is illustrated by the marked difference in pathogenesis between the organisms. Invasin causes invasion and translocation of the bacterium across the intestinal epithelium deeper into host tissue. Intimin also promotes adherence to epithelial cells, but not internalization; instead, it induces the A/E lesion. Invasin recognizes members of the integrin super-family from mammalian cells in order to facilitate bacterial adherence to and invasion of host cells. Moreover, two aspartic acid residues, reminiscent of the integrin-binding and synergy regions of fibronectin, are required for recognition (Hamburger et al., 1999). It has also been established that homo-multimerization of invasin from Y.pseudotuberculosis facilitates receptor clustering, tight adherence and the subsequent invasion signal (Dersch and Isberg, 1999). In contrast, intimin binds the Tir, which is introduced into the host cell membrane by the bacterium via a type III protein secretion/translocation system. For both intimin and invasin a C-terminal fragment of ∼190 amino acids is sufficient for function in which no significant sequence homology is present (<10%).
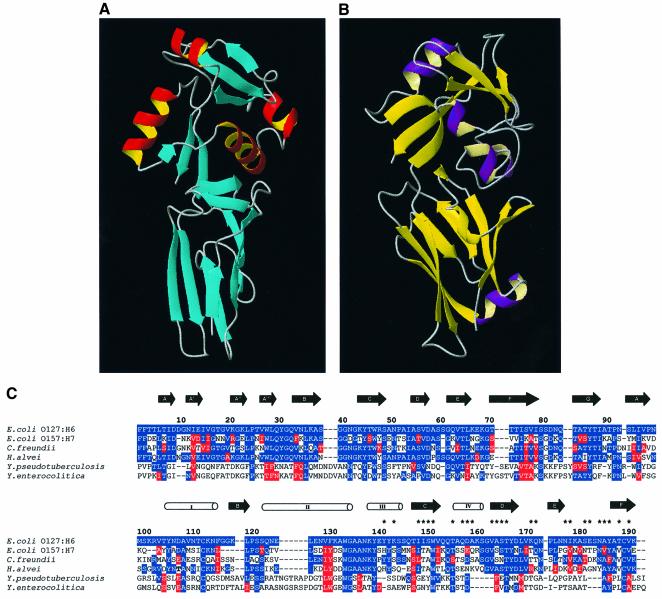
Remarkable likenesses can be revealed between the structures of intimin and invasin (Figure 4). Hamburger et al. determined the crystal structure of an extracellular portion of invasin (Inv497) (Hamburger et al., 1999). Inv497 is ∼180 Å in length and composed of five distinct domains (D1, D2, D3, D4 and D5), the first four of which resemble eukaryotic members of the IgSF. D3–D5 in invasin are analogous to the domains of Int280 (280 amino acid C-terminal fragment of intimin), which was revealed by earlier NMR studies (Kelly et al., 1998, 1999). It has also been reported that sequence alignment indicates the existence of a further IgSF domain within intimin. Also, secondary structure algorithms suggest that residues 445–558 contain a high β-sheet content but sequence alignment with known Ig domains (maximum of 20% identity, 25% conservation) falls below the cut-off for definitive assignment of an Ig-like fold. Therefore the extracellular portion of intimin contains at least four Ig-like domains arranged in a rod similar to the C-terminus of invasin, which conveys a highly accessible ‘adhesive tip’ to the target cell. The tip in both intimin and invasin is composed of the C-terminal 190 amino acids, which forms a super-domain. Despite no significant sequence homology, Int190 and D4/D5 from invasin are similar: 73 equivalent Cα atoms superimpose with an r.m.s.d. of 2.9 Å for D3 in intimin with D4 in invasin and 77 equivalent Cα atoms superimpose with an r.m.s.d. of 3.5 Å for in D4 intimin with D5 in invasin. The most notable differences occur within the divergent C-terminal domain (D4 in intimin and D5 in invasin); these include two additional helices in Int190 (helix III and IV), the positions of some of the secondary structure elements and the relative orientations of C-terminal domains. Despite these differences, the C-terminal 100 amino acids of both intimin and invasin possess a folding topology related to that of the CTLDs.
Fig. 4. The structural comparison of Int188 topology with D4/D5 from Invasin (Hamburger et al., 1999). (A) Schematic representation of Int188. Helices are shown as red tubes and strands as blue arrows. (B) Schematic representation of D4/D5 from invasin. Helices are shown as purple tubes, strands as yellow arrows. (C) Sequence alignment and topology for Int188 and invasin. The approximate location of secondary structure elements is also indicated; helices are delineated as open tubes and β-strands as black arrows. The amino acids positions are shown for Int190 from EPEC O127:H6 and correspond to residues 750–939 in full-length intimin. The residues highlighted in blue and red represent identity and conservation with the EPEC O127:H6 sequence, respectively. Intimins from E.coli O127:H6, E.coli O26:H- and E.coli O157:H7 have been immunologically categorized into types α, β and γ, respectively. The two invasin sequences, Y.pseudotuberculosis and Yersinia enterocolitica, are aligned on the basis of the structural superimposition provided by DALI. Asterisks represent amide resonances that are significantly perturbed upon the addition of Tir55 peptide.
Considering the limited sequence similarity between Int190 and D4–D5 of invasin (∼10% identity), and their different binding specificities and biological activities, it is remarkable that these proteins adopt very similar structures. These findings raise intriguing questions regarding the origin and the parallel evolution of these two bacterial adhesion molecules.
