Abstract
Yeast SIR2, the founding member of a conserved gene family, acts to modulate chromatin structure in three different contexts: silent (HM) mating-type loci, telomeres and rDNA. At HM loci and telomeres, Sir2p forms a complex with Sir3p and Sir4p. However, Sir2p’s role in rDNA silencing is Sir3/4 independent, requiring instead an essential nucleolar protein, Net1p. We describe two novel classes of SIR2 mutations specific to either HM/telomere or rDNA silencing. Despite their opposite effects, both classes of mutations cluster in the same two regions of Sir2p, each of which borders on a conserved core domain. A surprising number of these mutations are dominant. Several rDNA silencing mutants display a Sir2p nucleolar localization defect that correlates with reduced Net1p binding. Although the molecular defect in HM/telomere-specific mutants is unclear, they mimic an age-related phenotype where Sir3p and Sir4p relocalize to the nucleolus. Artificial targeting can circumvent the silencing defect in a subset of mutants from both classes. These results define distinct functional domains of Sir2p and provide evidence for additional Sir2p-interacting factors with locus-specific silencing functions.
Keywords: aging/nucleolus/silencing/Sir2/telomere
Introduction
Transcriptional silencing in yeast is a form of chromatin modification with general effects, not only on transcription, but also on DNA repair and recombination (reviewed in Stone and Pillus, 1998). This type of repression, analogous in several respects to heterochromatin in more complex eukaryotes, was first uncovered at silent (HM) mating-type loci, where it plays an important role in the mating behavior of haploid cells. Stable repression of mating-type genes requires four trans-acting silent information regulator (Sir) proteins (Rine and Herskowitz, 1987). Three of these Sir proteins (Sir2/3/4) form a complex required for a similar, although less stable, form of silencing observed at telomeres, known as telomere position effect (TPE) (Aparicio et al., 1991). A large body of genetic, biochemical and cell biological data support a two-step model for the establishment of silent chromatin in yeast (reviewed in Grunstein, 1998; Cockell and Gasser, 1999). In the first step, specific combinations of silencer- or telomere-binding proteins (e.g. Rap1, Abf1, ORC or Yku proteins) recruit the Sir2/3/4 complex to the chromosome. This Sir silencing complex then assembles along adjacent chromatin through a network of protein–protein interactions involving the Sir proteins themselves, and Sir3/4 contacts with the highly conserved N-terminal tails of histones H3 and H4.
Until very recently, relatively little was known about the molecular role of SIR2 in silencing, despite the fact that it is the founding member of a gene family in yeast that is highly conserved throughout evolution (Brachmann et al., 1995; Derbyshire et al., 1996). Yeast has four SIR2 homologs (HST1–4), whose functions may partially overlap with those of SIR2. SIR2 homologs in other yeasts, such as Kluyveromyces lactis (Chen and Clark-Walker, 1994) and Candida albicans (Perez-Martin et al., 1999) may also play a role in organizing chromatin structure. However, the existence of bacterial Sir2-like proteins, and cytoplasmic homologs in both human (Afshar and Murnane, 1999) and Leishmania major (Zemzoumi et al., 1998), suggests that the function of Sir2p homologs may not be restricted to chromatin.
These apparently disparate observations can be reconciled by recent studies pointing to an enzymatic function for the Sir2p family. Following up on studies of bacterial (Tsang and Escalante-Semerena, 1998) and human (Frye, 1999) SIR2 homologs, Tanny et al. (1999) presented evidence that yeast Sir2p can ribosylate histones in vitro, and that this activity might be important for its in vivo function. More recently, Imai et al. (2000) showed that Sir2p has, in addition, an NAD-dependent histone deacetylase activity. This latter observation is consistent with a previous report that silent chromatin is hypoacetylated, and that overexpression of SIR2 leads to global histone deacetylation (Braunstein et al., 1993). Taken together, these and other data suggest that individual Sir2 family members might be protein-modifying enzymes with very specific sets of targets.
Interestingly, Sir2p itself has multiple functions within the yeast nucleus. In fact, a large fraction of Sir2 protein is found within the nucleolus, physically associated with the tandem 9 kb rDNA repeats (Gotta et al., 1997). Significantly, sir2 mutants display increased recombination within the rDNA repeats (Gottlieb and Esposito, 1989), alterations in rDNA chromatin structure and loss of a form of silencing that is seen when pol II genes are placed within the rDNA array (Bryk et al., 1997; Fritze et al., 1997; Smith and Boeke, 1997). Unlike HM and telomeric silencing, rDNA silencing requires neither SIR3 nor SIR4, consistent with the observation that neither Sir3 nor Sir4 protein is normally found within the nucleolus (Gotta et al., 1997; Kennedy et al., 1997). Instead, mutation of SIR4 actually improves rDNA silencing (Smith et al., 1998). This fact, together with the observation that alterations in SIR2 gene dosage dramatically affect the level of rDNA silencing (Smith et al., 1998), suggests that a limiting pool of Sir2 protein is distributed between telomeres, HM loci and rDNA, in a manner regulated at least in part by Sir4p. The localization of Sir2p to the nucleolus, and its function there, requires an essential nucleolar protein, Net1p, with an additional (SIR2 independent) role in regulating the exit from mitosis (Shou et al., 1999; Straight et al., 1999; Visintin et al., 1999). Sir2p itself has other, unexpected functions within the nucleolus. It regulates life span in yeast, possibly by controlling the generation of extrachromosomal rDNA circles within the nucleolus (Kaeberlein et al., 1999), and is also required for a meiotic checkpoint response (San-Segundo and Roeder, 1999).
Here we address the question of how Sir2p can carry out two apparently distinct silencing functions, one involving the Sir2/3/4 complex at HM loci and telomeres, the other working through a SIR3/4-independent pathway in the nucleolus. Our approach was to search for mutations in SIR2 that would genetically separate its HM/telomeric silencing functions from its silencing activity at the rDNA. We describe the isolation and initial characterization of a collection of such locus-specific SIR2 mutants. Surprisingly, mutations of these two different types fall in either adjacent or overlapping regions that flank either side of the highly conserved ‘core’ domain. In addition, a remarkable number of these mutations are dominant or semi-dominant to wild type, in a locus-specific fashion. The molecular defects in these mutants have been explored by a series of protein localization, interaction and targeting experiments. The results emphasize the importance of the Sir2p–Net1p interaction in rDNA silencing and point to the existence of an as yet unidentified Sir2p interaction that is required to localize stably the Sir2/3/4 protein complex at telomeres and HM loci.
Results
Isolation of SIR2 mutants with locus-specific silencing defects
We used a polymerase chain reaction (PCR) mutagenesis procedure and plasmid ‘gap repair’ (Rothstein, 1991) to generate a library of SIR2 mutants directly in yeast cells. The host strain was designed so that three different silencing functions could be tested by simple plate assays. The screening procedure is summarized in Figure 1. From ∼5 × 104 yeast transformants tested, ∼70% were indistinguishable from wild type, whereas ∼30% behaved like SIR2 null mutants (i.e. loss of silencing at all three loci) and were not pursued further. Instead, we focused our attention on a small fraction of mutants (<1% of transformants) that showed a differential effect on silencing.
