Abstract
Processing of the p105 precursor to form the active subunit p50 of the NF-κB transcription factor is a unique case in which the ubiquitin system is involved in limited processing rather than in complete destruction of the target substrate. A glycine-rich region along with a downstream acidic domain have been demonstrated to be essential for processing. Here we demonstrate that following IκB kinase (IκK)-mediated phosphorylation, the C-terminal domain of p105 (residues 918–934) serves as a recognition motif for the SCFβ-TrCP ubiquitin ligase. Expression of IκKβ dramatically increases processing of wild-type p105, but not of p105-Δ918–934. Dominant-negative β-TrCP inhibits IκK-dependent processing. Furthermore, the ligase and wild-type p105 but not p105-Δ918–934 associate physically following phosphorylation. In vitro, SCFβ-TrCP specifically conjugates and promotes processing of phosphorylated p105. Importantly, the TrCP recognition motif in p105 is different from that described for IκBs, β-catenin and human immunodeficiency virus type 1 Vpu. Since p105-Δ918–934 is also conjugated and processed, it appears that p105 can be recognized under different physiological conditions by two different ligases, targeting two distinct recognition motifs.
Keywords: IκB kinase (IκK)/NF-κB/p105/β-TrCP/ubiquitin
Introduction
The NF-κB transcription factors play key roles in basic processes such as regulation of the immune and inflammatory responses, development and differentiation, malignant transformation and apoptosis (Baeuerle and Baltimore, 1996; Baldwin, 1996; Barnes and Karin, 1997; Ghosh et al., 1998; Foo and Nolan, 1999). The precursor molecules p105 and p100 undergo ubiquitin- and proteasome-mediated limited processing to yield the respective active subunits p50 and p52 (Palombella et al., 1994; Orian et al., 1995; Betts and Nabel, 1996), which are derived from the N-terminal domain of the molecule. The C-terminal domain is degraded (Fan and Maniatis, 1991). These subunits typically heterodimerize with members of the rel family, such as p65, RelB or c-Rel, to generate the active transcription factor. In the resting cell, the heterodimer generates a ternary complex with a member of the IκB family of inhibitory proteins and is sequestered in the cytosol. Following stimulation, specific IκB kinases are activated (Mercurio et al., 1997; Woronicz et al., 1997; Zandi et al., 1997) and phosphorylate the protein on serine residues 32 and 36 (Brown et al., 1995). The phosphorylation leads to recognition of the molecule by the SCFβ-TrCP ubiquitin ligase complex (see, for example, Yaron et al., 1998; Winston et al., 1999), polyubiquitylation on Lys21 and/or Lys22 (Scherer et al., 1995) and subsequent degradation by the 26S proteasome (Alkalay et al., 1995; Chen et al., 1995). Following degradation of IκBα, the heterodimer is translocated into the nucleus where it initiates specific transcription.
The ubiquitin pathway is involved in the regulation of many basic cellular processes, such as cell cycle progression, differentiation and development, and the immune and inflammatory responses. Involvement of the system in these processes is mediated via specific proteolysis of key regulatory proteins such as cyclins, transcriptional activators, membrane receptors and major histocompatibility complex (MHC) class I-restricted antigens. Degradation of a protein by the system involves two discrete steps: (i) formation of a polyubiquitin chain that is covalently attached to the target substrate; and (ii) degradation of the tagged protein by the 26S proteasome. Conjugation of ubiquitin requires the sequential action of three enzymes: the ubiquitin-activating enzyme, E1; one of several ubiquitin-carrier proteins, E2s (known also as ubiquitin-conjugating enzymes, UBCs); and a member of the ubiquitin–protein ligase, E3, family. E3s serve as substrate-binding subunits and play an essential role in specific substrate recognition. Several classes of E3s have been described. Among them are the SCF complexes that recognize phosphorylated substrates. These tetrameric complexes are comprised of Skp1, cullin1 and Rbx1/Roc1, which are common to all SCFs, and a variable F-box protein (for recent reviews on the ubiquitin system, see, for example, Laney and Hochstrasser, 1999; Voges et al., 1999; Kornitzer and Ciechanover, 2000; for a recent review on SCF complexes, see Deshaies, 1999). The variable F-box protein serves as the substrate recognition subunit. For example, an SCF complex that contains β-TrCP (SCFβ-TrCP) as the F-box protein recognizes phosphorylated IκBα, β-catenin and human immunodeficiency virus type 1 (HIV-1) Vpu via the sequence [DS(P)GΨXS(P)]. Recognition of other substrates via different motifs has not been ruled out. SCFSkp2 recognizes the singly phosphorylated p27Kip1 (Carrano et al., 1999) and possibly E2F-1 (Marti et al., 1999) via an as yet unidentified motif.
Limited processing of the precursor proteins p105 and p100 is mediated by the ubiquitin system (Fan and Maniatis, 1991; Palombella et al., 1994; Orian et al., 1995; Coux and Goldberg, 1998; Heusch et al., 1999) and appears to be the only established case in which the system is involved in limited processing rather than in complete destruction of its target. The mechanisms involved in this process have been partially elucidated. Lin and Ghosh (1996) demonstrated that a glycine-rich region (GRR) that spans amino acid residues 372–394 in mouse p105 is required for processing. A GRR in human p105 serves a similar function (Orian et al., 1999). The GRR appears to serve as a digestion ‘stop’ signal for the 26S proteasome. Processing also requires an additional domain that contains two essential lysine residues (441 and 442) that are important for ubiquitylation, and a downstream acidic region (residues 446–454) that may function as an E3 recognition motif (Orian et al., 1999). These findings suggest that processing requires at least two motifs, a processing ‘stop’ signal and a ubiquitylation/E3 recognition site. Fan and Maniatis (1991) have shown that a truncated form of p105, p60, can be processed to p50. Lin et al. (1998) have shown that p105 can be processed co-translationally, and synthesis of the complete molecule is not required for generation of p50. Taken together, studies from all three groups suggest that all the motifs that are essential for basal processing are contained within the first ∼550 amino acid residues. However, other studies have suggested a role for the C-terminal domain in regulated processing/degradation of p105. Fujimoto et al. (1995) and McKichan et al. (1996) have shown that stimulation-induced phosphorylation of serine and possibly threonine residues at the C-terminal domain of the molecule increases processing. Belich et al. (1999) have shown that TPL-2-mediated phosphorylation of the C-terminal domain of p105 leads, although indirectly, to accelerated degradation of the molecule. Heissmeyer et al. (1999) have shown that IκK-mediated phosphorylation of serine residues localized to a region that spans moieties 920–936 also leads to rapid degradation of p105. Although these studies appear to be somewhat contradictory in their final conclusion with regard to whether signal-induced C-terminal phosphorylation leads to enhanced processing or complete degradation of p105 (see Discussion), they all conclude that signal-induced phosphorylation of serine residues in the C-terminal domain serves an important regulatory role in NF-κB activation. In all these cases, however, the possible involvement of the ubiquitin system in the process has not been established.
Here we show that IκKβ-mediated phosphorylation at the C-terminal domain results in accelerated processing of p105 to p50, a process that is mediated by the SCFβ-TrCP E3.
Results
Residues 446–454 and 918–936 are both required for efficient processing of p105
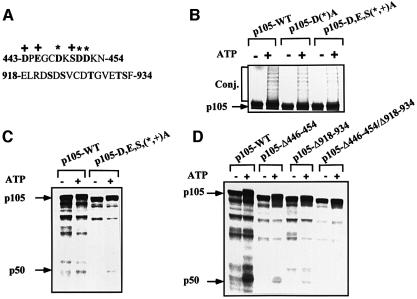
We have shown previously that residues 446–454, which are homologous to the IκBα E3 recognition motif, are required for processing of p105 (Orian et al., 1999). We postulated that the acidic residues in this region (Figure 1A) are important for ubiquitylation, possibly via binding of E3. Now we show that progressive replacement of these residues with alanine significantly decreases the efficiency of conjugation (Figure 1B) and subsequent processing (Figure 1C) of p105 in vitro, although it does not abolish them completely. Therefore, we predicted that an additional domain may be involved in recognition by the ubiquitin system. Since the C-terminal domain contains an IκK phosphorylation site that is involved in regulated processing/degradation of p105 (see Introduction), we decided to dissect the mechanism(s) that underlie its involvement in the process. As can be seen in Figure 1D, deletion of residues 446–454 reduces processing significantly (26% compared with processing of wild-type p105, as determined quantitatively). Deletion of residues 918–934, which comprise the IκK phosphorylation site, reduces processing even more (11% compared with wild type). Processing was reduced further in a p105 lacking both the acidic and the C-terminal domains. Since inhibition of processing is significant when any one of the domains is deleted, it is not clear whether they cooperate or act independently (see Figure 9B; Discussion).
