Abstract
Cytokines of the gp130 family exert their diverse biological effects by formation of stable high affinity transmembrane receptor complexes that are characterized by the presence of the shared transmembrane signalling receptor gp130. Different gp130 ligands form signalling complexes that vary in both composition and stoichiometry. Analysis of the three-dimensional structure of selected ligands and receptor elements indicates that ligands display three topologically conserved receptor recognition epitopes that interact with complementary ligand recognition elements. The composition of the signalling complex and downstream biological responses is defined by the relative affinity of different receptor components for these epitopes. The detailed structure of receptor recognition epitopes indicates that the generation of small molecule cytokine mimetics may be a feasible objective.
Keywords: cytokines/gp130/receptor recognition epitopes/receptor signalling complexes
Introduction
The formation of protein complexes is a central biochemical mechanism in signal transduction and cell regulation. This is epitomized by the interaction of cytokine ligands with transmembrane signalling receptors. The generation of a high affinity complex between a ligand and its target receptors outside the cell is the initiating step in the activation of signal transduction cascades inside the cell. An issue of fundamental biological significance is the biochemical specificity of this interaction. This dictates both the identity and kinetics of the resulting signal transduction processes and, accordingly, the associated biological responses. Important insights into the biochemical specificity of cytokine signalling have emerged from examining the structural details of the interaction between cytokines and their receptors. As a result, some general principles of receptor activation have begun to emerge. These ‘rules’ not only contribute to the general understanding of protein–protein interactions but also provide the basis for deliberate manipulation of cytokine signalling. In this paper we review the current understanding of ligand recognition by the cytokine superfamily of receptors (reviewed by Cosman, 1993; Wells and De Vos, 1996) with particular reference to ligands employing the shared transmembrane receptor gp130 (Hibi et al., 1990; reviewed by Taga and Kishimoto, 1997).
The cytokine family of receptors
The ‘cytokine’ superfamily of polypeptide regulators is defined by shared structural features of both ligands and receptors. The defining feature of cytokine ligands is the presence of a shared protein fold, ‘the four-helix bundle’. This consists of two pairs of anti-parallel α-helices connected by polypeptide loops. Detailed structural features further subdivide the family into three subsets (Boulay and Paul, 1993; Sprang and Bazan, 1993). ‘Short chain’ cytokines such as interleukins (IL)-2, -3 and -4 have relatively short α-helices, typically 8–10 residues in length. ‘Long chain’ cytokines such as growth hormone (GH), erythropoietin (EPO), granulocyte colony stimulating factor (GCSF) and the ‘gp130 cytokines’ discussed below have longer helices, from 10 to 20 residues in length. Finally, some cytokines, such as IL-5 (Milburn et al., 1993) and interferon γ (Walter et al., 1995), have eight α-helices forming a duplicated version of a four-helix bundle motif.
These ligands interact with receptors that share common structural motifs in the extracellular region. The most prominent is the cytokine receptor homology domain (CHD), which consists of two seven-stranded β-sandwich motifs connected by a proline-rich linker sequence (Bazan, 1990). The first, N-terminal, motif contains a characteristic pattern of cysteine residues that form inter-strand disulfide bonds. The signature feature of the CHD is the conserved amino acid sequence WSXWS in the second, C-terminal, motif. This feature can be exploited in the cloning and identification of novel cytokine family receptors (e.g. Elson et al., 1998; Sprecher et al., 1998).
Intracellular signalling processes are initiated by cytokine ligands forming a high affinity oligomeric complex with transmembrane receptors. This leads to the formation of a protein ‘signalling complex’ on the inside of the membrane that may include src family kinases (e.g. Ernst et al., 1994) as well as the JAK family of cytoplasmic tyrosine kinases, which associate with the cytoplasmic domain of the receptors, and the STAT family of transcription factors (reviewed by Ihle et al., 1994; Ihle, 1996; Heinrich et al., 1998). An important feature of this relationship is that individual JAK and STAT families exhibit preferential affinity for different receptors. This means that the composition of the ‘signalling complex’ and the particular repertoire of signals induced by receptor oligomerization are dictated by the identity and composition of transmembrane receptors complexed by the ligand. A central issue is, therefore, the molecular basis by which individual cytokines recognize receptors to cause the formation of an active signalling complex.
Receptor homodimerization: the growth hormone example
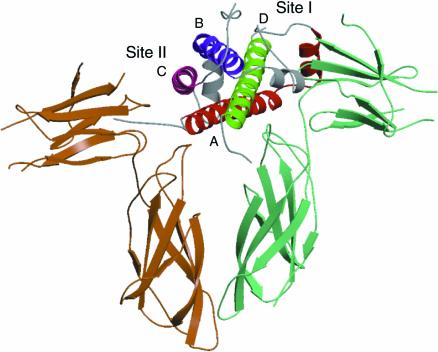
The pioneering study of the interaction between human growth hormone (hGH) and the growth hormone receptor (GHR) (De Vos et al., 1992; reviewed by Wells, 1996) has provided important initial insights into the structural basis of ‘long chain’ cytokine/receptor recognition. The crystal structure of the hGH–GHR complex (De Vos et al., 1992) reveals that a single molecule of hGH forms a homodimeric receptor complex by simultaneously binding two identical receptor subunits (Figure 1). Two discrete regions of the ligand, site I (located in the fourth α-helix) and site II (formed from regions in the first and third helices), each bind a receptor molecule via interaction with the CHD. As a result of this simultaneous interaction with two sites on the ligand (Cunningham et al., 1991) the cytoplasmic domains of the receptor are brought into close proximity and intracellular signalling processes are induced.
Fig. 1. Ribbon representation of the X-ray structure of the complex between GH and the GHR (De Vos et al., 1992). Two molecules of GHR bind one molecule of GH via sites I and II. The four α-helices of GH are colour coded: helix A, red; helix B, blue; helix C, purple; helix D, green.
Although large areas of protein surface are buried in formation of the hGH–GHR receptor–ligand complex, detailed mutagenesis and biophysical studies have shown that the affinity of the interaction is dominated by a few tightly clustered amino acid residues forming an energetic ‘hot spot’ (Clackson and Wells, 1995) in the centre of the binding interface. The identity of the residues that dominate the binding energy of a large protein–protein interface has been termed the ‘functional epitope’ (Cunningham and Wells, 1993). The affinity of the interaction (and hence biological specificity) is consequently defined by the structural complementarity of the functional epitopes of ligand and receptor.
Detailed analysis of the functional epitopes comprising the site I and site II interaction sites of hGH suggests a shared design (Cunningham and Wells, 1989; reviewed by Wells, 1996). The most significant residues, as defined by the consequences of mutation to alanine, are hydrophobic contacts involving either non-polar side chains such as tryptophan, the aliphatic components of non-polar side chains or the polypeptide backbone. The non-polar ‘core’ is surrounded by a halo of polar residues of lesser importance for binding energy.
Kinetic studies of the hGH–GHR interaction indicate that the primary role of the functional epitopes is to stabilize protein–protein interaction by decreasing the dissociation rate of the complex. These results have been used to suggest (Wells, 1996) that the initial contact between ligand and receptor is dominated by random diffusion or ‘rolling’ of protein surfaces producing many unstable contacts. The complex is then stabilized when the functional epitopes of the receptor and ligand engage.
Combinatorial receptor complexes
The hGH example described above represents the simplest case of protein complex formation in the cytokine receptor system: a single ligand engages with a single class of receptor. However, several cytokine family systems exhibit greater complexity. These are combinatorial systems that involve multiple ligands and receptors that are assembled to form signalling complexes with different compositions. Typically these involve one, or more, shared receptor components. Thus, IL-2, -4, -7 and -15 all associate with the common IL-2 receptor γ (reviewed by Kondo et al., 1993). GM-CSF, IL-5 and IL-3 all share the common IL-3 β receptor (reviewed by Guthridge et al., 1998). In this paper we consider a large family of ligands that associate with the common transducing receptor gp130 (Hibi et al., 1990). These systems raise the question: what is the molecular basis by which protein complexes of different compositions can be formed from shared elements?
gp130 cytokines
The gp130 system is a set of ligands and receptors that elicits biological responses by the formation of oligomeric signalling complexes that contain one or more copies of a common transmembrane transducing receptor gp130 (reviewed by Taga and Kishimoto, 1992, 1995). The gp130 family of cytokines therefore exhibits a very wide range of biological functions in vivo due both to the diversity of receptor complexes that can be formed and to developmental regulation of the components of intracellular signalling pathways with which receptor complexes engage. They can also exhibit common (or ‘redundant’) functions in vitro that are dependent upon the signalling functions of the shared gp130 receptor (reviewed by Taga and Kishimoto, 1997).
There are currently nine cytokine ligands defined that signal via formation of a receptor complex that contains one, or more, molecules of gp130 (Table I). They are all polypeptides of 180–200 amino acids that collectively exhibit relatively low overall sequence homology (∼20%). In some cases homologues have been isolated from different mammalian species that exhibit significant differences in biological specificity (see below). To date, gp130 cytokine homologues have not been identified in vertebrate species such as fish or amphibians although a sex pheromone protein from salamanders has recently been reported to exhibit sequence similarity to gp130 cytokines (Rollmann et al., 1999). It is likely that the currently defined gp130 cytokine family is not complete and other family members may emerge from genome sequencing efforts and other approaches.
Table I. The composition of gp130 cytokine receptor complexes.
| Ligand | Site I | Site II | Site III |
|---|---|---|---|
| LIFa | – | gp130 | LIF-R |
| OSMb | – | gp130 | LIF-R/OSM-R |
| CNTFc | CNTF-R | gp130 | LIF-R |
| CT-1d | ? | gp130 | LIF-R |
| IL-6e | IL-6R | gp130 | gp130 |
| IL-11f | IL-11R | gp130 | gp130 |
| KSV-IL-6g | IL-6R | gp130 | gp130 |
| Rm-IL-6h | IL-6R | gp130 | gp130 |
| CLC/NN-1i | ? | gp130 | LIF-R |
aLeukemia inhibitory factor: murine (Gearing et al., 1987); human (Gough et al., 1988).
bOncostatin M: human (Malik et al., 1989); murine (Yoshimura et al., 1996).
cCiliary neurotrophic factor (Stockli et al., 1989).
dCardiotrophin-1 (Pennica et al., 1995).
eInterleukin-6 (Hirano et al., 1986).
fInterleukin-11 (Paul et al., 1990).
gKaposi sarcoma associated human herpes virus 8 interleukin-6 (Neipel et al., 1997; Nicholas et al., 1997).
hRhesus macaque rhadinovirus interleukin-6 (Kaleeba et al., 1999).
iCardiotrophin-like cytokine/novel neurotrophin-1 (Senaldi et al., 1999; Shi et al., 1999).