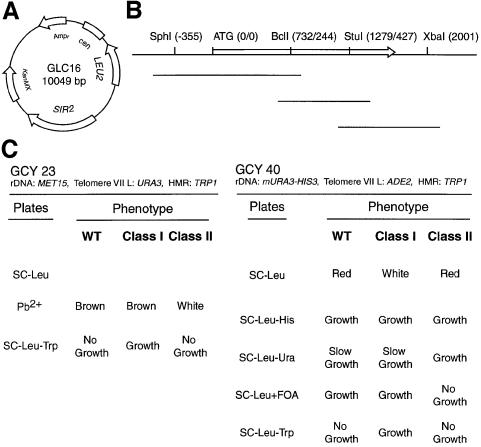
Fig. 1. PCR mutagenesis of SIR2 and screen for locus-specific mutants. (A) SIR2 plasmid GLC16 (CEN/LEU2/KanMX4 backbone) used both as a template for mutagenic PCR and as a backbone for transformation and ‘gap repair’. (B) Strategy for PCR-based mutagenesis of SIR2. Unique restriction sites (with nucleotide/amino acid positions given in parentheses) used to prepare three different linear plasmid molecules for ‘gap repair’ are indicated. The SIR2 open reading frame is indicated by the open arrow, and the location of the three different PCR products used for mutagenesis are shown by lines below. (C) Two strains (GCY23 and GCY40) used in the screen, their relevant reporter loci, and expected phenotypes for wild-type SIR2, class I and class II mutants.
Most locus-specific mutants fell into one of two different classes. Class I mutants were defective for both telomeric and HMR silencing but displayed normal rDNA silencing. Conversely, class II mutants had normal telomeric and HMR silencing but were defective in rDNA silencing. The silencing phenotypes of these mutants are shown in Figure 2 for a subset of the mutants. We also identified some alleles with a telomeric silencing defect but no loss of repression at either HMR or the rDNA. Given the fact that HMR silencing is stronger than TPE, these mutants might represent weak alleles of the class I type and have not been pursued further. In addition, we found a small number of mutants that were defective in both TPE and rDNA silencing, but normal at HMR. These might be weak alleles of SIR2 with a general defect, and therefore have not been characterized further. Finally, it is worth noting that in an independent screen we identified ∼2.0 × 104 mutants apparently defective in HMR silencing. None of these mutants displayed any detectable TPE, suggesting a tight association of these two forms of silencing with respect to SIR2 function.
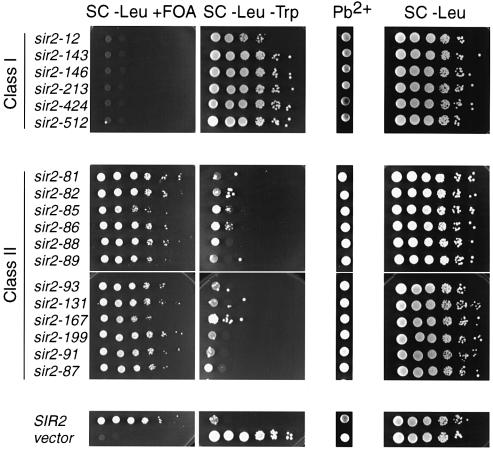
Fig. 2. Phenotypes of class I and class II mutants. Reporter strain GCY23 was transformed with plasmids (GLC16 or derivatives) containing the SIR2 mutant alleles indicated, SIR2 wild type or no SIR2 insert (vector). Ten-fold serial dilutions of cells were plated on SC-Leu+FOA (to measure TPE), SC-Leu-Trp (HMR silencing), Pb2+/G418 (rDNA silencing) and SC-Leu (total number of plasmid-containing cells).
Class II (loss of rDNA silencing) mutants were more common than class I mutants. Because rDNA silencing is extremely sensitive to SIR2 gene dosage (Smith et al., 1998) we were concerned that many of these mutants might result from even small effects on Sir2p stability in vivo. To test this possibility, epitope-tagged versions of the SIR2 mutations were constructed and their steady-state protein levels were measured by Western blotting. As might be expected, some mutants showed a decreased level of protein relative to wild type (data not shown). However, overexpression of class II mutant proteins does not restore wild-type function (data not shown). Moreover, the fact that most of these class II mutants are dominant to wild type (see below) argues against a simple protein level defect.
In order to determine the residues involved in locus-specific SIR2 function, a subset of class I and class II mutants was characterized by DNA sequencing (Figure 3). The results of this analysis are striking in several respects. To begin with, both class I and class II mutants are highly clustered within the same two regions of the protein, one relatively N-terminal, the other near the C-terminus. The C-terminal mutations of class I and class II types are actually overlapping (amino acids 485–502 and 437–511, respectively), and the N-terminal mutations of the two classes are immediately adjacent to each other on the linear sequence (223–235 and 154–220, respectively). There is a notable absence of locus-specific mutations within a large stretch of the most highly conserved part of the protein, termed the ‘core’ domain (Brachmann et al., 1995; Sherman et al., 1999).
Fig. 3. Mutations responsible for class I and class II phenotypes. (A) Graphical representation of Sir2p, depicting location of class I and class II mutations. The gray box indicates the highly conserved core domain of Sir2p (amino acids 254–498) and the mutated residues conferring class I and class II phenotypes are represented by black bars. (B) A list of mutations, divided according to class. Mutations in bold letters have been identified more than once, whereas those in parentheses do not contribute to the silencing phenotype of the mutant and have no effect on silencing when recloned by themselves. These changes are shown because some subsequent assays were performed with alleles containing these mutations. We observed an average of 2.8 mutations per clone for class I mutants, and 2 mutations per clone for class II mutants, including silent mutations. Because of the relative abundance of class II mutants, only those containing a minimum number of mutations were selected for subsequent analysis. The following pairs of mutants behaved similarly in all the subsequent assays, and thus only one mutant per group is shown in subsequent figures: sir2-12/sir2-423, sir2-146/sir2-472, sir2-213/sir2-404 and sir2-366/sir2-408/sir2-424. A summary of mutant phenotypes is shown in Supplementary Tables I and II, available at The EMBO Journal Online.
Certain amino acid changes were recurrent amongst the class I mutations. Interestingly, four of these were changes to arginine (R), whereas the two others each involved changes to glycine (G). The fact that nine of 11 sequenced class I alleles share at least one change in common with another allele suggests that examination of further alleles is unlikely to reveal many new mutations with this phenotype. In contrast, all but one of 12 class II single mutations are unique, suggesting that other single amino acid changes will give this phenotype. Furthermore, unlike the case for class I mutants, we failed to identify any common amino acid changes amongst class II mutants, and no changes to either glycine or arginine were observed. We also identified two extreme C-terminal truncations with a class II phenotype (sir2-87 and -91), both of which fall somewhat beyond the cluster of class II (or class I) C-terminal point mutations. Because these two truncation mutations map very close to each other, only the larger of the two (sir2-87) has been studied in all subsequent assays.
SIR2 class I and class II mutants are typically dominant to wild type
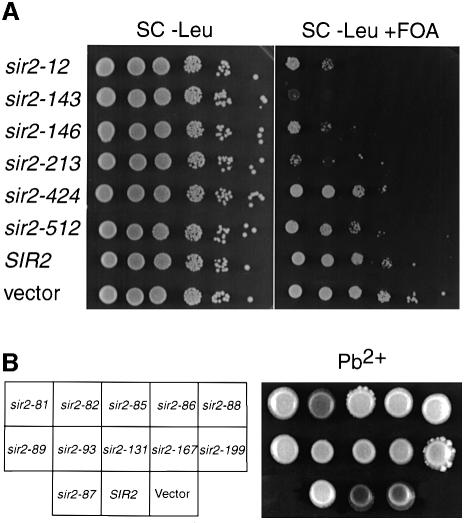
We first performed a simple genetic test to determine whether the class I or class II mutants are recessive or dominant to wild type. Mutant alleles were introduced on CEN/LEU2 plasmids in a SIR2 strain (GCY66). Surprisingly, almost all class II mutants caused a defect in rDNA silencing in the presence of the wild-type SIR2 gene (Figure 4B). Similarly, four of six class I mutants were either partially or completely defective in TPE (Figure 4A). As might be expected, class I mutants do not interfere with rDNA silencing, and class II mutants likewise have no effect on TPE in SIR2 cells (data not shown). Curiously, despite the strong dominant-negative effect of some class I mutants on TPE, none had any measurable effect on silencing of TRP1 at the HMR locus (data not shown).
