Abstract
The expression of Notch receptors on hematopoietic cells and of cognate ligands on bone marrow stromal cells suggests a possible role for Notch signalling in the regulation of hematopoiesis. In order to assess the involvement of Notch1 signalling in myelopoiesis, 32D myeloid progenitor cell lines were engineered to permit the conditional induction of the constitutively active intracellular domain of murine Notch1 (mN1IC) by the 4-hydroxytamoxifen-inducible system. The induction of mN1IC resulted in accelerated and increased granulocytic differentiation. These effects were observed under growth conditions that support differentiation and, to a lesser degree, under conditions that normally promote self-renewal. Transient transfection of mN1IC deletion mutants showed that the differentiation promoting activity correlated with RBP-J transactivation. Furthermore, expression of a transcriptionally active derivative of RBP-J (RBP-J–VP16) increased myeloid differentiation. To test further the role of Notch signalling in a physiological context, 32D cells expressing mNotch1 were cultured on fibroblast layers that either expressed or did not express the Notch ligand Jagged1. Similar to the induction of mN1IC, Jagged1 accelerated granulocytic differentiation of 32D cells. Taken together, our data suggest that activation of mNotch1 promotes myeloid differentiation via RBP-J transactivation.
Keywords: differentiation/hematopoiesis/mNotch1/RBP-J/signalling
Introduction
In invertebrates and vertebrates, intracellular signalling through the Notch transmembrane receptors has a role to play in proliferation and differentiation in many tissues throughout development (see reviews by Artavanis-Tsakonas et al., 1995; Simpson, 1995). Mechanisms through which Notch can influence cell fate include lateral inhibition and inductive signalling. Thus far, four Notch receptors (Notch1–4) have been identified in mammalian cells (Ellisen et al., 1991; Weinmaster et al., 1991, 1992; Del Amo et al., 1992; Reaume et al., 1992; Kopan and Weintraub, 1993; Lardelli and Lendahl, 1993; Lardelli et al., 1994; Uyttendaele et al., 1996), and consist of highly conserved transmembrane glycoprotein receptors with a single transmembrane domain (Figure 1). The extracellular domain of Notch contains a variable number of epidermal growth factor (EGF)-like repeats and lin-12/Notch repeats (LNR), which function in ligand binding and Notch activation (Rebay et al., 1991). The Notch intracellular domain includes the RAM domain, six cdc10/ankyrin repeats, motifs characteristic of molecules involved in protein–protein interactions, a nuclear localization signal and a C-terminal OPA (glutamine-rich) and PEST (proline-glutamate-serine-threonine-rich) region (Figure 1) (Del Amo et al., 1993; Kurooka et al., 1998). The RAM domain binds CSL proteins, identified in various species as CBF1 (also termed RBP-J recombination recognition sequence binding protein at the Jκ site) in mammals (Tamura et al., 1995); Suppressor of Hairless in Drosophila and Xenopus; and Lag-1 in Caenorhabditis elegans. Notch is activated by binding a member of the DSL (Drosophila Delta and Serrate and C.elegans Lag-2) family of cell surface proteins (Tax et al., 1994), which includes Delta and Jagged in mammals (Bettenhausen et al., 1995; Lindsell et al., 1995; Shawber et al., 1996a; Dunwoodie et al., 1997). Following activation, Notch is cleaved within the transmembrane domain, releasing the Notch intracellular domain (NIC) from the membrane (Kopan et al., 1996; Schroeter et al., 1998). The NIC then translocates to the nucleus where it can modulate gene expression via association with CSL proteins, and thereby may affect cell fate choice (Schroeter et al., 1998). This may not be the only mechanism, however, through which cell fate can be influenced by Notch since some aspects of Notch signalling do not apparently require the known CSL proteins (Shawber et al., 1996b; Bigas et al., 1998; Nofziger et al., 1999; see review by Weinmaster, 1997).
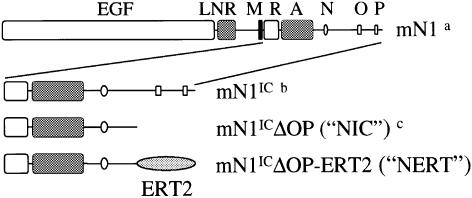
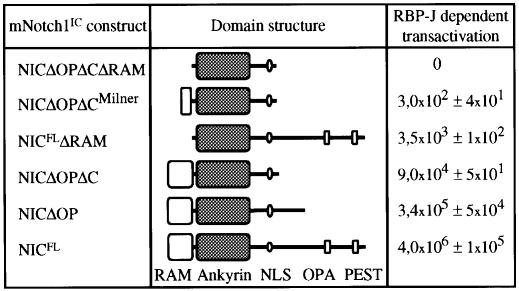
Fig. 1. Schematic diagram of the full-length mNotch1 receptor and the mNotch1 constructs used in this work. aaa 1–2531; baa 1747–2531; caa 1747–2293. mN1, mNotch1; IC, intracellular; EGF, EGF repeats; LNR, LIN repeats; M, transmembrane domain; R, RAM23 domain; A, ankyrin repeats; N, nuclear localization sequence; O, OPA sequence; P, PEST sequence; ERT2, hormone binding domain of human estrogen receptor (OHT-sensitive mutant).
The importance of this intracellular domain in Notch signalling was indicated by the observation that in some human T lymphoblastic leukemias, a chromosomal translocation resulted in a deletion of the extracellular domain of the human Notch1 gene (Ellisen et al., 1991). Further work then showed that expression of this truncated version can act as a constitutive activated Notch (Kopan et al., 1994; Nye et al., 1994; Milner et al., 1996).
Since Notch receptors and their ligands are expressed in hematopoietic tissues (Weinmaster et al., 1991, 1992; Milner et al., 1994, 1996; Shawber et al., 1996a; Jones et al., 1998; Li et al., 1998; Varnum-Finney et al., 1998; see review by Milner and Bigas, 1999) and, by analogy with other systems, it is likely that Notch receptors also play a role in the regulation of blood cell production. Notch1 mRNA and protein is found in immature CD34-positive hematopoietic progenitor cells, lymphoid, myeloid and erythroid precursor cell populations as well as in peripheral blood T and B lymphocytes, monocytes and neutrophils (reviewed by Milner and Bigas, 1999), indicating a general role for Notch in differentiation and/or proliferation of multiple hematopoietic lineages and at various stages of maturation. However, while several studies indicate that Notch1 is critically involved in T-cell lineage development (Robey et al., 1996; Washburn et al., 1997; Deftos et al., 1998; Pui et al., 1999; Radtke et al., 1999), a role for Notch signalling in regulating myelopoiesis remains unclear. Milner et al. (1996) and Carlesso et al. (1999), for example, have shown that ectopic expression of constitutively active forms of Notch1 inhibits differentiation of myeloid progenitor cell lines, whilst a study by Shelly et al. (1999) indicated that Notch is required for erythroid differentiation to proceed. Similarly, ambivalent results were found when primary hematopoietic progenitor cells were stimulated by the Notch receptor ligand Jagged1: in some studies a moderate increase in colony formation was observed (Jones et al., 1998; Varnum-Finney et al., 1998), whereas in others no effect or a decrease in colony formation was seen (Walker et al., 1999).
To investigate this further, we have now expressed the murine Notch1 intracellular domain (mN1IC; NIC, Figure 1), which is a constitutively active form of mNotch1, in the myeloid progenitor cell line 32D using an estrogen-inducible expression system, and looked at how mN1IC affects differentiation of the 32D cells under various growth and differentiation conditions. The results of this study show that activated mNotch1 promotes differentiation of 32D cells. Furthermore, using a fibroblast cell line engineered to express the Notch ligand Jagged1 (Lindsell et al., 1995) in a co-culture assay with 32D cells expressing the mNotch1 receptor, we demonstrate the physiological relevance of differentiation induction by activated mNotch1. In addition, to determine the signalling pathway(s) involved in the effect of mNotch1 on myeloid differentiation, we have expressed different parts of the mN1IC and a transcriptionally active form of RBP-J in 32D cells using a transient expression system. This allows the immediate analysis of cells expressing the respective target protein. Our data suggest that Notch signalling via RBP-J is responsible for the enhancement of myeloid differentiation by mNotch1.