Identification of the Tir-binding site within intimin
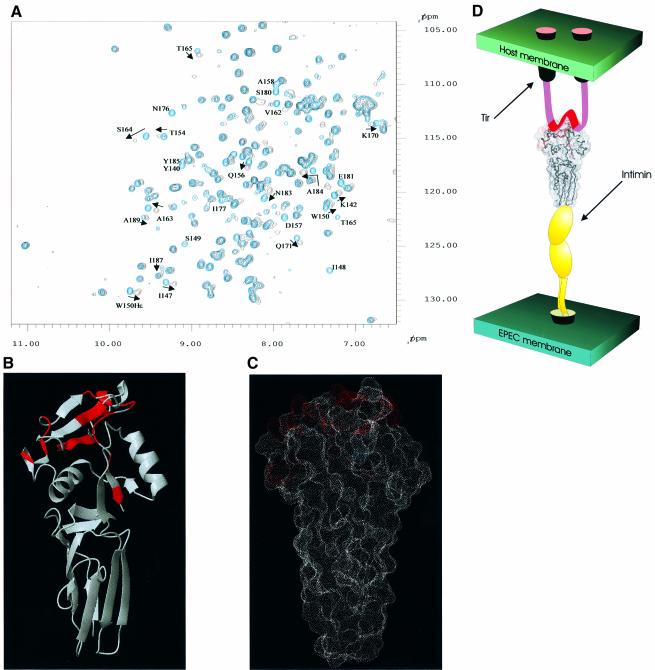
Recently, the intimin-binding region of Tir has been localized to a central region encompassing a 55 amino acid motif between two putative membrane-spanning helices (Kenny et al., 1997; de Grado et al., 1999; Hartland et al., 1999; Kenny, 1999). Analysis of line-widths and chemical shifts for Int188 amide resonances in the presence of the Tir55 peptide facilitated mapping of the Tir-binding surface. Amides, which exhibit large chemical shift changes and/or line broadening in the presence of Tir55, reveal residues likely to be directly involved in binding. A comparison of the 1H-15N HSQC spectra with and without saturating amounts of Tir55 is shown in Figure 5. A number of amide resonances move or broaden whilst the majority of the spectrum remains unchanged, which is indicative of a specific complex between Int188 and Tir55. The affected residues are concentrated within a region located in the C-terminal domain, D4, and are illustrated clearly in Figure 5, in which they are coloured red on schematic and solvent accessibility representations of Int188. Residues involved include Y140, K142, I147, I148, S149, W150, T154, Q156, D157, A158, V162, A163, S164, T165, K170, Q171, N176, I177, S180, E181, N183, A184, Y185, T187 and V189. For clarity these positions are numbered according to Int190 and marked by asterisks in the sequence alignment figure (Figure 4C). Principally, this region is located at the tip of the structure and forms a long thin surface that is centred on the upper, solvent exposed surface of the second β-sheet (strands C, D and E). This putative binding site is highly localized, covering an area on intimin that measures ∼20 × 8 Å. This data is also completely reproducible with Int280 (data not shown), which suggests that residues downstream of Int190 are unlikely to be involved in Tir55 binding. The highly localized nature of this region with the three-dimensional structure suggests that the portion of Tir in contact with intimin is likely to be shorter than the 55 amino acids currently proposed. Based on the structure, we hypothesize that the number of amino acid residues of Tir in contact with intimin may be as few as five if the conformation is completely extended, or as many as 14 if helical.
Fig. 5. (A) The 1H-15N HSQC NMR spectrum for Int188 (blue) with the identical region of the 1H-15N HSQC NMR spectrum for Int188 with Tir55 peptide overlaid (black). Peaks corresponding to amides that are affected upon addition of Tir55 are labelled with their residue number (for consistency these are numbered according to Int190). (B) Schematic representation of the structure of Int188. Residues shown in red indicate chemical shift/line-width perturbation in the presence of Tir55, namely Y140, K142, I147, I148, S149, W150, T154, Q156, D157, A158, V162, A163, S164, T165, K170, Q171, N176, I177, S180, E181, N183, A184, Y185, T187 and V189. These data are also summarized in Figure 3A. The data illustrate a possible binding site for Tir55, which is composed of highly concerted patch of residues in D4. For clarity Int188 is rotated by 180° with respect to Figure 3A. (C) A dot representation of the solvent-accessible surface for Int188. The residues that show chemical shift/line-width perturbation in the presence of Tir55 are shown in red. The data illustrate a possible binding site for Tir55, which is composed of a highly concerted patch of residues in D4. (D) Schematic representation of the quaternary structure of the entire extracellular region of intimin in complex with Tir peptide. IGSF domains D1 and D2 are shown in yellow. The Tir-binding fragment of intimin (i.e. Int190, D3 and D4) is illustrated by the space filling representation shown in (B). Tir is shown schematically: the two predicted transmembrane helices are shown as tubes and the intimin-binding peptide, which is manually docked onto the binding site, as a red ribbon.
Site-directed mutagenesis of intimin
Interestingly, based on the structural alignment provided by DALI (Figure 4), W150 (within Int190) is highly conserved in all intimin types, invasin and CTLDs. Although W150 is buried, it is located just below surface residues involved in binding and may therefore provide a structural scaffold for the Tir-binding site. One-dimensional 1H NMR spectra of the W150A mutant (data not shown) indicate that this protein is stable and fully folded, and therefore any effects of the modification are entirely due to localized structural effects and not global destabilization. Moreover, W150 shows chemical shift and line-width perturbations of its backbone and side-chain amide in the presence of Tir. This residue was therefore chosen for mutation and extensive functional analysis, in terms of intimin expression, A/E lesion formation and Tir binding. This residue in the intact intimin (W899) was subjected to site-directed mutagenesis using the cloned eae encoding intimin (pCVD438; Donnenberg and Kaper, 1991), generating plasmid pICC53 (Table I). The site-directed mutated eae clone was introduced into the eae mutant EPEC strain CVD206 (Donnenberg and Kaper, 1991), and its biological activities were characterized in terms of intimin expression, the ability to support A/E lesion formation and binding to Tir.