Fig. 4. Dominance test of class I and class II mutations. The SIR2 wild-type strain GCY66 was transformed with plasmids (GLC16 or derivatives) containing either class I or class II mutations, SIR2 wild type or no insert (vector). (A) Six different class I mutations (as indicated) assayed for telomeric silencing (adh4::URA3-TelVIIL). (B) Class II mutations assayed for rDNA silencing using the colony-color marker MET15. Overnight cultures of cells transformed with the plasmids indicated were spotted onto Pb2+/G418 plates.
Protein localization effects in sir2 locus-specific mutants
To ask whether the mutant phenotypes are correlated with specific changes in sub-nuclear protein localization, we examined cells containing Sir2 wild-type or mutant proteins fused to the green fluorescent protein (GFP). Sir2p–GFP fully complements a sir2 null mutation for HMR silencing and provides near wild-type silencing function at telomeres and rDNA (see Supplementary material, available at The EMBO Journal Online). Furthermore, the Sir2p–GFP nuclear localization pattern (Figure 5), a crescent-shaped nucleolar structure and several much weaker telomeric foci, is very similar to that previously reported for Sir2p by indirect immunofluorescence (IF) (Gotta et al., 1997).
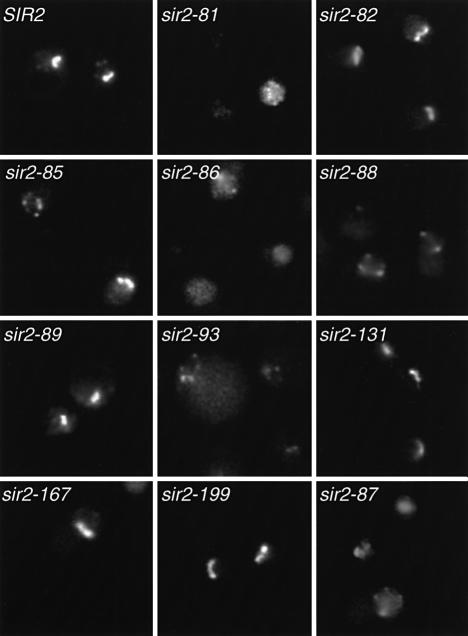
Fig. 5. Sub-nuclear localization of Sir2p and class II mutant proteins. A sir2Δ strain (GCY16) was transformed with a centromeric plasmid containing SIR2 or class II mutations (as indicated), in which the proteins are fused at their C-termini to GFP. Cultures were prepared and viewed by fluorescence microscopy as described in Materials and methods.
As expected, the Sir2p–GFP fluorescence patterns in all class I mutants examined are indistinguishable from wild type with respect to the bright nucleolar fluorescence (data not shown). In contrast, several class II mutants (sir2-81, -86, -88, -87 and -93) showed a clear loss of nucleolar Sir2p–GFP signal (Figure 5). Although these five class II mutants all lose Sir2p nucleolar localization, the redistribution of Sir2–GFP protein in these cells is not identical. Whereas sir2-81, -86 and -87 show general diffuse nuclear fluorescence with some brighter spots, sir2-88 and -93 have less diffuse staining and a few, more prominent telomeric foci. The sir2-85 mutant showed two distinct localization patterns: about half of the cells have normal nucleolar fluorescence, whereas the other half show a loss of nucleolar localization with weak foci remaining. Interestingly, the other class II mutants showed essentially normal Sir2p–GFP nucleolar fluorescence. These data indicate that the loss of rDNA silencing in half of the class II mutants can be explained, at least in part, by a failure of the mutant proteins to localize properly to the nucleolus. For the remainder of the class II mutants the loss of rDNA silencing may be due to a specific functional defect of the protein within the nucleolus.
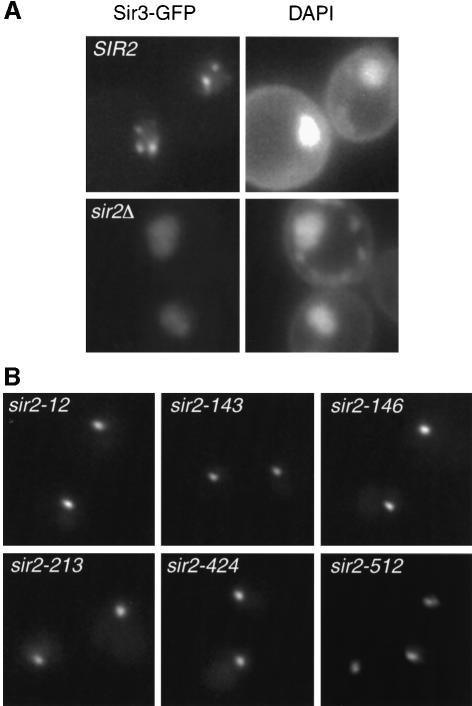
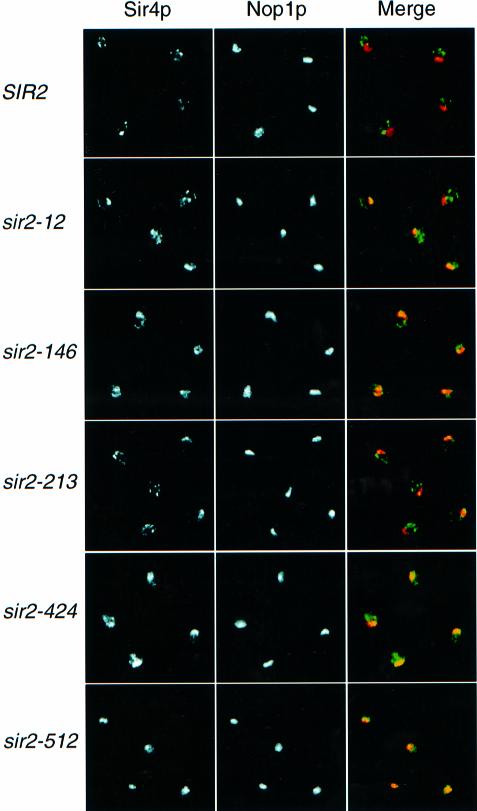
The punctate, presumably telomeric localization observed in Sir2p–GFP wild-type cells appeared to be affected in some class I mutants. However, loss of telomeric foci is difficult to assay since they are seen clearly in only ∼15% of wild-type cells. Consequently, we used a Sir3p–GFP fusion to monitor telomeric localization of the Sir protein complex in class I mutant cells. In SIR2 wild-type cells, Sir3p–GFP gives a clear punctate fluorescence pattern within the nucleus, in which 2–4 bright spots are typically observed above a low nuclear background (Figure 6A). This pattern presumably corresponds to telomere clusters (Gotta et al., 1996). In a sir2 null mutant, diffuse nuclear Sir3p–GFP fluorescence is seen (Figure 6A), consistent with a previous report using IF (Gotta et al., 1997). Interestingly, all class I mutants examined show a novel Sir3p–GFP localization phenotype in which a single, bright spot is observed within the nucleus (Figure 6B). Since it had been shown previously that Sir3p leaves telomeres and localizes to the nucleolus in a SIR2-dependent manner when SIR4 is mutated (Gotta et al., 1997), we wanted to test whether Sir3p localizes to the nucleolus in class I mutants. Using antibodies against Sir3p and Nop1p we found that Sir3p is mainly localized within the nucleolus in class I mutants, although some weak telomeric foci can sometimes be observed in sir2-12 and -213 strains (data not shown). To characterize further the integrity of the Sir complex at telomeres, we also examined Sir4p and Rap1p localization by IF. Interestingly, Sir4p also relocalizes to the nucleolus in all class I mutants tested, although not always to the same extent (Figure 7). Whereas in a sir2-512 strain Sir4p is almost exclusively concentrated at the nucleolus, it is still present at telomeres in sir2-12 and -213 mutants. Moreover, sir2-146 and -424 strains display a diffuse Sir4p nuclear staining in addition to nucleolar localization. Rap1p staining is also more diffuse in class I mutants (data not shown), consistent with the delocalization of the Sir complex from telomeres (Gotta et al., 1996).