Results
Expression of the activated intracellular domain of mNotch1 promotes granulocytic differentiation of 32D cells
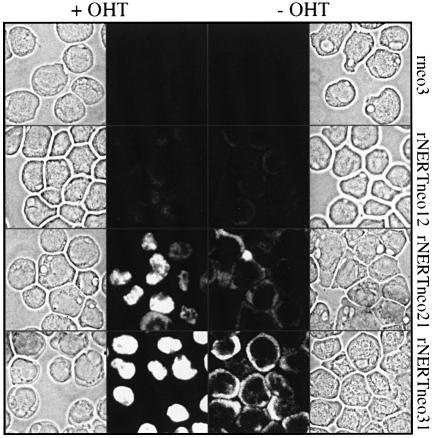
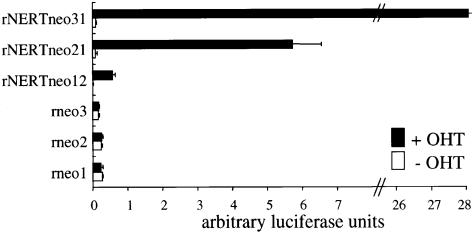
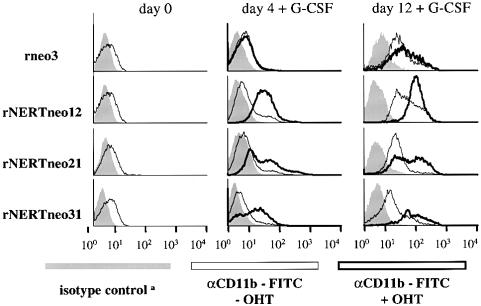
To obtain clones of 32D cells in which translocation to the nucleus of the activated intracellular domain of mNotch1 (mN1IC; Figure 1) can be regulated, 32D cells were transfected by electroporation with retroviral vectors carrying the mN1IC fused to the human estrogen receptor (rNERTneo), and then selected in the presence of geneticin and interleukin-3 (IL-3) and cloned in soft agar. As a control, cells were transfected with a vector that does not contain the mN1IC–estrogen receptor cDNA (rneo). Both vectors conferred geneticin resistance to transfected cells. Cultures of rNERTneo-transfected clones, in which expression of the correct sized NERT protein was confirmed by western analysis using an antibody against the human estrogen receptor α as probe (data not shown), were then tested for transactivation of the RBP-J pathway in the presence or absence of 4-hydroxytamoxifen (OHT). In the absence of OHT, when the mN1IC–estrogen receptor fusion protein was exclusively located in the cytoplasm (Figure 2), no transactivation of the RBP-J pathway was observed (Figure 3). After addition of OHT, the mN1IC–estrogen receptor fusion protein translocated to the nucleus (Figure 2) and the RBP-J pathway was transactivated in all clones used in this study (Figure 3). Transactivation of the RBP-J pathway in the different clones correlated with the amount of mN1IC–estrogen receptor fusion protein present in the nucleus of the cells, i.e. highest in clone 31 and lowest in clone 12 (Figures 2 and 3). As expected, control clones did not transactivate the RBP-J pathway in the presence or absence of OHT (Figure 3).
Fig. 2. OHT treatment induces translocation of the NERT protein from the cytoplasm to the nucleus in rNERTneo clones. The NERT protein was detected using immunofluorescence staining with a monoclonal anti-human estrogen receptor α antibody. No signal could be detected in rneo control cells. NERT protein levels correlate with the levels of OHT-inducible RBP-J-dependent transactivation in the rNERTneo clones (see also Figure 3). The cell at the lower side of panel rNERTneo21 +OHT is in mitosis. Magnification is ×630.
Fig. 3. Transactivation of the RBP-J pathway through mNotch1 is OHT-inducible in 32D rNERTneo clones. Cells were transfected with the reporter construct pGa981-6 (luciferase gene under the control of 12 RBP-J binding sites). Luciferase activity was measured 24 h after transfection and subsequent culture in the absence or presence of OHT. Each bar represents mean values from two independent transfections after correction for transfection efficiency ± SEM.
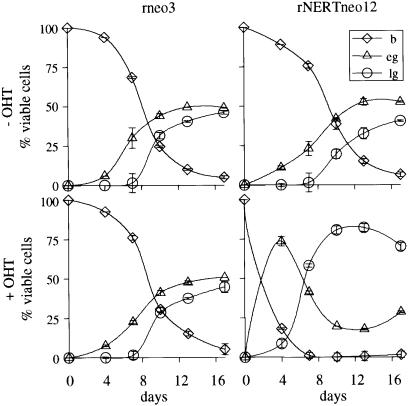
Differentiation of 32D cells into granulocytic cells may be modulated by modifying the concentrations of IL-3 and granulocyte colony stimulating factor (G-CSF) (Valtieri et al., 1987): When maintained in high IL-3, 32D cells proliferate as undifferentiated blasts. However, when G-CSF is added to the culture medium and IL-3 is reduced or absent, 32D cells differentiate into mature granulocytic cells. To assess the effect of activated mNotch1 on myeloid differentiation of 32D cells, rNERTneo-transfected 32D cells were thus cultured in the presence of G-CSF without IL-3 and in the presence or absence of OHT, and monitored for changes in morphology. Time course experiments revealed an accelerated onset of differentiation concomitant with an increase in immature and mature forms of granulocytic differentiation in the presence of OHT when compared with cells differentiated in the absence of OHT (Figures 4 and 5B). These results were further confirmed by fluorescence-activated cell sorter (FACS) analyses using an antibody directed against the neutrophil/macrophage cell surface markers Mac-1 (mCD11b; Figure 6) and Gr-1 (data not shown). Although cell proliferation of the rNERTneo 32D cells was reduced in the presence of OHT (data not shown), differentiating cultures of rNERTneo-transfected cells contained a significantly higher absolute number of differentiated cells in the presence of OHT (p <0.001; Table I). Control cultures of rneo-transfected 32D cells differentiated virtually identically to the parental 32D cells and did not differentiate significantly more in the presence of OHT, indicating that OHT itself has no significant effect on granulocytic differentiation of 32D cells (Figures 4 and 5B; Table I). Thus, our data show that under conditions that allow differentiation of 32D cells, activated mNotch1 both accelerates and enhances differentiation into granulocytic cells.
Fig. 4. Activated mN1IC accelerates and enhances G-CSF-induced granulocytic differentiation of 32D cells. rNERTneo and rneo cell clones were each differentiated (in quadruplicate) by removal of IL-3 and addition of G-CSF for 13 days. OHT was included in two of each set of four cultures from day 0. The experiment was repeated a total of nine times using three pairs of rNERTneo and rneo clones with virtually identical results. A representative experiment is shown. b, undifferentiated blast cells; eg, early granulocytes (promyelocytes and myelocytes); lg, late granulocytes (metamyelocytes and granulocytes).
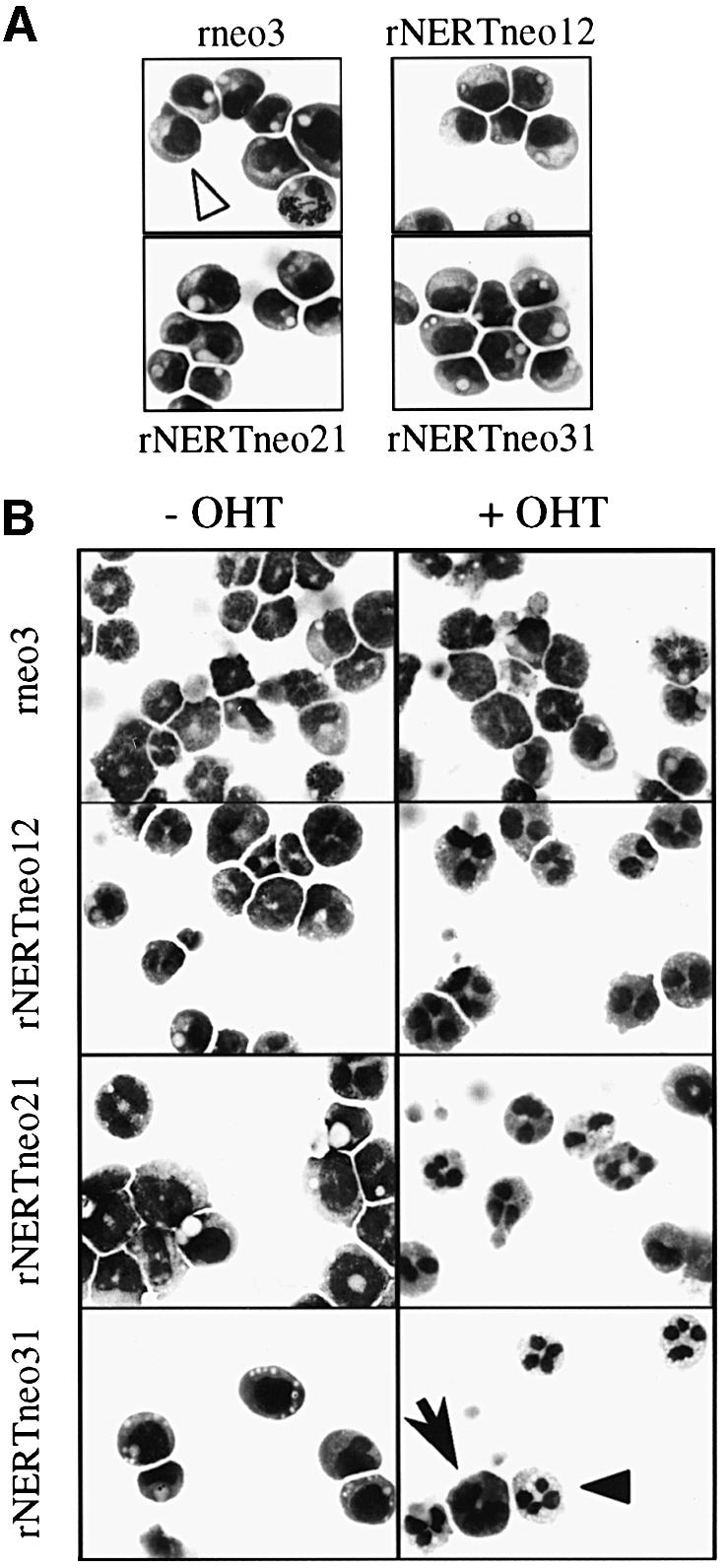
Fig. 5. Activated mN1IC promotes G-CSF-induced granulocytic differentiation of 32D cells. Cells were stained with May–Grünwald–Giemsa. Magnification is ×630. (A) Morphology of undifferentiated 32D clones. Under self-renewing conditions and in the absence of OHT, all clones consisted of >99% undifferentiated blast cells. The panel for rneo3 contains a mitotic cell. (B) Morphology of differentiated 32D clones. Cells were differentiated by removal of IL-3 and addition of G-CSF for 9 days in duplicates. One of each duplicate was treated with OHT from day 0. Each of three rneo clones analysed yielded identical results, and a representative experiment is shown here. Analysis after 6 or 12 days of differentiation showed the same effect of mN1IC on the differentiation of 32D cells. One undifferentiated blast cell (open arrowhead), one early granulocyte (arrow) and one late granulocyte (closed arrowhead) are marked. This experiment was repeated nine times yielding virtually identical results.
