Abstract
Using a sordarin derivative, an antifungal drug, it was possible to determine the structure of a eukaryotic ribosome⋅EF2 complex at 17.5 Å resolution by three-dimensional (3D) cryo-electron microscopy. EF2 is directly visible in the 3D map and the overall arrangement of the complex from Saccharomyces cerevisiae corresponds to that previously seen in Escherichia coli. However, pronounced differences were found in two prominent regions. First, in the yeast system the interaction between the elongation factor and the stalk region of the large subunit is much more extensive. Secondly, domain IV of EF2 contains additional mass that appears to interact with the head of the 40S subunit and the region of the main bridge of the 60S subunit. The shape and position of domain IV of EF2 suggest that it might interact directly with P-site-bound tRNA.
Keywords: elongation/GM193663/sordarin/translation/yeast
Introduction
The elongation cycle of protein synthesis consists of three basic steps: A-site occupation, peptidyl transfer and translocation. While the peptidyl transferase activity is an intrinsic activity of the large ribosomal subunit, the rates of A-site occupation and translocation are greatly enhanced by soluble elongation factors. EF-Tu in prokaryotes and the homologous EF1α in eukaryotes form a ternary complex with the aminoacylated tRNA (aa-tRNA) and GTP that delivers the aa-tRNA into the ribosomal A-site. The translocation reaction is mediated by EF-G in prokaryotes and the homologous EF2 in eukaryotes. All these factors belong to the G-protein superfamily (Bourne et al., 1991). For a long time, it was generally accepted that these elongation factors follow the classical scheme of G-proteins. They successively bind to the ribosome in the GTP conformation, catalyze their respective step on the ribosome, subsequently hydrolyze GTP and dissociate in the GDP conformation. More recently, this view has been challenged in the case of the translocation reaction and it was suggested that EF-G and possibly EF2 may act like force-generating motor proteins (Abel and Jurnak, 1996; Rodnina et al., 1997).
Because of the high degree of evolutionary conservation of the protein synthesis machinery (rRNA, ribosomal proteins, translation factors), the general framework of protein synthesis is believed to be the same in prokaryotes and eukaryotes. However, eukaryotic 80S ribosomes are considerably larger than the prokaryotic 70S ribosomes and also protein synthesis appears to be more complex in eukaryotes (see Hershey et al., 1996). Apart from translation initiation, which is far more complicated in eukaryotes, there are also significant differences in the ribosomal elongation phase. In addition to the GTP molecules hydrolyzed by EF-Tu/EF1α and EF-G/EF2, eukaryotes consume ATP during the elongation cycle. In fungi this ATP hydrolysis takes place on a third elongation factor, EF3 (Skogerson and Wakatama, 1976). Polypeptide chain elongation in fungi requires EF3-dependent ATP hydrolysis to release deacylated tRNA from the ribosomal E-site (Triana-Alonso et al., 1995).
Cryo-electron microscopy (cryo-EM) has been used in recent years to study the structure of the ribosome and its binding to functional ligands. Even at moderate resolutions, in the range 15–20 Å, binding positions and conformational changes of elongation factors could be deduced from the resulting density maps (Stark et al., 1997; Agrawal et al., 1998, 1999).
It has recently been found that the interaction of EF2 with the ribosome is the cellular target for members of the sordarin family of antifungal compounds (Capa et al., 1998; Dominguez and Martin, 1998; Dominguez et al., 1998; Gomez-Lorenzo and Garcia-Bustos, 1998; Justice et al., 1998, 1999). Even though there is a high degree of homology between the fungal and mammalian protein synthesis machineries, these inhibitors are highly specific for the fungal elongation step (Kinsman et al., 1998). This study was prompted by the possibility that sordarin compounds could stabilize the yeast ribosome⋅EF2 complex and allow its imaging by cryo-EM, as has been previously accomplished with fusidic acid and the Escherichia coli ribosome⋅EF-G complex (Agrawal et al., 1998). Sordarin derivative GM193663 was used to stabilize the ribosome⋅EF2 complex and to obtain the first three-dimensional (3D) reconstruction of a eukaryotic 80S ribosome in complex with one of several protein factors of the protein synthesis machinery. The resolution of the map, 17.5 Å, is the highest obtained so far for a eukaryotic ribosome. It has allowed us to determine the binding position of EF2 and to discuss the similarities and possible differences in the mechanism of translocation between prokaryotes and eukaryotes.