Fig. 6. Determination of the integrity of the Sir complex at telomeres by Sir3p–GFP localization. A sir2Δ SIR3–GFP strain (GCY150) was transformed with LEU2 integrative plasmids coding for SIR2 or class I mutations, as indicated. (A) Sir3p–GFP delocalizes from telomeres in the absence of Sir2p. Comparison of GFP fluorescence in SIR2 and sir2Δ strains. DAPI (4′,6-diamidine-2-phenylindole) staining of DNA is shown on the right. (B) Sir3p–GFP shows a novel localization pattern in the presence of all class I mutations.
Fig. 7. Co-localization of Sir4p and Nop1p by indirect immunofluorescence in sir2 class I mutants. Cells (SIR2 and the five sir2 class I mutants indicated) were stained with both anti-Sir4p (detected by a DTAF (5-([4,6-dichlorotriazin-2-yl] amino)-fluorescein) conjugated antibody, green) and anti-Nop1p (detected by a Cy5-conjugated antibody, red). Independent signals from Sir4p and Nop1p are shown in the first two columns. The merge of these two images for each strain is shown on the right, where the coincidence of Sir4p and Nop1p signals is yellow.
Interactions with Sir4p and Net1p
Sir2p is known to interact with several other proteins, which are themselves essential either for HM and telomeric silencing, or for rDNA silencing. Sir2p interacts with Sir4p both in vitro and in vivo (Moazed and Johnson, 1996; Moazed et al., 1997; Strahl-Bolsinger et al., 1997; P.Moretti and D.Shore, unpublished results), and loss of Sir4p abolishes both TPE and HM silencing. Likewise, Sir2p interacts with Net1p, a nucleolar protein that is itself required for rDNA silencing (Shou et al., 1999; Straight et al., 1999). One might expect, therefore, that class I mutations could result from defects in the interaction with Sir4p, and class II mutants from an inability to interact properly with Net1p.
We have developed two-hybrid assays to measure both of these interactions in the context of class I and class II mutations. To test Sir2p–Sir4p interactions, Sir2p or Sir2p mutants were fused at amino acid 78 to the LexA protein, and a Gal4p activation domain (GAD)–Sir4p hybrid was constructed with the C-terminal half of Sir4p (amino acids 737–1358). As expected, none of the class II alleles was defective in the Sir4p interaction. Surprisingly, though, all of the class I mutants also appeared to interact normally with GAD–Sir4p (data not shown).
To measure the Sir2p–Net1p interaction, we found that the combination of LexA–Net1p(566–801) and GAD–Sir2p(1–562) gave the strongest signal, thus narrowing down the part of Net1p sufficient for Sir2p interaction to a 236 amino acid region. As expected from the nucleolar function of Net1p, none of the SIR2 class I mutant proteins was defective in interacting with LexA–Net1p. In contrast, three of the class II mutants (sir2-81, -86 and -87) were completely defective in this Net1p interaction (Table I). Interestingly, these three mutants display a complete loss of nucleolar localization for the corresponding Sir2p–GFP hybrid proteins (Figure 5). It should be noted, though, that three other mutants with a defect in Sir2p nucleolar localization (sir2-85, -88 and -93) interact normally with Net1p in this assay.
Table I. Two-hybrid analysis of GAD–Sir2p/LexA–Net1p interactions.
| Class I mutants | Class II mutants | ||
|---|---|---|---|
| Sir2-12 | + | Sir2-81 | – |
| Sir2-143 | + | Sir2-82 | + |
| Sir2-146 | + | Sir2-85 | + |
| Sir2-213 | + | Sir2-86 | – |
| Sir2-424 | + | Sir2-88 | + |
| Sir2-512 | + | Sir2-89 | + |
| Sir2-93 | + | ||
| Sir2-131 | + | ||
| Sir2-167 | + | ||
| Sir2-199 | + | ||
| Sir2-87 | – |
(+) Strong blue color detected after ∼2–4 h in β-galactosidase assay on nitrocellulose filters. (–) No signal detected after 24 h of incubation.
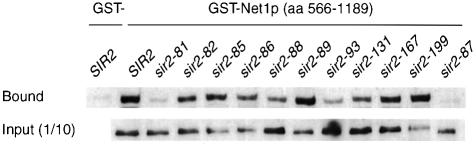
To confirm these two-hybrid results with Net1p, a glutathione S-transferase (GST) pull-down assay was performed using a GST–Net1(566–1189) hybrid protein and yeast whole-cell extracts made from strains expressing Myc-tagged versions of Sir2p wild-type or class II mutant proteins (Figure 8). The results of the two-hybrid and GST pull-down assays were qualitatively similar, except for the sir2-86 mutant, which fails to interact with LexA–Net1p in the two-hybrid assay but does still bind to GST–Net1p. Moreover, the sir2-88 and -93 mutants show a slight but reproducible decrease in Net1p interaction that was not detected in the two-hybrid assay. These differences could be due to the different fragment sizes of Net1p used in the LexA and GST hybrid assays, or to a different sensitivity of the two assays. Nonetheless, these data are consistent with the Sir2p–GFP localization results, and confirm the importance of Net1p in recruiting Sir2p to the nucleolus. Every mutant with a reduced Net1p interaction, whether detected in the two-hybrid or in the GST pull-down assay, has a loss of Sir2p nucleolar localization.
Fig. 8. In vitro interaction of wild-type and class II mutant proteins with Net1p. Binding of Myc epitope-tagged Sir2 proteins from whole-cell yeast extracts (wild type and class II mutants, as indicated) to GST–Net1(566–1189) protein bound to glutathione beads. Bound protein was detected by Western blotting using a monoclonal antibody against the Myc tag.
Targeting Sir2 mutant proteins to silencers, telomeres and rDNA
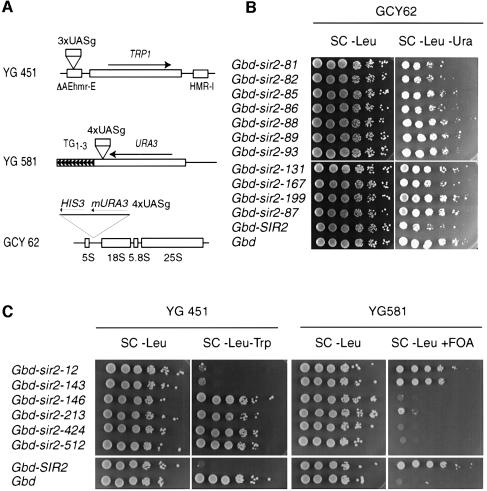
Sir2 protein does not appear to bind directly to DNA and thus is presumably localized to its sites of action by interactions with other proteins. To distinguish between a protein localization problem and a different functional defect, we fused both class I and class II mutant proteins to the Gal4p DNA-binding domain (Gbd) and targeted the hybrids to modified HM, telomeric and rDNA loci containing Gal4p DNA-binding sites (UASg) adjacent to suitable reporter genes (Buck and Shore, 1995; Marcand et al., 1996) (Figure 9A).
Fig. 9. Targeted silencing at telomeres, HMR and the rDNA locus. (A) Schematic representation of targeting silencing loci: HMR (YG451), adh4::URA3 on Chr. VII-L (YG581) or downstream of a 5S rRNA gene (GCY62). All of the targeting strains are also sir2Δ::kanMX4. (B) Effect of Gbd–Sir2p class II mutants (as indicated) targeted to the rDNA, with Gbd–Sir2p and Gbd alone for comparison. (C) Effect of Gbd–Sir2p class I mutants (as indicated) targeted to either a telomere or HMR. Gbd–Sir2p and Gbd alone serve as controls.