Fig. 1. The acidic domain (residues 443–454) and the C-terminal IκK phosphorylation site of p105 are both required for processing of p105 in vitro. (A) Sequence of amino acid residues 443–454 and 918–934 of p105. The asterisks denote aspartate residues 448, 451 and 452, which were replaced by alanine in the experiments described in (B) and (C). The plus signs denote Asp443, Glu445 and Ser450, which were replaced by alanine in the same experiments. Acidic and serine residues in the 443–454 sequence and serine and threonine residues in the 918–934 sequence are in bold. (B) ATP-dependent conjugation of wild-type-p105, p105-D(*)A and p105-D,E,S(*,+)A. Conjugation of the p105 acidic domain mutants was carried out in a crude HeLa cell extract as described in Materials and methods. (C) ATP-dependent processing of wild-type p105 and p105-D,E,S(*,+)A. Processing of wild-type p105 and p105-D,E,S(*,+)A was carried out in a crude HeLa cell extract as described in Materials and methods. (D) ATP-dependent processing of wild-type p105, p105-Δ446–454, p105-Δ918–934 and p105-Δ446–454/Δ918–934. Processing of the different p105 deletion mutants was carried out as described in (C). Conj. denotes conjugates. p50 and p105 denote the site of migration of the two molecules, respectively.
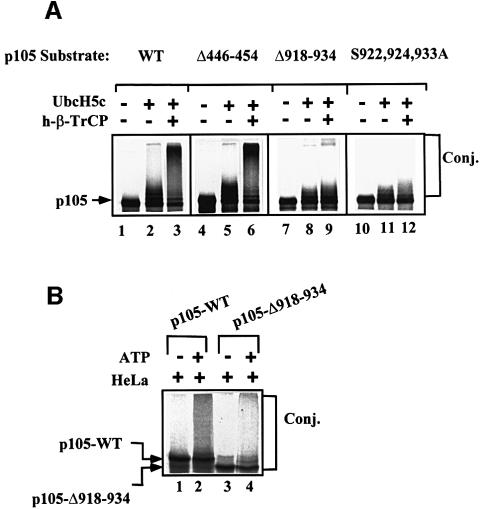
Fig. 9. Two ubiquitin ligases are involved in p105 processing, one of them, SCFβ-TrCP, requires the presence of residues 918–934 and, in particular, serine residues 922, 924 and 933. (A) SCFβ-TrCP conjugates wild-type and Δ446–454, but not Δ918–934 and Δ446–454-S922,924,933A p105s. In vitro translated and kinase-phosphorylated wild-type (lanes 1–3), Δ446–454 (lanes 4–6), Δ918–934 (lanes 7–9) and Δ446–454-S922,924,933A (lanes 10–12) p105s were subjected to in vitro conjugation in a reconstituted cell-free system as described in Materials and methods. When indicated, Ubc5Hc and SCFh-β-TrCP were added as described. (B) Conjugation of wild-type and Δ918–934 p105s in crude HeLa extract. In vitro translated and kinase-phosphorylated wild-type (lanes 1 and 2) and Δ918–934 (lanes 3 and 4) p105s were subjected to in vitro conjugation as described in Materials and methods. The cell-free system contained crude HeLa cell extract as a source of the conjugating enzymes. Proteins were resolved and visualized as described in Materials and methods.
IκKβ is required for processing of p105 via the precursor’s C-terminal domain
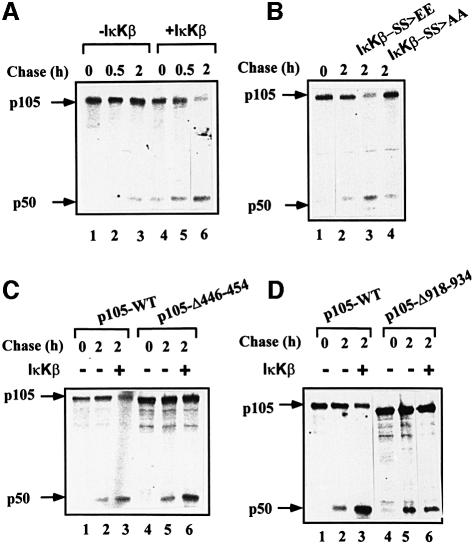
Since IκKs can modify the C-terminal domain of p105 (Heissmeyer et al., 1999), it was important to test whether the modification affects processing. As IκKβ is the kinase involved in the pro-inflammatory response (Li et al., 1999), we decided to test initially the role of this kinase. As can be seen in Figure 2A, expression of constitutively active IκKβ significantly stimulates processing of p105 (compare lanes 2 and 3 with lanes 5 and 6). As can be seen in lane 6, after 2 h of chase in the presence of the kinase, almost all the precursor protein disappeared. While most of it was processed to p50, it appears that a significant part was completely degraded (see also Discussion). Stimulation is probably due to the catalytic activity of IκKβ, as an inactive enzyme did not stimulate processing (Figure 2B, compare lane 3 with lane 4). At this point, it was important to confirm the role of the C-terminal domain in IκK-dependent stimulated processing. As can be seen in Figure 2C, the kinase stimulated processing of wild-type p105 and p105-Δ446–454 to the same extent, ruling out a role for the acidic domain in the process (compare lanes 2 and 3 with lanes 5 and 6). (The apparent similar quantity of p50 generated from the wild-type and the Δ446–454 proteins is due to an excess of the expressed mutant in this experiment. The efficiency of generation of p50 from p105-Δ446–454 is significantly lower than that from the wild type; see, for example, Figure 1 and Orian et al., 1999.) In contrast, the kinase did not have any effect on processing of p105-Δ918–936 (Figure 2D, compare lanes 2 and 3 with lanes 5 and 6). Thus, it appears that IκKβ stimulates p105 processing via its activity on the C-terminal domain of the molecule.
Fig. 2. IκKβ-mediated processing of p105 requires the C-terminal domain of the precursor molecule. (A) Transfection of cells with constitutively active IκKβ significantly stimulates processing of p105. Cos-7 cells were transiently transfected with a cDNA coding for wild-type p105 with or without a cDNA coding for IκKβ. Processing was monitored in a pulse–chase labeling and immunoprecipitation experiment as described in Materials and methods. (B) Enhanced processing of p105 requires active IκKβ. Cos-7 cells were transiently transfected with a cDNA coding for wild-type p105 without or with cDNAs coding for either constitutively active (IκKβ-SS>EE) or inactive (IκKβ-SS>AA) IκKβs (see Materials and methods). Processing was monitored as described in (A). (C) The acidic domain of p105 (residues 446–454) is not required for IκKβ-mediated processing of the molecule. Cos-7 cells were transiently transfected with cDNAs coding for either wild-type p105 or p105-Δ446–454, with or without a cDNA coding for constitutively active IκKβ. Processing was monitored as described in (A). (D) The C-terminal phosphorylation domain of p105 (residues 918–934) is required for IκKβ-mediated processing of the precursor molecule. Cos-7 cells were transiently transfected with cDNAs coding for either wild-type p105 or p105-Δ918–934, with or without a cDNA coding for IκKβ. Processing was monitored as described in (A).
p105 is phosphorylated in its C-terminal domain by IκKβ
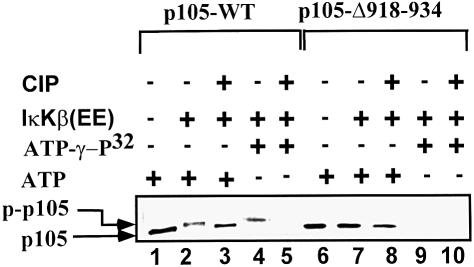
To demonstrate that IκKβ phosphorylates the C-terminal domain of p105, we incubated in vitro translated, 35S-labeled wild-type p105 or p105-Δ918–934 in the presence of active IκKβ and ATP. As can be seen in Figure 3 (lanes 1–3), the wild-type protein is converted into a more slowly migrating form following incubation in the presence of the kinase and ATP. This change in molecular mass is probably due to phosphorylation, as the slower migrating form can be converted into a faster migrating form following addition of alkaline phosphatase (Figure 3, lane 3). In contrast, migration of p105-Δ918–934 is not affected following incubation in the presence of the kinase and ATP (Figure 3, compare lanes 6 and 7). It should be noted that at times, a small change can still be seen in the migration of the Δ918–934 mutant following its incubation in the presence of ATP; however, it is much smaller than the change observed in the molecular weight of the wild-type protein (see, for example, Figure 1D). This small change is probably due to weak phosphorylation of serine and threonine residues localized to the 850–891 region (Heissmeyer et al., 1999). IκKβ-mediated phosphorylation at the C-terminal domain could also be observed when we utilized unlabeled p105 incubated in the presence of the kinase and [α-32P]ATP: as can be seen in Figure 3 (lane 4), wild-type p105 is phosphorylated, and the label disappears following addition of the phosphatase (lane 5). In contrast, the Δ918–934 mutant is not phosphorylated by IκKβ (Figure 3, lane 9). These in vitro experiments show that IκKβ modifies p105 on serine and/or threonine residues in the region that spans moieties 918–934 (see, however, Figure 9).