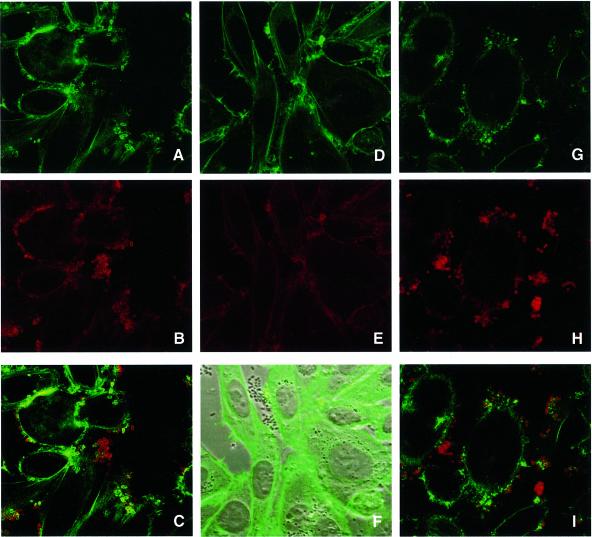
Western blots of whole bacterial cell extracts (data not shown) and immunofluorescence microscopy of infected HEp-2 cells confirmed that the mutation introduced into the cloned eae gene did not affect intimin expression or association with the bacterial outer membrane (Figure 6). The ability of CVD206(pICC53) to induce A/E lesions on cultured HEp-2 cells was investigated using the fluorescent actin staining (FAS) test as a marker for lesion formation. Replacement of W899 with alanine (pICC53) did not affect surface expression of intimin but resulted in adherent bacteria that are unable to initiate host cytoskeletal rearrangements (Figure 6). CVD206(pCVD438) produced both FAS and intimin positive staining while CVD206 alone gave a double-negative reaction (Figure 6).
Fig. 6. FAS reaction (A, D, G), intimin staining (B, E, H) and overlaid images (C, F and I) of infected HEp-2 cells. CVD206(pCVD438) (A–C) showed both FAS and intimin staining. No staining was observed using strain CVD206 alone (D, E) although adhering bacteria could be observed by phase contrast (F). Substitution of W899 in strain CVD206(pICC54) resulted in FAS negative staining although surface intimin expression was not affected (G–I).
To ascertain whether or not this FAS-negative phenotype was due to impaired binding to Tir, gel overlay and yeast two-hybrid system binding assays were employed. In contrast to the fusion protein MBP–Int190, which showed a clear interaction with Tir-M in gel overlay experiments (Figure 7A), no interaction could be detected with MBP–Int190 or MBP–Int280 containing the modification (MBP–Int190W150A, Figure 7A; MBP–Int280W240A, data not shown). For the yeast two-hybrid system, DNA encoding Int190W150A and Int280W240A were cloned into pGAD424 (plasmid pICC44 and pICC62, respectively), while plasmid pICC10, expressing Tir, was used as the bait. Replica plating onto selective media produced vigorously growing colonies in yeast stains co-transformed with pICC10 and pICC39 (encoding the wild-type Int190). However, co-transformation of pICC10 with pICC44 (encoding Int190W150A) or pICC62 (encoding Int280W240A) did not support growth on the selective media. Measuring the β-galactosidase activities in these strains revealed background levels (Figure 7B), while strains harbouring pICC10 and pICC39 produced elevated levels of β-galactosidase activity (Figure 7B).
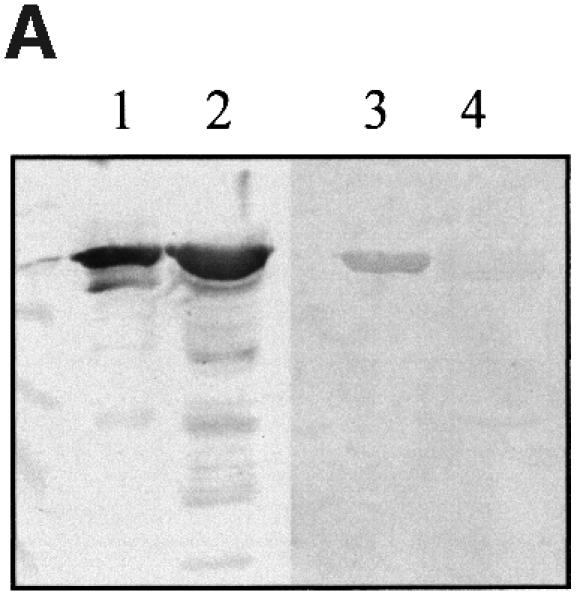
Fig. 7. (A) Detection of Int190–Tir interactions using gel overlays. Western blots of MBP–Int mutants were reacted with a rabbit MBP antiserum or overlayed with Tir-M. Similar levels of MBP–Int190 (lane 1) and MBP–Int190W150A (lane 2) fusion proteins were detected with polyclonal antiserum, while Tir-M only bound to MBP–Int190 (lane 3). (B) Detection of Int–Tir interactions using the yeast two hybrid system. β-galactosidase assays showing a 7-fold increase in enzymatic activity in strains co-expressing the whole Tir polypeptide and Int190 compared with Tir–Int190A150, Tir–Int280A240 and single plasmid transformants.
The results taken together are entirely consistent and provide further evidence for the conclusions of the NMR titration experiments. In summary, altering W150 in Int190 or W240 in Int280 (W899 in intact intimin) prevents EPEC from forming actin pedestals by specifically disrupting the intimin–Tir interaction. This mutation lies proximal to the Tir-recognition region, which comprises a 20 × 8 Å patch of residues within the C-terminal 100 amino acids (D4).
This work represents the first report of high-resolution structure information for intimin. We also provide compelling evidence for a Tir-binding site at the extremity of intimin, which extends from the bacterial membrane. Figure 5D illustrates a schematic model for the structure of the intimin–Tir peptide complex. The results provide a useful insight that will aid the development of preventative therapies against EPEC.
Materials and methods
Bacterial strains and plasmids
Bacterial strains used in this study included wild-type EPEC strain E2348/69 (O127:H6), eae deletion mutant derivative CVD206 (Donnenberg and Kaper, 1991) and E.coli strains TG1 and BL21. Bacterial strains were grown in L-broth. Media were supplemented with 50 µg/ml kanamycin or 100 µg/ml ampicillin where appropriate. The plasmids are listed in Table I.