Upon targeting, only two class I mutants (sir2-12 and -143) were able to restore strong silencing at both HMR and the marked telomere (Figure 9C). As expected, all of the class I hybrids were proficient in targeted rDNA silencing (data not shown). Gbd–Sir2p hybrids of the class II type also displayed different phenotypes, in this case when targeted to a (4×UASg)-mURA3 reporter placed within the rDNA repeats (Figure 9B). Three mutant proteins (Sir2-81, -82 and -86) restored silencing to almost wild-type levels when targeted, whereas the others displayed little or no activity compared with the Gbd alone control, or with the strain lacking UASg sites (Figure 9B; data not shown). As expected, all of the class II hybrids were proficient in targeted telomeric and HM silencing (data not shown). It is interesting to compare these results with the in vivo localization data described above. Although the simplest model would predict that localization-defective mutants could be rescued by Gbd-mediated targeting to the rDNA, this is not what was observed. Of the five mutants completely defective in nucleolar localization, only two restored rDNA silencing upon targeting (sir2-81 and -86). This suggests that the four remaining mutants are not only defective in nucleolar localization, but are also unable to form an active silencing complex at the rDNA. It is also worth noting that the sir2-82 mutant, in which the mutant Sir2p appeared to be localized normally in the nucleolus, was rescued by targeting. These data indicate that Gbd-mediated targeting can rescue defects in some of the SIR2 mutants that are not apparent at the level of sub-nuclear localization of Sir2p–GFP.
Discussion
The genetic demonstration presented here that Sir2 protein has separable, locus-specific silencing functions is consistent with previous studies showing that the protein collaborates with different sets of factors in rDNA and HM/telomeric silencing. Therefore, mutations that disrupt a specialized Sir2p interaction, for example with Sir4p or Net1p, would be predicted to result in class I or class II phenotypes, respectively. Such mutations might be expected to identify independent interaction surfaces on Sir2p and to behave as recessive, loss-of-function mutations. The data presented here, however, reveal a more complex picture.
Evidence for a common recognition surface on Sir2p for both HM/telomeric- and rDNA-specific silencing factors
One striking feature of both classes of mutations is that they cluster in two distinct, short regions of the protein that are separated from each other by an ∼250 amino acid region that constitutes most of the highly conserved ‘core’ domain of the SIR2 family. Remarkably, the C-terminal clusters of both classes are overlapping, whereas the N-terminal mutations map immediately adjacent within an ∼80 amino acid stretch. We speculate that these two core-flanking regions form interaction surfaces for proteins (or protein complexes) involved in locus-specific silencing functions. One implication of this model is that a competition between rDNA and HM/telomeric sites for limiting Sir2p (Smith et al., 1998; Cockell et al., 2000) might be played out, at least in part, at the level of Sir2p-interacting proteins specific to one or another silencing function.
It is interesting to note that the collection of class I mutations is restricted to a relatively small number of residues, and the changes themselves are often to either arginine or glycine. In contrast, all class II mutations identified different residues, with no obvious pattern to either the wild-type or mutant amino acids. One possible explanation for these different properties is that Sir2p-interacting proteins important for HM/telomeric silencing rely on a relatively restricted set of critical contacts whose loss (replacement by glycine) or substitution by a bulky, positively charged residue (arginine) is necessary to abolish function. Protein–protein interactions important for rDNA silencing, on the other hand, might rely on a more disperse set of weaker interactions.
The identification of two distinct regions where both class I and class II mutants map raises the possibility that two different proteins (or complexes) interact with Sir2p at each of the two types of silent loci (HM/telomere and rDNA). However, the fact that mutations in the two different regions cannot be distinguished phenotypically might argue against this model. An alternative hypothesis would be that the two regions are located close to each other on the surface of the folded protein and interact with a single protein or complex of proteins at a given silencing locus. This idea is consistent with the identification of class II mutations in both regions that weaken Net1p binding. However, the dominance of both types of mutations is difficult to understand in terms of simple models that involve only one Sir2p-interacting factor at each silencing site (see below).
A corollary to the observed clustering of the locus-specific mutations is the absence of such alleles throughout most of the conserved ‘core’ region (amino acids 254–498). We suggest that this region performs a common, essential enzymatic function required for both rDNA and HM/telomeric silencing, either mono-ADP ribosylation of specific target proteins (Frye, 1999; Tanny et al., 1999), deacetylation (Imai et al., 2000) or both. Consistent with this idea, mutation of several different conserved motifs within the core domain abolishes all SIR2 silencing activity (Sherman et al., 1999). According to this model, most of the core domain either contains no information relevant to target protein specificity, or it is difficult to obtain mutations in this region that alter target specificity without simultaneously weakening or abolishing the protein’s essential enzymatic activity.
Because our screen asked for loss-of-function mutations, one might expect that they would be recessive to wild type. Instead, we found many dominant alleles in both classes, with respect to rDNA and telomeric silencing. Perhaps the fact that we have selected mutations that retain function for at least one site of action has vastly increased the likelihood that they will exert a dominant effect at another locus. Nonetheless, the prevalence of dominant mutations in our collection appears to be unusual, and may provide important clues regarding the mechanism of Sir2p action at rDNA and telomeres (see below). It is worth noting that other groups have recently generated dominant SIR2 mutations by more directed approaches. Sherman et al. (1999) have described a yeast/human Sir2 chimera in which the core of a human SIR2 homolog replaces the Sir2p core, which has a dominant-negative phenotype. Similarly, a core domain point mutation (Sir2394L) is dominant, at least with respect to rDNA and telomeric silencing (S.Perrod and S.Gasser, personal communication).
Despite the prevalence of class I mutants with a dominant effect on TPE, none caused any derepression of HMR. In their original genetic analysis of SIR function at HM loci, Rine and Herskowitz (1987) also found only recessive SIR2 alleles (13 in total). The absence of dominant alleles in our collection with respect to HMR may reflect important mechanistic differences between silencing at this locus as opposed to either telomeres or the rDNA. The special role of Sir1p at HM loci (Pillus and Rine, 1989; Chien et al., 1993; Fox et al., 1997), and its interaction with Sir4p (Triolo and Sternglanz, 1996), might underlie this difference (see below).
Class I mutants: evidence for additional HM/telomeric silencing factors
Biochemical data (Moazed et al., 1997) suggest that the Sir2p interaction with Sir4p might be critical for its function in HM/telomeric silencing. It was thus surprising to find that all class I mutations interact normally with Sir4p in a two-hybrid assay. Furthermore, the relocalization of Sir4p to the nucleolus in all class I mutants also suggests that the Sir2p–Sir4p interaction is intact in these mutants. The failure to identify Sir2 mutant proteins with a clear Sir4p interaction defect might indicate that Sir4p and a protein involved in rDNA silencing may interact with the same region of Sir2p such that mutations abolishing only the Sir4p interaction (thus leading to a class I phenotype) cannot be obtained. Consistent with this idea, Cockell et al. (2000) have shown that an N-terminal fragment of Sir2p binds to Sir4p and is required for rDNA silencing.