Fig. 6. Activated mN1IC accelerates expression of CD11b (Mac-1) in differentiating 32D cells. aIsotype controls of cells with or without OHT treatment were identical. Cells were induced to differentiate by removal of IL-3 and addition of G-CSF. On days 0, 4, 8 and 12, respectively, the cells were analysed by FACS analysis for expression of CD11b as a marker for granulocytic differentiation. On day 0, only cells without OHT treatment were analysed. By day 4 of differentiation in the presence of OHT, CD11b expression levels were equivalent to those seen after 12 days of differentiation in the absence of OHT. This experiment was repeated twice with the same results.
Table I. Activated mN1IC enhances differentiation of 32D cells in the absence of G-CSF.
| 32D clone | Percentage of cell typesa |
Total numberd of differentiated cells ×105 | pe | ||
|---|---|---|---|---|---|
| Blasts | EGb | LGc | |||
| rneo3 | |||||
| –OHT | 99 ± 1 | 1 ± 1 | 0 ± 0 | 2 ± 2 | |
| +OHT | 97 ± 1 | 3 ± 1 | 0 ± 0 | 5 ± 2 | |
| rNERTneo12 | |||||
| –OHT | 100 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | |
| +OHT | 13 ± 1 | 56 ± 3 | 31 ± 4 | 48 ± 4 | ≤0.001 |
| rNERTneo21 | |||||
| –OHT | 95 ± 1 | 5 ± 1 | 0 ± 0 | 16 ± 3 | |
| +OHT | 53 ± 2 | 44 ± 1 | 3 ± 1 | 42 ± 2 | ≤0.001 |
aCells were cultured in low concentrations of IL-3 (1–10 U IL-3 per ml) for 7 days. Mean values ± SEM of two representative experiments using 2 U IL-3 per ml are shown. The experiment under low IL-3 conditions was repeated nine times yielding virtually identical results.
bEarly granulocytes (promyelocytes and myelocytes).
cLate granulocytes (metamyelocytes and granulocytes).
dCalculated by multiplication of the absolute cell number with the percentage of differentiated cells (EG and LG).
eThe increase in total number of differentiated cells after OHT treatment is statistically significant.
To test whether activated mNotch1 also promotes differentiation of 32D cells under conditions that favour self-renewal, rNERTneo-transfected 32D cell clones were analysed for their morphology in the presence of high concentrations of IL-3 and in the presence or absence of OHT. In the presence of a high concentration of IL-3 and in the absence of OHT, all cells showed blast cell morphology (Figure 5A). In contrast, in the presence of a high concentration of IL-3 and OHT, rNERTneo-transfected 32D cell clones also contained a low number (5%) of differentiated granulocytic cells, whereas in control clones all cells remained undifferentiated (data not shown).
This indicates that activated Notch may be influencing the probability of self-renewal versus differentiation. To analyse this further, the clonogenicity of these cells was assayed by cloning in semi-solid media containing high concentrations of IL-3. After 4 days of culture in the presence or absence of OHT, the colony-forming ability of rNERTneo transfected 32D cells and control rneo transfected 32D cells was evaluated. Under culture conditions that allow differentiation, i.e. in the presence of G-CSF and absence of IL-3, the number of clonogenic cells of rNERTneo-transfected 32D cells grown in the presence of OHT for 4 days was markedly reduced when compared with rNERTneo-transfected cells grown in the absence of OHT (p <0.02; Table II); whereas control rneo-transfected 32D cells showed no significant difference in the clonogenicity in the presence or absence of OHT (Table II). Under conditions that normally promote self-renewal, i.e. in the presence of high concentrations of IL-3, the number of clonogenic cells of rNERTneo-transfected 32D cells was reduced ∼2- to 4-fold after culture for 4 days in the presence of OHT (p <0.02 for rNERTneo 21 and rNERTneo 12), whereas the number of clonogenic cells of rneo-transfected 32D control cells remained unaffected by OHT. These results suggest that activated Notch1 decreases self-renewal and induces differentiation of 32D cells even in the presence of high concentrations of IL-3.
Table II. Activated mN1IC reduces clonogenicity of 32D cells.
| 3D clone | Percentage clonogenicitya | Absolute number of clonogenic cellsb | Relative clonogenicity (%)c |
|---|---|---|---|
| rneo3 | |||
| –OHT | 69 ± 6 | 1.7 × 105 | |
| +OHT | 61 ± 14 | 1.5 × 105 | 88 |
| rNERTneo 21 | |||
| –OHT | 58 ± 6 | 1.5 × 105 | |
| +OHT | 14 ± 1 | 1.4 × 104 | 9 |
aThe number of clonogenic cells grown in the presence or absence of OHT and in the presence of G-CSF for 4 days was determined by plating the cells in soft agar in the presence of 150 U/ml IL-3 as described in Materials and methods. Values represent mean values ± SEM.
bThe total number of clonogenic cells per culture on day 4 was calculated using percentage clonogenicity and total viable cell numbers. On day 0, each culture was started with 3.5 × 104 cells.
cRatio of the absolute number of clonogenic cells in the presence of OHT to the number in the absence of OHT. Relative clonogenicity is significantly reduced for rNERTneo 21 cells (p <0.02), but not for rneo3 cells. Values represent averages from three independent experiments. A similar reduction in clonogenicity of rNERTneo cells was observed after 6 days of culture in the presence of OHT (data not shown).
These effects of Notch become even more apparent as the IL-3 concentration is reduced. When rNERTneo-transfected 32D cells were cultured in the presence of reduced amounts of IL-3 (1–10 U IL-3 per ml) and monitored for changes in morphology, rNERTneo-transfected 32D cells, in the absence of OHT, showed few differentiating cells and no terminally differentiated cells at day 7 in the presence of low concentrations of IL-3 (Table I). In the presence of OHT, however, up to 87% of the rNERTneo-transfected cells were differentiated into granulocytic cells at day 7 (Table I). Control rneo-transfected 32D cells retained a predominantly immature blast cell morphology until day 7 when cultured in the presence of low concentrations of IL-3, regardless of the addition of OHT (Table I).
The Jagged/Notch pathway accelerates granulocytic differentiation of 32D cells
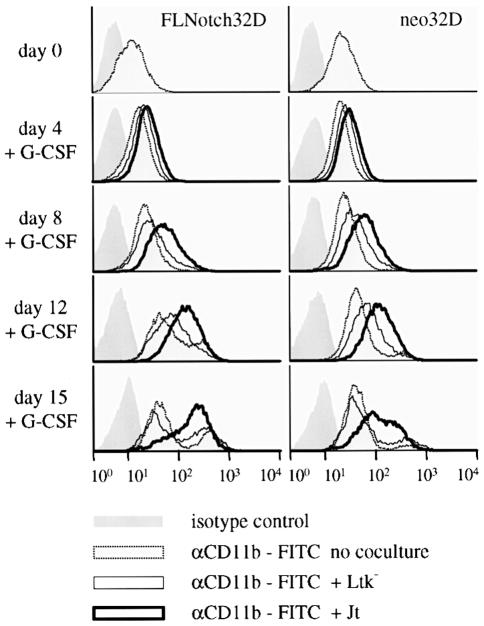
Since the activated form of mNotch1 promoted granulocytic differentiation of 32D cells, we asked whether this same phenotype could be induced by Notch as a result of ligand binding. To test this hypothesis, we used a fibroblast cell line engineered to express the Notch ligand Jagged1 (JT cells; Lindsell et al., 1995). JT cells can activate Notch in co-cultured C2C12 myoblasts and thereby prevent muscle cell differentiation (Lindsell et al., 1995). Expression of Jagged1 on JT cells but not on the parental mouse Ltk– cells was confirmed by western blot analysis (data not shown). Our assay involved co-culturing the JT cells or the parental Ltk– cells with either 32D cells stably transfected to express full-length mNotch1 (FLNotch32D), control transfected 32D cells (neo32D) or parental untransfected 32D cells. Western blot analysis using antibodies directed against mNotch1 confirmed that FLNotch32D cells expressed high levels of the mNotch1 receptor but also revealed that the parental 32D cells do already express the mNotch1 receptor (data not shown). To analyse the influence of the Jagged/Notch pathway on 32D cell differentiation, FLNotch32D, neo32D or parental 32D were cultured in differentiation medium in the presence of JT or Ltk– fibroblasts and monitored for expression of the mature myeloid cell surface marker Mac-1 (mCD11b). As shown in Figure 7, co-culture of FLNotch32D cells with JT cells resulted in an accelerated onset of differentiation when compared with FLNotch32D cells co-cultured with the parental Ltk– fibroblasts or with 32D cells in the presence of G-CSF alone. Neo32D and the untransfected parental 32D cells showed a virtually identical increase in Mac-1 expression as FLNotch32D cells when differentiated on JT cells (Figure 7), suggesting that the endogenous Notch expression in 32D cells is sufficient for biological activity of the Jagged/Notch pathway. The promotion of granulocytic differentiation of 32D cells by co-culture with Jagged1-expressing cells was similar to the effects of activated mN1IC on granulocytic differentiation of 32D cells (Figures 6 and 7). Thus, these results indicate a physiological role for the activated mN1IC in promoting granulocytic differentiation of 32D cells.