Receptor binding sites are based on published experimental data or conservation of key residues in receptor binding epitopes. (?), the identity of the receptor is currently uncertain.
The gp130 cytokine family collectively exhibits a very broad range of biological activities in vivo (reviewed by Taga and Kishimoto, 1997). The most important information on their biological function derives from targeted gene ablation studies (reviewed by Heinrich et al., 1998). These reveal that each member of the family that has been studied yields a unique phenotype upon inactivation. For example, ablation of IL-6 results in impairment of acute phase and anti-viral responses (Kopf et al., 1994; Poli et al., 1994), whereas deletion of ciliary neurotrophic factor (CNTF) results in late-onset motor neuron degeneration (Masu et al., 1993) and deletion of the leukaemia inhibitory factor (LIF) gene results in female infertility due to failure of embryo implantation (Stewart et al., 1992; Escary et al., 1993). These data imply that, despite the use of overlapping receptor signalling components, each cytokine has a characteristic repertoire of biological functions in vivo.
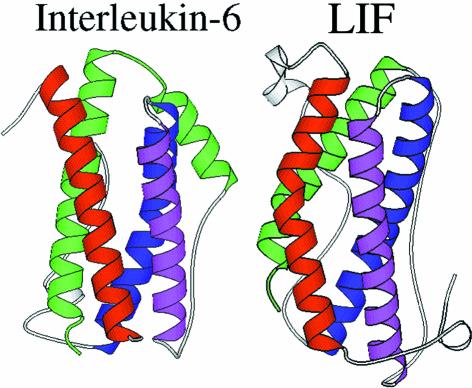
The structure of gp130 cytokines has been the subject of extensive experimental investigation. The tertiary structures of mouse and human LIF (Robinson et al., 1994; Hinds et al., 1998), CNTF (McDonald et al., 1995), IL-6 (Somers et al., 1997; Xu et al., 1997) and oncostatin M (OSM) (Hoffman et al., 1996; M.Deller, K.R.Hudson, E.Y.Jones and J.K.Heath, in preparation) have been defined by either X-ray crystallography or nuclear magnetic resonance and, in some cases, by both techniques. These studies reveal that each ligand exhibits the predicted (Bazan, 1991) ‘long chain’ four-helix bundle topology. This comprises four tightly packed α-helices (termed A, B, C and D) ranging from 15 to 22 amino acids in length. The helices are connected in an up-up-down-down arrangement by three polypeptide loops (Figure 2). The A–B and C–D loops are relatively long as they connect parallel helices whereas the B–C loop is shorter as it connects a pair of antiparallel helices.
Fig. 2. Ribbon representation of the crystal structures of IL-6 (Somers et al., 1997) and LIF (Robinson et al., 1994). The four helices are colour coded: helix A, red; helix B, blue; helix C, purple; helix D, green.
Closer inspection of the available structures and comparison of NMR with crystallographic data reveal differences and similarities between different family members. First, the polypeptide loops connecting the helices are most probably flexible in solution. They exhibit different lengths and topologies in different molecules and, in some cases, cannot be fully traced in the electron density maps. Sequence alignments also suggest that the inter-helical loops can also vary in length between homologues from different species. A feature of divergence between family members is the form of the A and D helices. A comparison of IL-6 and LIF (Figure 2) reveals that in IL-6 these helices are straight, whereas in LIF, CNTF and hOSM they exhibit a marked kink in their structure. This feature allows the gp130 cytokine family to be split into two sub-classes: (i) the ‘straight’ ligands such as IL-6 and (from modelling studies) IL-11; and (ii) the viral IL-6 proteins and the ‘kinked’ ligands, which include LIF, CNTF and hOSM, and (from modelling studies) cardiotrophin-1 (CT-1) and cardiotrophin-like cytokine/novel neurotrophin-1 (CLC/NN-1) (Table I). This divergence in structure may, as discussed below, reflect a functional difference in receptor recognition.
gp130 family receptors
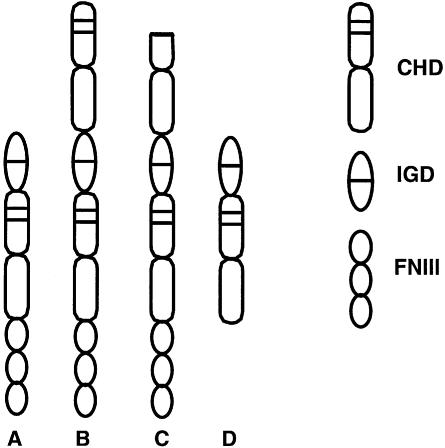
The signalling functions of gp130 cytokines are executed by association with a specific set of receptors (Figure 3). These are of two varieties. The first are ‘signalling receptors’: transmembrane proteins that have cytoplasmic domains that engage with the intracellular JAK/STAT signalling machinery. This class currently encompasses gp130 itself (Hibi et al., 1990), the LIF-receptor (LIF-R; Gearing et al., 1991; Tomida et al., 1994) and the OSM-receptor (OSM-R; Mosley et al., 1996; Lindberg et al., 1998). The second, ‘non-signalling’ class of receptors interact with ligands but do not participate directly in activation of intracellular signalling via association with intracellular mediators. They either have abbreviated cytoplasmic domains: IL-6R (Yamasaki et al., 1988) and IL-11R (Hilton et al., 1994), or are linked to the external face of the plasma membrane by a lipid anchor e.g. CNTF-R (Davis et al., 1991). Non-signalling receptors can, nevertheless, exert biological functions when added to target cells in a soluble form (e.g. Karow et al., 1996).
Fig. 3. The organization of protein modules in the extracellular domains of gp130 family receptors (see Table I). The receptors are generated from three modules: the cytokine receptor homology domain (CHD), the IG-like domain (IGD) and three copies of a fibronectin type III repeat (FNIII). (A) gp130; (B) the LIF-R; (C) the OSM-R; (D) soluble receptors (CNTF-R, IL-6R, IL-11R).
Biological functions of gp130 family receptors
The biological functions mediated by gp130 family receptors have also been analysed by the generation of loss-of-function mutations in mice. These studies, although currently incomplete, indicate that each receptor mediates characteristic signalling processes. Thus, ablation of gp130 results in a complex fatal phenotype including defects in cardiac function (Yoshida et al., 1996; Betz et al., 1998; Hirota et al., 1999), whereas inactivation of LIF-R results in a complex phenotype that includes loss of motor neurons (Li et al., 1995; Ware et al., 1995). Genetic inactivation of IL-11R results in female sterility due to defective decidualization (Bilinski et al., 1998; Robb et al., 1998). There is currently incomplete concordance of phenotypes between ligand knockouts and receptor knockouts e.g. CNTF (Masu et al., 1993) and CNTF-R (DeChiara et al., 1995), which suggests that further members of the gp130 family of receptors remain to be discovered. Indeed there are indications that CT-1 may interact with a lipid-anchored non-signalling receptor (Robledo et al., 1997) and several ‘orphan’ receptors with strong sequence similarities to gp130 family receptors have recently been identified (e.g. Elson et al., 1998; Sprecher et al., 1998).
Functional anatomy of gp130 receptors
The gp130 family of receptors can be considered essentially modular in form (Figure 3). Three structural motifs are combined to form the extracellular region. The canonical CHD is found in all gp130 family receptors. This occurs once in gp130 and the three non-signalling receptors. The LIF-R and OSM-R have two copies of the CHD. The second motif is an ‘immunoglobulin-like domain’ (IGD) that is found in all gp130 receptors. The presence of the IGD distinguishes gp130 family receptors from the hGH-R prototype. The IGD is always located at the C-terminus of the most membrane-proximal CHD and, as the name suggests, is predicted to comprise seven β-strands linked by inter-strand loops. In the absence of direct information, the possible structure of the IGDs can be inferred from modelling studies (Horsten et al., 1997; Simpson et al., 1997). These indicate that the IGD is composed of seven short β-strands linked by solvent-exposed loops of varying lengths. There is relatively poor sequence conservation between the IGDs of different receptors. Those residues that are conserved appear to be located within regions of β-strand, indicative of a primary role in receptor structure. The third element of the extracellular domain comprises multiple copies of seven-stranded ‘fibronectin type III’ (FNIII) motifs. Comparison of these repeats to known structures of FNIII ‘strings’ (Huber et al., 1994) predicts that this region adopts an extended rod-like structure. A significant feature of the membrane-proximal FNIII repeat motifs is that they occur in all the transmembrane signalling receptors but are absent from the ‘non-signalling’ receptors. This indicates that the FNIII domain may have some role in transmembrane signalling such as stabilization and/or orientation of the transmembrane receptor dimers.
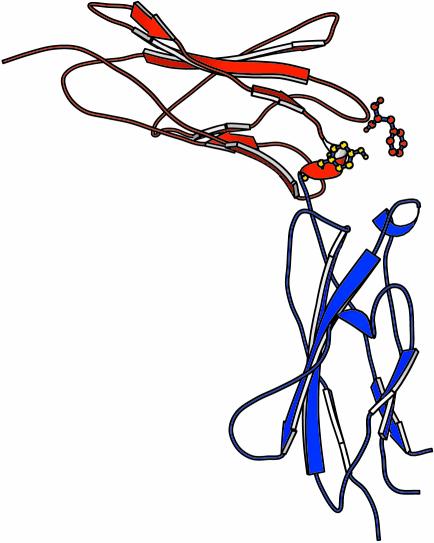
There is, as yet, no complete structure for any member of the gp130 family of receptors. The second, C-terminal, component of the gp130 CHD has been studied by NMR (Kernebeck et al., 1999), and the complete CHD of the common receptor gp130 has been characterized by crystallographic techniques (Bravo et al., 1998). This structure (Figure 4) reveals a strong general similarity to the CHD of the hGH, GCSF-R (Aritomi et al., 1999), IL-4 (Hage et al., 1999) and Epo-R receptors (Syed et al., 1998). It is composed of two seven-stranded β-barrels connected by a proline-rich linker at approximately right angles. As a result of this configuration the inter-strand loops are exposed at the ‘elbow’ between the two domains. The principal point of divergence between current CHD structures is the angle formed between the two β-barrel domains, which results in subtle differences in the relative exposure of inter-strand loops. There are many inter-domain hydrogen bonds, indicating that the overall structure of the gp130 CHD is relatively rigid and unlikely to undergo major structural rearrangements upon ligand engagement.