Since the Sir2/3/4 complex appears to remain intact in class I mutants, it seems likely that these Sir2 mutant proteins are defective in interacting with another (still unidentified) factor that is itself essential either for targeting or stabilizing the Sir complex at HM loci and telomeres. Several additional lines of evidence support this idea. Perhaps the most compelling argument for the existence of additional Sir2p-interacting factors, at least at telomeres, is the finding that many class I mutants are dominant to wild-type Sir2p for telomeric silencing. We propose two related models to explain this dominant effect. In the first model, Sir2p class I mutant proteins are defective in an interaction with protein X and fail to localize stably to either HM loci or telomeres. They exert their dominant effect by interacting with a second factor (protein Y), which is limiting for telomeric silencing and is effectively titrated away from these sites. Protein Y in this case might be either Sir4p or Sir3p, or it could be an unidentified factor critical for TPE. Alternatively, mutant Sir2p might be recruited to HM loci and telomeres by protein Y, where it directly disrupts the function of wild-type Sir2p by incorporating into and destabilizing Sir protein complexes (because of its inability to interact properly with protein X). The stabilizing effect of Sir1p interactions at HM loci might be sufficient to overcome either the titration or ‘poisoning’ effects of mutant Sir2 proteins. These ideas will become testable when we know more about the protein–protein interactions that are defective in class I mutants.
The dramatic relocalization of Sir3p and Sir4p in class I mutants also provides an important clue regarding their molecular defect. This effect requires the presence of the mutant Sir2 protein, which suggests that the Sir2/3/4 complex is intact in these cells but simply re-localized to the nucleolus. This observation strongly implies the existence of an additional Sir2p-interacting factor (factor X, above) required specifically for telomeric (and probably HM) localization of the Sir complex. The movement of Sir3p and Sir4p that we observe is reminiscent of the SIR2- and UTH4-dependent relocalization of Sir3p to the nucleolus seen in aging cells (Kennedy et al., 1997). Interestingly, aging is also associated with the simultaneous loss of both TPE and HM silencing (Kennedy et al., 1995; Smeal et al., 1996). All of our class I mutants thus mimic an effect seen in old wild-type cells in which the nucleolus apparently becomes a ‘sink’ for Sir proteins. It is tempting to speculate, then, that the loss of the hypothesized Sir2p–factor X interaction in class I mutants occurs normally in old cells. It is important to point out that the role of the Sir2/3/4 complex in the nucleolus, if any, is still unclear (Kaeberlein et al., 1999).
The targeting experiments with class I mutant proteins reveal a clear difference between Sir2-12p and -143p, which restore strong repression at both HMR and a telomere when tethered there, and the remaining mutants that have no effect whatsoever in this assay. Although the underlying cause of this phenotype is still not clear, it emphasizes the complexity of Sir2p function even at a single site of action. One interpretation of the Sir2-12p and -143p mutants is that they are capable of forming stable Sir2/3/4 complexes that can assemble along silent chromatin (Strahl-Bolsinger et al., 1997), but are defective in an interaction that initiates, or ‘nucleates’ this reaction. This postulated nucleation interaction might involve as yet unidentified factors, or might simply require different or tighter contacts with Sir4 and/or Sir3 proteins. The apparent loss of an initiation function in Sir2-12p and -143p proteins may be related to the observations of Grunstein and coworkers (Strahl-Bolsinger et al., 1997), who proposed distinct roles for Sir2p in ‘core’ and ‘extended’ heterochromatin.
Class II mutations: the importance of Net1p and additional factors in rDNA silencing
Class II mutants clearly emphasize a central role of Net1p in Sir2p localization to the nucleolus. Every Sir2p mutant with a lower affinity for Net1p (as measured either by the two-hybrid assay, GST pull-down or both methods) has a partial or complete loss of nucleolar localization. The converse is also true, apart from one notable exception, Sir2-85p, which seems to interact with Net1p normally, yet localizes to the nucleolus only in some cells. Apart from its central role in Sir2p localization, though, Net1p might not be essential for rDNA silencing per se. Sir2-81 is defective in a Net1p interaction, as measured both in the two-hybrid and GST–Net1p pull-down assays, yet it can restore rDNA silencing upon targeting.
Despite our ability to assign a plausible molecular defect to several class II mutants (a defective Net1p interaction), others still escape explanation. Again we are led to invoke the existence of unidentified Sir2p-interacting factor(s), in this case with specific nucleolar functions. With regard to the dominance of all but one class II mutation, we would also suggest that this is due either to the titration of a locus-specific factor or to the entry of mutant protein into a specific complex and the ‘poisoning’ of a subsequent assembly reaction leading to silencing.
In summary, the mutants described here provide new molecular information regarding locus-specific Sir2p function, and point to the existence of as yet uncharacterized factors controlling the sub-nuclear localization of Sir protein complexes. We anticipate that further study of these novel SIR2 mutants, particularly their biochemical properties, will yield important new molecular insights into silencing and aging.
Materials and methods
Media, strains and plasmids
Yeast strains used in this study are listed in Table II. Growth and manipulation of yeast was carried out according to standard procedures (Rose et al., 1990). Further details, including all plasmid constructions, can be found in the Supplementary material.
Table II. Yeast strains used in this study.
| Strain | Genotype | Reference |
|---|---|---|
| W303-1A | MATa ade2-1 trp1-1 can1-100 leu2-3.112 his3-11,15 ura3-1 | Thomas and Rothstein (1989) |
| GCY16 | MATa sir2::KanMX4 | this study |
| GCY23 | MATa RDN1::MET15 ΔAhmr::TRP1 adh4::URA3 Tel VIIL sir2Δ met15Δ ADE2 | this study |
| GCY40 | MATα RDN1::mURA3-HIS3 ΔAhmr::TRP1 adh4::ADE2 Tel VIIL sir2::KanMX4 | this study |
| GCY62 | MATα RDN1::4×UASg-mURA3-HIS3 sir2::KanMX4 | this study |
| GCY66 | MATα RDN1::MET15 ΔAhmr::TRP1 adh4::URA3 Tel VIIL met15Δ ADE2 | this study |
| GCY150 | MATa SIR3–GFP sir2::KanMX4 | this study |
| YG451 | MATa ΔAEhmr::3×UASg-TRP1 sir2::KanMX4 | Irina Serdobova |
| YG581 | MATa adh4::4×UASg-URA3 Tel VIIL sir2::KanMX4 | Irina Serdobova |
| CTY10-5D | MATa ade2-1 trp1-901 leu2-3 his3-200 gal4 gal80 URA3::lexA op-LacZ | Bartel and Fields (1995) |
All YG and GCY strains are isogenic derivatives of W303 (Thomas and Rothstein, 1989).
Mutagenesis and screening
The SIR2 gene was mutagenized by PCR in three separate, partially overlapping segments (Figure 1B). PCR conditions (25 ng of template, 100 pmol of primer, 3 mM MgCl2, 0.5 mM MnCl2, 0.2 mM dATP, 0.2 mM dGTP, 1 mM dTTP, 1 mM dCTP, 1 U of Taq polymerase, in a final volume of 50 µl with 30 cycles of amplification) were optimized to yield an average of 1–3 mutations in the final product. The PCR products were mixed separately with a SIR2-containing plasmid, in which most of the corresponding fragment of the gene had been removed by restriction digestion, and transformed into yeast. ‘Gap-repair’ (Rothstein, 1991) of the linearized plasmids by the PCR products generated a high frequency of transformants that were then screened directly for silencing defects. Omission of the PCR product resulted in a severe drop (>100-fold) in transformation frequency.
Either of two different strains (GCY23 and GCY40) was used to screen for silencing defects after transformation. In both strains the TRP1 gene was used to measure repression at HMR (Brand et al., 1985; Sussel and Shore, 1991). In one strain (GCY23) the URA3 gene was used as a TPE reporter (Gottschling et al., 1990), which allowed us to measure silencing by the ability to grow in the presence of the drug 5-fluoroorotic acid (5-FOA). Silencing within the rDNA in this strain was measured using the MET15 gene, which provides a sensitive colony-color assay for expression on plates containing lead nitrate (Smith and Boeke, 1997). In GCY40 the ADE2 gene was used as a colony-color reporter for telomeric silencing (Gottschling et al., 1990) and a modified URA3 gene within the rDNA repeats was used to monitor rDNA silencing (Smith and Boeke, 1997). Both reporter strains contained a complete deletion of the SIR2 open reading frame.