Results
Overall structure of the yeast 80S ribosome
Using micrographs at several defoci, contrast transfer function (CTF) correction, and a much larger dataset, we were able to drastically improve the resolution of the cryo-EM map of the yeast 80S ribosome from Saccharomyces cerevisiae. Previous reconstructions of the yeast 80S ribosome yielded resolutions of 35 Å for the empty ribosome (Verschoor et al., 1998) and of 26 Å for an 80S–Sec61 complex (Beckmann et al., 1997). Here we present the structure of the S.cerevisiae 80S ribosome in complex with elongation factor EF2. The complex was stabilized by using the sordarin derivative GM193663, a translational inhibitor that is specific for fungi and probably acts in a way similar, although not identical, to fusidic acid in prokaryotes (Capa et al., 1998; Dominguez et al., 1998, 1999; Justice et al., 1998; Kinsman et al., 1998). Approximately 60% of the 80S ribosomes carried EF2 (see Table I). This is lower than the corresponding binding of EF-G to the E.coli 70S ribosomes (80%), but compares very well with the reported 60% activity of yeast ribosomes in tRNA binding (Triana-Alonso et al., 1995).
Table I. EF2 and sordarin binding to 80S ribosomes.
| Background | 80S | EF2 | 80S + EF2 | |
|---|---|---|---|---|
| Sordarin (pmol) | 0.2 | 0.2 | 0.9 | 2.3 |
| v | – | – | – | 0.58 |
In a total volume of 50 µl, 5 pmol of 80S ribosomes and 51 pmol of EF2 were incubated in the presence of [3H]sordarin (20 µM, f.c.). Controls were performed using only sordarin, 80S ribosomes and sordarin, or EF2 and sordarin. After the binding reaction (see Materials and methods) the sample was passed through a PD10 gel filtration column. The amount of sordarin present in the flow-through was determined by measuring the radioactivity in the flow-through. The occupancy v (pmol sordarin/pmol 80S ribosomes) was calculated taking the value of EF2 + sordarin as the background value and taking into account that only 2.4 pmol of 80S were present in the flow-through.
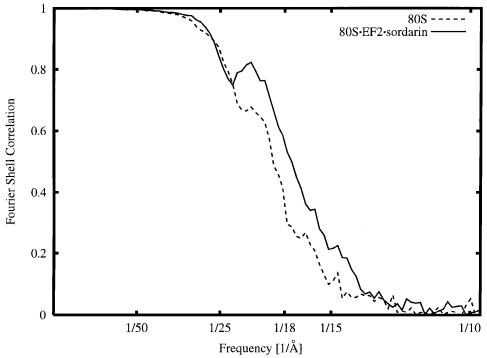
The resolution curve for the final reconstruction is shown in Figure 1. With the conservative cut-off value of 0.5 (see Malhotra et al., 1998 for discussion), the resolution achieved is 17.5 Å, but the reconstruction still contains information of lower accuracy beyond this point (13.4 Å with the 3-σ criterion; see Orlova and van Heel, 1997). A control reconstruction of the empty ribosome yielded a resolution of 18.9 Å (see Figure 1), confirming that, among other factors, the stabilization of the conformation by binding of the ligand is an important factor in the improvement of resolution.
Fig. 1. Fourier shell correlation curve, indicating a resolution of 17.5 Å with a 0.5 cut-off criterion for the 80S⋅EF2⋅sordarin complex (solid line) and 18.9 Å for the vacant 80S ribosome (dashed line).