Fig. 4. Ribbon representation of the crystal structure of the gp130 CHD (Bravo et al., 1998). The locations of the two principal residues involved in ligand recognition via site II are presented in ball-and-stick format: Phe73 (red) and Tyr100 (blue). Residues are numbered according to the crystal structure.
A prominent unsolved issue is the relative orientation of the three modules in the complete structure of the extracellular domain of these receptors. It is likely that the connection between the IGD and the C-terminal CHD involves a right-angle turn. In all cases, the junction between the IGD and CHD involves a proline-rich sequence comparable to the region linking the two components of the CHD. Taken together the existing structural and modelling data suggest that the extracellular domains of the gp130 receptors most closely resemble a chain of varying numbers of seven-stranded β-sandwich modules connected by linkers that impose specific turns between individual modules. As will be discussed below, an important consequence of these proposed turns is the exposure of inter-strand loops that have the potential to interact with ligands.
Ligand recognition
The gp130 family of receptors collectively exhibits differences in affinity for gp130 ligands that are absolutely central to understanding their function. The preferences for association with different gp130 receptors dictate the composition of the signalling complex formed by an individual ligand (Table I). The structural basis of these differences is accordingly a central problem in defining the specificity of signalling via gp130 cytokines.
These preferences fall into three categories. The first is where a ligand binds to a receptor with high (typically nano-molar) affinity: thus, hOSM binds to gp130, LIF binds to LIF-R, IL-6 binds to IL-6R, IL-11 binds to IL-11R and CNTF binds to CNTF-R. The second case is where a ligand only binds to a receptor with high affinity when it is bound to another receptor: thus, IL-6 binds gp130 with high affinity in the presence of IL-6R, IL-11 binds gp130 when bound to IL-11R and CNTF binds gp130 when bound to CNTF-R. In each of these cases a receptor of the non-signalling class is required to enable the interaction with gp130. As a result the formation of the high affinity signalling complex is dominated by the interaction between the ligand and a non-signalling receptor.
The third form of preference involves the case in which species differences exist between homologous ligands in their ability to bind receptors. For example, human OSM can bind both the human LIF-R and OSM-R, whereas murine OSM binds the murine OSM-R but not LIF-R (Ichihara et al., 1997). In a similar fashion, murine LIF binds the murine LIF-R with high affinity and the human LIF-R with low affinity (Owczarek et al., 1993; Layton et al., 1994; Robinson et al., 1994). This preference has proved useful for defining the sites of interaction between ligands and receptors and needs to be taken into account when analysing the activity of ligands between species (e.g. Clegg et al., 1996).
Receptor recognition epitopes in ligands
The availability of high resolution structural information, combined with predictions from molecular modelling, has allowed the detailed definition by site-directed mutagenesis of the functional epitopes of gp130 cytokines involved in receptor recognition. These data reveal an important departure from the classic ‘two site’ models defined above. gp130 cytokines have three topologically discrete sites of interaction with receptors (Inoue et al., 1995; Paonessa et al., 1995; Di Marco et al., 1996; Hudson et al., 1996; Barton et al., 1999). The first two sites, site I and site II, are analogous in location to their hGH counterparts: site I is located at the end of the D helix and site II is composed of residues in the A and C helices. However, a defining feature of the gp130 family is the existence of an additional interaction epitope, site III, which is located at the N-terminal tip of the D helix.
Matching the three interaction epitopes for each cytokine with the cognate receptor (Table I) reveals a combinatorial pattern of receptor engagement in which some key themes become apparent. Site I, if used, is always engaged by a non-signalling receptor: IL-6R, IL-11R or CNTF-R. Site II is always engaged by gp130. Site III is always occupied by a second signalling receptor: gp130, LIF-R or OSM-R, depending upon the identity of the ligand. This pattern represents an important departure in that signalling receptors in this system engage via sites II and III rather than sites I and II of the hGH prototype. Combining this information with the ‘preferences’ described above leads to a further rule. The association with signalling receptors via site III is always relatively high affinity. The association with non-signalling receptor via site I is also of relatively high affinity whereas the association with ligands with site II may be either high affinity or low affinity depending upon the requirement for a non-signalling receptor bound to site I. The use of a non-signalling receptor by some ligands therefore compensates for the absence of a high affinity site II gp130 binding site. It seems very likely that ligand-dependent formation of the complex between non-signalling receptors and gp130 involves the formation of a receptor–receptor interface, since the introduction of mutations into the C-terminal half of the CHD of the IL-6R inhibits IL-6 signalling without affecting ligand binding (Yawata et al., 1993; Salvati et al., 1995).
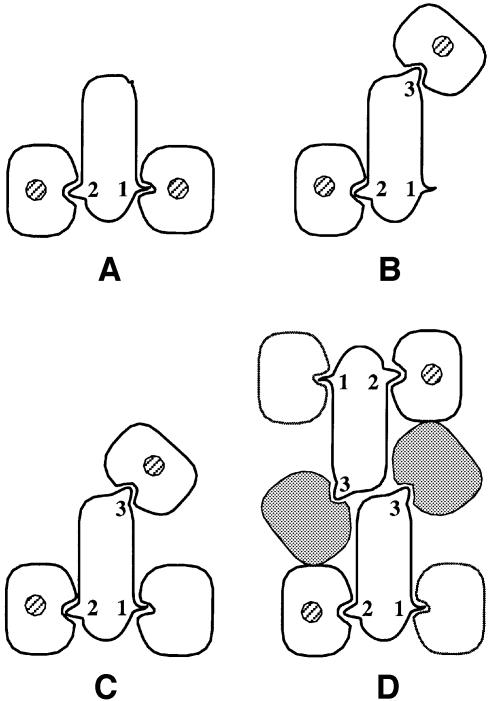
This three-site design has a further twist: in some cases, such as IL-6 and IL-11, gp130 binds to two different regions of the ligand, whereas in other cases, such as LIF, CNTF and OSM, gp130 only binds one site. If (as indeed proves to be the case) the interaction between gp130 and sites II and III involves physically distinct regions of the receptor, it follows that different classes of higher order complexes can be formed by different ligands. In particular, it would be predicted (Figure 5) that ligands which bind two copies of gp130 will form a hexameric complex comprising two copies of gp130, two copies of ligand and (where appropriate) two copies of the cognate non-signalling receptor. Ligands that bind a single copy of gp130 are predicted (Figure 5) to form a ‘trimeric’ complex composed of one copy of gp130, one copy of ligand, one copy of the second signalling receptor (LIF-R or OSM-R) and, where appropriate, one copy of a non-signalling receptor. The existence of these two different types of higher order complexes has been confirmed by studies of the stoichiometry and composition of receptor ligand complexes in solution (Ward et al., 1994; De Serio et al., 1995; Paonessa et al., 1995; Zhang et al., 1997).
Fig. 5. Schematic representation of the differing compositions and stoichiometries of cytokine receptor complexes. The transmembrane domains of ‘signalling receptors’ are indicated by the presence of hatched circles. This shows that the relative orientation of signalling receptors may differ between the ‘dimeric’ type of complex (A) and the higher order complexes (B), (C) and (D). Ligand recognition epitopes are numbered as in Table I. (A) Two receptors/one ligand illustrated by the GH–GHR prototype (Figure 1). In this case, sites I and II are occupied by signalling receptor. (B) Two receptors/one ligand illustrated by the LIF–LIF-R–gp130 and OSM–OSM-R–gp130 complexes. In this case, sites II and III are occupied by signalling receptors. gp130 occupies site II. (C) Three receptors/one ligand illustrated by the predicted CNTF complex. The non-signalling CNTF-R binds via site I of the ligand, gp130 binds via site II and LIF-R binds via site III. (D) Four receptors/two ligands (‘hexameric complexes’) illustrated by the predicted IL-11R–gp130–IL-11 and IL-6R–gp130–IL-6 complexes. In this case, one molecule of gp130 simultaneously binds two molecules of ligand. Site II binding is mediated by the CHD and site III binding is mediated by the IGD (hatched). The non-signalling receptor binds site I. As a result of two molecules of gp130 binding ligand there are two molecules of ligand in the complex.
The employment of three distinct receptor binding sites by ligands of the gp130 family therefore results in two types of complexes: those that differ in composition and those that differ in stoichiometry.