Fig. 3. IκKβ-dependent phosphorylation of p105 requires residues 918–934. The effect of IκKβ was monitored using wild-type p105 (lanes 1–5) or p105-Δ918–934 (lanes 6–10). Translated [35S]methionine-labeled (lanes 1–3 and 6–8) or unlabeled (lanes 4 and 5, and 9 and 10; an amount equivalent to the labeled protein) p105s were incubated in the presence of unlabeled (lanes 1–3 and 6–8) or [γ-32P]ATP (lanes 4 and 5, and 9 and 10) as described in Materials and methods. IκKβ was added as indicated. Following incubation for 25 min, calf intestine alkaline phosphatase (10 U) was added where indicated, and the incubation continued for an additional 5 min. p105 was precipitated using anti-p50 antibody and immobilized protein A. Samples were resolved via SDS–PAGE and proteins visualized via phosphoimaging. p-p105 denotes phosphorylated p105.
ΔF-box β-TrCP inhibits processing of p105, the precursor’s IκKβ phosphorylation domain
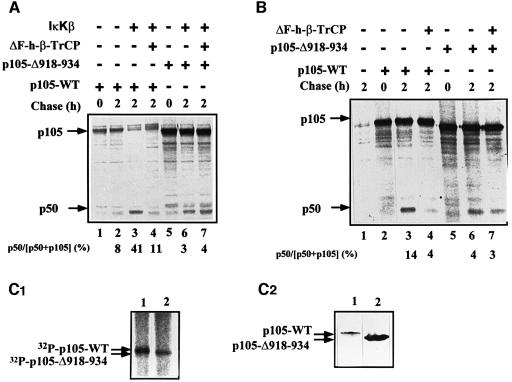
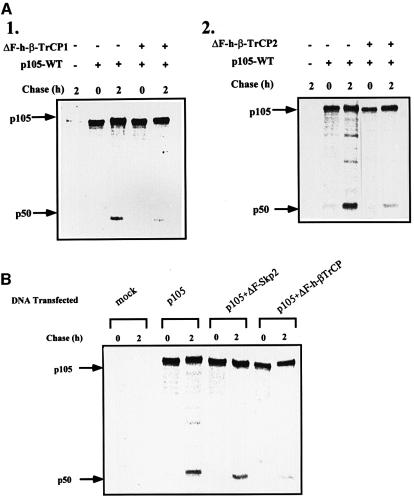
At this stage, it was important to identify the E3 involved in recognition of the phosphorylated C-terminal region of p105. Obvious candidates are members of the SCF family of ligases such as SCFβ-TrCP and SCFSkp2 that recognize phosphorylated substrates. Transfection of cells with the dominant-negative E3, ΔF-box β-TrCP1, significantly inhibits IκKβ-dependent processing of wild-type p105 (Figure 4A, compare lanes 3 and 4). In contrast, processing of p105-Δ918–934 is not affected (compare lanes 6 and 7). It should be noted that this experiment was carried out in the presence of the active form of IκKβ. An obvious question involves the role of the C-terminal domain under basal conditions. To our surprise, we found that the dominant-negative TrCP inhibits processing of p105 in the absence of the active kinase as well (Figure 4B, compare lanes 3 and 4). This is probably due to basal phosphorylation of p105 that occurs in the absence of exogenous stimulation, or to stress-induced activation caused by the transfection of the DNA probes. Indeed, as can be seen in Figure 4C1, p105 is also phosphorylated in non-stimulated cells, and the wild-type protein is modified to a significantly greater extent compared with its p105-Δ918–934 counterpart, suggesting a role for C-terminal phosphorylation in p105 processing under these conditions. It is possible that the difference in phosphorylation between the wild-type and the Δ918–934 p105s is even larger than that observed, as in most experiments the expression of the mutated p105 was much greater than that of the wild-type protein (Figure 4C2; compare also, for example, lanes 5 and 1 in Figure 4A). Quantitatively, however, the amount of p50 generated following IκKβ phosphorylation is significantly greater than that formed under basal conditions and, therefore, the inhibition by the mutant E3 is more striking [30% (41–11) reduction in p50 generation in the presence of the kinase versus 10% (14–4) in its absence; Figure 4A and B, respectively]. The striking effect of IκKβ is also reflected in the finding that most of the p105 precursor disappears in the presence of the kinase (Figure 4A, lane 3), and this disappearance is strongly inhibited by the dominant-negative E3 (lane 4). In the absence of the kinase, there is almost no disappearance of p105, while slow processing occurs under these conditions, reflecting the low efficiency of the process under basal conditions (Figure 4B, lane 3). Based on these findings, we concluded that TrCP may serve as the E3 that recognizes the phosphorylated C-terminal domain of p105, and is involved in phosphorylation-mediated processing of the molecule. To determine the specificity of β-TrCP1, we monitored the effect of the second isoform of TrCP, TrCP2. The two proteins are highly homologous, but nevertheless distinct. While both recognize IκBα (Suzuki et al., 1999), TrCP2 does not recognize β-catenin (Hart et al., 1999). As can be seen in Figure 5A1 and A2, the two ΔF-box derivatives inhibit p105 processing in vivo to the same extent. In contrast, the dominant-negative species of a different F-box protein, ΔF-box Skp2, does not affect p105 processing (Figure 5B).
Fig. 4. ΔF-box β-TrCP1 inhibits processing of wild-type p105 but not p105-Δ918–934. (A) Effect of ΔF-box β-TrCP1 on processing of wild-type p105 and p105-Δ918–934 in the presence of IκKβ. Cos-7 cells were transiently transfected with cDNAs coding for either wild-type p105 or p105-Δ918–934. cDNAs coding for ΔF-box β-TrCP1 and IκKβ were co-transfected as indicated. Processing was monitored in a pulse–chase labeling and immunoprecipitation experiment as described in Materials and methods. (B) Effect of ΔF-box β-TrCP1 on processing of wild-type p105 and p105-Δ918–934 in the absence of IκKβ. Cos-7 cells were transiently transfected with cDNAs coding for either wild-type p105 or p105-Δ918–934. A cDNA coding for ΔF-box β-TrCP was co-transfectd as indicated. Processing was monitored as described in (A). Quantitative analysis of processing ([p50/p50 + p105] × 100) is presented (bottom of panels) following subtraction of background radioactivity (time 0). (C) The C-terminal domain of p105 is phosphorylated in vivo under non-stimulated conditions. (C1) Phosphorylation of wild-type and Δ918–934 p105s in vivo. Cos-7 cells were transiently transfected with cDNAs coding for either wild-type p105 or p105-Δ918–934. Cells were labeled with H3[32P]O4, and the labeled p105 was precipitated and visualized as described in Materials and methods. (C2) Western blot analysis of expressed wild-type p105 and p105-Δ918–934. Cos-7 cells were transfected with cDNAs coding for either wild-type p105 or p105-Δ918–934 as described in (A). Proteins were analyzed using anti-p50 antibody and western blot analysis.
Fig. 5. ΔF-box β-TrCP1 and ΔF-box β-TrCP2, but not ΔF-box Skp2, inhibit processing of p105. (A) Effect of ΔF-box β-TrCP1 and 2 on the processing of p105. Cos-7 cells were transiently transfected with a cDNA coding for wild-type p105. A cDNA coding for ΔF-box β-TrCP1 (A1) or ΔF-box β-TrCP2 (A2) was co-transfected where indicated. Processing of p105 was monitored in a pulse–chase labeling and immunoprecipitation experiment as described in Materials and methods. (B) Effect of ΔF-box Skp2 on processing of wild-type p105. Cos-7 cells were transiently transfected with a cDNA coding for wild-type p105. cDNAs coding for ΔF-box Skp2 or ΔF-box β-TrCP1 were co-transfected where indicated. Processing of p105 was monitored as described in (A).