Fig. 7. The Jagged/Notch pathway accelerates expression of CD11b (Mac-1) in differentiating 32D cells. Cells were induced to differentiate by removal of IL-3 and addition of G-CSF and cultured in the presence of JT, control Ltk– fibroblasts or G-CSF alone. On days 0, 4, 8, 12 and 15, respectively, the cells were analysed by FACS analysis for expression of CD11b as a marker for granulocytic differentiation. This experiment was repeated three times with virtually identical results.
Increased granulocytic maturation of 32D cells correlates with activation of the RBP-J signalling pathway
Notch signalling regulates expression of downstream genes by activation of the RBP-J transcription factor. This requires the RAM domain and ankyrin repeats of Notch (Kato et al., 1997). However, some Notch-dependent processes occur by RBP-J-independent mechanisms (Shawber et al., 1996b; Nofziger et al., 1999). To dissect the signalling pathway(s) that mediates the enhancing effect of activated mNotch1 on granulocytic differentiation, we designed several deletion mutants of mN1IC (Figure 8) and analysed these mutants for their influence on differentiation of 32D cells in the presence of G-CSF. The deletion constructs were cloned into a plasmid-based transient expression system in which a target protein is co-expressed with the eGFP. This system allows the analysis of cells expressing the respective target protein directly after transfection and sorting for GFP-positive cells without altering their phenotype (Z.McIvor, C.M.Heyworth, B.A.Johnson, S.Pearson, H.Fiegler, L.Hampson, T.M.Dexter and M.A.Cross, submitted). 32D cells were transfected by electroporation with the plasmid constructs depicted in Figure 8, respectively. As a control, 32D cells were transfected with vector DNA without an insert. Western blot analysis using antibodies directed against the Flag- or myc-tag, respectively, confirmed expression of the correct sized Notch1 mutant proteins in all transfected 32D cell cultures (data not shown). Transactivation of the RBP-J-dependent signalling pathway by the mN1IC mutants was then tested by co-transfection of a reporter construct consisting of a luciferase gene under the control of 12 RBP-J binding sites (Strobl et al., 1997). Notch mutants that contained the complete RAM23 domain strongly transactivated the reporter construct (Figure 8). Transactivation, however, was more pronounced when the Notch mutants also included the C-terminal OPA and PEST sequences (Figure 8). To assess the effect of the different Notch mutant proteins on granulocytic differentiation of 32D cells, 32D cells were cultured in the presence of G-CSF without IL-3 for 9 days directly after transfection, and GFP-positive 32D cells sorted 48 h after transfection and induction of differentiation were monitored for changes in morphology and cell surface differentiation markers. As shown in Table III, all 32D cell cultures that expressed mN1IC mutant proteins yielded significantly increased granulocytic differentiation as compared with control transfected 32D cells. The increase in granulocytic differentiation by the Notch mutant proteins did not correlate with the level of transactivation of the RBP-J-dependent signalling pathway (Figure 8 and Table III), suggesting that low levels of transactivation of the RBP-J-dependent signalling pathway already result in a maximal biological response.
Fig. 8. Transactivation of the RBP-J-dependent signalling pathway by mN1IC deletion constructs. Each of the mN1IC deletion constructs was co-transfected with pGA981-6 into 32D cells and luciferase activity was determined in two to six independent transfections. Values shown are the mean ± SEM from independent transfections after correction for transfection efficiency.
Table III. Activation of the RBP-J signalling pathway enhances G-CSF-induced differentiation of 32D cells.
| Transfected construct | Increase in the percentage of differentiated cellsa after 9 days of differentiationb |
||
|---|---|---|---|
| Differentiated cellsc | nd | p | |
| (control) GFP | n.a. | ||
| NICΔOPΔCΔRAM–GFP | +4 (±0.9) | 3 | ≤0.05 |
| NICFLΔRAM–GFP | +9 (± 2.9) | 3 | ≤0.05 |
| NICΔOPΔCMilner–GFP | +13 (± 1.5) | 4 | ≤0.001 |
| NICΔOPΔC–GFP | +27 (± 3.7) | 3 | ≤0.01 |
| NICΔOP–GFP | +26 (± 4.3) | 4 | ≤0.01 |
| NICFL–GFP | +21 (± 2.0) | 5 | ≤0.001 |
| RBP-J–VP16–GFP | +17 (± 5.3) | 7 | ≤0.001 |
| tTa–GFPe | +1 (± 4.5) | 3 | not significant |
aValues shown were corrected for the respective values from CMVeGFP control transfected cells of each experiment.
bDifferentiation was induced directly after transfection of the cells by culturing the cells in 1000 U/ml G-CSF without IL-3. GFP positive cells were sorted 2 days later.
cEarly and late granulocytes. Mean values ± SEM of all experiments are shown. n.a., not applicable.
dNumber of independent experiments in which the respective constructs were tested.
etTa, which contains the functional VP16 transactivation domain, was used as a control for RBP-J–VP16.
A fusion protein of RBP-J with the viral transactivation domain VP16, RBP-J–VP16, has been used as a transcriptionally active form of RBP-J (Waltzer et al., 1995). We showed above that activation of the Notch signalling pathway led to increased granulocytic differentiation of 32D cells. To examine whether the increased differentiation of 32D cells by activated Notch is mediated through RBP-J, we transfected RBP-J–VP16 using the GFP sorting system into 32D cells and monitored the RBP-J–VP16-expressing 32D cells for changes in morphology. RBP-J–VP16-expressing 32D cells showed strong transactivation that was comparable with transactivation of the RBP-J pathway mediated by the activated full-length mN1IC construct [5 × 106 and 4 × 106 relative light units (RLU), respectively] and granulocytic differentiation was significantly increased as compared with cells transfected with the eGFP vector alone (p <0.001; Table III). Transfection of the tTa control construct containing a functional VP16 transactivation domain did not alter 32D cell differentiation (Table III), showing that the effect of RBP-J–VP16 is specific for the RBP-J signalling pathway. Thus, activated RBP-J also promotes granulocytic differentiation of 32D cells.
Constitutively active mNotch1 does not block G-CSF-induced differentiation in 32D cells
Recently, it has been described that constitutive ectopic expression of a truncated intracellular form of mNotch1 in 32D cells inhibits G-CSF-induced differentiation and allows continued proliferation of undifferentiated cells (Milner et al., 1996). Since the mN1IC construct used by Milner et al. lacked a complete functional RAM domain and showed very little transactivation capacity of the RBP-J pathway (see Figure 8 and Table III), we reasoned that RBP-J signalling might be responsible for the opposing effects observed in our study. However, when we transiently expressed the MT-mN1-ICΔOP construct (NICΔOPΔCMilner–GFP) in 32D cells using the GFP sorting system described above, no block in differentiation in response to G-CSF was observed. In contrast, although the transactivation of the RBP-J pathway was very low, 32D cells expressing the MT-mN1-ICΔOP protein showed a significantly higher degree of terminal granulocytic differentiation in the presence of G-CSF and in the absence of IL-3 than control vector transfected cells (p <0.001; Table III). These findings support the contention that Notch signalling mediated by RBP-J promotes granulocytic differentiation and does not block G-CSF-induced differentiation of 32D cells.
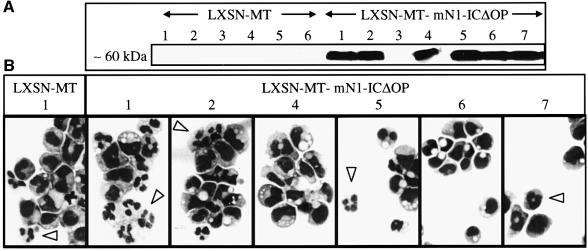
To determine whether the block in G-CSF-induced differentiation observed by Milner et al. could be attributed to the differences in the viral constructs and experimental procedures used, we infected our 32D cells with a retroviral vector expressing MT-mN1-ICΔOP constitutively (Milner et al., 1996). As a control, 32D cells were infected with a vector expressing only the myc epitope tag (LXSN-MT). Both vectors conferred geneticin resistance to infected cells. After selection and cloning in the presence of G418 and IL-3, three control infected clones and six MT-mN1-ICΔOP clones were cultured in the presence of G-CSF without IL-3 and monitored for changes in morphology. Eleven days after induction of differentiation ∼50% of the control 32D cells (three clones analysed) were terminally differentiated into granulocytic cells (Figure 9B). Of the six MT-mN1-ICΔOP clones, however, two clones differentiated similarly to the control clones, two clones were blocked in differentiation, and one clone showed a delayed and one an accelerated differentiation pattern when compared with the control infected cells (Figure 9B). Expression of the correct sized MT-mN1-ICΔOP protein in all clones was confirmed by western blot analysis using an anti-myc-tag antibody (Figure 9A). These data show that although ectopic expression of active mN1IC can lead to the production of cell lines that are blocked in differentiation, this is not a consistent phenomenon, indicating that other changes, in addition to activated Notch, are required.