Structure of the functional epitopes of ligands
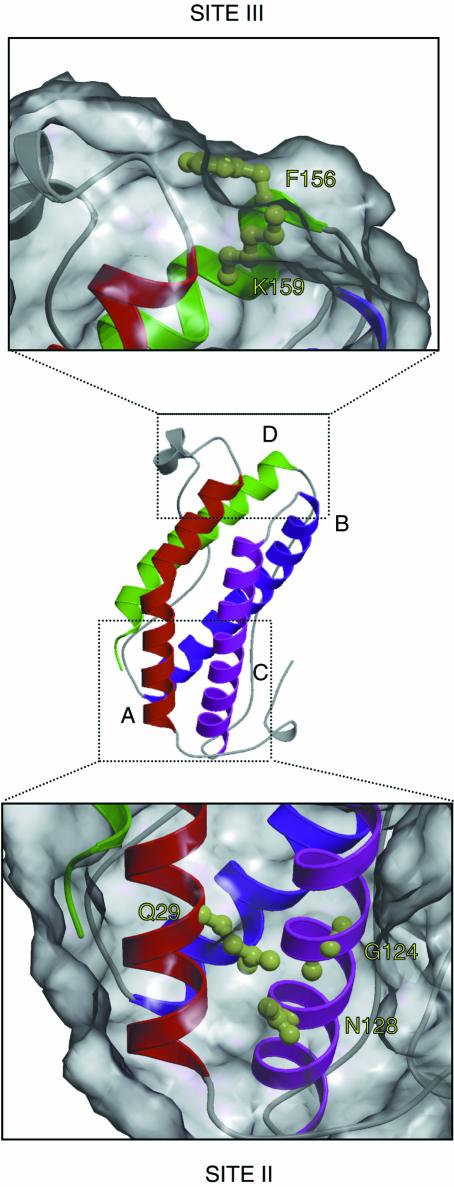
The availability of high resolution structural data combined with functional analysis of receptor recognition epitopes by mutagenesis studies leads to the definition of the structural features involved in receptor recognition by the gp130 family of cytokines. This reveals that receptor interaction, like the hGH model, involves a recognition ‘hot spot’ that is dominated by a small number of amino acid residues located close together in space. Thus, mutagenesis of LIF (Hudson et al., 1996) and OSM (K.R.Hudson and J.K.Heath, unpublished) has shown that the interaction with gp130 via site II involves a small cluster of residues in the A and C helices (Figure 6). This functional epitope is dominated by a conserved glycine residue that, as a consequence of the absence of a side chain, forms a hydrophobic concavity in helix C with the Cα backbone. The neighbouring asparagine residue is exposed at the next turn of the α-helix. Members of the gp130 cytokine family that do not interact with gp130 with high affinity alone, and require a non-signalling receptor, such as CNTF, do not show conservation of residues in the site II epitope.
Fig. 6. High resolution structure of the site II and site III receptor recognition epitopes of LIF. The location of the four helices is indicated as in the ribbon diagram of Figure 2. The definition of the principal components of the functional receptor recognition epitopes is derived from the mutagenesis data of Hudson et al. (1996). Site II is composed of residues in the A and C helices (Q29, G124 and N128). Site III is composed of F156 and K159.
Site III in LIF, OSM and CNTF (all of which interact with LIF-R) is dominated by two residues (Di Marco et al., 1996; Hudson et al., 1996): a conserved solvent-exposed phenylalanine and an adjacent arginine (Figure 6). Parallel mutagenesis studies of IL-6 and IL-11, which interact with gp130 via site III (Paonessa et al., 1995; Barton et al., 1999), reveal a similar arrangement of a solvent-exposed hydrophobic residue (in this case tryptophan) and an adjacent basic residue (in this case arginine). These findings strongly indicate that, as with hGH, the specificity of receptor recognition is dictated by the identity of a relatively small number of amino acids that share common chemical features.
Site I residues involved in the interaction with non-signalling receptors have been defined for IL-6 (reviewed by Simpson et al., 1997) and IL-11 (Barton et al., 1999). In both cases the dominating feature is an arginine residue located towards the end of the D helix. A potential prototype for the arrangement of the site I interaction has recently been provided by the high resolution crystal structure of the short chain cytokine IL-4 bound to the specific high affinity cytokine receptor IL-4R α (Hage et al., 1999). This reveals that the functional epitope involved in the contact between ligand and receptor is formed from a mosaic of three clusters of trans-interacting residues. These ‘avocado’ clusters are composed of a core of polar residues surrounded by an outer region of residues with hydrophobic character. The centre of this cluster includes a prominent arginine residue in the ligand, which forms a salt bridge with an aspartate residue in the N-terminal half of the CHD of IL-4R α. This configuration is the inverse of the site II form described above.
Ligand recognition epitopes in receptors
The availability of the three-dimensional structure of the gp130 CHD has allowed the complementary gp130 recognition epitope for site II in ligands to be defined by mutagenesis (Horsten et al., 1997). These studies reveal that the interaction with site II is also dominated by a small number of physically adjacent residues. In particular, mutation of the prominent solvent-exposed phenylanine and tyrosine in the N-terminal module (Figure 4) elicits a significant loss of affinity for ligands that interact with gp130 with high affinity such as LIF and OSM and IL-6–IL-6R complexes. Comparison of these findings with the well defined site II interaction of hGH and GHR (reviewed by Wells, 1996) suggests that the site II interaction in both cases may exhibit strong conservation in which a prominent non-polar residue from the CHD of the receptor interacts with a hydrophobic cleft in the C helix of the ligand formed from the polypeptide backbone. This interaction is stabilized by further interactions between residues in the ‘halo’ of the functional epitope as defined by Wells and colleagues (reviewed by Wells, 1996). It is interesting to note that a non-polar residue is found in the equivalent position of the N-terminal module of the CHD in virtually every cytokine family receptor known. This implies that the hGH model may provide a precedent for the entire spectrum of site II CHD ligand interactions.
The feature of the gp130 system that is so far unique is the existence of the site III receptor binding epitope that interacts with a second signalling receptor in the complex. The site III binding epitope of gp130 proves to be located in the IGD. This has been demonstrated by analysis of the interaction of gp130 mutants in which the IGD has been deleted (Hammacher et al., 1998; Kurth et al., 1999; Moritz et al., 1999). These mutants exhibit impaired signalling functions in the presence of IL-6–IL-6R but retain the ability to interact with LIF and IL-6–IL-6R with high affinity. In addition, a monoclonal antibody that exhibits the ability to block responses mediated by IL-6–IL-6R, but not LIF or OSM, binds to an epitope located in the IGD of gp130 (Liautard et al., 1997).
Similar data show that the recognition site for ligands that interact with LIF-R via site III also lies in the IGD of LIF-R. Analysis of a series of chimeric receptors constructed between mouse and human LIF-R (which differ in their ligand recognition properties) reveals that the major determinant for species-specific recognition of LIF lies in the IGD of LIF-R (Owczarek et al., 1997). A point mutation in LIF-R that blocks LIF-R binding is also located in the IGD of LIF-R (Taupin et al., 1999). Alanine-scanning mutagenesis of the IGD of LIF-R has revealed a short sequence, thought to be located in a putative inter-strand loop, which is required for the binding of OSM to LIF-R (K.Chobotova, K.R.Hudson and J.K.Heath, manuscript submitted). The same region also contains a recognition epitope for a monoclonal antibody that inhibits signalling mediated via LIF-R (Pitard et al., 1997). Finally, it has proved possible experimentally to convert a site III gp130 recognition epitope into a site III LIF-R recognition epitope by exchange of residues in the site III region (Kallen et al., 1999). This result indicates that the site III recognition sites in gp130 and LIF-R are functionally analogous but exhibit preferences for different ligands. This is supported by the finding that OSM can interact with the IGD of gp130 with low affinity in solution (Staunton et al., 1998).
Collectively these results show that the IGD of signalling receptors of the gp130 family is involved in binding ligands through the site III recognition motif. At the present time there is no direct information on the three-dimensional structure of any gp130 family IGD or, of equal significance, of the structural interrelationship between the IGD and the following CHD. These structures are important goals of future research. As described above, however, it seems likely that there is a bend or turn in the region linking the IGD and CHD. It is also interesting that the site III recognition motif of gp130 ligands is, in structural terms, very similar to the site II recognition motif of the gp130 receptor. This suggests that the interaction of site III with the IGD of signalling receptors may involve the characteristic exposed non-polar site III residue of the ligand associating with a hydrophobic patch or cleft in the receptor.
The final significance of the identification of the IGD as a prominent recognition element is that it means that gp130 has two ligand recognition motifs: site II elements in the CHD and site III elements in the IGD. Thus, gp130 has the ability to bind simultaneously two molecules of ligand where the LIF-R can only bind one. This difference between the two signalling receptors is the central distinction between the hexameric and trimeric modes of signalling complex formation described above.
Conformations of signalling receptors
The principal doctrine of ligand-mediated receptor activation is that the function of ligand is to dimerize two transmembrane receptors. The consequent association of the intracellular domains activates intracellular signalling. But, ‘do the intracellular domains care how the extracellular domains are brought together by the cytokine?’ (Ballinger and Wells, 1998). In other words, is the elaborate architecture of extracellular signalling complexes merely a means to bring receptors into close proximity or is a specific orientation imposed upon the interacting cytoplasmic domains as a result of extracellular complex formation?
At first sight it would appear that there is abundant evidence that receptor dimerization is sufficient to elicit intracellular signalling. It has been shown many times that the extracellular and intracellular domains of cytokine family receptors are interchangeable between related receptors. Thus, chimeras between the gp130 intracellular domain and the extracellular domain of the GCSF-R can emulate many gp130-dependent signalling events in the presence of GCSF (e.g. Niwa et al., 1998). Extensive studies with the EPO-R, and other receptors, have shown that a variety of different strategies for eliciting receptor dimerization can result in activation of intracellular signalling (Qui et al., 1998).
This idea has recently been challenged by the crystal structures of the extracellular domain of the EPO-R bound either to EPO or two different EPO-mimetic peptides that exhibit either agonist or antagonist activity in biological assays (Livnah et al., 1996, 1999). These structures reveal a structure–activity relationship in which all three ligands bind two molecules of EPO-R. However, the two EPO-mimetic peptides formed complexes in which interreceptor orientation differed from that of the authentic EPO ligand (Syed et al., 1998). This is predicted to result in an increased physical separation of receptors at the interface with the plasma membrane. The largest departure from the conformation of the native EPO–EPO-R complex was observed with the agonist peptide, whereas the antagonistic peptide formed a complex of intermediate orientation. This indicates that ‘inactive’ conformations of receptor dimers might exist.
This idea has been given further support from a study (Remy et al., 1999) in which varying lengths of flexible amino acid linker sequence have been introduced into the EPO-R between the transmembrane domain and two complementary fragments of the enzyme dihydrofolate reductase (DHFR) as a ‘proximity reporter’. In this experimental design, DHFR activity is reconstituted if the two fragments are brought into close physical proximity. It was found that insertion of short (5–10 amino acid) linkers only permitted reconstitution of DHFR activity in the presence of ligand whereas long (30 amino acid) linkers were able to reconstitute DHFR activity in the absence of ligands. Moreover, fusion of the DHFR reporters to JAK kinase resulted in activation of DHFR activity in the presence of EPO and full-length EPO-R. These findings indicate that the function of the normal EPO ligands is to bring receptor cytoplasmic domains into close vicinity upon association with the receptor and, in the absence of ligand, the receptor cytoplasmic domains are separated by a distance equal to the 30 amino acid linker. A corollary of this proposal is that the cytoplasmic domains of cytokine receptors do not interact directly in the absence of ligand. This result shows that dimerization can be necessary, but not sufficient, to induce receptor activation, and that receptors may form non-signalling dimers in the absence of ligand.