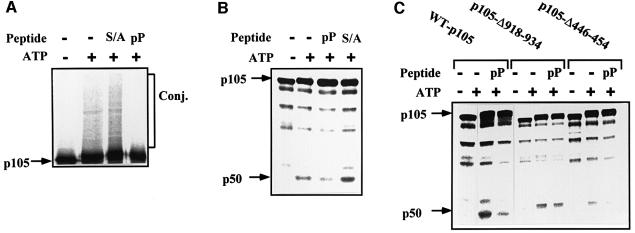
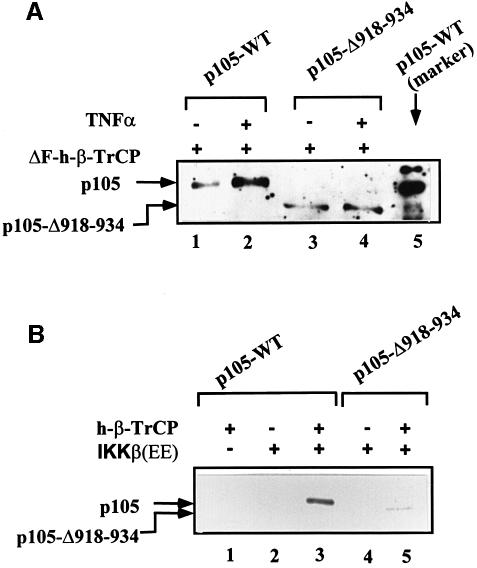
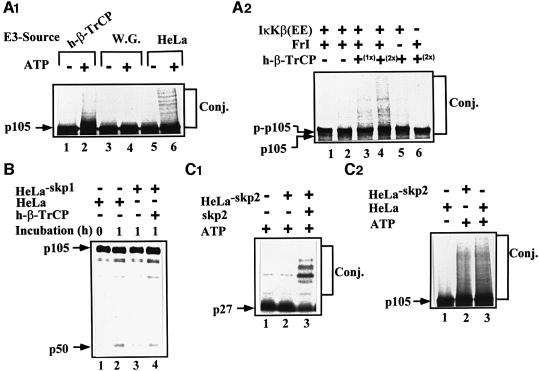
To explore further the role of β-TrCP in p105 recognition, we utilized the phosphopeptide that spans the phosphorylation domain of IκBα. This peptide binds specifically to TrCP and inhibit its activity towards IκBα and β (Yaron et al., 1997, 1998). As can be seen in Figure 6A, the peptide significantly inhibits conjugation of p105 in vitro. In contrast, a similar peptide in which the two phosphorylated serine residues were substituted with alanine was inactive. The peptides had similar effects on processing of p105 (Figure 6B). At this point, it was important to demonstrate that the phosphopeptide acts via inhibition of recognition of the C-terminal domain of p105 by TrCP. As is shown in Figure 6C, the peptide inhibits processing of the wild-type and Δ446–454 p105s, but not of the Δ918–934-p105. This finding complements our initial finding that IκKβ stimulates, in the intact cell, processing of the wild-type and Δ446–454 proteins, but not of the Δ918–934 deletion mutant (Figure 2C and D, respectively). It should be stressed again that processing of any of the single mutants is significantly less efficient than that of the wild-type protein (see also Figure 1D). The series of experiments presented in Figures 4–6 is based on the utilization of inhibitory E3 and peptides. Therefore, more direct evidence is necessary in order to establish firmly the role of SCFβ-TrCP in p105 processing. If TrCP is indeed the E3 involved in recognition of the phosphorylated C-terminus of p105, it is expected that it will associate physically with the substrate, and that the association will be enhanced following stimulation. Indeed, as can be seen in Figure 7A, physical association between TrCP1 and p105 is increased several fold following tumor necrosis factor-α (TNF-α) stimulation of cells. Deletion of the IκKβ phosphorylation site (residues 918–934) abrogates the enhanced binding. The fact that TrCP was able to bind the Δ918–934 p105 is probably due to the minor phosphorylation sites still available in this mutant (see above; Heissmeyer, 1999). It should be noted that TrCP transfected alone did not pull down any p105 cross-reactive material (not shown). Similarly, p105 transfected alone did not bind to the Ni beads (not shown). Further supporting the notion that TrCP associates directly with p105 is the experiment presented in Figure 7B. Here, in a reconstituted cell-free system, only p105 that was phosphorylated by IκKβ could be precipitated by TrCP. Wild-type p105 that was incubated in the absence of the kinase or p105-Δ918–934 that was incubated in the presence of the kinase failed to associate with TrCP. Lastly, it was important to demonstrate directly that β-TrCP can conjugate ubiquitin to C-terminally phosphorylated p105 and promote its processing. As can be seen in Figure 8A1, tetrameric SCFβ-TrCP complex conjugates an in vitro phosphorylated wild-type p105 (lanes 1 and 2). In contrast, a crude wheat germ extract that lacks this complex cannot reconstitute conjugation (lanes 3 and 4). As a control, we used a crude HeLa extract that, not surprisingly, catalyzes efficient conjugation (lanes 5 and 6). The experiment described in Figure 8A2 clearly demonstrates that TrCP-mediated conjugation requires the prior activity of IκKβ. In the absence of the kinase, TrCP cannot promote conjugation (compare lanes 3 and 4 with lane 6). It is of note that a substrate incubated in the absence of the kinase migrates more rapidly (lane 6) than that incubated in its presence (lanes 1–5). To demonstrate a role for TrCP in p105 processing, we depleted HeLa cell extract of SCFβ-TrCP using an antibody specific to Skp1. Utilization of the anti-Skp1 antibody was necessary, as depletion with anti-TrCP was not efficient due to the low affinity of the antibody we have. As can be seen in Figure 8B, Skp1-depleted HeLa cell extract demonstrated a lower processing activity (compare lanes 2 and 3). The inhibited activity could be restored following addition of purified recombinant SCFβ-TrCP. Addition of recombinant Skp1 alone was inefficient (not shown). Similarly, addition of an irrelevant antibody (anti-MyoD) had no effect on processing (not shown). The ability to reconstitute a cell-free conjugation assay enabled us to test the specificity of TrCP and to compare its activity with that of a different F-box protein, Skp2. As can be seen in Figure 8C1, an Skp2-depleted HeLa cell extract was unable to conjugate p27Kip1 (lane 2). Activity could be restored following addition of purified Skp2. In contrast, the Skp2-depleted extract could conjugate phosphorylated p105 efficiently (Figure 8C2). All these experiments firmly establish the role of β-TrCP in recognition of the phosphorylated C-terminal domain of p105. This is particularly important as this domain does not resemble the binding site for TrCP [S(P)GΨXS(P)] in the three previously identified substrates of this enzyme: HIV-1 Vpu, IκBα and β-catenin (see above).
Fig. 6. The IκBα phosphopeptide specifically inhibits conjugation and processing of p105 in vitro. (A) The IκBα phosphopeptide inhibits conjugation of p105. Conjugation of p105 in a cell-free system was monitored as described in Materials and methods in the absence or presence of ATP, the IκBα-derived phosphopeptide (pP) or the S32,36A control peptide (S/A) as indicated. (B) The IκBα phosphopeptide inhibits processing of p105. Processing of p105 in a cell-free system was monitored as described in Materials and methods and in (A). (C) Effect of the IκBα-derived phosphopeptide on processing of wild-type p105, p105-Δ918–934 and p105-Δ446–454. Processing of wild-type p105, p105-Δ918–934 and p105-Δ446–454 was monitored in a cell-free system as described in Materials and methods in the absence or presence of ATP and the IκBα-derived phosphopeptide (pP), as indicated.
Fig. 7. Wild-type p105, but not p105-Δ918–934, associates physically with ΔF-box β-TrCP following phosphorylation. (A) TNF-α stimulates physical association between p105 and β-TrCP in cells. Cos-7 cells were transiently transfected with cDNAs coding for His6-tagged ΔF-box β-TrCP and either wild-type p105 or p105-Δ918–934 as indicated. After 40 h, TNF-α was added as described in Materials and methods. Cells were disrupted, His-TrCP was immobilized, and proteins were visualized via western blot analysis using anti-p50 antibody as described in Materials and methods. The site of migration of wild-type p105 is marked by separation of in vitro translated labeled p105 (lane 5). (B) Association between p105 and β-TrCP in vitro requires prior IκK-mediated phosphorylation. In vitro translated and labeled wild-type p105 (lanes 1–3) or p105-Δ918–934 (lanes 4 and 5) were incubated in the absence (lane 1) or presence (lanes 2–5) of IκK and ATP as described in Materials and methods. His-tagged β-TrCP was added when indicated. Following incubation, the complex was immobilized and proteins resolved and visualized as described in Materials and methods.