Yeast assays
HMR, telomeric and rDNA silencing assays were performed as described previously (Gottschling et al., 1990; Sussel and Shore, 1991; Smith and Boeke, 1997). Two-hybrid assays in strain CTY10-5D (Bartel and Fields, 1995) were performed as before (Moretti et al., 1994), except in the case of Sir2–Net1 interactions, which were measured by a nitrocellulose filter assay (Breeden and Nasmyth, 1987) due to poor growth in liquid culture.
In vitro binding assays
GST and GST–Net1p(566–1189) fusion proteins were expressed in Escherichia coli strain BL21 and purified essentially as described (Moretti et al., 1994), except that 1% Triton X-100 was added to improve solubilization. Typical binding reactions used 1 ml of crude bacterial extract (250 µl for GST alone) and 50 µl of glutathione–agarose slurry. Incubation was carried out at 4°C for 1 h on a rotating wheel. The beads where washed three times with phosphate-buffered saline (PBS) and resuspended in 1 ml of PBS. Crude whole-cell yeast extract was prepared by vortexing in the presence of glass beads. Samples were incubated as above, but for 2 h. The agarose beads were washed three times in PBS, resuspended in 50 µl of SDS sample buffer, and boiled for 5 min before SDS gel electrophoresis and Western blotting. Input and bound Sir2-9×Myc were detected with the 9E10 monoclonal antibody (hybridoma cell line kindly provided by G.Evan) using the ECL detection system (Amersham). Protein levels were quantified using a phosphoimager (Bio-Rad).
Microscopy
For Sir2p– and Sir3p–GFP localization studies, overnight cultures in selective media supplemented with 6 mg/l adenine were used. Microscopy was performed on a Zeiss Axiovert S100 microscope with a 100× oil-immersion objective and an FITC filter set. Images were captured on a Hamamatsu C4742-95 digital camera using Openlab 2.0.4 software. Indirect immunofluorescence was performed as described (Gotta et al., 1996), with antibodies against Rap1p, Sir3p and Sir4p (Gotta et al., 1997). Confocal microscopy was performed on a Zeiss Axiovert 100 microscope using a 63× Plan-Apochromat objective (1.4 oil) as described (Gotta et al., 1996). In all cases, no signal from one fluorochrome could be detected on the other filter set.
Supplementary material
The supplementary material for this paper is available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank S.Perrod and S.Gasser for invaluable help with indirect immunofluorescence confocal microscopy, and R.Sternglanz, J.Smith, J.Boeke, W.Zaccharaie and P.Silver for gifts of DNAs. We also thank A.Bianchi, M.Cockell, S.Gasser and K.Mishra for their thoughtful comments on the manuscript, all other members of the Shore laboratory for helpful discussions, I.Serdobova for strains YG451 and YG581, and N.Roggli for help in preparing the figures. Support from the Swiss National Science Fund, the Swiss Cancer League and the Canton of Geneva is gratefully acknowledged. This work was also supported by a grant allocated by AETAS, Foundation for Research into Ageing, Geneva, with the financial support of the Foundation A. R. & J.Leenaards, Lausanne.
References
- Afshar G. and Murnane,J.P. (1999) Characterization of a human gene with sequence homology to Saccharomyces cerevisiae SIR2. Gene, 234, 161–168. [DOI] [PubMed] [Google Scholar]
- Aparicio O.M., Billington,B.L. and Gottschling,D.E. (1991) Modifiers of position effect are shared between telomeric and silent mating-type loci in S.cerevisiae. Cell, 66, 1279–1287. [DOI] [PubMed] [Google Scholar]
- Bartel P.L. and Fields,S. (1995) Analyzing protein–protein interactions using two-hybrid system. Methods Enzymol., 254, 241–263. [DOI] [PubMed] [Google Scholar]
- Brachmann C.B., Sherman,J.M., Devine,S.E., Cameron,E.E., Pillus,L. and Boeke,J.D. (1995) The SIR2 gene family, conserved from bacteria to humans, functions in silencing, cell cycle progression and chromosome stability. Genes Dev., 9, 2888–2902. [DOI] [PubMed] [Google Scholar]
- Brand A.H., Breeden,L., Abraham,J., Sternglanz,R. and Nasmyth,K. (1985) Characterization of a ‘silencer’ in yeast: a DNA sequence with properties opposite to those of a transcriptional enhancer. Cell, 41, 41–48. [DOI] [PubMed] [Google Scholar]
- Braunstein M., Rose,A.B., Holmes,S.G., Allis,C.D. and Broach,J.R. (1993) Transcriptional silencing in yeast is associated with reduced nucleosome acetylation. Genes Dev., 7, 592–604. [DOI] [PubMed] [Google Scholar]
- Breeden L. and Nasmyth,K. (1987) Cell cycle control of the yeast HO gene: cis- and trans-acting regulators. Cell, 48, 389–397. [DOI] [PubMed] [Google Scholar]
- Bryk M., Banerjee,M., Murphy,M., Knudsen,K.E., Garfinkel,D.J. and Curcio,M.J. (1997) Transcriptional silencing of Ty1 elements in the RDN1 locus of yeast. Genes Dev., 11, 255–269. [DOI] [PubMed] [Google Scholar]
- Buck S.W. and Shore,D. (1995) Action of a RAP1 carboxy-terminal silencing domain reveals an underlying competition between HMR and telomeres in yeast. Genes Dev., 9, 370–384. [DOI] [PubMed] [Google Scholar]
- Chen X.J. and Clark-Walker,G.D. (1994) Sir2 mutants of Kluyveromyces lactis are hypersensitive to DNA-targeting drugs. Mol. Cell. Biol., 14, 4501–4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien C.T., Buck,S., Sternglanz,R. and Shore,D. (1993) Targeting of SIR1 protein establishes transcriptional silencing at HM loci and telomeres in yeast. Cell, 75, 531–541. [DOI] [PubMed] [Google Scholar]
- Cockell M.M. and Gasser,S.M. (1999) Nuclear compartments and gene regulation. Curr. Opin. Genet. Dev., 9, 199–205. [DOI] [PubMed] [Google Scholar]
- Cockell M.M., Perrod,S. and Gasser,S.M. (2000) Analysis of Sir2p domains required for rDNA and telomeric silencing in Saccharomyces cerevisiae. Genetics, 154, 1069–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derbyshire M.K., Weinstock,K.G. and Strathern,J.N. (1996) HST1, a new member of the SIR2 family of genes. Yeast, 12, 631–640. [DOI] [PubMed] [Google Scholar]
- Fox C.A., Ehrenhofer-Murray,A.E., Loo,S. and Rine,J. (1997) The origin recognition complex, SIR1 and the S phase requirement for silencing. Science, 276, 1547–1551. [DOI] [PubMed] [Google Scholar]
- Fritze C.E., Verschueren,K., Strich,R. and Easton Esposito,R. (1997) Direct evidence for SIR2 modulation of chromatin structure in yeast rDNA. EMBO J., 16, 6495–6509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye R.A. (1999) Characterization of five human cDNAs with homology to the yeast SIR2 gene: Sir2-like proteins (sirtuins) metabolize NAD and may have protein ADP-ribosyltransferase activity. Biochem. Biophys. Res. Commun., 260, 273–279. [DOI] [PubMed] [Google Scholar]