Another way of looking at this problem would be to say that signalling receptors can form different types of complexes that can be either competent or incompetent to engage the intracellular signalling machinery. This idea has been applied to the gp130 system by Grotzinger et al. (1999), who have proposed, on the basis of modelling studies, that the formation of the hexameric IL-6–IL-11 signalling complex proceeds from a tetrameric complex with two gp130 molecules to a hexameric complex involving the addition of another subunit of IL-6–IL-6R. The hexameric complex is completed in this model by the addition of a second IL-6–IL-6R subunit. An important feature of this model is the proposal that the ‘relative position of the two gp130 molecules’ in the tetrameric complex is different from that of the hexameric complex and this feature is associated with signalling activity. In particular, it is proposed that the tetrameric, but not the hexameric, conformation represents the active signalling complex. This would predict that high concentrations of ligand (which should favour the hexameric form) should inhibit signalling, leading to ‘bell-shaped’ dose response curves. This latter prediction is certainly not seen in at least some manifestations of homodimeric gp130 signalling (e.g. Karow et al., 1996). In addition, studies of the formation of complexes derived from non-signalling ectodomains in solution indicate that the hexameric complex is formed by means of an unstable trimeric intermediate (Ward et al., 1996). It is unlikely that this model can be decisively tested until some means are devised to visualize the composition of active signalling complexes in live cells.
An alternative representation of the mechanism of receptor activation has been outlined by Ballinger and Wells (1998). This emphasizes kinetic aspects of receptor complex formation and proposes that activation of signalling requires a prolonged period of receptor dimerization that consequently requires the formation of a stable receptor complex. In this view many receptor conformations may exist but only stable complexes will result in signal activation. Based upon the detailed understanding of the mechanisms of ligand engagement described above, this model proposes that signalling is determined by the rate at which receptor complexes dissociate. In the presence of ligand, receptors are ‘locked’ into a complex that is sufficiently stable to engage the signal transduction machinery. This would indicate that the crystal structure of the antagonist EPO peptidomimetic complexed to receptor has captured an essentially unstable form of the receptor complex. In this light it is notable that virtually all artificial mechanisms of eliciting receptor dimerization, such as the introduction of interreceptor covalent bonds, result in the formation of highly stable receptor dimers. This model also accounts for the observation that every mutation that modifies receptor complex stability has a concomitant effect on intracellular signalling and biological activity. Indeed it is this very mechanism that lies behind the development of ligands with altered functionality, as discussed below. This model is also open to decisive experimental testing in that it predicts that it is impossible to activate signalling by reducing the stability of receptor complexes.
Altered ligands and the prospect for therapy
As discussed above, one reason for seeking to understand the structural features of cytokine–receptor interactions is to use the information gained to devise means of intervening in cytokine signalling pathways. These intervention methods can take two forms: the derivation of mutant proteins with altered functionality and the identification of chemical compounds that interact with either ligands or receptors.
Mutant proteins
Based upon the preceding analysis three strategies can be proposed to develop mutant ligands whose biological activities are different from ‘wild-type’ ligands. The first would be to make mutants that are defective in their ability to bind specific signalling receptor components. In principle, these mutants would exhibit ‘dominant-negative’ or antagonistic biological properties since they would be predicted to form ‘sterile’ complexes that lack an essential signalling component. In reality, the practical utility of such mutants is dependent upon the affinity of the site that is destroyed, since any mutant ligand has to retain the ability to form a stable receptor complex in order to compete effectively with wild-type ligands. In the case of LIF, an effective antagonist has been generated by mutation of site II residues involved in gp130 recognition. This yields a protein that is still able to bind LIF-R with high affinity and therefore has the ability to antagonize biological effects that are mediated by known, or unknown, cytokines that associate with LIF-R (Wollert et al., 1996; Vernallis et al., 1997). Similarly, mutants of IL-6 that are defective in binding gp130 via site III are able to antagonize signalling mediated by IL-6–IL6R complexes (Brakenhoff et al., 1994; Demartis et al., 1996; Renne et al., 1998).
The second strategy involves the opposite approach: in this case mutations are generated that increase the affinity of interaction with specific signalling receptors. These molecules would be expected to exhibit ‘super-agonist’ activity with increased potency compared with their wild-type counterparts. This method has been employed with some success for IL-6, where the selection of mutants with enhanced affinity for gp130 has been generated, either by selection for high affinity binding using ligands subjected to random mutation in site II or site III residues, or by covalent fusion of the ligand to IL-6R using a flexible linker (Savino et al., 1994; Cabibbo et al., 1995; Toniatti et al., 1996; Fischer et al., 1997; Pflanz et al., 1999).
The third approach involves generating ligands that exhibit qualitatively distinct biological specificity compared with their wild-type progenitors. In principle, this can be achieved by altering the identity of a receptor bound at a particular site. This approach has so far been confined to exchanging biological specificities between related cytokines. Thus, Kallen et al. (1999) exchanged the site III epitope of IL-6 for its CNTF counterpart. This hybrid molecule acquired a requirement for LIF-R to activate signalling. An interesting extension of this idea would be to alter the repertoire of signalling receptors so that entirely novel signalling complexes are generated. It is possible to imagine, for example, the generation of mutants in which the association with gp130 is substituted for that of a closely related receptor such as GCSF-R. This class of mutant is predicted to exhibit different biological specificities to the parent ligands as a result of the formation of novel receptor complexes. A second example would be a case where site I, which is usually occupied by non-signalling receptors, was modified to be occupied by a second signalling receptor.
Small molecules
It has generally been thought that protein–protein interactions of the type involved in cytokine receptor signalling would represent poor targets for the development of small molecule agonists and antagonists. This belief largely derives from consideration of the large interfaces formed on the formation of a complex. However, as described above, extensive structure–function studies have shown that in many, if not all cases, the affinity of the complex is dominated by the identity of a few key residues in the ligand and receptor. This suggests that the production of small molecule mimetics of cytokines should be possible. Such mimetics could either bind to, or emulate, a specific functional epitope. Significant progress has been made by screening compound libraries for chemical entities that can activate receptor signalling. Compounds have recently been identified that can act as agonists or antagonists of GCSF-R (Tian et al., 1998) and EPO-R signalling (Qureshi et al., 1999). Notably, in the latter study a chemical antagonist was converted into an agonist by oligomerization. This finding is in accord with what might be predicted from the behaviour of bona fide polypeptide ligands and indicates that more sophisticated chemical mimetics might be generated from dendromeric compounds containing different functional groups. Despite these promising beginnings some important obstacles remain. The current generation of compounds exhibits greatly reduced potency compared with their natural protein partners. Moreover, as described above, it is not clear whether truly effective chemical mimetics of cytokine signalling would need to elicit specific changes in the conformation of receptor complexes as well as merely bind appropriate partners.
Conclusions
The gp130 cytokines are an example of a family of ligands that employ a combinatorial mechanism of receptor engagement to achieve biological specificity and diversity of function. The central feature of their mechanism of action is the use of a common transmembrane transducing receptor gp130. The accumulated evidence indicates that the diversity of gp130-dependent receptor signalling complexes produced by the engagement of different ligands involves simple structural rules. Each ligand displays three stereotypical binding sites, or epitopes, with differential affinities for cognate receptors. Receptors display single or multiple corresponding ligand recognition sites, which results in the formation of signalling complexes that can differ in both composition and stoichiometry. Receptor recognition epitopes involve a small number of key solvent-exposed residues that may provide a target for the generation of small molecule cytokine mimetics.
Acknowledgments
Acknowledgements
We are grateful to the Cancer Research Campaign for continued support of this work. We thank Yvonne Jones, Laura Grey, Robert Robinson, Keith Hudson, Ann Vernallis, Mark Hall, Marc Deller and Victoria Barton for discussion and collaboration.