Preparation of MBP–Int280 derivatives
DNA segments encoding Int190, Int146, Int235, Int134, IntD1&2 and IntD2 (Figure 1; Table I) were amplified by PCR (primers are listed in Table III) using E2348/69 DNA as template (Frankel et al., 1995). The truncated Int280 fragments were cloned into pMal-c vector in E.coli TG1 and the MBP–Int280 derivatives purified and analysed by 12% SDS–PAGE as described previously (Frankel et al., 1995).
Table III. List of primers.
| Primer | Sequence |
|---|---|
| 280-Fa | 5′-GGAATTCATTACTGAGATTAAGGCT |
| 190-F | 5′-GGAATTCTTTTTTACAACGCTTACAATT |
| 190-Rb | 5′-CGGGATCCTTATTTTACACAAGTGGC |
| 188-F | 5′-AAACATATGACGACGCTTACAATTGATG |
| 146-F | 5′-GGAATTCGGTAATATTGAAATTGTTGGAACC |
| 146-R | 5′-CGGGATCCTTAAGCTGTTTGTTGTACCCATGAAAT |
| D2+D3-F | 5′-GGAATTCATTACTGAGATTAAGGCT |
| D2+D3-R | 5′-CGGGATCCTAATGCAGCCCCCCATGC |
| D4-F | 5′-GGAATTCCTTACAATTGATGACGGT |
| D4-R | 5′-CGGGATCCTAATGCAGCCCCCCATGC |
aF, forward primer.
bR, reverse primer.
Gel overlays
Purified MBP–Int280 derivatives or MBP alone were separated by SDS–PAGE, blotted onto a nitrocellulose membrane and blocked with 10% skim-milk in phosphate-buffered saline (PBS), 0.1% Tween-20 overnight as described previously (Frankel et al., 1995; Hartland et al., 1999). His-Tir-M was purified as described previously (Hartland et al., 1999). The nitrocellulose membranes were reacted with 5 µg/ml of His-Tir-M in PBS, 0.1% Tween-20 for 2 h and washed twice for 5 min in PBS, 0.1% Tween-20. His-Tir-M binding to the different MBP–Int280 derivatives was detected with anti-His antiserum (1:2000 for 2 h) and then anti-rabbit antibodies conjugated to alkaline phosphatase (1:2000 for 1 h). Binding of the individual MBP–Int280 derivatives to immobilized Tir-M was performed as described previously (Hartland et al., 1999).
Yeast two-hybrid system
The yeast two-hybrid host PJ69-4A (MATa trp1-901 leu2-3,112 ura3-52 his3-200 gal4D gal80D LYS2::GAL1-HIS3 GAL2-ADE1 met2::GAL7-lacZ) was selected for use in this study, which confers the advantage of three independent reporter genes under the control of three different GAL promoters (James et al., 1996). DNA fragments encoding the Int280 derivatives and Tir derivatives were cloned, following PCR amplification (Table I), into the ADH1 driven fusion vectors pGBT9 (carrying the GAL4-binding domain, BD) and pGAD424 (carrying the GAL4 activation domain, AD) respectively (Clontech). The constructs were used to transform PJ69-4A with initial selection for the plasmid encoded TRP1 and LEU2 genes (Hartland et al., 1999). The resulting transformants were replica plated onto 3-aminotriazole-containing medium to select for the HIS3 reporter, and onto SC minus Trp, Leu, Ade to select for the ADE2 reporter. The function of the lacZ reporter was quantified in cell extracts by assaying for β-galactosidase activity using O-nitrophenyl β-d-galactopyranoside as a substrate (Miller, 1972).
Site-directed mutagenesis
Site-directed mutagenesis of the eae gene was performed using the QuickChange Site-directed mutagenesis kit (Stratagene) following the manufacturer’s instructions, using double-stranded pCVD438 as template. Complementary mutagenesis oligonucleotide pairs incorporating single amino acid substitutions are as follows:
Sense oligonucleotides: 5′-GACTATAATTTCAGCTGTACAACAAACAGC (A150).
Antisense oligonucleotides: 5′-GCTGTTTGTTGTACAGCTGAAATTATAGTC (A150).
Mutated plasmid containing staggered nicks was generated by extension of primers annealed to opposite strands of the denatured plasmid by temperature cycling (1 cycle at 95°C for 30 s; then 16 cycles at 95°C for 30 s, 55°C for 1 min, 68°C for 18 min) in the presence of the high fidelity Pfu DNA polymerase. Synthesized DNA containing the desired mutation was selected from the original DNA template by incubation with Dpn1 at 37°C for 1 h, on the basis that dam-methylated parental DNA template would be susceptible to digestion whereas the newly synthesized unmethylated mutated plasmid would not. Nicks in the plasmid were repaired following transformation of 1 µl of the synthesized products into competent E.coli XL1-Blue cells.
Chloramphenicol-resistant transformants were randomly selected and inoculated to overnight L-broths for preparation of plasmid mini-preps (Qiagen). Correct incorporation of each mutation was monitored by DNA sequencing using an automated DNA sequencer. Mutated pCVD438 derivatives were also used as PCR templates for subcloning the DNA fragments encoding Int190 and Int280 into pMAL-c (pICC52 and pICC63, respectively) and pGAD424 (pICC44 and pICC62, respectively) for expression as MBP fusion in E.coli and for the yeast two hybrid-system, respectively (Table I).
Mutated plasmid was transformed to competent CVD206 cells (eae–; Donnenberg and Kaper, 1991) for analysis of phenotypic effects. The mutated pCVD438 derivative was also used as a PCR template for subcloning the DNA fragments encoding Int190 into pMAL-c2 (pICC52), and pGAD424 (pICC44) for expression as MBP fusion in E.coli and for the yeast two hybrid-system, respectively (Table I).