- Gotta M., Laroche,T., Formenton,A., Maillet,L., Scherthan,H. and Gasser,S.M. (1996) The clustering of telomeres and colocalization with Rap1, Sir3 and Sir4 proteins in wild-type Saccharomyces cerevisiae. J. Cell Biol., 134, 1349–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotta M., Strahl-Bolsinger,S., Renauld,H., Laroche,T., Kennedy,B.K., Grunstein,M. and Gasser,S.M. (1997) Localization of Sir2p: the nucleolus as a compartment for silent information regulators. EMBO J., 16, 3243–3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb S. and Esposito,R.E. (1989) A new role for a yeast transcriptional silencer gene, SIR2, in regulation of recombination in ribosomal DNA. Cell, 56, 771–776. [DOI] [PubMed] [Google Scholar]
- Gottschling D.E., Aparicio,O.M., Billington,B.L. and Zakian,V.A. (1990) Position effect at S.cerevisiae telomeres: reversible repression of pol II transcription. Cell, 63, 751–762. [DOI] [PubMed] [Google Scholar]
- Grunstein M. (1998) Yeast heterochromatin: regulation of its assembly and inheritance by histones. Cell, 93, 325–328. [DOI] [PubMed] [Google Scholar]
- Imai S., Armstrong,C.M., Kaeberlein,M. and Guarente,L. (2000) Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature, 403, 795–800. [DOI] [PubMed] [Google Scholar]
- Kaeberlein M., McVey,M. and Guarente,L. (1999) The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev., 13, 2570–2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy B.K., Austriaco,N.R.,Jr, Zhang,J. and Guarente,L. (1995) Mutation in the silencing gene SIR4 can delay aging in S.cerevisiae. Cell, 80, 485–496. [DOI] [PubMed] [Google Scholar]
- Kennedy B.K. et al. (1997) Redistribution of silencing proteins from telomeres to the nucleolus is associated with extension of life span in S.cerevisiae. Cell, 89, 381–391. [DOI] [PubMed] [Google Scholar]
- Marcand S., Buck,S.W., Moretti,P., Gilson,E. and Shore,D. (1996) Silencing of genes at nontelomeric sites in yeast is controlled by sequestration of silencing factors at telomeres by Rap1 protein. Genes Dev., 10, 1297–1309. [DOI] [PubMed] [Google Scholar]
- Moazed D. and Johnson,D. (1996) A deubiquitinating enzyme interacts with SIR4 and regulates silencing in S.cerevisiae. Cell, 86, 667–677. [DOI] [PubMed] [Google Scholar]
- Moazed D., Kistler,A., Axelrod,A., Rine,J. and Johnson,A.D. (1997) Silent information regulator protein complexes in Saccharomyces cerevisiae: a SIR2/SIR4 complex and evidence for a regulatory domain in SIR4 that inhibits its interaction with SIR3. Proc. Natl Acad. Sci. USA, 94, 2186–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti P., Freeman,K., Coodly,L. and Shore,D. (1994) Evidence that a complex of SIR proteins interacts with the silencer and telomere-binding protein RAP1. Genes Dev., 8, 2257–2269. [DOI] [PubMed] [Google Scholar]
- Perez-Martin J., Uria,J.A. and Johnson,A.D. (1999) Phenotypic switching in Candida albicans is controlled by a SIR2 gene. EMBO J., 18, 2580–2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillus L. and Rine,J. (1989) Epigenetic inheritance of transcriptional states in S.cerevisiae. Cell, 59, 637–647. [DOI] [PubMed] [Google Scholar]
- Rine J. and Herskowitz,I. (1987) Four genes responsible for a position effect on expression from HML and HMR in Saccharomyces cerevisiae. Genetics, 116, 9–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose M.D., Winston,F. and Hieter,P. (1990). Methods in Yeast Genetics: A Laboratory Course Manual. Plainview, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Rothstein R. (1991) Targeting, disruption, replacement and allele rescue: integrative DNA transformation in yeast. Methods Enzymol., 194, 281–301. [DOI] [PubMed] [Google Scholar]
- San-Segundo P.A. and Roeder,G.S. (1999) Pch2 links chromatin silencing to meiotic checkpoint control. Cell, 97, 313–324. [DOI] [PubMed] [Google Scholar]
- Sherman J.M., Stone,E.M., Freeman-Cook,L.L., Brachmann,C.B., Boeke,J.D. and Pillus,L. (1999) The conserved core of a human SIR2 homologue functions in yeast silencing. Mol. Biol. Cell, 10, 3045–3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shou W., Seol,J.H., Shevchenko,A., Baskerville,C., Moazed,D., Chen,Z.W., Jang,J., Charbonneau,H. and Deshaies,R.J. (1999) Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell, 97, 233–244. [DOI] [PubMed] [Google Scholar]
- Smeal T., Claus,J., Kennedy,B., Cole,F. and Guarente,L. (1996) Loss of transcriptional silencing causes sterility in old mother cells of S.cerevisiae. Cell, 84, 633–642. [DOI] [PubMed] [Google Scholar]
- Smith J.S. and Boeke,J.D. (1997) An unusual form of transcriptional silencing in yeast ribosomal DNA. Genes Dev., 11, 241–254. [DOI] [PubMed] [Google Scholar]
- Smith J.S., Brachmann,C.B., Pillus,L. and Boeke,J.D. (1998) Distribution of a limited Sir2 protein pool regulates the strength of yeast rDNA silencing and is modulated by Sir4p. Genetics, 149, 1205–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone E.M. and Pillus,L. (1998) Silent chromatin in yeast: an orchestrated medley featuring Sir3p. BioEssays, 20, 30–40. [DOI] [PubMed] [Google Scholar]
- Strahl-Bolsinger S., Hecht,A., Luo,K. and Grunstein,M. (1997) SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev., 11, 83–93. [DOI] [PubMed] [Google Scholar]
- Straight A.F., Shou,W., Dowd,G.J., Turck,C.W., Deshaies,R.J., Johnson,A.D. and Moazed,D. (1999) Net1, a Sir2-associated nucleolar protein required for rDNA silencing and nucleolar integrity. Cell, 97, 245–256. [DOI] [PubMed] [Google Scholar]
- Sussel L. and Shore,D. (1991) Separation of transcriptional activation and silencing functions of the RAP1-encoded repressor/activator protein 1: isolation of viable mutants affecting both silencing and telomere length. Proc. Natl Acad. Sci. USA, 88, 7749–7753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanny J.C., Dowd,G.J., Huang,J., Hilz,H. and Moazed,D. (1999) An enzymatic activity in the yeast Sir2 protein that is essential for gene silencing. Cell, 99, 735–745. [DOI] [PubMed] [Google Scholar]
- Thomas B.J. and Rothstein,R. (1989) Elevated recombination rates in transcriptionally active DNA. Cell, 56, 619–630. [DOI] [PubMed] [Google Scholar]
- Triolo T. and Sternglanz,R. (1996) Role of interactions between the origin recognition complex and SIR1 in transcriptional silencing. Nature, 381, 251–253. [DOI] [PubMed] [Google Scholar]
- Tsang A.W. and Escalante-Semerena,J.C. (1998) CobB, a new member of the SIR2 family of eukaryotic regulatory proteins, is required to compensate for the lack of nicotinate mononucleotide:5,6-dimethylbenzimidazole phosphoribosyltransferase activity in cobT mutants during cobalamin biosynthesis in Salmonella typhimurium LT2. J. Biol. Chem., 273, 31788–31794. [DOI] [PubMed] [Google Scholar]
- Visintin R., Hwang,E.S. and Amon,A. (1999) Cfi1 prevents premature exit from mitosis by anchoring Cdc14 phosphatase in the nucleolus. Nature, 398, 818–823. [DOI] [PubMed] [Google Scholar]
- Zemzoumi K., Sereno,D., Francois,C., Guilvard,E., Lemesre,J.L. and Ouaissi,A. (1998) Leishmania major: cell type dependent distribution of a 43 kDa antigen related to silent information regulatory-2 protein family. Biol. Cell, 90, 239–245. [DOI] [PubMed] [Google Scholar]