References
- Aritomi M., Kunishima,N., Okamoto,T., Kuroki,R., Ota,Y. and Morikawa,K. (1999) Atomic structure of the GCSF–receptor complex showing a new cytokine-receptor recognition scheme. Nature, 401, 713–717. [DOI] [PubMed] [Google Scholar]
- Ballinger M.D. and Wells,J.A. (1998) Will any dimer do? Nature Struct. Biol., 5, 938–940. [DOI] [PubMed] [Google Scholar]
- Barton V.A., Hudson,K.R. and Heath,J.K. (1999) Identification of three distinct receptor binding sites of murine interleukin-11. J. Biol. Chem., 274, 5755–5761. [DOI] [PubMed] [Google Scholar]
- Bazan J.F. (1990) Structural design and molecular evolution of a cytokine receptor superfamily. Proc. Natl Acad. Sci. USA, 87, 6934–6938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazan J.F. (1991) Neuropoietic cytokines in the hematopoietic fold. Neuron, 7, 197–208. [DOI] [PubMed] [Google Scholar]
- Betz U.A.K., Bloch,W., van den Broek,M., Yoshida,K., Taga,T., Kishimoto,T., Addicks,K., Rajewsky,K. and Muller,W. (1998) Postnatally induced inactivation of gp130 in mice results in neurological, cardiac, hematopoietic, immunological, hepatic and pulmonary defects. J. Exp. Med., 188, 1955–1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilinski P., Roopenian,D. and Gossler,A. (1998) Maternal IL-11Rα function is required for normal decidual and fetoplacental development in mice. Genes Dev., 12, 2234–2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulay J. and Paul,W.E. (1993) Hemapoietin sub-family classification based on size, gene organization and sequence homology. Curr. Biol., 3, 573–581. [DOI] [PubMed] [Google Scholar]
- Brakenhoff J.P., de Hon,F.D., Fontaine,V., ten Boekel,E., Schooltink,H., Rose-John,S., Heinrich,P.C., Content,J. and Aarden,L.A. (1994) Development of a human interleukin-6 receptor antagonist. J. Biol. Chem., 269, 86–93. [PubMed] [Google Scholar]
- Bravo J., Staunton,D., Heath,J.K. and Jones,E.Y. (1998) Crystal structure of a cytokine-binding region of gp130. EMBO J., 17, 1665–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabibbo A., Sporeno,E., Toniatti,C., Altamura,S., Savino,R., Paonessa,G. and Ciliberto,G. (1995) Monovalent phage display of human interleukin (hIL)-6: selection of superbinder variants from a complex molecular repertoire in the hIL-6 D-helix. Gene, 167, 41–47. [DOI] [PubMed] [Google Scholar]
- Clackson T. and Wells,J.A. (1995) A hot spot of binding energy in a hormone–receptor interface. Science, 267, 383–386. [DOI] [PubMed] [Google Scholar]
- Clegg C.H., Rulffes,J.T., Wallace,P.M. and Haugen,H.S. (1996) Regulation of an extrathymic T-cell development pathway by oncostatin M. Nature, 384, 261–263. [DOI] [PubMed] [Google Scholar]
- Cosman D. (1993) The hematopoietin receptor superfamily. Cytokine, 5, 95–106. [DOI] [PubMed] [Google Scholar]
- Cunningham B.C. and Wells,J.A. (1989) High-resolution epitope mapping of hGH–receptor interactions by alanine-scanning mutagenesis. Science, 244, 1081–1085. [DOI] [PubMed] [Google Scholar]
- Cunningham B.C. and Wells,J.A. (1993) Comparison of a structural and a functional epitope. J. Mol. Biol., 234, 554–563. [DOI] [PubMed] [Google Scholar]
- Cunningham B.C., Ultsch,M., De Vos,A.M., Mulkerrin,M.G., Clauser,K.R. and Wells,J.A. (1991) Dimerization of the extracellular domain of the human growth hormone receptor by a single hormone molecule. Science, 254, 821–825. [DOI] [PubMed] [Google Scholar]
- Davis S., Aldrich,T.H., Valenzuela,D.M., Wong,V.V., Furth,M.E., Squinto,S.P. and Yancopoulos,G.D. (1991) The receptor for ciliary neurotrophic factor. Science, 253, 59–63. [DOI] [PubMed] [Google Scholar]
- DeChiara T.M., Vejsada,R., Poueymirou,W.T., Acheson,A. and Suri,C. (1995) Mice lacking the CNTF receptor, unlike mice lacking CNTF, exhibit profound motor neuron deficits at birth. Cell, 83, 313–322. [DOI] [PubMed] [Google Scholar]
- Demartis A., Bernassola,F., Savino,R., Melino,G. and Ciliberto,G. (1996) Interleukin 6 receptor superantagonists are potent inducers of human multiple myeloma cell death. Cancer Res., 15, 4213–4218. [PubMed] [Google Scholar]
- De Serio A., Graziani,R., Laufer,R., Ciliberto,G. and Paonessa,G. (1995) In vitro binding of ciliary neurotrophic factor to its receptors: evidence for the formation of an IL-6-type hexameric complex. J. Mol. Biol., 254, 795–800. [DOI] [PubMed] [Google Scholar]
- De Vos A.M., Ultsch,M. and Kossiakoff,A.A. (1992) Human growth hormone and extracellular domain of its receptor: crystal structure of the complex. Science, 255, 306–312. [DOI] [PubMed] [Google Scholar]
- Di Marco A., Gloaguen,I., Graziani,R., Paonessa,G., Saggio,I., Hudson,K.R. and Laufer,R. (1996) Identification of ciliary neurotrophic factor (CNTF) residues essential for leukemia inhibitory factor receptor binding and generation of CNTF receptor antagonists. Proc. Natl Acad. Sci. USA, 93, 9247–9252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elson G.C., Graber,P., Losberger,C., Herren,S., Gretener,D., Menoud,L.N., Wells,T.N., Kosco-Vilbois,M.H. and Gauchat,J.-F. (1998) Cytokine-like factor-1, a novel soluble protein, shares homology with members of the cytokine type I receptor family. J. Immunol., 161, 1371–1379. [PubMed] [Google Scholar]
- Ernst M., Gearing,D.P. and Dunn,A.R. (1994) Functional and biochemical association of Hck with the LIF/IL-6 receptor signal transducing subunit gp130 in embryonic stem cells. EMBO J., 13, 1574–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escary J.L., Perreau,J., Dumenil,D., Ezine,S. and Brulet,P. (1993) Leukaemia inhibitory factor is necessary for maintenance of haematopoietic stem cells and thymocyte stimulation. Nature, 363, 361–364. [DOI] [PubMed] [Google Scholar]
- Fischer M., Goldschmitt,J., Peschel,C., Brakenhoff,J.P., Kallen,K.J., Wollmer,A., Grotzinger,J. and Rose-John,S. (1997) I. A bioactive designer cytokine for human hematopoietic progenitor cell expansion. Nature Biotechnol., 15, 142–145. [DOI] [PubMed] [Google Scholar]
- Gearing D.P., Gough,N.M., King,J.A., Hilton,D.J., Nicola,N.A., Simpson,R.J., Nice,E.C., Kelso,A. and Metcalf,D. (1987) Molecular cloning and expression of cDNA encoding a murine myeloid leukaemia inhibitory factor (LIF). EMBO J., 6, 3995–4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gearing D.P., Thut,C.J., VandeBos,T., Gimpel,S.D., Delaney,P.B., King,J., Price,V., Cosman,D. and Beckmann,M.P. (1991) Leukemia inhibitory factor receptor is structurally related to the IL-6 signal transducer, gp130. EMBO J., 10, 2839–2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gough N.M., Gearing,D.P., King,J.A., Willson,T.A., Hilton,D.J., Nicola,N.A. and Metcalf,D. (1988) Molecular cloning and expression of the human homologue of the murine gene encoding myeloid leukemia-inhibitory factor. Proc. Natl Acad. Sci. USA, 85, 2623–2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotzinger J., Kernebeck,T., Kallen,K.J. and Rose-John,S. (1999) IL6 type cytokine receptor complexes: hexamer, tetramer or both? Biol. Chem., 380, 803–813. [DOI] [PubMed] [Google Scholar]
- Guthridge M.A., Stomski,F.C., Thomas,D., Woodcock,J.M., Bagley,C.J., Berndt,M.C. and Lopez,A.F. (1998) Mechanism of activation of the GM-CSF, IL-3, and IL-5 family of receptors. Stem Cells, 16, 301–313. [DOI] [PubMed] [Google Scholar]
- Hage T., Sebald,W. and Reinemer,P. (1999) Crystal structure of the interleukin-4/receptor α chain complex reveals a mosaic binding interface. Cell, 97, 271–281. [DOI] [PubMed] [Google Scholar]
- Hammacher A., Richardson,R.T., Layton,J.E., Smith,D.K., Angus,L.J., Hilton,D.J., Nicola,N.A., Wijdenes,J. and Simpson,R.J. (1998) The immunoglobulin-like module of gp130 is required for signaling by interleukin-6, but not by leukemia inhibitory factor. J. Biol. Chem., 273, 22701–22707. [DOI] [PubMed] [Google Scholar]
- Heinrich P.C., Behrmann,I., Muller-Newen,G., Schaper,F. and Graeve,L. (1998) Interleukin-6-type cytokine signalling through the gp130/Jak/STAT pathway. Biochem. J., 334, 297–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibi M., Murakami,M., Saito,M., Hirano,T., Taga,T. and Kishimoto,T. (1990) Molecular cloning and expression of an IL-6 signal transducer, gp130. Cell, 63, 1149–1157. [DOI] [PubMed] [Google Scholar]
- Hilton D.J. et al. (1994) Cloning of murine IL-11 receptor α-chain: requirement for gp130 for high affinity binding and signal transduction. EMBO J., 13, 4765–4775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinds M.G., Maurer,T., Zhang,J.G., Nicola,N.A. and Norton,R.S. (1998) Solution structure of leukemia inhibitory factor. J. Biol. Chem., 273, 13738–13745. [DOI] [PubMed] [Google Scholar]
- Hirano T. et al. (1986) Complementary DNA for a novel human interleukin (BSF-2) that induces B lymphocytes to produce immunoglobulin. Nature, 324, 73–76. [DOI] [PubMed] [Google Scholar]
- Hirota H., Chen,J., Betz,U.A., Rajewsky,K., Gu,Y., Ross,J.,Jr, Müller,W. and Chien,K.R. (1999) Loss of a gp130 cardiac muscle cell survival pathway is a critical event in the onset of heart failure during biomechanical stress. Cell, 97, 189–198. [DOI] [PubMed] [Google Scholar]
- Hoffman R.C. et al. (1996) Resonance assignments for oncostatin M, a 24-kDa α-helical protein. J. Biomol. NMR, 7, 273–282. [DOI] [PubMed] [Google Scholar]