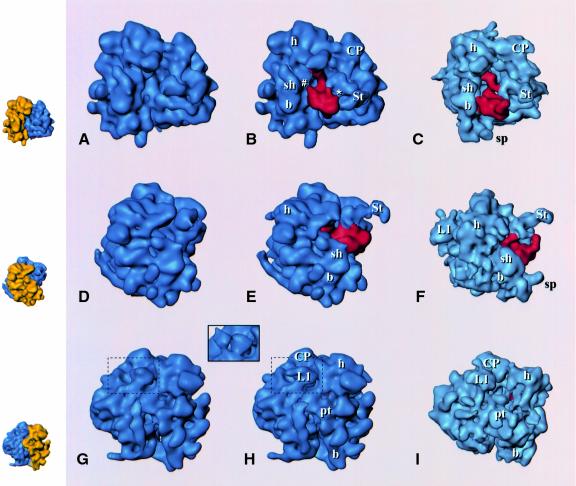
Since the resolution is in the same range as for published 3D cryo-EM reconstructions of the E.coli 70S ribosome (Agrawal et al., 1998, 1999), a direct comparison of both structures is possible (Figure 2). The similarity between the two structures observed at lower resolution (Verschoor et al., 1998) has become more pronounced. Landmarks observed in the large subunit (central protuberance, stalk, L1 protuberance, tunnel through the subunit) and the small subunit (head, body, platform, shoulder) all bear a detailed resemblance. However, the yeast ribosome has additional elements distributed over the periphery of the structure, which are due to the expansion segments in the rRNA (Dube et al., 1998b; Spahn et al., 1999) and 24 additional proteins (Warner, 1999). Another difference is in the relative arrangement of the subunits (Figure 2). This difference is more complicated than a mere rotation of the 40S subunit, found by Dube et al. (1998a) for the mammalian ribosome. To analyze the changes, we aligned several landmark features of the large subunits of the E.coli 70S and the S.cerevisiae 80S ribosome, i.e. the central protuberance, the L7/L12 stalk, the tunnel and the main bridge B2 (see Frank et al., 1995a for a definition). This analysis shows that when compared with the 70S ribosome, the shoulder of the small subunit in the 80S ribosome is shifted away from the large subunit, while its head is located closer to the central protuberance and its platform is closer to the large subunit. Furthermore, one striking difference is observed in the arrangement of components within the 60S subunit: the L1 protuberance is shifted towards the central protuberance of the 60S subunit when compared with its position in the 50S subunit (Figure 2G–I).
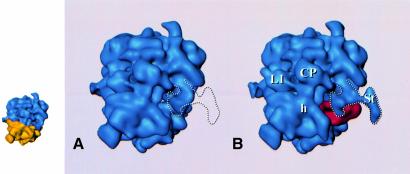
Fig. 2. Reconstructions of the 80S ribosome from S.cerevisiae without (A, D, G) and with (B, E, H) EF2 bound, and the EF-G-bound 70S ribosome from E.coli (C, F, I; adapted from Agrawal et al., 1999), presented in three equivalent views. Upper row (A–C): side view, with small subunit on the left and large subunit on the right, showing the binding sites of the elongation factors; middle row (D–F): view from the small subunit solvent side, with (E) and (F) showing the extended stalk (St); bottom row (G–I): view from the L1 protein side. The inset above (G) and (H) shows a comparison of the L1 region [within the dashed boundary in (G) and (H)] with the ‘split’ appearance of L1 in the yeast ribosome in a previous reconstruction (Beckmann et al., 1997). EF2 and EF-G are shown in red. Small insets on the left depict the 80S control map with 40S and 60S colored in yellow and blue, respectively, in corresponding orientations as an interpretation aid. Landmarks, small subunit: h, head; b, body; pt, platform; sh, shoulder (for the designation of subunit body, see Figure 3; large subunit: CP, central protuberance; L1, L1 protuberance; St, extended stalk.
Localization and interaction of EF2 with the ribosome
An analysis of the difference map between the 80S⋅EF2⋅sordarin complex and the vacant ribosome gives a distinct density corresponding to EF2 (see the supplementary data at The EMBO Journal Online) and reveals a large conformational shift of the stalk region upon EF2 binding, with EF2 located close to the base of the stalk and the A-site (see below). The EF2 occupancy of ribosomes was at least 60%, sufficient to observe EF2 directly in the 3D map (Figure 2B and E). The fraction of vacant 80S ribosomes present in the preparation (<40%) should weaken the EF2 density and should make it more difficult to observe conformational changes. However, EF2 and some conformational changes (see below) can be clearly observed, showing that the EF2-bound fraction of 80S ribosomes is dominant in the data used for the reconstruction. Thus, the main effect of the fraction of vacant ribosomes present is probably a reduction in resolution.