Detection of intimin expression by western blotting and FAS
Expression of the intimin derivatives was determined by western blotting. Briefly, stationary L-broth cultures were diluted 1:100 in Dulbecco’s modified Eagle’s medium (DMEM) and incubated for 3 h at 37°C. An equivalent of an OD600 of 0.5 was loaded onto 7.5% SDS–PAGE as described previously (Adu-Bobie et al., 1998). The electrophoresed polypeptides were transferred to a nitrocellulose membrane and immunodetection of intimin was performed using an intimin-α antiserum, diluted 1:500. FAS test (using FITC–phalloidin) (Knutton et al., 1989) and intimin immunostaining (using anti-Int280 and TRITC-conjugated anti-rabbit antibodies) were employed to detect A/E lesion formation and intimin expression on infected HEp-2 cells by confocal microscopy.
Recombinant expression of Int190 and Int188
The 570 bp (encoding Int190) and 564 bp (encoding Int188) fragments of the eae gene from EPEC E2348/69 (and a TAA stop condon) were cloned, following PCR amplification (the primer sequences are listed in Table III), into the prokaryotic expression vector pET3a using restriction sites NdeI and BamHI, generating plasmid pICC54 and pICC59. The plasmid was transformed into E.coli strain BL21 containing pLysS and grown in Luria–Bertani broth supplemented with 100 µg/ml ampicillin and 30 µg/ml chloramphenicol. For NMR, the cultures were grown in minimal media containing 0.07% 15NH4Cl and 0.4% [13C]glucose supplemented with 100 µg/ml ampicillin and 30 µg/ml chloramphenicol. Cultures were grown until mid-log phase, and then protein expression induced by the addition of 0.4 mM isopropyl-β-d-thiogalactopyranoside (IPTG). Growth was continued overnight at 30°C after which the cultures were harvested by centrifugation. Since Int188 was found to be insoluble, the cell pellets were resuspended in lysis buffer [20 mM Tris pH 8.0, 1 mM EDTA, 0.5 mM phenylmethylsulfonyl fluoride (PMSF)], sonicated and centrifuged to pellet the insoluble and inclusion body material. The supernatant fraction was discarded.
Refolding and purification of Int188
The pellets were washed (50 mM Tris pH 8.0), and spun for 15 min at 20 000 g. This was repeated twice, firstly washing in Triton buffer (50 mM Tris pH 8.0, 1% Triton), and secondly guanidine (50 mM Tris pH 8.0, 0.5 mM guanidine). Following the final wash, the inclusion body pellets were dissolved in 50 ml of solubilization buffer (50 mM Tris pH 8.0, 1 mM EDTA, 6 M guanidine) by stirring overnight at room temperature. The solubilized inclusion bodies were then passed slowly by peristaltic pump into 1 l of refolding buffer [50 mM Tris pH 8.0, 1 mM EDTA, 1 M arginine, 1 mM glutathione (reduced), 0.5 mM glutathione (oxidized)] at 4°C overnight, stirring. The refolded protein was then concentrated to 50 ml using an amicon apparatus with a 10 kDa cut-off membrane. This was then dialysed against 50 mM Tris pH 8.0, 1 mM EDTA. The refolded protein was purified by cation exchange chromatography using sulfopropyl Sepharose in 20 mM sodium acetate pH 5.2. Pure intimin was eluted in 15% NaCl, the samples pooled, dialysed into 20 mM sodium acetate pH 5.2 and concentrated for NMR.
NMR spectroscopy
For NMR spectroscopy, 1 mM samples of [13C/15N]Int188 were prepared in 0.5 ml sodium acetate buffer at pH 5.2. Approximately 10% v/v D2O was added to the sample. All NMR spectra were recorded at 500 MHz proton frequency on a four-channel Bruker DRX500 spectrometer equipped with a z-shielded gradient triple resonance probe. The temperature was maintained at 310 K throughout the experiments. The sequence-specific backbone 1HN, 15N, 13Cα, 13Cβ and 13C′ assignments were completed using HNCA, HN(CO)CA, CBCA(CO)NH and HBHA(CO)NH experiments (Grzesiek and Bax, 1992a,b; Kay et al., 1994; Muhandiram and Kay, 1994). The secondary structure and side chain assignments were determined using 1H-15N nuclear Overhauser (NOESY) HSQC and HCCH-total correlation (TOCSY) spectroscopy (Bax et al., 1990). Distance restraints for the structure calculation were obtained from 1H-15N NOESY HSQC (Marion et al, 1989; Norwood et al., 1990) (80 ms mixing time), 1H-13C NOESY HSQC (80 ms mixing time) and double 13C-edited HMQC NOESY HSQC (Vuister et al., 1993) (80 ms mixing time) spectra recorded on a fully exchanged D2O sample. All the experiments used gradients for coherence selection together with the sensitivity enhancement protocol (Kay et al., 1994).
Structure calculation
NOE cross peak intensities were measured at 80 ms mixing time. The structures were calculated from random starting coordinates on the basis of 1528 distance restraints, comprising 771 short range NOEs (residue i to residue 1 < i < 4), 455 long range NOEs (residue i to residue i >4), 151 hydrogen bonds and one disulfide bond. The calculation also included 307 dihedral angle restraints, composed of 112 φ and 111 ψ angles. A hybrid torsion angle and Cartesian co-ordinate dynamics protocol was executed within the program X_PLOR (Nilges et al., 1988; Brünger, 1993). The distance restraints were calibrated internally using known distances. NOEs observed at 80 ms mixing time were placed in three categories on the basis of estimated cross peak intensities: strong (<2.8 Å), medium (<3.5 Å) and weak (<5.0 Å). The dihedral angle restraints were estimated using the backbone torsion angle prediction package TALOS (Cornilescu et al., 1999). No distance violation >0.2 Å and no dihedral violation >4.0° was found.