Fig. 8. β-TrCP, but not Skp2, is the ubiquitin ligase involved in conjugation and processing of phosphorylated p105 in vitro. (A1) h-β-TrCP reconstitutes p105 conjugation. 35S-labeled p105 translated in wheat germ extract was subjected to in vitro kinase assay in the presence of IκKβ, and conjugation was carried out as described in Materials and methods. Recombinant and purified SCFh-β-TrCP (lanes 1 and 2), wheat germ extract (75 µg of protein; lanes 3 and 4) or crude HeLa extract (75 µg of protein; lanes 5 and 6) were used as the source of E3. (A2) IκKβ-dependent phosphorylation of p105 is required for its β-TrCP-mediated conjugation. 35S-labeled p105 (as in A1) was subjected to in vitro kinase assay in the presence (lanes 1–5) or absence (lane 6) of IκKβ as described in Materials and methods, and conjugation was monitored as described. Purified recombinant tetrameric SCFβ-TrCP was used at 0.75 (1×; lane 3) and 1.5 (2×; lanes 4 and 6) µg. (B) SCFβ-TrCP reconstitutes processing of p105. 35S-labeled p105 (as in A1) was subjected to in vitro kinase assay as described in Materials and methods. Processing was carried out in a complete (lanes 1 and 2) or Skp1-depleted (lanes 3 and 4) HeLa cell extract in the absence (lane 3) or presence (lane 4) of purified SCFβ-TrCP as described in Materials and methods. (C) Skp2 is not required for conjugation of phosphorylated p105. (C1) Conjugation of p27Kip1 requires Skp2. In vitro trans lated 35S-labeled p27Kip1 was incubated in the presence of Skp2-immunodepleted synchronized HeLa extract as described in Materials and methods. Recombinant and purified Skp2 (0.5 µg) were added when indicated. (C2) Conjugation of p105 is Skp2 independent. 35S-labeled p105 (as in A1) was subjected to in vitro kinase assay in the presence of IκKβ as described in Materials and methods. Conjugation was monitored in the presence of complete or Skp2-immunodepleted HeLa cell extract as indicated and described in Materials and methods. All notes are as described in the legend to Figure 1.
p105 is conjugated by two distinct E3s, one of which, β-TrCP, recognizes the phosphorylated C-terminal serine residues of the molecule
To analyze the C-terminal motif of p105 targeted by TrCP, we initially monitored conjugation of wild-type, Δ446–454 and Δ918–934 p105s in a reconstituted cell-free system. As can be seen in Figure 9A, the wild-type (lanes 1–3) and Δ446–454 (lanes 4–6) proteins are conjugated efficiently by the SCF complex. In contrast, p105-Δ918–934 does not generate high molecular weight adducts (lanes 7–9). To identify specifically the residues that play a role in recognition of phosphorylated p105, we substituted serine residues 922, 924 and 933 with alanine. As can be seen in Figure 9A (lanes 10–12), these replacements abolished the ability of TrCP to catalyze conjugation, strongly suggesting that these residues are targeted by the kinase. While these findings clearly support the notion that IκK is mostly a serine kinase, they further corroborate the notion that the TrCP recognition motif in p105 is novel. We noted that the E2 used, UbcH5c (see also Discussion), generates low molecular weight adducts with all three proteins (Figure 8A, lanes 2, 5, 8 and 11). This reaction may be catalyzed by the E2 alone, or along with a non-specific E3 present in the crude wheat germ extract.
While recognition of the phosphorylated C-terminal domain is mediated by SCFβ-TrCP, it is not clear whether this E3 is also involved in recognition of the upstream acidic domain. In cells, dominant-negative ΔF-box β-TrCP has a minor effect on processing of p105-Δ918–934 (Figure 4A and B, compare lanes 6 and 7 with lanes 3 and 4). Also, processing of the C-terminally deleted mutant is not affected by the inhibitory phosphopeptide (Figure 6C). Furthermore, p105-Δ446–454 and the wild-type proteins are conjugated equally by the SCF complex in vitro (Figure 9A, lanes 3 and 6), suggesting that the acidic domain is not recognized by this ligase. To test a possible role for an additional, as yet unidentified, E3 in targeting the acidic domain, wild-type and the C-terminally deleted p105s were incubated in the presence of crude HeLa extract. Unlike the SCF complex that cannot conjugate p105-Δ918–934, here both proteins were conjugated efficiently (Figure 9B), suggesting that the HeLa extract contains an additional, acidic domain-recognizing E3. Similar to its processing (Figure 1D), conjugation of the Δ918–934 species is less effective than that of the wild type. This is probably due to the elimination of the TrCP conjugation motif. While indirect, these findings strongly suggest that p105 is conjugated by at least two ligases, recognizing two distinct motifs. It is not known whether Lys441 and Lys442 are shared by the two E3s as ubiquitylation sites.
Discussion
We have shown that, in addition to the GRR and the downstream acidic domain, p105 processing also depends on residues 918–934 (Figure 1). Mechanistic analysis shows that IκKβ utilizes this region to promote accelerated processing (Figure 2). This processing is probably mediated via phosphorylation of the C-terminal domain (Figure 3). Similar data, though not identical (see below), were also reported by Heissmeyer et al. (1999). Since phosphorylation by IκKβ leads to recruitment of SCFβ-TrCP to the pIκBα, we thought to test the potential role of this ligase in p105 processing. Expression of the dominant-negative ΔF-box species of TrCP significantly inhibited processing of wild-type but not Δ918–934 p105s (Figure 4). The effect was specific, as the dominant-negative form of Skp2, a different F-box protein, was inactive (Figure 5). To corroborate further the notion that TrCP is the p105 C-terminal E3, we utilized a specific phosphopeptide that spans the IκBα signaling domain and inhibits IκBα conjugation. The phosphopeptide inhibits, in a specific manner, conjugation and processing of p105 (Figure 6). An additional experiment has shown that TrCP associates with phosphorylated wild-type but not with the C-terminally deleted p105 both in vivo and in vitro (Figure 7). Lastly, we have shown that purified recombinant SCFβ-TrCP can reconstitute specifically conjugation and processing of phosphorylated wild-type p105 (Figure 8). Also, TrCP cannot reconstitute conjugation of either p105-Δ918–934 or p105-S922,924,933A, strongly suggesting that phosphorylation of these specific serine residues is essential for recognition by the E3 (Figure 9). In the SCF reconstitution experiment, we used UbcH5c as the E2 enzyme. Interestingly, this E2 is also the enzyme that catalyzes ubiquitylation, along with SCFβ-TrCP, of signal-induced and phosphorylated IκBα (Gonen et al., 1999). The possible role of other E2 enzymes, and in particular that of Ubc3/Cdc34, which also acts along with SCF complexes, has not been studied. Since, unlike purified SCF, crude HeLa extract conjugated p105-Δ918–934 (Figure 9B), we concluded that p105 is targeted by at least two distinct E3s that recognize different structural motifs.
Several proteins are targeted via recognition of two distinct domains, probably by different E2s and E3s. Among them are p53, which is targeted under different conditions by Mdm2 (Honda et al., 1997) and E6-AP (Scheffner et al., 1993). The yeast mating type transcriptional regulator MATα2 is also targeted by two signals, Deg1 and Deg2, and two E2 enzymes, Ubc6 and Ubc7 (Chen et al., 1993); however, the physiological significance and identity of the E3s involved in recognition of the two signals have remained obscure. The model protein lysozyme is targeted following recognition of its α-NH2 lysine residue by the N-end rule Ubc-14 kDa and E3α, and of a downstream ‘body’ site by E2-F1 (UbcH7) and an unidentified E3 (Gonen et al., 1996).
Several mechanisms can explain the complex recognition of p105. One simple explanation is that the acidic domain is involved in basal processing, whereas the function of the C-terminal domain is to enable accelerated, signal-induced processing. Indeed, it has been shown that stimulation of cells leads to phosphorylation of serine and possibly threonine residues in the C-terminal domain, with subsequent increased processing (Fujimoto et al., 1995; MacKichan et al., 1996). Basal processing can occur co-translationally from a nascent polypeptide chain that does not contain the C-terminal domain (Lin et al., 1998). However, even here, the minimally required chain that can undergo processing must include the acidic recognition domain (Lin et al., 1998; Orian et al., 1999). An alternative explanation involves the putative function of the seven ankyrin repeat domain of p105 that spans residues 544–803. It has been shown that different transcriptional regulators such as p50 (Liou et al., 1992), c-rel, p65 (Rice et al., 1992) and Bcl-3 (Hatada et al., 1992) bind specifically to this region. Under basal conditions, binding sequesters them in the cytosol. Their release and subsequent translocation to the nucleus will be possible only following destruction of the ankyrin repeat domain. It is possible that following synthesis of one or more ankyrin repeats and binding of the transcription factors, the acidic domain involved in basal processing is not accessible to the ligase. Therefore, a novel recognition domain must be synthesized, preferably at the C-terminal region of the molecule. This domain will enable binding of ligase, ubiquitylation and partial (processing) or complete degradation of p105. This will release the active bound factors and, in the case of processing, will generate an additional molecule of p50 or p52. Synthesis of complete p105 that contains a signal-induced phosphorylation domain for processing/degradation and that is resistant to co-translational processing ensures sequestration of an inactive set of transcription factors that are ‘ready to go’ following stimulation. Indeed, Belich et al. (1999) and Heissmeyer et al. (1999) reported that phosphorylation at the C-terminal domain of p105 by TPL-2 and IκK, respectively, stimulates proteolysis of p105. It should be noted, however, that immunoprecipitated TPL-2 does not phosphorylate p105 in vitro, suggesting that it may act as an upstream kinase, possibly for IκKβ, as is NIK in the case of IκBα. Heissmeyer et al. (1999) showed that the accelerated degradation leads to activation of the Bcl-3⋅p50 transcriptional complex that follows release of sequestered p50. We have shown that, in addition, IκK phosphorylation leads to accelerated processing and increased formation of p50, thus amplifying the signal even further by generating a larger quantity of the active transcriptional factors. It should be noted that even in this case, a large part of p105 is completely degraded, as the generated p50 cannot account for all the lost p105 (see, for example, Figure 2A, lanes 4 and 6, or Figure 2B, lanes 1 and 3). While processing appears to be an intrinsic property of p105, complete degradation probably reflects the inefficiency of the GRR as a processing ‘stop’ signal. It is of note that the C-terminal recognition pathway also operates under basal conditions (Figure 4B). This is probably due to phosphorylation of p105 (Figure 4C) that may result, for example, from stress-induced activation of the internal cellular signaling pathway that follows transfection. The malignantly transformed cells utilized may also have partially activated signaling pathways. A constitutively active kinase that is different from IκKβ or activation of IκKβ by a kinase that is different from IκKα (O’Mahony et al., 2000) may also be involved in basal phosphorylation. Li et al. (1994) have already shown that p105 is phosphorylated in unstimulated cells, and becomes hyperphosphorylated following stimulation. We demonstrated that a significant part of this modification occurs on the C-terminal domain. It should be emphasized, however, that even on that background, signaling significantly increases processing, as reported by Fujimoto et al. (1995) and McKichan et al. (1996). Our data showing that IκKβ increases processing (Figure 2) and that TrCP associates physically with phosphorylated p105 (Figure 7) strongly support the notion that signaling plays an important role in p105 processing/degradation that results in enhanced transcription.