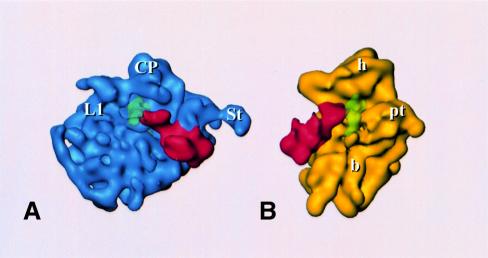
The additional density corresponding to EF2 (Figures 2B, E, H and 3) was isolated by threshold analysis. The overall shape and location of the EF2 mass so determined (Figure 3) resemble those for the EF-G mass from the 70S⋅EF-G⋅GDP⋅fusidic acid complex (Agrawal et al., 1998). This similarity strongly suggests that EF2 is present in its natural binding site, even though it was bound to the ribosome in the absence of mRNA and tRNA and in the presence of sordarin. We note that in the E.coli system the position and conformation of EF-G were not affected by the presence of mRNA and tRNA (Agrawal et al., 1998, 1999). The resolution of the yeast complex is high enough to allow the tentative identification of the different EF2 domains by homology with EF-G (Ævarsson et al., 1994; Czworkowski et al., 1994). There is a good match in overall shape and size between the density mass of EF2 and a theoretical model (Capa et al., 1998; not shown), although the placement of additional structural elements in the model of domain IV, accounting for 36 additional residues, does not show detailed agreement. The best match between the model and the EM density is obtained by fitting domains I and II to the density map, and turning domains III, IV and V together as a unit by 25° towards the central protuberance, around a pivoting point between domains II and III. This is similar to the operation that was necessary in the case of EF-G to obtain an optimum fit between the X-ray structure of EF-G and the extra density attributed to EF-G in the 70S⋅EF-G⋅GDP⋅fusidic acid complex (Agrawal et al., 1998).
Fig. 3. Reconstruction of EF2-bound 80S ribosome, split into the 60S (A) and 40S (B) subunit, each shown with EF2; P-site-bound tRNA is also shown (green, transparent) in a position inferred by analogy to its experimental localization in E.coli (Malhotra et al., 1998). Landmarks are the same as in Figure 2.
There are several sites where the EF2 density is fused with the 80S density. These sites represent contacts between the factor and the ribosome or at least positions of close approach. In general, the sites of interaction between the elongation factor and the ribosome appear to be the same as in the E.coli system (Agrawal et al., 1998). Domain II of EF2 interacts with the shoulder of the 40S subunit close to the site (Figure 3b) where protein S4 has been localized in a recent X-ray map of the 30S subunit of a prokaryotic ribosome (Clemons et al., 1999). Domain III of EF2 forms an apparent contact with the body of the 40S subunit closer to the decoding site (hidden in Figure 3B), and both domain V and the G domain interact with the stalk base region of the 60S subunit (Figures 2B, E and 3A). The GTP face of the G domain interacts with the 60S subunit in the lower portion of the stalk base, at the place where the α-sarcin–ricin stem–loop has been placed in cryo-EM maps of the 70S ribosome from E.coli (Agrawal et al., 2000; Gabashvili et al., 2000) and a 5 Å X-ray map of the 50S subunit from Thermus thermophilus (Ban et al., 1999). Finally, domain IV of EF2 is larger and more complex than its bacterial counterpart and makes extensive contacts with the 40S subunit and also the 60S subunit (see below). Specifically, these contacts involve the head of the 40S subunit and one of the intersubunit bridges. Most importantly, it reaches into the region immediately bordering the decoding site (Figures 2B and 3B).
With the exception of domain III, these interactions remain visible for a wide range of threshold levels, strongly indicating that physical contacts are made. Although the overall picture of the ribosome–factor interaction is very similar in the E.coli complex (Agrawal et al., 1998, 1999) and the yeast complex, the mode of interaction appears to be different in important regions. The interaction between domain V of EF2 and the stalk region seems to be the same, but there is a marked change in the contact between the stalk base region and the G/G′ domain of the factor. Instead of a clear ‘arc-like’ connection seen in E.coli (Agrawal et al., 1998, 1999; Figure 2F), the corresponding contact in the yeast system is much broader and fused with the domain V contact. The contacts that build the arc are also visible, but here the connection is shorter, broader and more straight. Furthermore, there is an additional, weaker contact between EF2 and the stalk base region located closer to the base of the extended portion of the stalk (marked by an asterisk in Figure 2B). Domain II of EF2 is larger than the corresponding domain in EF-G, and an additional contact is formed between the extra mass of domain II and the 40S subunit close to its site of interaction with domain III (marked by a hash in Figure 2B).