NMR-binding studies
The 55 amino acid peptide encompassing the intimin-binding region of Tir was synthesized using Fmoc chemistry on a Perseptive Biosystems 9050+ automatic peptide synthesizer with customized protocols. Coupling was performed using HBTU (benzotriazol-N,N,N′,N′-tetramethyluronium hexafluorophosphate) chemistry. After removal of the final Fmoc group, the resin was washed with methanol and dichloromethane and dried under high vacuum. The peptide was released from the resin (500 mg) during 2 h with a cocktail (50 ml) of trifluoroacetic acid (TFA) 92.5%, phenol 2%, water 2%, ethanedithiol 2%, anisole 1% and triisobutylsilane 1%. Characterization was performed using reversed-phase HPLC on a C18 silica wide-pore column (15 cm × 2.1 mm) with an elution gradient of 10–50% acetonitrile containing 0.1% TFA over 40 min. Further purification where necessary was performed on a 25 cm × 22 mm column using the same conditions. The molecular weight was confirmed by electrospray mass spectrometry. The final peptide sample was assessed to be >75% pure. Two 0.5 mM Int188 samples were prepared in acetate buffer at pH 5.2; one was left untouched and to the other freeze-dried Tir55 was added to obtain a 10:1 Tir55:Int188 molar ratio. 1H-15N HSQC spectra were recorded on the two starting samples and on sequential mixtures in order to follow the titration of the amide resonances, which facilitated their assignment. The experiment was repeated twice: again with Int188 and once with Int280.
Atomic co-ordinates
The atomic co-ordinates have been deposited in the Brookhaven Protein Data Bank and can be obtained from S.Matthews (s.j.matthews@ic.ac.uk).
Acknowledgments
Acknowledgements
We are grateful to Dr Xuemei Yuan for help with constructing the figures and Chris Hale for the microscopy work. S.M. is indebted for the financial support of The Wellcome Trust and the BBSRC. G.D. and G.F. would also like to acknowledge the support of The Wellcome Trust. In addition, G.D. would like to acknowledge financial support from Action Research.
References
- Adu-Bobie J., Frankel,G., Bain,C., Goncaleves,A.G., Trabulsi,L.R., Douce,G., Knutton,S. and Dougan,G. (1998) Detection of intimin α, β, γ and δ, four intimin derivatives expressed by attaching and effacing microbial pathogens. J. Clin. Microbiol., 36, 662–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bax A., Clore,G.M. and Gronenborn,A.M.J. (1990) 1H-1H correlation via isotropic mixing of 13C magnetization, a new three-dimensional approach for assigning 1H and 13C spectra of 13C enriched proteins. Magn. Res., 88, 425–431. [Google Scholar]
- Boyington J.G., Riaz,A.N., Patamawam,A., Coligan,J.E., Brook,A.G. and Sun,P.D. (1999) Structure of the CD94 reveals a novel C-type lectin fold: implications for the NK cell-associated CD94/NKG2 receptors. Immunity, 10, 75–82. [DOI] [PubMed] [Google Scholar]
- Brünger A.T. (1993) X_PLOR Manual. Version 3.1. Yale University Press, New Haven, CT. [Google Scholar]
- Cornilescu G., Delaglio,F. and Bax,A. (1999) Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR, 13, 289–302. [DOI] [PubMed] [Google Scholar]
- de Grado M., Abe,A., Gauthier,A., Steele-Mortimer,O., DeVinney,R. and Finlay,B.B. (1999) Identification of the intimin binding domain of Tir of enteropathogenic Escherichia coli.Cell. Microbiol., 1, 7–18. [DOI] [PubMed] [Google Scholar]
- Deibel C., Kramer,S., Chakraborty,T. and Ebel,F. (1998) EspE, a novel secreted protein of attaching and effacing bacteria, is directly translocated into infected host cells, where it appears as a tyrosine-phosphorylated 90 kDa protein. Mol. Microbiol., 28, 463–474. [DOI] [PubMed] [Google Scholar]
- Dersch P. and Isberg,R.R. (1999) A region of Yersinia pseudo tuberculosis invasin protein enhance integrin-mediated uptake into mammalian cells and promotes self-association. EMBO J., 18, 1199–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnenberg M.S. and Kaper,J.B. (1991) Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect. Immun., 59, 4310–4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnenberg M.S., Tacket,C.O., James,S.P., Losonsky,G., Nataro,J.P., Wasserman,S.S., Kaper,J.B. and Levine,M.M. (1993a) Role of the eaeA gene in experimental enteropathogenic Escherichia coli infection. J. Clin. Invest., 92, 1412–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnenberg M.S., Tzipori,S., McKee,M.L., O’Brien,A.D., Alroy,J. and Kaper,J.B. (1993b) The role of the eae gene of enterohemorrhagic Escherichia coli in intimate attachment in vitro and in a porcine model. J. Clin. Invest., 92, 1418–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnenberg M.S., Yu,J. and Kaper,J.B. (1993c) A second chromosomal gene necessary for intimate attachment of enteropathogenic Escherichia coli to epithelial cells. J. Bacteriol., 175, 4670–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebel F., Podzadel,T., Rohde,M., Kresse,A.U., Kramer,S., Deibel,C., Guzman,C.A. and Chakraborty,T. (1998) Initial binding of Shiga toxin-producing Escherichia coli to host cells and subsequent induction of actin rearrangements depend on filamentous EspA-containing surface appendages. Mol. Microbiol., 30, 147–161. [DOI] [PubMed] [Google Scholar]