Materials and methods
Materials
Materials for SDS–PAGE and Bradford reagent were from Bio-Rad. A mixture of l-35S-labeled methionine and cysteine for metabolic labeling, [35S]methionine for in vitro translation, [α-32P]ATP, [γ-32P]ATP, H3[32P]O4, as well as pre-stained molecular weight markers and immobilized protein A were obtained from Amersham Pharmacia Biotech. Tissue culture sera and media were from Biological Industries, Bet Haemek, Israel or from Sigma. Antibodies (polyclonal) against NF-κB1 p50 and Skp2 were from Santa Cruz. Anti His-tag and Ni-NTA resin were from Qiagen. Anti-hemagglutinin (HA), anti-FLAG and anti-T7 were from Roche, Sigma and Novagen, respectively. Ubiquitin, dithiothreitol (DTT), ATP, phosphocreatine, creatine phosphokinase, 2-deoxyglucose and Tris buffer [Tris(hydroxymethyl)aminomethane] were from Sigma. Hexokinase and Fugene™ 6 transfection reagent were from Roche. HEPES and protease inhibitor cocktail were from Calbiochem. The wheat germ extract-based transcription–translation coupled kit (TNT®) was from Promega. Restriction and modifying enzymes were from New England Biolabs. Reagents for ECL were from Pierce. Oligonucleotides were synthesized by Biotechnology General, Rehovot, Israel. IkBα-derived synthetic peptides (doubly phosphorylated and S32,36A, both spanning residues 28–39) were synthesized by SynPep (Dublin, CA). TNF-α was from PeproTech EC Ltd, UK. All other reagents were of high analytical grade.
Cell lines
Cos-7 and HeLa cells were grown at 37°C in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum (FCS). HI-5 insect cells were grown at 27°C in Grace medium supplemented with 10% FCS, lactalbumin hydrolysate and yeastolate. All transfections were carried out using the Fugene reagent.
Plasmids and construction of mutants
Wild-type human p105 cDNAs used for in vitro translation (pT7β105) and for transient transfection (in pCI-neo) were described previously (Orian et al., 1995, 1999). The different p105 mutants were generated in these two vectors by site-directed mutagenesis using the QuikChange™ kit (Stratagene). cDNAs coding for the constitutively active (S176,180E) and dominant-negative (S176,180A) IκBβ kinase were as described (Mercurio et al., 1997). cDNAs coding for h-β-TrCP1 (Margottin et al., 1998) and β-TrCP2 (Suzuki et al., 1999) were generated by RT–PCR of RNA derived from 293 and HeLa cells, respectively. Using two-step PCR (Higuchi et al., 1988), the F-box deletion mutants of the two proteins were generated by removing nucleotides 93–536 and 1–511 from the cDNAs of h-β-TrCP1 and h-β-TrCP2, respectively. Using PCR, His6 tags were fused to the N-terminal residues of the TrCPs. The cDNAs were subcloned into the pCAGGS expression vector (Niwa, 1991). ΔF-box Skp2 for cell expression was as described (Carrano et al., 1999). The sequence of all constructs was confirmed using ABI 310 or 377 autosequencers.
Expression of recombinant proteins in insect cells
h-β-TrCP1 was cloned via RT–PCR of 293 cell RNA. Similarly, Skp1 and RBX1/Roc1 cDNAs were cloned from HeLa cells. To discriminate between the three proteins, His6, FLAG and T7 tags were fused to their N-terminal residues, respectively. All cDNAs were subcloned into the pVL1393 vector (Invitrogen). Recombinant baculovirus constructs were generated using the Bac-PAK6 baculovirus expression system (Clontech). Recombinant baculovirus vector expressing C-terminally HA-tagged cullin1 was provided by Dr Arnim Pause. For the genera tion of a tetrameric His-TrCP1-containing SCF complex (h-β-TrCP1⋅Skp1⋅cullin1⋅Rbx1/Roc1), cells were co-infected with all four viruses. After 60 h, cells were harvested and lysed in a buffer containing 1% Triton X-100, 50 mM Tris–HCl pH 8.0, 150 mM NaCl, 10% glycerol, 0.1 mM DTT and protease inhibitor cocktail. Solubilized cells were centrifuged at 14 000 g for 20 min at 4°C, and the supernatant collected. SCFβ-TrCP complex was purified using Ni-NTA–agarose (Qiagen). The resin-recovered complex contained all four components as determined via western blot analysis using the appropriate anti-tag antibodies. Baculovirus-expressed constitutively active Skp2 and IκKβ were as described (Carrano et al., 1999; Mercurio et al., 1999, respectively).
Preparation of cell extracts
HeLa cell extract was prepared by hypotonic lysis as described previously (Orian et al., 1995). Cell cycle-synchronized/Skp2-depleted HeLa extract was prepared using a specific anti-Skp2 antibody as described (Carrano et al., 1999). Skp1-depleted HeLa extract was prepared using a specific antibody (Transduction Laboratories) and immobilized protein A.
In vitro processing of p105
All p105 proteins and p27kip were translated in vitro using a wheat germ extract-based transcription–translation coupled kit in the presence of l-[35S]methionine according to the manufacturer’s instructions. Processing of labeled p105 to p50 was monitored as described (Orian et al., 1995, 1999). To inhibit SCFβ-TrCP-dependent processing, a doubly phosphorylated peptide that spans the IκBα recognition domain [28-DRHDS32(P)GLDS36(P)MKD-39] and the inactive S32,36A peptide were added at 40 µM at the pre-incubation step along with bestatin (20 µg/ml) as described (Gonen et al., 1999). Following incubation, reaction mixtures were resolved via SDS–PAGE (10%). Gels were dried and analyzed by phosphoimager (Fuji, Japan).
In vitro kinase assays
In vitro translated labeled p105 was phosphorylated in a cell-free system in a reaction mixture that contained in 25 µl: 20 µl of wheat germ extract containing the labeled p105 (see above), 5 mM MgCl2 and 0.5 mM ATP. When indicated, unlabeled p105 was used instead of the labeled protein, and [γ-32P]ATP (2.5 µCi) substituted for the unlabeled nucleotide. Baculovirus-expressed IκKβ (∼0.4 µg) was added to the cell-free system when indicated. Reaction mixtures were incubated for 20 min at 30°C.
In vitro conjugation assays
Ubiquitin–p105 conjugates were generated in a crude extract in an assay similar to that described for processing, but with the following modifications: (i) ubiquitinaldehyde (UbAl; 0.5 µg), a specific inhibitor of certain isopeptidases (Hershko and Rose, 1987), was present in the reaction mixture; (ii) okadaic acid was added at 1 µM; and (iii) the proteasome inhibitor MG132 was added at a final concentration of 20 µM. Where indicated, the IκBα-derived peptides were added to the reaction mixture as described for processing of p105. Reconstitution of conjugation with purified SCFβ-TrCP1 complex was carried out using 2.5 µl (∼20 000 c.p.m.) of the [35S]methionine-labeled p105 that was phosphorylated in vitro by IκKβ as described above. E1 was provided by the wheat germ extract that contained the substrate, whereas E2 was provided by the addition of reticulocyte fraction I (2.5 µl; Blumenfeld et al., 1995), or purified recombinant UbcH5c (0.5 µg; Gonen et al., 1999). Purified SCF complex was added at ∼1.5 µg. Reactions were incubated at 37°C for 30 min, resolved via SDS–PAGE (7.5%) and analyzed as described above. Conjugation of p27Kip1 in HeLa cell extract was carried out as described (Carrano et al., 1999).