Fig. 9. Constitutive ectopic expression of mN1IC does not necessarily block G-CSF-induced differentiation of 32D cell clones. 32D cells were infected with LXSN-MT control or LXSN-MT-mN1-ICΔOP virus (Milner et al., 1996), respectively, and clonal cell lines were analysed 37 days after infection. (A) Myc-tagged mN1-ICΔOP protein was detected by western blot analysis using the 9E10 monoclonal antibody against the myc-tag. Under the experimental conditions used, the myc-tags of control vector infected clones could not be detected. Lanes 5–7 were run on a separate gel. (B) Morphology of different 32D cell clones 11 days after induction of differentiation. Cells were induced to differentiate by removal of IL-3 and addition of G-CSF. Differentiation was virtually identical for all LXSN-MT virus-infected control clones analysed. Some representative differentiated cells are marked by arrowheads. Cells were stained with May–Grünwald–Giemsa. Magnification is ×630.
Discussion
Based on the expression of activated mNotch1 derivatives from inducible or transiently transfected constructs and on the presentation of the Notch ligand Jagged1 to mNotch1-expressing cells we provide evidence that Notch signalling can accelerate and enhance granulocytic differentiation. While our results add to a growing body of evidence that Notch signalling influences hematopoiesis, the nature of the consequences described here differ in some respects to those reported previously. Most strikingly, we find that the induction of mNotch1 activity in 32D cells promotes differentiation, in contrast to previous studies that demonstrated a differentiation block (Milner et al., 1996; Bigas et al., 1998; Jones et al., 1998; Li et al., 1998; Varnum-Finney et al., 1998; Carlesso et al., 1999; Han et al., 2000).
How can this discrepancy be explained? Several of the studies indicating a block in differentiation were based on the observation that co-culture of hematopoietic progenitor cells with stromal cell lines expressing the Notch ligand Jagged led to an increase in the number of immature hematopoietic progenitor cells (Jones et al., 1998; Li et al., 1998; Varnum-Finney et al., 1998; Carlesso et al., 1999). In contrast, however, and in line with our work, a separate study recently reported a decrease in myeloid colony formation after co-culture of immature progenitor cells with Jagged-expressing cell lines (Walker et al., 1999). The outcome of co-culture experiments may be influenced by many factors, including additional growth factors added, inherent functional differences among the different Notch ligands, and possible differences between different ligand-expressing cell lines and the parental cell lines in the type and amount of cytokines made, adhesion molecules expressed or other factors unrelated to Jagged expression. In other words, it is difficult to conclude with any certainty that the differentiation block observed in some studies is solely attributable to Notch signalling. Interestingly, constitutive Notch1 activity did not block or delay myeloid differentiation in vivo (Pui et al., 1999).
The proposition that Notch1 activation by Jagged1 inhibits differentiation of myeloid progenitor cells was based on several studies with truncated soluble forms of the hJagged1 or hDelta-like1 protein (Li et al., 1998; Varnum-Finney et al., 1998; Han et al., 2000). In contrast to our results with Jagged-expressing cells, a peptide consisting of the DSL domain of hJagged1 (Li et al., 1998) did not have an influence on differentiation of 32D cells expressing full-length mNotch1 (T.Schroeder, M.Eulitz and U.Just, unpublished data). Since in Drosophila truncated forms of Delta and Serrate have been shown to have dominant negative effects (Hukriede et al., 1997; Sun and Artavanis-Tsakonas, 1997), a likely explanation for this discrepancy is that the soluble forms of the Notch ligands cannot substitute for Notch ligands bound on the cell surface of stromal cells but may act as a dominant negative form of Jagged or Delta.
Most evidence for a block in myeloid differentiation by activated Notch is derived from studies that, like ours, used the myeloid progenitor cell line 32D (Milner et al., 1996; Bigas et al., 1998). However, in contrast to our data that activated NIC accelerates and enhances granulocytic differentiation, these studies demonstrated that differentiation-blocked cells emerged after constitutive expression of activated NIC. While it is formally possible that this discrepancy is due to differences in the 32D populations used (Migliaccio et al., 1989), it seems more likely to reflect differences between the short term (this study) and long term (previous studies) manipulation of expression in the 32D system. In support of this, we found that 32D populations infected with retrovirus constitutively expressing activated Notch1 yielded some clones that were blocked in G-CSF-induced differentiation following several weeks of culture, but this was by no means universal. Considering that we observed the differentiation-promoting effects of Notch1 signalling even under high IL-3 conditions, which normally favour self-renewal, it seems likely that extended culture of a 32D population expressing Notch1 may favour the outgrowth of genetic variants that are blocked in differentiation.
In previous studies, 32D cells expressing constitutively active mNIC were refractory to the differentiation-promoting action of G-CSF (Milner et al., 1996; Bigas et al., 1998). This activity of mNotch1 was attributed to a specific region of mNotch1, termed Notch cytokine response (NCR) region (Bigas et al., 1998). Since we have shown in this study that mN1IC promotes granulocytic differentiation of 32D cells, it seems possible that the inhibition of G-CSF-induced differentiation reported previously was due to the selection of cells that are resistant to the differentiation-promoting activities of both mN1IC and G-CSF. Further studies will elucidate how the signal transduction pathways for mNotch1 and G-CSF are linked, and if and how the proposed NCR of mNotch1 is involved.
Constitutive active forms of Notch that contain the RAM domain have been shown to interact both physically and functionally with CSL (RBP-J in mammals) proteins (Jarriault et al., 1995; Tamura et al., 1995; Hsieh et al., 1996). In accordance with previously published work (Kurooka et al., 1998), our data with different mN1IC constructs show that three functional domains, the RAM domain, the ankyrin repeats and C-terminal TAD domain, are involved in RBP-J transactivation. Furthermore, we demonstrate in this study that Notch signalling via RBP-J clearly correlates with myeloid differentiation. A transcriptionally active form of RBP-J (RBP-J–VP16) also increased granulocytic differentiation of 32D cells. Taken together, our results suggest that activated mNotch1 induces granulocytic differentiation of 32D cells and that this action of mNotch1 is mediated by RBP-J signalling. The RBP-J protein is commonly used by the Notch family members (Kato et al., 1996) and directly and uniquely targeted by Notch signalling (Oka et al., 1995; de la Pompa et al., 1997). Thus, although our observations do not exclude the possibility that additional, RBP-J- independent signalling is also involved in myeloid differentiation, RBP-J may function as a master protein in differentiative decisions in hematopoiesis.
Based on the evidence presented in this study, we propose that the primary effect of mNotch1 on 32D cells is to promote rather than to inhibit granulocytic differentiation. The Notch effect appears to be mediated via RBP-J activation. Our initial studies using multipotent hematopoietic progenitors suggest that activated mNotch1 also promotes differentiation of other myeloid lineages (T.Schroeder and U.Just, manuscript in preparation). Previous studies have indicated a role for Notch signalling in the regulation of T lymphoid versus B lymphoid lineage decisions as well as αβ versus γδ T cell fate and CD4 versus CD8 αβ T cell fate (Robey et al., 1996; Washburn et al., 1997; Pui et al., 1999). Bearing this and the widespread expression of Notch throughout hematopoietic progenitors in mind, it is tempting to speculate that Notch may be involved in a range of lineage decisions, possibly extending back to divergence of the myeloid and lymphoid compartments.
Materials and methods
Plasmid constructions
All mNotch1 amino acids (aa) positions used in this publication correspond to the DDBJ/EMBL/GenBank accession number Z11886.
Construction of OHT-inducible mN1IC retroviral vectors
An open reading frame (ORF) for an OHT-inducible, Flag-tagged intracellular mN1IC protein was generated. A N-terminal Flag-tag (Sigma Europe) was fused in-frame to aa 1751–2290 of the intracellular domain of mNotch1 (Hsieh et al., 1996; Höfelmayr et al., 1999) and the construct was sequenced for verification. Subsequently, aa 282–595 of the human estrogen receptor were C-terminally fused in-frame. A mutant form of the human estrogen receptor (ERT2), which is responsive to OHT, was used (Feil et al., 1997). The complete reading frame was cloned into the multiple cloning site of a derivative of the murine embryonal stem cell virus (MESV)-based retroviral vector, p50-X-neo (Grez et al., 1990; Laker et al., 1998) carrying the neomycin resistance gene (rneo). This plasmid was called rNERTneo. After transfection, transcription was driven by a 5′ MESV-LTR and neo was translated after splicing out the Notch-ERT2 gene. The empty neomycin resistance plasmid, containing only neo, was used for control transfections.