- Horsten U., Muller-Newen,G., Gerhartz,C., Wollmer,A., Wijdenes,J., Heinrich,P.C. and Grotzinger,J. (1997) Molecular modeling-guided mutagenesis of the extracellular part of gp130 leads to the identification of contact sites in the interleukin-6 (IL-6).IL-6 receptor.gp130 complex. J. Biol. Chem., 272, 23748–23757. [DOI] [PubMed] [Google Scholar]
- Huber A.H., Wang,Y.M., Bieber,A.J. and Bjorkman,P.J. (1994) Crystal structure of tandem type III fibronectin domains from Drosophila neuroglian at 2.0 Å. Neuron, 12, 717–731. [DOI] [PubMed] [Google Scholar]
- Hudson K.R., Vernallis,A.B. and Heath,J.K. (1996) Characterization of the receptor binding sites of human leukemia inhibitory factor and creation of antagonists. J. Biol. Chem., 271, 11971–11978. [DOI] [PubMed] [Google Scholar]
- Ichihara M., Hara,T., Kim,H., Murate,T. and Miyajima,A. (1997) Oncostatin M and leukemia inhibitory factor do not use the same functional receptor in mice. Blood, 90, 165–173. [PubMed] [Google Scholar]
- Ihle J.N. (1996) STATs: signal transducers and activators of transcription. Cell, 84, 331–334. [DOI] [PubMed] [Google Scholar]
- Ihle J.N., Witthuhn,B.A., Quelle,F.W., Yamamoto,K., Thierfelder,W.E., Kreider,B. and Silvennoinen,O. (1994) Signaling by the cytokine receptor superfamily: JAKs and STATs. Trends Biochem. Sci., 19, 222–227. [DOI] [PubMed] [Google Scholar]
- Inoue M., Nakayama,C., Kikuchi,K., Kimura,T., Ishige,Y., Ito,A., Kanaoka,M. and Noguchi,H. (1995) D1 cap region involved in the receptor recognition and neural cell survival activity of human ciliary neurotrophic factor. Proc. Natl Acad. Sci. USA, 92, 8579–8583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaleeba J.A., Bergquam,E.P. and Wong,S.W. (1999) A rhesus macaque rhadinovirus related to Kaposi’s sarcoma-associated herpesvirus/human herpesvirus 8 encodes a functional homologue of interleukin-6. J. Virol., 73, 6177–6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallen K.J. et al. (1999) Receptor recognition sites of cytokines are organized as exchangeable modules. J. Biol. Chem., 274, 11859–11867. [DOI] [PubMed] [Google Scholar]
- Karow J., Hudson,K.R., Hall,M.A., Vernallis,A.B., Taylor,J.A., Gossler,A. and Heath,J.K. (1996) Mediation of interleukin-11-dependent biological responses by a soluble form of the interleukin-11 receptor. Biochem. J., 318, 489–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kernebeck T. et al. (1999) The signal transducer gp130: solution structure of the carboxy terminal domain of the cytokine receptor homology region. Protein Sci., 8, 5–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo M., Takeshita,T., Ishii,N., Nakamura,M., Watanabe,S.J., Arai,K. and Sugamura,K. (1993) Sharing of the interleukin-2 (IL-2) receptor γ chain between receptors for IL-2 and IL-4. Science, 262, 1874–1877. [DOI] [PubMed] [Google Scholar]
- Kopf M., Baumann,H., Freer,G., Freudenberg,M., Lamers,M., Kishimoto,T., Zinkernagel,R., Bluethmann,H. and Kohler,G. (1994) Impaired immune and acute-phase responses in interleukin-6-deficient mice. Nature, 368, 339–342. [DOI] [PubMed] [Google Scholar]
- Kurth I., Horsten,U., Pflanz,S., Dahmen,H., Kuster,A., Grotzinger,J., Heinrich,P.C. and Muller-Newen,G. (1999) Activation of the signal transducer glycoprotein 130 by both IL-6 and IL-11 requires two distinct binding epitopes. J. Immunol., 162, 1480–1487. [PubMed] [Google Scholar]
- Layton M.J., Lock,P., Metcalf,D. and Nicola,N.A. (1994) Cross-species receptor binding characteristics of human and mouse leukemia inhibitory factor suggest a complex binding interaction. J. Biol. Chem., 269, 17048–17055. [PubMed] [Google Scholar]
- Li M., Sendtner,M. and Smith,A. (1995) Essential function of LIF receptor in motor neurons. Nature, 378, 724–727. [DOI] [PubMed] [Google Scholar]
- Liautard J., Sun,R.X., Cotte,N., Gaillard,J.P., Mani,J.C., Klein,B. and Brochier,J. (1997) Specific inhibition of IL-6 signalling with monoclonal antibodies against the gp130 receptor. Cytokine, 9, 233–241. [DOI] [PubMed] [Google Scholar]
- Lindberg R.A., Juan,T.S., Welcher,A.A., Sun,Y., Cupples,R., Guthrie,B. and Fletcher,F.A. (1998) Cloning and characterization of a specific receptor for mouse oncostatin M. Mol. Cell. Biol., 18, 3357–3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livnah O., Stura,E.A., Johnson,D.L., Middleton,S.A., Mulcahy,L.S., Wrighton,N.C., Dower,W.J., Jolliffe,L.K. and Wilson,I.A. (1996) Functional mimicry of a protein hormone by a peptide agonist: the EPO receptor complex at 2.8 Å. Science, 273, 464–471. [DOI] [PubMed] [Google Scholar]
- Livnah O., Stura,E.A., Middleton,S.A., Johnson,D.L., Jolliffe,L.K. and Wilson,I.A. (1999) Crystallographic evidence of preformed dimers of erythropoietin receptor before ligand activation. Science, 283, 987–990. [DOI] [PubMed] [Google Scholar]
- Malik N. et al. (1989) Molecular cloning, sequence analysis and functional expression of a novel growth regulator, oncostatin M. Mol. Cell. Biol., 9, 2847–2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masu Y., Wolf,E., Holtmann,B., Sendtner,M. and Brem,G. (1993) Disruption of the CNTF gene results in motor neuron degeneration. Nature, 365, 27–32. [DOI] [PubMed] [Google Scholar]
- McDonald N.Q., Panayotatos,N. and Hendrickson,W.A. (1995) Crystal structure of dimeric human ciliary neurotrophic factor determined by MAD phasing. EMBO J., 14, 2689–2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milburn M.V., Hassell,A.M., Lambert,M.H., Jordan,S.R., Proudfoot,A.E., Graber,P. and Wells,T.N. (1993) A novel dimer configuration revealed by the crystal structure at 2.4 Å resolution of human interleukin-5. Nature, 363, 172–176. [DOI] [PubMed] [Google Scholar]
- Moritz R.L., Ward,L.D., Tu,G.F., Fabri,L.J., Ji,H., Yasukawa,K. and Simpson,R.J. (1999) The N-terminus of gp130 is critical for the formation of the high affinity interleukin-6 receptor complex. Growth Factors, 16, 265–278. [DOI] [PubMed] [Google Scholar]
- Mosley B., De Imus,C., Friend,D., Boiani,N., Thoma,B., Park,L.S. and Cosman,D. (1996) Dual oncostatin M (OSM) receptors. Cloning and characterization of an alternative signaling subunit conferring OSM specific receptor activation. J. Biol. Chem., 271, 32635–32643. [DOI] [PubMed] [Google Scholar]
- Neipel F., Albrecht,J.C., Ensser,A., Huang,Y.Q., Li,J.J., Friedman-Kien,A.E. and Fleckenstein,B. (1997) Human herpesvirus 8 encodes a homolog of interleukin-6. J. Virol., 71, 839–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas J. et al. (1997) Kaposi’s sarcoma-associated human herpesvirus-8 encodes homologues of macrophageinflammatory protein-1 and interleukin-6. Nature Med., 13, 287–292. [DOI] [PubMed] [Google Scholar]
- Niwa H., Burdon,T., Chambers,I. and Smith,A.G. (1998) Self-renewal of pluripotent embryonic stem cells is mediated via activation of STAT3. Genes Dev., 12, 2048–2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owczarek C.M., Layton,M.J., Metcalf,D., Lock,P., Willson,T.A., Gough,N.M. and Nicola,N.A. (1993) Inter-species chimeras of leukaemia inhibitory factor define a major human receptor-binding determinant. EMBO J., 12, 3487–3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owczarek C.M., Zhang,Y., Layton,M.J., Metcalf,D., Roberts,B. and Nicola,N.A. (1997) The unusual species cross-reactivity of the leukemia inhibitory factor receptor α chain is determined primarily by the immunoglobulin-like domain. J. Biol. Chem., 272, 23976–23985. [DOI] [PubMed] [Google Scholar]
- Paonessa G., Graziani,R., De Serio,A., Savino,R., Ciapponi,L., Lahm,A., Salvati,A.L., Toniatti,C. and Ciliberto,G. (1995) Two distinct and independent sites on IL-6 trigger gp130 dimer formation and signalling. EMBO J., 14, 1942–1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul S.R. et al. (1990) Molecular cloning of a cDNA encoding interleukin 11, a stromal cell-derived lymphopoietic and hematopoietic cytokine. Proc. Natl Acad. Sci. USA, 87, 7512–7516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennica D. et al. (1995) Expression cloning of cardiotrophin 1, a cytokine that induces cardiac myocyte hypertrophy. Proc. Natl Acad. Sci. USA, 92, 1142–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pflanz S., Tacken,I., Grotzinger,J., Jacques,Y., Dahmen,H., Heinrich,P.C. and Muller-Newen,G. (1999) A fusion protein of interleukin-11 and soluble interleukin-11 receptor acts as a superagonist on cells expressing gp130. FEBS Lett., 450, 117–122. [DOI] [PubMed] [Google Scholar]
- Pitard V. et al. (1997) Production of monoclonal antibodies against the leukemia inhibitory factor low affinity receptor, gp190 and their characterisation. J. Immunol. Methods, 205, 177–190. [DOI] [PubMed] [Google Scholar]
- Poli V., Balena,R., Fattori,E., Markatos,A., Yamamoto,M., Tanaka,H., Ciliberto,G., Rodan,G.A. and Costantini,F. (1994) Interleukin-6 deficient mice are protected from bone loss caused by estrogen depletion. EMBO J., 13, 1189–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qui H., Belanger,A., Yoon,H.-W.P. and Bunn,H.F. (1998) Homodimerization restores biological activity to an inactive erythropoietin mutant. J. Biol. Chem., 273, 11173–11176. [DOI] [PubMed] [Google Scholar]