Transient transfections and processing of p105 in cells
Cos-7 cells were transiently transfected with 3–5 µg of wild-type or the various p105 mutant cDNAs. Where indicated, the p105 cDNAs were co-transfected along with 2–3 µg of cDNAs coding for the different species of IκKβ, ΔF-box TrCP or ΔF-box Skp2. Processing of p105 was monitored in pulse–chase and immunoprecipitation experiments 40 h after transfection as described (Orian et al., 1999). When indicated, cells were incubated for 30 min in phosphate-free medium followed by 30 min labeling with 0.5 mCi/ml of H3[32P]O4 (pulse only). Following lysis, labeled proteins were precipitated with anti-p105 antibody. Immune complexes were collected using immobilized protein A. Following SDS–PAGE (10%), proteins were visualized by a phosphoimager.
Physical association between p105 and His-ΔFbox β-TrCP1
Complex formation between p105 and His-ΔFbox β-TrCP in cells was monitored as follows. Cells were transfected with the indicated constructs. At 40 h post-transfection, cells were stimulated with 20 ng/ml TNF-α for 15 min. Immediately following stimulation, cells were harvested, washed twice with phosphate-buffered saline (PBS) and lysed using a hypotonic lysis buffer containing 20 mM Tris–HCl pH 7.6, 10 mM β-mercaptoethanol and protease inhibitor cocktail. Cells were disrupted by three cycles of freezing (liquid N2) and thawing. Following additional douncing, the broken cells were centrifuged at 14 000 g for 20 min at 4°C. Ni-NTA resin was added to the supernatant (200 µg of protein), and binding was performed at 4°C for 1 h followed by three washes with TENGEN buffer (50 mM Tris–HCl pH. 7.4, 1 mM EDTA, 100 mM NaCl, 10% glycerol, 0.1% NP-40). Following SDS–PAGE (10%), proteins were visualized via ECL using anti-p50 antibody. For monitoring association in a cell-free system, in vitro translated and 35S-labeled p105 was subjected to kinase assay in the presence or absence of IκKβ as described above. Purified baculovirus-expressed recombinant His-tagged β-TrCP (∼0.5 µg) was added as indicated and, following 30 min incubation at 30°C, the complex was immobilized as described above. Proteins were resolved via SDS–PAGE and visualized by phosphoimager.
Acknowledgments
Acknowledgements
The authors thank Dr Arnim Pause (Boehringer Ingelheim, Canada) for the baculovirus cullin-1 expression vector, Dr Michele Pagano from NYU Medical Center for the ΔF-box Skp2 mammalian cell expression vector, and Drs Eyal Bengal (Faculty of Medicine, Technion, Haifa, Israel) and Chaim Kahana (Weizmann Institute, Rehovot, Israel) for helpful discussions. This research was supported by grants from the Israel Science Foundation founded by the Israeli Academy of Sciences and Humanities—Centers of Excellence Program, the German–Israeli Foundation for Scientific Research and Development (GIF), the Israel Cancer Society, the German–Israeli Project Cooperation (DIP), the Foundation for Promotion of Research at the Technion, a research grant administered by the Vice President of the Technion for Research (to A.C.), a grant from the National Institutes of Health (NIH; to A.L.S.), a grant from the US–Israel Binational Science Foundation (to A.C. and A.L.S.) and a TMR grant from the European Community (to A.I. and A.C.). Partial support was also obtained from Signal Pharmaceuticals, Inc., San Diego, CA. A.O. was supported by a Fellowship from the Clore Foundation. Purchasing of the ABI 310 autosequencer was supported partially by a grant from the Israel Science Foundation founded by the Israeli Academy of Sciences and Humanities.
References
- Alkalay I., Yaron,A., Hatzubai,A., Orian,A., Ciechanover,A. and Ben Neriah,Y. (1995) Stimulation-dependent IκBα phosphorylation marks the NF-κB inhibitor for degradation via the ubiquitin–proteasome pathway. Proc. Natl Acad. Sci. USA, 92, 10599–10603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baeuerle P.A. and Baltimore,D. (1996) NF-κB: ten years after. Cell, 87, 13–20. [DOI] [PubMed] [Google Scholar]
- Baldwin A.S. (1996) The NF-κB and IκB proteins: new discoveries and insights. Annu. Rev. Immunol., 14, 649–683. [DOI] [PubMed] [Google Scholar]
- Barnes P.J. and Karin,M. (1997) Nuclear factor-κB: a pivotal transcription factor in chronic inflammatory diseases. N. Engl. J. Med., 336, 1066–1071. [DOI] [PubMed] [Google Scholar]
- Belich M.P., Salmerón,A., Johnston,L.H. and Ley,S.C. (1999) TPL-2 kinase regulates the proteolysis of the NF-κB-inhibitory protein NF-κB1 p105. Nature, 397, 363–368. [DOI] [PubMed] [Google Scholar]
- Betts J.C. and Nabel,G.J. (1996) Differential regulation of NF-κB2 (p100) processing and control by amino-terminal sequences. Mol. Cell. Biol., 16, 6363–6371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenfeld N., Gonen,H., Mayer,A., Smith,C.E., Siegel,N.R., Schwartz,A.L. and Ciechanover,A. (1994) Purification and characterization of a novel species of ubiquitin-carrier protein, E2, that is involved in degradation of non-‘N-end rule’ protein substrates. J. Biol. Chem., 269, 9574–9581. [PubMed] [Google Scholar]
- Brown K., Gerstberger,S., Carlson,L., Franzoso,G. and Siebenlist,U. (1995) Control of IκBα proteolysis by site specific signal-induced phosphorylation. Science, 267, 1485–1488. [DOI] [PubMed] [Google Scholar]
- Carrano A.C., Eytan,E., Hershko,A. and Pagano,M. (1999) SKP2 is required for ubiquitin-mediated degradation of the CDK inhibitor p27. Nature Cell Biol., 1, 193–199. [DOI] [PubMed] [Google Scholar]
- Chen P., Johnson,P., Sommer,T., Jentsch,S. and Hochstrasser,M. (1993) Multiple ubiquitin-conjugating enzymes participate in the in vivo degradation of the yeast MATα2 repressor. Cell, 74, 357–369. [DOI] [PubMed] [Google Scholar]
- Chen Z., Hagler,J., Palombella,V., Melandri,F., Scherer,D., Ballard,D. and Maniatis,T. (1995) Signal-induced site-specific phosphorylation targets IκBα to ubiquitin–proteasome pathway. Genes Dev., 9, 1586–1597. [DOI] [PubMed] [Google Scholar]
- Coux O. and Goldberg,A.L. (1998) Enzymes catalyzing ubiquitination and proteolytic processing of the p105 precursor of nuclear factor κB1. J. Biol. Chem., 273, 8820–8828. [DOI] [PubMed] [Google Scholar]
- Deshaies R.J. (1999) SCF and cullin/ring H2-based ubiquitin ligases. Annu. Rev. Cell. Dev. Biol., 15, 435–467. [DOI] [PubMed] [Google Scholar]
- Fan C.M. and Maniatis,T. (1991) Generation of the p50 subunit of NF-κB by processing of p105 through an ATP-dependent pathway. Nature, 354, 395–398. [DOI] [PubMed] [Google Scholar]
- Foo S.Y. and Nolan,G.P. (1999) NF-κB to the rescue: RELs, apoptosis and cellular transformation. Trends Genet., 15, 229–235. [DOI] [PubMed] [Google Scholar]
- Fujimoto K., Yasuda,H., Sato,Y. and Yamamoto,K. (1995) A role for phosphorylation in the proteolytic processing of the human NF-κB1 precursor. Gene, 165, 183–189. [DOI] [PubMed] [Google Scholar]
- Ghosh S., May,M.J. and Kopp,E.B. (1998) NF-κB and Rel proteins: evolutionarily conserved mediators of immune responses. Annu. Rev. Immunol., 16, 225–260. [DOI] [PubMed] [Google Scholar]
- Gonen H. et al. (1996) Isolation, characterization and partial purification of a novel ubiquitin–protein ligase, E3: targeting of protein substrates via multiple and distinct recognition signals and conjugating enzymes. J. Biol. Chem., 271, 302–310. [DOI] [PubMed] [Google Scholar]
- Gonen H., Bercovich,B., Orian,A., Carrano,A., Takizawa,C., Yamanaka,K., Pagano,M., Iwai,K. and Ciechanover,A. (1999) Identification of the ubiquitin-carrier proteins, E2s, involved in signal-induced conjugation and subsequent degradation of IκBα. J. Biol. Chem., 274, 14823–14830. [DOI] [PubMed] [Google Scholar]