Construction of Notch deletion mutant and RBP-J–VP16 expression vectors
For the generation of different mN1IC–GFP and RBP-J–VP16–GFP constructs, the vector CMVeGFP (Z.McIvor, C.M.Heyworth, B.A.Johnson, S.Pearson, H.Fiegler, L.Hampson, T.M.Dexter and M.A.Cross, submitted) was used as a backbone. Briefly, it carries a human cytomegalovirus (hCMV) promotor, a multiple cloning site (MCS), an internal ribosomal entry site (IRES) and an eGFP ORF from 5′ to 3′. ORFs containing the following mN1IC parts were cloned into the MCS of CMVeGFP (name of the constructs and position of tags in parentheses): aa 1751–2531 (NICFL–GFP, myc-tag between aa 2293 and 2294, from plasmid pEFBos neo Notch; Kato et al., 1997); aa 1804–2531 (NICFLΔRAM–GFP, myc-tag between aa 2293 and 2294); aa 1751–2293 (NICΔOP–GFP, N-terminal Flag-tag); aa 1751–2183 (NICΔOPΔC–GFP, N-terminal Flag-tag); aa 1810–2183 (NICΔOPΔCMilner–GFP, six N-terminal myc-tags; Milner et al., 1996); aa 1804–2183 (NICΔOP ΔCΔRAM–GFP). RBP-J–VP16–GFP was constructed by cloning a PCR product containing a RBP-J–VP16 ORF without poly(A) signal (Waltzer et al., 1995) into the CMVeGFP. tTa–GFP was constructed by cloning the ORF for the tetracycline-dependent transactivator from the plasmid pUHD15–1 (Gossen and Bujard, 1992) into CMVeGFP. Functionality of the VP16 transactivation domain in the tTa protein was confirmed by co-transfection of tTa–GFP and the reporter plasmid pUHC13-3 (Gossen and Bujard, 1992) into 32D cells. In the absence of tetracycline, tTa induced high expression of luciferase in 32D cells.
Cell culture
32D cells were maintained in Iscove’s modified Dulbecco’s medium (IMDM) supplemented with 10% fetal calf serum (FCS) and mouse IL-3 conditioned medium (mIL-3 CM; Karasuyama and Melchers, 1988) at a concentration that stimulated optimal cell growth (which corresponds to 150 U rIL-3 per ml). Cells were kept in a density between 5 × 104 and 106 cells per ml and were regularly checked to be free of mycoplasma contamination using a Mycoplasma PCR Elisa Kit (Roche, Germany). Cells were carefully controlled for normal growth rates, factor dependency and differentiation (Valtieri et al., 1987). All experiments shown were conducted with recombinant mouse IL-3 (Roche, Germany). For activation of the OHT-inducible mN1IC 32D cells, 4-hydroxy-tamoxifen (RBI, USA) was added to the medium at a final concentration of 50 nM.
Differentiation of 32D cells was induced by washing the cells once in IMDM containing 10% FCS and plating 2 × 105 cells per ml in IMDM containing 10% FCS and 1000 U/ml hG-CSF (Neupogen, Amgen, USA). Aliquots were removed at the time points indicated for analysis. To ensure optimal growth and differentiation conditions, the cells were split to constant density and fed with fresh differentiation medium every fourth day. Viable cells were counted by Trypan blue dye exclusion. Differentiation of 32D cells was monitored by morphological scoring of May–Grünwald–Giemsa stained cytospins. The cell types were classified according to the following criteria. Undifferentiated 32D cells (blasts): scant basophilic (dark blue) cytoplasma and a single large relatively round nucleus with fine, dense chromatin structure containing several nucleoli; early granulocytes (promyelocytes and myelocytes): azurophilic or neutrophilic granularity, ring-shaped or indented nucleus, few or no nucleoli, coarser nuclear chromatin and light-end, acidophilic cytoplasma with increased cytoplasma/nucleus ratio; late granulocytes (metamyelocytes and neutrophils): banded or segmented nucleus with coarse, clumped chromatin structure, light and acidophilic cytoplasma and increased cytoplasma/nucleus ratio. Considerable care was taken to validate accurate differential counts. All differential counts were done on 100–200 cells in a blinded fashion by T.Schroeder and U.Just.
For assessment of colony forming cell capacity (clonogenicity), aliquots of the rneo or rNERTneo 32D cells were taken after 4 days of culture in the presence or absence of OHT, and plated in triplicates at various cell densities (30, 100, 300 and 1000 cells/ml) in culture medium (IMDM, 10% FCS) containing 10% mIL-3 CM (which corresponds to 150 U rIL-3 per ml) and 0.3% agar, in the presence or absence of OHT (triplicate wells in 6-well plates). Colonies (>50 cells) were counted after 7 days.
Virus-producing, Ltk– and JT cell lines were kept in IMDM supplemented with 10% FCS.
For co-culture experiments of Ltk– or JT cells with 32D cells, Ltk– or JT cells were seeded in 6-well plates at a density such that they would be 50% confluent the next day. After 12 h, 10532D cells were seeded onto the Ltk– or JT cells in differentiation medium. Every 3 days the 32D cells were transferred to freshly prepared Ltk– or JT cell layers.
Transient transfections, luciferase assays and reporter plasmids
32D cells were transfected by electroporation using a Bio-Rad Gene PulserII at 320 V, 1050 µFa and infinite resistance. Cells (4 × 106) were transfected in 400 µl of normal growth medium in Bio-Rad 4-mm cuvettes, respectively. Transfections for GFP sorting assays were done using between 20 and 50 µg of DNA. For RBP-J-dependent reporter assays, 32D cells were transfected in duplicates with 10 µg of the different Notch constructs and 5 µg reporter plasmid pGa981-6, as described above. Reporter plasmid pGa981-6 contains 12 RBP-J binding sites in front of the minimal β-globin promotor driving the expression of a luciferase gene (Strobl et al., 1997). Cells were cultured in the presence or absence of 50 nM OHT and harvested after 24 h in phosphate-buffered saline (PBS) containing 1% Triton X-100 and 1 mM dithiothreitol. Cells were spun for 10 min at 20 000 g at 4°C and luciferase activity of the supernatants was measured using standard protocols. Transfection efficiency in different clones was measured by parallel, separate transfection of CMV-Luc, a plasmid carrying a luciferase gene under the control of a CMV promotor and enhancer. To control the specificity of the RBP-J transactivation, reporter transfections using plasmid pGa50-7 were done in parallel. pGa50-7 contains a luciferase gene under the control of the minimal β-globin promotor (Strobl et al., 1997). None of the Notch constructs tested influenced the luciferase expression from this plasmid.
Stable transductions and selection procedures
On day 0, 32D cells were electroporated in triplicate with 10 µg of the plasmid rNERTneo or the control plasmid rneo, respectively, and flushed into 5 ml growth medium. On day 2, selection was started by addition of 0.5 mg/ml G418 (Gibco, Germany). On day 8, the G418 concentration was increased to 1 mg/ml, and was further increased to 1.5 mg/ml on day 13. Eighteen days after transfection, all cells were resistant to G418 and were frozen. After re-thawing, single cell lines were generated by cloning in soft agar. One hundred cells were plated per 6-well plate in 3 ml growth medium containing 1.5 mg/ml G418 and 0.33% Bacto agar (Difco, Germany). After 9 days, single colonies were picked, transferred to liquid culture, expanded for 11 days and single cell lines were frozen. From the transfection to freezing of single cell lines, cells were in culture for 38 days. Viral supernatants were harvested from the producer cell lines for LXSN-MT and LXSN-MT-mNotch1-ICΔOP (Milner et al., 1996) using standard procedures. 32D cells were infected by incubation with viral supernatant for 12 h in the presence of IL-3. Cells were selected and cloned as described above and analysed 37 days after infection.
FLNotch32D were generated by transfection with the plasmid pEFBos-neo-Notch (Kato et al., 1997). As controls, 32D cells were transfected with the control plasmid pEFBos-neo (neo32D). Cells were transfected by electroporation and geneticin-resistant clones were selected and cloned as described above.
Western blot analysis
Cells were lysed in PBS containing 10 mM EDTA, 1% Triton X-100, sodium fluoride (50 mM), vanadate (1 mM), aprotinin (9.5 µg/ml), phenylmethylsulfonyl fluoride (PMSF, 1 mM), antipain (2 µg/ml), leupeptin (2 µg/ml), pepstatinA (2 µg/ml) and chymostatin (2 µg/ml) (Sigma Europe). Cell extracts were collected and centrifuged at 20 000 g for 10 min at 4°C. For western blot analysis, protein extracts of 4 × 105 cells were electrophoresed by polyacrylamide gel electrophoresis (PAGE) and blotted to polyvinylidene fluoride membranes (Millipore, Germany) using standard protocols. Rainbow marker (10 µg) (Amersham, Europe) was used as a size standard. The NERT protein was detected with a monoclonal murine antibody directed against the human estrogen receptor α (#sc-8002, Santa Cruz, USA). Tagged Notch proteins were detected with antibodies directed against Flag-tag (#F3165, Sigma, Europe) or against myc-tag (#sc-40, Santa Cruz Biotechnology, USA), respectively. HA-tagged rJagged1 in JT cells was detected with an antibody against the HA-tag (#3808–1, Clontech, USA). Full-length mNotch1 was detected with a monoclonal antibody against the intracellular domain of mNotch1. The detected proteins were visualized by the enhanced chemiluminescence system (Amersham, Europe).