- Qureshi S.A. et al. (1999) Mimicry of erythropoietin by a nonpeptide molecule. Proc. Natl Acad. Sci. USA, 96, 12156–12161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remy I., Wilson,I.A. and Michnick,S.W. (1999) Erythropoietin receptor activation by a ligand induced conformation change. Science, 283, 990–993. [DOI] [PubMed] [Google Scholar]
- Renne C., Kallen,K.J., Mullberg,J., Jostock,T., Grotzinger,J. and Rose-John,S. (1998) A new type of cytokine receptor antagonist directly targeting gp130. J. Biol. Chem., 273, 27213–27219. [DOI] [PubMed] [Google Scholar]
- Robb R.L., Li,R., Hartley,L., Nandurkar,H.H., Koentgen,F. and Begley,C.G. (1998) Infertility in female mice lacking the receptor for interleukin 11 is due to a defective uterine response to implantation. Nature Med., 4, 303–308. [DOI] [PubMed] [Google Scholar]
- Robinson R.C., Grey,L.M., Staunton,D., Vankelecom,H., Vernallis,A.B., Moreau,J.F., Stuart,D.I., Heath,J.K. and Jones,E.Y. (1994) The crystal structure and biological function of leukemia inhibitory factor: implications for receptor binding. Cell, 77, 1101–1116. [DOI] [PubMed] [Google Scholar]
- Robledo O., Fourcin,M., Chevalier,S., Guillet,C., Auguste,P., Pouplard Barthelaix,A., Pennica,D. and Gascan,H. (1997) Signaling of the cardiotrophin-1 receptor. Evidence for a third receptor component. J. Biol. Chem., 272, 4855–4863. [DOI] [PubMed] [Google Scholar]
- Rollmann S.M., Houck,L.D. and Feldhoff,R.C. (1999) Proteinaceous pheromone affecting female receptivity in a terrestrial salamander. Science, 285, 1907–1909. [DOI] [PubMed] [Google Scholar]
- Salvati A.L., Lahm,A., Paonessa,G., Ciliberto,G. and Toniatti,C. (1995) Interleukin-6 (IL-6) antagonism by soluble IL-6 receptor α mutated in the predicted gp130-binding interface. J. Biol. Chem., 270, 12242–12249. [DOI] [PubMed] [Google Scholar]
- Savino R., Ciapponi,L., Lahm,A., Demartis,A., Cabibbo,A., Toniatti,C., Delmastro,P., Altamura,S. and Ciliberto,G. (1994) Rational design of a receptor super-antagonist of human interleukin-6. EMBO J., 13, 5863–5870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senaldi G. et al. (1999) Novel neurotrophin-1/B cell-stimulating factor-3: a cytokine of the IL-6 family. Proc. Natl Acad. Sci. USA, 96, 11458–11463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Wang,W., Yourey,P.A., Gohari,S., Zukauskas,D., Zhang,J., Ruben,S. and Alderson,R.F. (1999) Computational EST database analysis identifies a novel member of the neuropoietic cytokine family. Biochem. Biophys. Res. Commun., 262, 132–138. [DOI] [PubMed] [Google Scholar]
- Simpson R.J., Hammacher,A., Smith,D.K., Matthews,J.M. and Ward,L.D. (1997) Interleukin-6: structure–function relationships. Protein Sci., 6, 929–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers W., Stahl,M. and Seehra,J.S. (1997) 1.9 Å crystal structure of interleukin 6: implications for a novel mode of receptor dimerization and signaling. EMBO J., 16, 989–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprang S.R. and Bazan,J.F. (1993) Cytokine structural taxonomy and mechanisms of receptor engagement. Curr. Opin. Struct. Biol., 3, 815–827. [Google Scholar]
- Sprecher C.A., Grant,F.J., Baumgartner,J.W., Presnell,S.R., Schrader, S.K., Yamagiwa,T., Whitmore,T.E., O’Hara,P.J. and Foster,D.F. (1998) Cloning and characterization of a novel class I cytokine receptor. Biochem. Biophys. Res. Commun., 246, 82–90. [DOI] [PubMed] [Google Scholar]
- Staunton D., Hudson,K.R. and Heath,J.K. (1998) The interactions of the cytokine-binding homology region and immunoglobulin-like domains of gp130 with oncostatin M: implications for receptor complex formation. Protein Eng., 11, 1093–1102. [DOI] [PubMed] [Google Scholar]
- Stewart C.L., Kaspar,P., Brunet,L.J., Bhatt,H., Gadi,I., Kontgen,F. and Abbondanzo,S.J. (1992) Blastocyst implantation depends on maternal expression of leukaemia inhibitory factor. Nature, 359, 76–79. [DOI] [PubMed] [Google Scholar]
- Stockli K.A., Lottspeich,F., Sendtner,M., Masiakowski,P., Carroll,P., Gotz,R., Lindholm,D. and Thoenen,H. (1989) Molecular cloning, expression and regional distribution of rat ciliary neurotrophic factor. Nature, 342, 920–923. [DOI] [PubMed] [Google Scholar]
- Syed R.S. et al. (1998) Efficiency of signalling through cytokine receptors depends critically on receptor orientation. Nature, 395, 511–516. [DOI] [PubMed] [Google Scholar]
- Taga T. and Kishimoto,T. (1992) Cytokine receptors and signal transduction. FASEB J., 6, 3387–3396. [DOI] [PubMed] [Google Scholar]
- Taga T. and Kishimoto,T. (1995) Signaling mechanisms through cytokine receptors that share signal transducing receptor components. Curr. Opin. Immunol., 7, 17–23. [DOI] [PubMed] [Google Scholar]
- Taga T. and Kishimoto,T. (1997) Gp130 and the interleukin-6 family of cytokines. Annu. Rev. Immunol., 15, 797–819. [DOI] [PubMed] [Google Scholar]
- Taupin J.-L., Miossec,V., Pitard,V., Blanchard,F., Daburon,S., Raher,S., Jacques,Y., Godard,A. and Moreau,J.-F. (1999) Binding of leukemia inhibitory factor (LIF) to mutants of its low affinity receptor, gp190, reveals a LIF binding site outside and interactions between the two cytokine binding domains. J. Biol. Chem., 274, 14482–14489. [DOI] [PubMed] [Google Scholar]
- Tian S.S. et al. (1998) A small, nonpeptidyl mimic of granulocyte-colony-stimulating factor. Science, 281, 257–259. [DOI] [PubMed] [Google Scholar]
- Tomida M., Yamamoto-Yamaguchi,Y. and Hozumi,M. (1994) Three different cDNAs encoding mouse D-factor/LIF receptor. J. Biochem., 115, 557–562. [DOI] [PubMed] [Google Scholar]
- Toniatti C., Cabibbo,A., Sporena,E., Salvati,A.L., Cerretani,M., Serafini,S., Lahm,A., Cortese,R. and Ciliberto,G. (1996) Engineering human interleukin-6 to obtain variants with strongly enhanced bioactivity. EMBO J., 15, 2726–2737. [PMC free article] [PubMed] [Google Scholar]
- Vernallis A.B., Hudson,K.R. and Heath,J.K. (1997) An antagonist for the leukemia inhibitory factor receptor inhibits leukemia inhibitory factor, cardiotrophin-1, ciliary neurotrophic factor and oncostatin M. J. Biol. Chem., 272, 26947–26952. [DOI] [PubMed] [Google Scholar]
- Walter M.R., Windsor,W.T., Nagabhushan,T.L., Lundell,D.J., Lunn,C.A., Zauodny,P.J. and Narula,S.K. (1995) Crystal structure of a complex between interferon-γ and its soluble high-affinity receptor. Nature, 376, 230–235. [DOI] [PubMed] [Google Scholar]
- Ward L.D., Howlett,G.J., Discolo,G., Yasukawa,K., Hammacher,A., Moritz,R.L. and Simpson,R.J. (1994) High affinity interleukin-6 receptor is a hexameric complex consisting of two molecules each of interleukin-6, interleukin-6 receptor and gp-130. J. Biol. Chem., 269, 23286–23289. [PubMed] [Google Scholar]
- Ward L.D., Hammacher,A., Howlett,G.J., Matthews,J.M., Fabri,L., Moritz,R.L., Nice,E.C., Weinstock,J. and Simpson,R.J. (1996) Influence of interleukin-6 (IL-6) dimerization on formation of the high affinity hexameric IL-6.receptor complex. J. Biol. Chem., 271, 20138–20144. [DOI] [PubMed] [Google Scholar]
- Ware C.B. et al. (1995) Targeted disruption of the low-affinity leukemia inhibitory factor receptor gene causes placental, skeletal, neural and metabolic defect. Development, 121, 1283–1299. [DOI] [PubMed] [Google Scholar]
- Wells J.A. (1996) Binding in the growth hormone receptor complex. Proc. Natl Acad. Sci. USA, 93, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells J.A. and De Vos,A.M. (1996) Hematopoietic receptor complexes. Annu. Rev. Biochem., 65, 609–634. [DOI] [PubMed] [Google Scholar]
- Wollert K.C. et al. (1996) Cardiotrophin-1 activates a distinct form of cardiac muscle cell hypertrophy. Assembly of sarcomeric units in series VIA gp130/leukemia inhibitory factor receptor-dependent pathways. J. Biol. Chem., 271, 9535–9545. [DOI] [PubMed] [Google Scholar]
- Xu G.Y., Yu,H.A., Hong,J., Stahl,M., McDonagh,T., Kay,L.E. and Cumming,D.A. (1997) Solution structure of recombinant human interleukin-6. J. Mol. Biol., 268, 468–481. [DOI] [PubMed] [Google Scholar]
- Yamasaki K., Taga,T., Hirata,Y., Yawata,H., Kawanishi,Y., Seed,B., Taniguchi,T., Hirano,T. and Kishimoto,T. (1988) Cloning and expression of the human interleukin-6 (BSF-2/IFN β 2) receptor. Science, 241, 825–828. [DOI] [PubMed] [Google Scholar]
- Yawata H., Yasukawa,K., Natsuka,S., Murakami,M., Yamasaki,K., Hibi,M., Taga,T. and Kishimoto,T. (1993) Structure–function analysis of human IL-6 receptor: dissociation of amino acid residues required for IL-6-binding and for IL-6 signal transduction through gp130. EMBO J., 12, 1705–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K. et al. (1996) Targeted disruption of gp130, a common signal transducer for the interleukin 6 family of cytokines, leads to myocardial and hematological disorders. Proc. Natl Acad. Sci. USA, 93, 407–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura A., Ichihara,M., Kinjyo,I., Moriyama,M., Copeland,N.G., Gilbert,D.J., Jenkins,N.A., Hara,T. and Miyajima,A. (1996) Mouse oncostatin M: an immediate early gene induced by multiple cytokines through the JAK–STAT5 pathway. EMBO J., 15, 1055–1063. [PMC free article] [PubMed] [Google Scholar]
- Zhang J.G., Owczarek,C.M., Ward,L.D., Howlett,G.J., Fabri,L.J., Roberts,B.A. and Nicola,N.A. (1997) Evidence for the formation of a heterotrimeric complex of leukaemia inhibitory factor with its receptor subunits in solution. Biochem. J., 325, 693–700. [DOI] [PMC free article] [PubMed] [Google Scholar]