- Hart M. et al. (1999) The F-box protein β-TrCP associates with phosphorylated β-catenin and regulates its activity in the cell. Curr. Biol., 9, 207–210. [DOI] [PubMed] [Google Scholar]
- Hatada E.N., Nieters,A., Wulczyn,F.G., Naumann,M., Meyer,R., Nucifora,G., McKeithan,T.W. and Scheidereit,C. (1992) The ankyrin repeat domains of the NF-κB precursor p105 and the protooncogene Bcl-3 act as specific inhibitors of NF-κB DNA binding. Proc. Natl Acad. Sci. USA, 89, 2489–2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heissmeyer V., Krappmann,D., Wulczyn,F.G. and Scheidereit,C. (1999) NF-κB p105 is a target of IκB kinases and controls signal induction of Bcl-3–p50 complexes. EMBO J., 18, 4766–4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A. and Rose,I.A. (1987) Ubiquitin-aldehyde: a general inhibitor of ubiquitin-recycling processes. Proc. Natl Acad. Sci. USA, 84, 1829–1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heusch M., Lin,L., Geleziunas,R. and Greene,W.C. (1999) The generation of NF-κB2 p52: mechanism and efficiency. Oncogene, 18, 6201–6208. [DOI] [PubMed] [Google Scholar]
- Higuchi R., Krummel,B. and Saiki,R.K. (1988) A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res., 16, 7351–7367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda R., Tanaka,H. and Yasuda,H. (1997) Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett., 420, 25–27. [DOI] [PubMed] [Google Scholar]
- Kornitzer D. and Ciechanover,A. (2000) Modes of regulation of ubiquitin-mediated protein degradation. J. Cell Physiol., 182, 1–11. [DOI] [PubMed] [Google Scholar]
- Laney J.D. and Hochstrasser,M. (1999) Substrate targeting in the ubiquitin system. Cell, 97, 427–430. [DOI] [PubMed] [Google Scholar]
- Li C.C., Korner,M., Ferris,D.K., Chen,E., Dai,R.M. and Longo,D.L. (1994) NF-κB/Rel family members are physically associated phosphoproteins. Biochem J., 303, 499–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., van Antwerp,D., Mercurio,F., Lee,K.F. and Verma,I.M. (1999) Severe liver degeneration in mice lacking the IκB kinase 2 gene. Science, 284, 321–325. [DOI] [PubMed] [Google Scholar]
- Lin L. and Ghosh,S. (1996) A glycine-rich region in NF-κB p105 functions as a processing signal for the generation of the p50 subunit. Mol. Cell. Biol., 16, 2248–2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L., DeMartino,G.N. and Greene,W.C. (1998) Cotranslational biogenesis of NF-κB p50 by the 20S proteasome. Cell, 92, 819–828. [DOI] [PubMed] [Google Scholar]
- Liou H.C., Nolan,G.P., Ghosh,S., Fujita,T. and Baltimore,D. (1992) The NF-κB p50 precursor, p105, contains internal IκB-like inhibitor that preferentially inhibits p50. EMBO J., 8, 3003–3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKichan M.L., Logeat,F. and Israël,A. (1996) Phosphorylation of p105 PEST sequence via a redox insensitive pathway up-regulates processing of p50 NF-κB. J. Biol. Chem., 271, 6084–6091. [DOI] [PubMed] [Google Scholar]
- Margottin F., Bour,S.P., Durand,H., Selig,L., Benichou,S., Richard,V., Thomas,D., Strebel,K. and Benarous,R. (1998) A novel human WD protein, h-β-TrCP, that interacts with HIV-1 Vpu, connects CD4 to the ER degradation pathway through an F-box motif. Mol. Cell, 1, 565–574. [DOI] [PubMed] [Google Scholar]
- Marti A., Wirbelauer,C., Scheffner,M. and Krek,W. (1999) Interaction between ubiquitin–protein ligase SCFSKP2 and E2F-1 underlies the regulation of E2F-1 degradation. Nature Cell Biol., 1, 14–19. [DOI] [PubMed] [Google Scholar]
- Mercurio F. et al. (1997) IKK-1 and IKK-2: cytokine-activated IκB kinases essential for NF-κB activation. Science, 278, 860–866. [DOI] [PubMed] [Google Scholar]
- Mercurio F. et al. (1999) IκB kinase (IKK)-associated protein 1, a common component of the heterogeneous IKK complex. Mol. Cell. Biol., 19, 1526–1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H., Yamamura,K. and Miyazaki,J. (1991) Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene, 108, 193–199. [DOI] [PubMed] [Google Scholar]
- O’Mahony A., Lin,X., Geleziunas,R. and Greene,W.C. (2000) Activation of the heterodimeric IκB kinase α (IKKα)–IKKβ complex is directional: IKKα regulates IKKβ under both basal and stimulated conditions. Mol. Cell. Biol., 20, 1170–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orian A., Whiteside,S., Israël,A., Stancovski,I., Schwartz,A.L. and Ciechanover,A. (1995) Ubiquitin-mediated processing of the NF-κB transcriptional activator precursor p105: reconstitution of a cell-free system and identification of the ubiquitin-carrier protein, E2, and a novel ubiquitin–protein ligase, E3, involved in conjugation. J. Biol. Chem., 270, 21707–21714. [DOI] [PubMed] [Google Scholar]
- Orian A., Schwartz,A.L., Israël,A., Whiteside,S., Kahana,C. and Ciechanover,A. (1999) Structural motifs involved in ubiquitin-mediated processing of the NFκB precursor p105: roles of the glycine-rich region and a downstream ubiquitination domain. Mol. Cell. Biol., 19, 3664–3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palombella V., Rando,O., Goldberg,A.L. and Maniatis,T. (1994) The ubiquitin–proteasome pathway is required for processing the NF-κB1 precursor protein and the activation of NF-κB. Cell, 78, 773–785. [DOI] [PubMed] [Google Scholar]
- Rice N.R., MacKichan,M.L. and Israël,A. (1992) The precursor of NF-κB p50 has IκB-like functions. Cell, 71, 243–253. [DOI] [PubMed] [Google Scholar]
- Scheffner M., Huibregtse,J.M., Vierstra,R.D. and Howley,P.M. (1993) The HPV-16 E6 and E6–AP complex functions as a ubiquitin–protein ligase in the ubiquitination of p53. Cell, 75, 495–505. [DOI] [PubMed] [Google Scholar]
- Scherer D.C., Brockman,J.A., Chen,Z., Maniatis,T. and Ballard,D.W. (1995) Signal-induced degradation of IκBα requires site-specific ubiquitination. Proc. Natl Acad. Sci. USA, 92, 11259–11263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H. et al. (1999) IκBα ubiquitination is catalyzed by an SCF-like complex containing Skp1, Cullin-1 and two F-box/WD40-repeat proteins, β-TrCP1 and β-TrCP2. Biochem. Biophys. Res. Commun., 256, 127–132. [DOI] [PubMed] [Google Scholar]
- Voges D., Zwickl,P. and Baumeister,W. (1999) The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu. Rev. Biochem., 68, 1015–1068. [DOI] [PubMed] [Google Scholar]
- Winston J.T., Strack,P., Beer-Romero,P., Chu,C.Y., Elledge,S.J. and Harper,J.W. (1999) The SCFβ-TRCP-ubiquitin ligase complex associates specifically with phosphorylated destruction motifs in IκBα and β-catenin and stimulates IκBα ubiquitination in vitro. Genes Dev., 13, 270–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woronicz J.D., Gao,X., Cao,Z., Rothe,M. and Goeddel,D.V. (1997) IκB kinase-β: NF-κB activation and complex formation with IκB kinase-α and NIK. Science, 278, 866–869. [DOI] [PubMed] [Google Scholar]
- Yaron A. et al. (1997) Inhibition of NF-κB cellular function via specific targeting of the IκBα-ubiquitin ligase. EMBO J., 16, 6486–6494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaron A. et al. (1998) Identification of the receptor component of the IκBα-ubiquitin ligase. Nature, 396, 590–594. [DOI] [PubMed] [Google Scholar]
- Zandi E., Rothwarf,D.M., Delhase,M., Hayakawa,M. and Karin,M. (1997) The IκB kinase complex (IKK) contains two kinase subunits, IKKα and IKKβ, necessary for IκB phosphorylation and NF-κB activation. Cell, 91, 243–252. [DOI] [PubMed] [Google Scholar]