FACS analysis
Fluorescein isothiocyanate (FITC)-conjugated monoclonal antibodies directed against CD11b (Mac-1) and Gr-1 were used to analyse granulocytic differentiation of 32D cells. Cells were harvested by centrifugation and resuspended in PBS containing 3% FCS. Fc-Block (#01241D, Pharmingen, Europe) was added at a dilution of 1:100 for 5 min at room temperature. Subsequently, a FITC-labelled antibody directed against mCD11b (#01714D, Pharmingen Europe), or a FITC-labelled antibody directed against Gr-1 (#01214A, Pharmingen, Europe), or a FITC-labelled isotype-matched control antibody (#11184C, Pharmingen Europe) was added at a dilution of 1:100. After an incubation for 45 min at room temperature in the dark, cells were washed twice and resuspended in PBS containing 1% FCS and 1 µg/ml propidium iodide. FACS analysis was done with a Becton Dickinson FACScan machine and Cell Quest software, using standard procedures.
FACS sorting
Two days after transfection, cells were harvested by centrifugation and resuspended in PBS containing 3% FCS, 2 µg/ml propidium iodide and 25 µg/ml DNase (Roche, Germany). Clumps of cells were removed by filtering through a nylon mesh and living, GFP-positive cells were sorted with a Becton Dickinson FACS Star Plus machine, using standard procedures. Untransfected cells were used to set gates for GFP-negative cells.
Immunofluorescent staining
32D clones were cultured in IL-3 in the presence or absence of OHT for 12 h. Cytospins were prepared, air dried and fixed in methanol for 7 min at room temperature. The fixed cells were incubated with 10% FCS in PBS for 30 min at room temperature and then incubated with the primary antibody for 1 h at 37°C. A monoclonal murine antibody against the human estrogen receptor α (#sc-8002, Santa Cruz, USA) was used at a dilution of 1:100 in 1% bovine serum albumin (BSA) in PBS. After three washes in 1% BSA in PBS, the cells were incubated with the secondary antibody for 1 h at 37°C. A Cy3-conjugated goat anti-mouse IgG antibody (Becton Dickinson, USA) was used at a dilution of 1:1000 in 1% BSA in PBS. Subsequently, the cells were washed three times in PBS and mounted in fluorescent mounting medium (DAKO, Germany). Microphotographic slides were taken with an Axiophot microscope (Zeiss, Germany), using identical exposure times for all immunofluorescent pictures. Slides were scanned with a Minolta Sprint Scan 35 slide scanner using identical settings for all slides and compiled to one figure using the Microsoft Powerpoint software.
Statistical analysis
Statistical differences were assessed using the Student’s t-test for paired data.
Acknowledgments
Acknowledgements
We are very grateful to Dr M.Brielmeier, Dr M.Cross, Prof. P.Chambon, Prof. T.Honjo, Dr Z.McIvor, Dr L.Milner, Dr C.Stocking, Dr L.Strobl, Dr G.Weinmaster and Dr U.Zimber-Strobl for providing reagents. We thank Dr J.Ellwart for FACS sorting and Prof. G.Bornkamm, Dr M.Cross and Prof. T.M.Dexter for helpful suggestions and critical reading of the manuscript. This work was supported by the Deutsche Forschungsgemeinschaft (DFG Research Group ‘Regulatory Protein Networks’ WI 319/11-3 project 9 to U.J.). This report represents a part of the doctoral thesis by T.S.
References
- Artavanis-Tsakonas S., Matsuno,K. and Fortini,M. (1995) Notch signalling. Science, 268, 225–232. [DOI] [PubMed] [Google Scholar]
- Bettenhausen B., Hrabe de Angelis,M., Simon,D., Guenet,J. and Gossler,A. (1995) Transient and restricted expression during mouse embryogenesis of Dll1, a murine gene closely related to Drosophila Delta. Development, 121, 2407–2418. [DOI] [PubMed] [Google Scholar]
- Bigas A., Martin,D. and Milner,L. (1998) Notch1 and Notch2 inhibit myeloid differentiation in response to different cytokines. Mol. Cell. Biol., 18, 2324–2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlesso N., Aster,J., Sklar,J. and Scadden,D. (1999) Notch1-induced delay of human haemopoietic progenitor cell differentiation is associated with altered cell cycle kinetics. Blood, 93, 838–848. [PubMed] [Google Scholar]
- Deftos M., He,Y.-W., Ojala,E. and Bevan,M. (1998) Correlating Notch signaling with thymocyte maturation. Immunity, 9, 777–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Amo F., Smith,D., Swiatek,P., Gendron-Maguire,M., Greenspan,R., McMahon,A. and Gridley,T. (1992) Expression pattern of Motch, a mouse homolog of Drosophila Notch, suggests an important role in early postimplantation mouse development. Development, 115, 737–744. [DOI] [PubMed] [Google Scholar]
- Del Amo F., Gendron,M., Swiatek,P., Jenkins,N., Copeland,N. and Gridley,T. (1993) Cloning, analysis, and chromosomal localization of Notch-1, a mouse homolog of Drosophila Notch. Genomics, 15, 259–264. [DOI] [PubMed] [Google Scholar]
- de la Pompa J. et al. (1997) Conservation of the Notch signalling pathway in mammalian neurogenesis. Development, 124, 1139–1148. [DOI] [PubMed] [Google Scholar]
- Dunwoodie S., Henrique,D., Harrison,S. and Beddington,S. (1997) Mouse Dll3: a novel divergent Delta gene which may complement the function of other Delta homologues during early pattern formation in the mouse embryo. Development, 124, 3065–3076. [DOI] [PubMed] [Google Scholar]
- Ellisen L., Bird,J., West,D., Soreng,A., Reynolds,T., Smith,S. and Sklar,J. (1991) TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell, 66, 649–661. [DOI] [PubMed] [Google Scholar]
- Feil R., Wagner,J., Metzger,D. and Chambon,P. (1997) Regulation of Cre recombinase activity by mutated estrogen receptor ligand-binding domains. Biochem. Biophys. Res. Commun., 237, 752–757. [DOI] [PubMed] [Google Scholar]
- Gossen M. and Bujard,H. (1992) Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl Acad. Sci. USA, 89, 5547–5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grez M., Akgün,E., Hilberg,F. and Ostertag,W. (1990) Embryonic stem cell virus, a recombinant murine retrovirus with expression in embryonic stem cells. Proc. Natl Acad. Sci. USA, 87, 9202–9206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han W., Ye,Q. and Moore,M. (2000) A soluble form of human Delta-like-1 inhibits differentiation of hematopoietic progenitor cells. Blood, 95, 1616–1625. [PubMed] [Google Scholar]
- Höfelmayr H., Strobl,L., Stein,C., Laux,G., Marschall,G., Bornkamm,G. and Zimber-Strobl,U. (1999) Activated mouse Notch1 transactivates Epstein–Barr virus nuclear antigen 2-regulated viral promoters. J. Virol., 73, 2770–2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh J., Henkel,T., Salmon,P., Robey,E., Peterson,M. and Hayward,S. (1996) Truncated mammalian Notch1 activates CBF1/RBPJk-repressed genes by a mechanism resembling that of Epstein–Barr virus EBNA2. Mol. Cell. Biol., 16, 952–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hukriede N., Gu,Y. and Fleming,R. (1997) A dominant-negative form of Serrate acts as a general antagonist of Notch activation. Development, 124, 3427–3437. [DOI] [PubMed] [Google Scholar]
- Jarriault S., Brou,C., Logeat,F., Schroeter,E., Kopan,R. and Israel,A. (1995) Signalling downstream of activated mammalian Notch. Nature, 377, 355–358. [DOI] [PubMed] [Google Scholar]
- Jones P., May,G., Healy,L., Brown,J., Hoyne,G., Delassus,S. and Enver,T. (1998) Stromal expression of Jagged 1 promotes colony formation by fetal haemopoietic progenitor cells. Blood, 92, 1505–1511. [PubMed] [Google Scholar]
- Karasuyama H. and Melchers,F. (1988) Establishment of mouse cell lines which constitutively secrete large quantities of interleukin 2, 3, 4 or 5 using cDNA expression vectors. Eur. J. Immunol., 18, 97–104. [DOI] [PubMed] [Google Scholar]
- Kato H., Sakai,T., Tamura,K., Minoguchi,S., Shirayoshi,Y., Hamada,Y., Tsujimoto,Y. and Honjo,T. (1996) Functional conservation of mouse Notch receptor family members. FEBS Lett., 395, 221–224. [DOI] [PubMed] [Google Scholar]
- Kato H., Taniguchi,Y., Kurooka,H., Minoguchi,S., Sakai,T., Nomura-Okazaki,S., Tamura,K. and Honjo,T. (1997) Involvement of RBP-J in biological functions of mouse Notch1 and its derivatives. Development, 124, 4133–4141. [DOI] [PubMed] [Google Scholar]