- Frankel G., Candy,D.C., Everest,P. and Dougan,G. (1994) Characterization of the C-terminal domains of intimin-like proteins of enteropathogenic and enterohemorrhagic Escherichia coli, Citrobacter freundii and Hafnia alvei. Infect. Immun., 62, 1835–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel G., Candy,D.C., Fabiani,E., Adu-Bobie,J., Gil,S., Novakova,M., Phillips,A.D. and Dougan,G. (1995) Molecular characterization of a carboxy-terminal eukaryotic-cell-binding domain of intimin from enteropathogenic Escherichia coli.Infect. Immun., 63, 4323–4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel G., Lider,O., Hershkoviz,R., Mould,A.P., Kachalsky,S.G., Candy,D.C.A., Cahalon,L., Humphries,M.J. and Dougan,G. (1996) The cell-binding domain of intimin from enteropathogenic Escherichia coli binds to β1 integrins. J. Biol. Chem., 271, 20359–20364. [DOI] [PubMed] [Google Scholar]
- Frankel G., Phillips,A.D., Rosenshine,I., Dougan,G., Kaper,J.B. and Knutton,S. (1998a) Enteropathogenic and enterohaemorrhagic Escherichia coli: more subversive elements. Mol. Microbiol., 30, 911–921. [DOI] [PubMed] [Google Scholar]
- Frankel G., Philips,A.D., Novakova,M., Batchelor,M., Hicks,S. and Dougan,G. (1998b) Generation of Escherichia coli intimin-derivatives with differing biological activities using site-directed mutagenesis of the intimin C-terminus domain. Mol. Microbiol., 29, 559–570. [DOI] [PubMed] [Google Scholar]
- Graves B.J. et al. (1994). Insight into e-selectin ligand interaction from the crystal-structure and mutagenesis of the lec EGF domains. Nature, 367, 532–538. [DOI] [PubMed] [Google Scholar]
- Gronwald W., Loewen,M.C., Lix,B., Daugualis,A.J., Sonnichsen,F.D., Davies,P.L. and Sykes,B.D. (1998) The solution structure of type II antifreeze protein reveals a new member of the lectin family. Biochemistry, 37, 4712–4721. [DOI] [PubMed] [Google Scholar]
- Grzesiek S. and Bax,A. (1992a) An efficient experiment for sequential backbone assignment of medium-sized isotopically enriched proteins. J. Magn. Reson., 99, 201–207. [Google Scholar]
- Grzesiek S. and Bax,A. (1992b) Improved 3D triple resonance NMR techniques applied to a 31 kDa protein. J. Magn. Reson., 96, 432–440. [Google Scholar]
- Hamburger Z.A., Brown,M.S., Isberg,R.R. and Bjorkman,P.J. (1999) Crystal structure of invasin: a bacterial integrin-binding protein. Science, 286, 291–295. [DOI] [PubMed] [Google Scholar]
- Hartland E.L., Batchelor,M., Delahay,R.M., Hale,C., Matthews,S., Dougan,G., Knutton,S., Connerton,I. and Frankel,G. (1999) Binding of intimin from enteropathogenic Escherichia coli to Tir and to host cells. Mol. Microbiol., 32, 151–158. [DOI] [PubMed] [Google Scholar]
- Hicks S., Frankel,G., Kaper,J.B., Dougan,G. and Phillips,A.D. (1998) Role of intimin and bundle foming pili in enteropathgenic Escherichia coli adhesion to paediatric intestine in vitro.Infect. Immun., 66, 1570–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins L.M., Frankel,G., Connerton,I., Goncalves,N.S., Dougan,G. and MacDonald,T.T. (1999a) Role of bacterial intimin in colonic hyperplasia and inflammation. Science, 285, 588–591. [DOI] [PubMed] [Google Scholar]
- Higgins L.M., Frankel,G., Douce,G., Dougan,G. and MacDonald,T.T. (1999b) Citrobacter rodentium infection in mice elicits a mucosal Th1 cytokine response and lesions similar to those in murine inflammatory bowel disease. Infect. Immun., 67, 3031–3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isberg R.R., Voorhis,D.L. and Falkow,S. (1987) Identification of invasin: a protein that allows enteric bacteria to penetrate cultured mammalian cells. Cell, 50, 769–778. [DOI] [PubMed] [Google Scholar]
- James P., Halladay,J. and Craig,E.A. (1996) Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics, 144, 1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis K.G. and Kaper,J.B. (1996) Secretion of extracellular proteins by enterohemorrhagic Escherichia coli via a putative type III secretion system. Infect. Immun., 64, 4826–4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis K.G., Giron,J.A., Jerse,A.E., McDaniel,T.K., Donnenberg,M.S. and Kaper,J.B. (1995) Enteropathogenic Escherichia coli contains a putative type III secretion system necessary for the export of proteins involved in attaching and effacing lesion formation. Proc. Natl Acad. Sci. USA, 92, 7996–8000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerse A.E., Yu,J., Tall,B.D. and Kaper,J.B. (1990) A genetic locus of enteropathogenic Escherichia coli necessary for the production of attaching and effacing lesions on tissue culture cells. Proc. Natl Acad. Sci. USA, 87, 7839–7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay L.E., Xu,G.Y. and Yamazaki,T. (1994) Enhanced-sensitivity triple-resonance spectroscopy with minimal water saturation. J. Magn. Reson., 109, 129–133. [Google Scholar]
- Kelly G., Prasannan,S., Daniell,S., Frankel,G., Dougan,G., Connerton,I. and Matthews,S. (1998) Sequential assignment of the triple labelled 30.1 kDa cell adhesion domain of intimin from enteropathogenic E.coli.J. Biomol. NMR, 12, 189–191. [DOI] [PubMed] [Google Scholar]
- Kelly G., Prasannan,S., Daniell,S., Fleming,K., Frankel,G., Dougan,G., Connerton,I. and Matthews,S. (1999) Structure of the cell-adhesion fragment of intimin from enteropathogenic Escherichia coli.Nature Struct. Biol., 6, 313–318. [DOI] [PubMed] [Google Scholar]
- Kenny B. (1999) Phosphorylation of tyrosine 474 of the enteropathogenic Escherichia coli (EPEC) Tir receptor molecule is essential for actin nucleating activity and is preceded by additional host modifications. Mol. Microbiol., 33, 1229–1241. [DOI] [PubMed] [Google Scholar]
- Kenny B., Lai,L.C., Finlay,B.B. and Donnenberg,M.S. (1996) EspA, a protein secreted by enteropathogenic Escherichia coli, is required to induce signals in epithelial cells. Mol. Microbiol., 20, 313–323. [DOI] [PubMed] [Google Scholar]