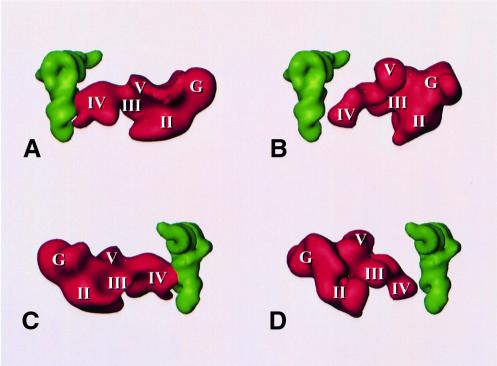
The most unexpected difference between the E.coli complex and the yeast complex is found in domain IV of the elongation factor, the domain implicated in tRNA mimicry (Nissen et al., 1995). Instead of ending in a single tip, domain IV of S.cerevisiae EF2 ends in a broad, fork-shaped structure with three prongs (Figures 3 and 4). The extra mass, which apparently corresponds to the sequence insertions in the EF-G domain IV homolog, interacts strongly with the ribosome. One side prong of the structure interacts with the head of the 40S subunit, the other with the bridge of the 60S subunit, which is homologous to the bridge B2a region of the 50S subunit of prokaryotes (Cate et al., 1999; Gabashvili et al., 2000), the most massive connection between the large and small subunit. [An analysis of the maps of the EF-G-bound ribosome, in the light of the detailed study of bridging connections between the subunits by Gabashvili et al. (2000), revealed that EF-G domain IV also contacts this bridge, but in a different geometric constellation.] The central prong of EF2 domain IV might be the homolog of the tip of that domain in EF-G as it is found in a similar position.
Fig. 4. Relative arrangements of (A and C) P-site tRNA and EF2 in the 80S ribosome (with tRNA placed into the 80S density map in a position equivalent to its observed position in the fMet-tRNAfMet–70S ribosome map of Malhotra et al., 1998) and (B and D) P-site tRNA (Malhotra et al., 1998) and EF-G (Agrawal et al., 1998) in the 70S ribosome. Roman numerals I–V and G mark the domains in EF-G and homologous domains in EF2 according to the model of Capa et al. (1998).
A possible interaction of domain IV of EF2 with P-site-bound tRNA
Domain IV of EF2 reaches deeper into the ribosome than the homologous domain of EF-G, which implies that it comes closer to the anticodon stem of P-site-bound tRNA. As yet, there has been no experimental localization of the P-site-bound tRNA in the yeast ribosome, and therefore we have superimposed the yeast structure with a model of P-site-bound tRNA derived from the E.coli ribosome (Malhotra et al., 1998). This appears to be justified since the functionally important regions of the ribosome are evolutionarily conserved and, accordingly, the structures of the ribosomes from the two species in this region (including the bridge B2 region) are very similar and well aligned. In this arrangement, domain IV of EF2 comes to lie very close to the tRNA. Two of the prongs of the fork-shaped end of domain IV that were identified above wrap around the anticodon stem–loop, roughly following the major groove of the tRNA (Figure 4). This fork is extended by the interaction of one of its prongs with the bridge B2a.
Conformational changes in the ribosome upon EF2 binding
In the vacant ribosome the extended stalk was not observed, probably due to its high mobility, but it became visible in the 80S⋅EF2⋅sordarin complex (Figure 2). Since the same 80S ribosome preparation was used for both reconstructions, this finding indicates that, as in prokaryotic ribosomes (Agrawal et al., 1998), the conformation of the yeast ribosome stalk becomes fixed in an extended configuration following factor binding. The extended stalk in yeast appears to be less strong, probably due to the lower occupancy of the elongation factor (60% in the yeast system versus >80% for E.coli). However, it can be clearly seen that the stalk extends outwards from the ribosome towards the cytosol, in agreement with its proposed role as an early anchoring point for soluble factors (van Agthoven et al., 1977; Möller and Maassen, 1986; Moazed et al., 1988; Uchiumi et al., 1990).