- Kopan R. and Weintraub,H. (1993) Mouse notch: expression in hair follicles correlates with cell fate determination. J. Cell Biol., 121, 631–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopan R., Nye,J. and Weintraub,H. (1994) The intracellular domain of mouse Notch: a constitutively activated repressor of myogenesis directed at the basic helix–loop–helix region of MyoD. Development, 120, 2385–2396. [DOI] [PubMed] [Google Scholar]
- Kopan R., Schroeter,E., Weintraub,H. and Nye,J. (1996) Signal transduction by activated mNotch: importance of proteolytic processing and its regulation by the extracellular domain. Proc. Natl Acad. Sci. USA, 93, 1683–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurooka H., Kuroda,K. and Honjo,T. (1998) Roles of the ankyrin repeats and C-terminal region of the mouse Notch1 intracellular region. Nucleic Acids Res., 26, 5448–5455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laker C., Meyer,J., Schopen,A., Friel,J., Heberlein,C., Ostertag,W. and Stocking,C. (1998) Host cis-mediated extinction of a retrovirus permissive for expression in embryonal stem cells during differentiation. J. Virol., 72, 339–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lardelli M. and Lendahl,U. (1993) Motch A and motch B—two mouse Notch homologues coexpressed in a wide variety of tissues. Exp. Cell Res., 204, 364–372. [DOI] [PubMed] [Google Scholar]
- Lardelli M., Dahlstrand,J. and Lendahl,U. (1994) The novel Notch homologue mouse Notch 3 lacks specific epidermal growth factor repeats and is expressed in proliferating neuroepithelium. Mech. Dev., 46, 123–136. [DOI] [PubMed] [Google Scholar]
- Li L. et al. (1998) The human homolog of rat Jagged1 expressed by marrow stroma inhibits differentiation of 32D cells through interaction with Notch1. Immunity, 8, 43–55. [DOI] [PubMed] [Google Scholar]
- Lindsell C., Shawber,C., Boulter,J. and Weinmaster,G. (1995) Jagged: a mammalian ligand that activates Notch1. Cell, 80, 909–917. [DOI] [PubMed] [Google Scholar]
- Migliaccio G., Migliaccio,A., Kreider,B., Rovera,G. and Adamson,J. (1989) Selection of lineage-restricted cell lines immortalized at different stages of haemopoietic differentiation from the murine cell line 32D. J. Cell Biol., 109, 833–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milner L. and Bigas,A. (1999) Notch as a mediator of cell fate determination in haemopoiesis: evidence and speculation. Blood, 93, 2431–2448. [PubMed] [Google Scholar]
- Milner L., Kopan,R., Martin,D. and Bernstein,I. (1994) A human homologue of the Drosophila developmental gene, Notch, is expressed in CD34+ haemopoietic precursors. Blood, 83, 2057–2062. [PubMed] [Google Scholar]
- Milner L., Bigas,A., Kopan,R., Brashem-Stein,C., Bernstein,I. and Martin,D. (1996) Inhibition of granulocytic differentiation by mNotch1. Proc. Natl Acad. Sci. USA, 93, 13014–13019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nofziger D., Miyamoto,A., Lyons,K. and Weinmaster,G. (1999) Notch signaling imposes two distinct blocks in the differentiation of C2C12 myoblasts. Development, 126, 1689–1702. [DOI] [PubMed] [Google Scholar]
- Nye J., Kopan,R. and Axel,R. (1994) An activated Notch suppresses neurogenesis and myogenesis but not gliogenesis in mammalian cells. Development, 120, 2421–2430. [DOI] [PubMed] [Google Scholar]
- Oka C. et al. (1995) Disruption of the mouse RBP-J κ gene results in early embryonic death. Development, 121, 3291–3301. [DOI] [PubMed] [Google Scholar]
- Pui J. et al. (1999) Notch1 expression in early lymphopoiesis influences B versus T lineage determination. Immunity, 11, 299–308. [DOI] [PubMed] [Google Scholar]
- Radtke F., Wilson,A., Stark,G., Bauer,M., van Meerwijk,J., MacDonald,H. and Aguet,M. (1999) Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity, 10, 547–558. [DOI] [PubMed] [Google Scholar]
- Reaume A., Conlon,R., Zirngibl,R., Yamaguchi,T. and Rossant,J. (1992) Expression analysis of a Notch homologue in the mouse embryo. Dev. Biol., 154, 377–387. [DOI] [PubMed] [Google Scholar]
- Rebay I., Fleming,R., Fehon,R., Cherbas,L., Cherbas,P. and Artavanis-Tsakonas,S. (1991) Specific EGF repeats of Notch mediate interactions with Delta and Serrate: implications for Notch as a multifunctional receptor. Cell, 67, 687–699. [DOI] [PubMed] [Google Scholar]
- Robey E., Chang,D., Itano,A., Cado,D., Alexander,H., Lans,D., Weinmaster,G. and Salmon,B. (1996) An activated form of Notch influences the choice between CD4 and CD8 T cell lineages. Cell, 87, 483–492. [DOI] [PubMed] [Google Scholar]
- Schroeter E., Kisslinger,J. and Kopan,R. (1998) Notch-1 signaling requires ligand-induced proteolytic release of intracellular domain. Nature, 393, 382–386. [DOI] [PubMed] [Google Scholar]
- Shawber C., Boulter,J., Lindsell,C. and Weinmaster,G. (1996a) Jagged2: a serrate-like gene expressed during rat embryogenesis. Dev. Biol., 180, 370–376. [DOI] [PubMed] [Google Scholar]
- Shawber C., Nofziger,D., Hsieh,J., Lindsell,C., Bogler,O., Hayward,D. and Weinmaster,G. (1996b) Notch signaling inhibits muscle cell differentiation through a CBF1-independent pathway. Development, 122, 3765–3773. [DOI] [PubMed] [Google Scholar]
- Shelly L., Fuchs,C. and Miele,L. (1999) Notch-1 inhibits apoptosis in murine erythroleukemia cells and is necessary for differentiation induced by hybrid polar compounds. J. Cell. Biochem., 73, 164–175. [DOI] [PubMed] [Google Scholar]
- Simpson P. (1995) The Notch connection. Nature, 375, 736–737. [DOI] [PubMed] [Google Scholar]
- Strobl L., Höfelmayr,H., Stein,C., Marschall,G., Brielmeier,M., Laux,G., Bornkamm,G. and Zimber-Strobl,U. (1997) Both Epstein–Barr viral nuclear antigen 2 (EBNA2) and activated Notch1 transactivate genes by interacting with the cellular protein RBP-J κ. Immunobiology, 198, 299–306. [DOI] [PubMed] [Google Scholar]
- Sun X. and Artavanis-Tsakonas,S. (1997) Secreted forms of DELTA and SERRATE define antagonists of Notch signaling in Drosophila. Development, 124, 3439–3448. [DOI] [PubMed] [Google Scholar]
- Tamura K., Taniguchi,Y., Minoguchi,S., Sakai,T., Tun,T., Furukawa,T. and Honjo,T. (1995) Physical interaction between a novel domain of the receptor Notch and the transcription factor RBP-J κ/Su (H). Curr. Biol., 5, 1416–1423. [DOI] [PubMed] [Google Scholar]
- Tax F., Yeargers,J. and Thomas,J. (1994) Sequence of C.elegans Lag-2 reveals a cell signaling domain shared with Delta and Serrate of Drosophila. Nature, 368, 150–154. [DOI] [PubMed] [Google Scholar]
- Uyttendaele H., Marazzi,G., Wu,G., Yan,Q., Sassoon,D. and Kitajewski,J. (1996) Notch/int-3, a mammary proto-oncogene, is an endothelial cell-specific mammalian Notch gene. Development, 122, 2251–2259. [DOI] [PubMed] [Google Scholar]
- Valtieri M., Tweardy,D., Caracciolo,D., Johnson,K., Mavilio,F., Altmann,S., Santoli,D. and Rovera,G. (1987) Cytokine-dependent granulocytic differentiation: regulation of proliferative and differentiative responses in a murine progenitor cell line. J. Immunol., 138, 3829–3835. [PubMed] [Google Scholar]
- Varnum-Finney B. et al. (1998) The Notch ligand, Jagged-1, influences the development of primitive haemopoietic precursor cells. Blood, 91, 4084–4091. [PubMed] [Google Scholar]
- Walker L., Lynch,M., Silverman,S., Fraser,J., Boulter,J., Weinmaster,G. and Gasson,J. (1999) The Notch/Jagged pathway inhibits proliferation of human haemopoietic progenitors in vitro. Stem Cells, 17, 162–171. [DOI] [PubMed] [Google Scholar]
- Waltzer L., Bourillot,P., Sergeant,A. and Manet,E. (1995) RBP-J κ repression activity is mediated by a co-repressor and antagonized by the Epstein–Barr virus transcription factor EBNA2. Nucleic Acids Res., 23, 4939–4945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washburn T., Schweighoffer,E., Gridley,T., Chang,D., Fowlkes,B., Cado,D. and Robey,E. (1997) Notch activity influences the αβ versus γδ T cell lineage decision. Cell, 88, 833–843. [DOI] [PubMed] [Google Scholar]
- Weinmaster G. (1997) The ins and outs of notch signaling. Mol. Cell. Neurosci., 9, 91–102. [DOI] [PubMed] [Google Scholar]
- Weinmaster G., Roberts,V. and Lemke,G. (1991) A homolog of Drosophila Notch expressed during mammalian development. Development, 113, 199–205. [DOI] [PubMed] [Google Scholar]
- Weinmaster G., Roberts,V. and Lemke,G. (1992) Notch2: a second mammalian Notch gene. Development, 116, 931–941. [DOI] [PubMed] [Google Scholar]