- Kenny B., DeVinney,R., Stein,M., Reinscheid,D.J., Frey,E.A. and Finlay,B.B. (1997) Enteropathogenic E.coli (EPEC) transfers its receptor for intimate adherence into mammalian cells. Cell, 91, 511–520. [DOI] [PubMed] [Google Scholar]
- Kohda D., Morton,G., Parkar,A.A., Hatanaka,H., Inagaki,F.M., Campbell,I. and Day,A. (1996) Solution structure of the link module: a hyaluronan-binding domain involved in extracellular matrix stability and cell migration. Cell, 86, 767–775. [DOI] [PubMed] [Google Scholar]
- Knutton S., Baldwin,P.H., Williams,P.H. and McNeish,A.S. (1989) Actin accumulation at site of bacterial adhesion to tissue culture cells: basis of a new diagnostic test for enteropathogenic and enterohaemorrhagic Escherichia coli.Infect. Immun., 57, 1290–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutton S., Rosenshine,I., Pallen,M.J., Nisan,I., Neves,B.C., Bain,C., Wolff,C., Dougan,G. and Frankel,G. (1998) A novel EspA-associated surface organelle of enteropathogenic Escherichia coli involved in protein translocation into epithelial cells. EMBO J., 17, 2166–2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai L.C., Wainwright,L.A., Stone,K.D. and Donnenberg,M.S. (1997) A third secreted protein that is encoded by the enteropathogenic Escherichia coli pathogenicity island is required for transduction of signals and for attaching and effacing activities in host cells. Infect. Immun., 65, 2211–2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong J.M., Fournier,R.S. and Isberg,R.R. (1990) Identification of the integrin binding domain of the Yersinia pseudotuberculosis invasin protein. EMBO J., 9, 1979–1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong J.M., Morrissey,P.E. and Isberg,R.R. (1993) A 76-amino acid disulfide loop in the Yersinia pseudotuberculosis invasin protein is required for integrin receptor recognition. J. Biol. Chem., 268, 20524–20532. [PubMed] [Google Scholar]
- Liu H., Magoun,L., Luperchio,S., Schauer,D.B. and Leong,J.M. (1999) The Tir-binding region of enterohaemorrhagic Escherichia coli intimin is sufficient to trigger actin condensation after bacterial-induced host cell signalling. Mol. Microbiol., 34, 67–81. [DOI] [PubMed] [Google Scholar]
- Marion D., Driscoll,P.C., Kay,L.E., Wingfield,P.T., Bax,A., Gronenborn,A.M. and Clore,G.M. (1989) Overcoming the overlap problem in the assignment of 1H NMR spectra of larger proteins by use of three-dimensional heternuclear 1H-15N Hartmann–Hahn multiple quantum coherence and nuclear Overhauser-multiple quantum coherence spectroscopy: application to interleukin 1. Biochemistry, 28, 6150–6156. [DOI] [PubMed] [Google Scholar]
- McDaniel T.K., Jarvis,K.G., Donnenberg,M.S. and Kaper,J.B. (1995) A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc. Natl Acad. Sci. USA, 92, 1664–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J.H. (1972) Experiments in Molecular Genetics. Cold Spring Harbor Laborator Press, Cold Spring Harbor, NY. [Google Scholar]
- Moon H.W., Whipp,S.C., Argenzio,R.A., Levine,M.M. and Giannella,R.A. (1983) Attaching and effacing activities of rabbit and human enteropathogenic Escherichia coli in pig and rabbit intestines. Infect. Immun., 41, 1340–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhandiram D.R. and Kay,L.E. (1994) Gradient-enhanced triple-resonance 3-dimensional NMR experiments with improved sensitivity. J. Magn. Reson., B103, 203–216. [Google Scholar]
- Nataro J.P. and Kaper,J.B. (1998) Diarrheagenic Escherichia coli.Clin. Microbiol. Rev., 11, 142–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilges M., Gronenborn,A.M. and Clore,G.M. (1988) Determination of 3-dimensional structures of proteins by simulated annealing with interproton distance restraints—application to crambin, potato carboxypeptidase inhibitor and barley serine proteinase inhibitor-2. Protein Eng., 2, 27–38. [DOI] [PubMed] [Google Scholar]
- Norwood T.J., Boyd,J., Heritage,J.E., Soffe,N. and Campbell,I.D. (1990) Comparison of techniques for 1H-detected heteronuclear 1H-15N spectroscopy. J. Magn. Reson., 87, 488–501. [Google Scholar]
- Oswald E., Schmidt,H., Morabito,S., Karch,H., Marches,O. and Caprioli,A. (2000) Typing of intimin genes in human and animal enterohemorrhagic and enetropathogenic Escherichia coli: characterisation of a new intimin variant. Infect. Immun., 68, 64–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perna N.T., Mayhew,G.F., Posfai,G., Elliott,S., Donnenberg,M.S., Kaper,J.B. and Blattner,F.R. (1998) Molecular evolution of a pathogenicity island from enterohemorrhagic Escherichia coli O157:H7. Infect. Immun., 66, 3810–3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuister G.W., Clore,G.M., Gronenborn,A.M., Powers,R., Garrett,D.S., Tschudin,R. and Bax,A. (1993) Increased resolution and improved spectral quality in 4-dimensional 13C/13C-separated HMQC-NOESY-HMQC spectra using pulsed field gradients. J. Magn. Reson., B101, 210–213. [Google Scholar]
- Weis W.I. and Drickamer,K. (1996) Structural basis of lectin–carbohydrate recognition. Annu. Rev. Biochem., 65, 441–473. [DOI] [PubMed] [Google Scholar]
- Wolff C., Nisan,I., Hanski,E., Frankel,G. and Rosenshine,I. (1998) Protein translocation into HeLa cells by infecting enteropathogenic Escherichia coli.Mol. Microbiol., 28, 143–155. [DOI] [PubMed] [Google Scholar]
- Yu J. and Kaper,J.B. (1992) Cloning and characterization of the eae gene of enterohaemorrhagic Escherichia coli O157:H7. Mol. Microbiol., 6, 411–417. [DOI] [PubMed] [Google Scholar]