In addition, a second conformational change can be observed in the stalk region: the whole stalk region moves towards the central protuberance (Figure 5). This movement is indicated by a comparison of the positive and negative masses in the difference map between the 80S⋅EF2 complex and the vacant 80S ribosome, and can also be observed directly (Figure 5). Such a drastic movement in response to EF2 binding has not been observed in the bacterial complex (Agrawal et al., 1998, 1999).
Fig. 5. Top view of the 80S ribosome (A) without and (B) with EF2 bound, indicating the structural rearrangement of the stalk region. The outline of the stalk region in the 80S⋅EF2⋅sordarin complex is indicated by the white dashed line. Small inset on the left: control map (A) with 40S and 60S colored in yellow and blue, respectively, as an interpretation aid.
Discussion
The reconstruction of the sordarin-stabilized 80S⋅EF2 complex represents the highest resolution density map of a eukaryotic ribosome obtained so far. We believe that the improvement from 26 Å (Beckmann et al., 1997) to 17.5 Å (Figure 1) is due to three main factors: (i) conformational homogeneity and stability; (ii) increased cryo-EM dataset; and (iii) the use of multiple defocus groups.
In the case of the fMet-tRNAfMet⋅ribosome complex of E.coli (Malhotra et al., 1998), higher resolution could be obtained than with the vacant ribosome, and it was conjectured that as a rule, functionally meaningful complexes might be better defined conformationally than their vacant counterparts. The large jump in resolution in the current study seems to be due, at least in part, to the stabilizing effect of EF2 binding, in agreement with the earlier observations.
There is no doubt that another reason for the improvement in resolution is the increase in the number of particles, now at 17 716, especially as it is combined with the strategy of collecting images over a wide range of defoci and CTF correction. As a result, the density map is well defined, with high-resolution features such as thin intersubunit bridges observed for the E.coli ribosome (Frank et al., 1995a,b), and without a noticeable prevalence of features in a particular size range familiar from previous non-CTF-corrected reconstructions of the eukaryotic ribosome.
Mobility of ribosomal components
The overall structure of the yeast 80S ribosome is quite similar to that of the 70S ribosome from E.coli. Features such as the stalk of the large subunit, the tunnel through the large subunit and the main bridge B2 (Frank et al., 1995a; Cate et al., 1999; Gabashvili et al., 2000) can be well aligned. This is in line with the interpretation that bacterial ribosomes and the inner core of the eukaryotic ribosomes are well conserved during evolution (Dube et al., 1998a) and that eukaryotic ribosomes contain additional elements, such as the expansion segments in the rRNA, at the surface (Dube et al., 1998b; Spahn et al., 1999). However, the relative arrangement of the subunits can change from organism to organism (Dube et al., 1998a; this study) and additional secondary structure elements can occupy different positions (Spahn et al., 1999).
A striking example for such a displacement in this study is the L1 protuberance. Compared with its position in the E.coli 70S ribosome, L1 is shifted towards the intersubunit space. A comparison in the L1 region of our new reconstructions of the 80S ribosome from S.cerevisiae with our previous one (Beckmann et al., 1997) reveals that the previous reconstruction was probably based on a mixed population of ribosomes. There, the L1 region has a complex shape and appears to be the result of the superposition of L1 proteins in two different positions: one in a position identical to that of the L1 protuberance observed in the current study and another, clearly observed at slightly lower threshold, in an ∼50 Å shifted position. The angle between the strands that connect L1 with the ribosome in either position is ∼90°. The position of L1 in E.coli, by comparison, appears to be in between the extreme positions observed for yeast, except that it is also shifted in a perpendicular direction, towards the small subunit (Malhotra et al., 1998). Also of interest in this context are earlier observations of smaller scale movements of L1 in response to changes in buffer conditions and functional states of the E.coli ribosome (R.K.Agrawal and J.Frank, unpublished results).
Most likely, the dramatic changes in L1 position in the S.cerevisiae 80S ribosome are in response to preparation conditions, with the presence or absence of tRNA and elongation factors being the most decisive determinants. The 80S preparation used by Beckmann et al. (1997) probably contained some quantities of bound tRNAs as well as elongation factors, as also indicated by the appearance of the P stalk in extended form in their reconstruction (by analogy with E.coli, where the extended state of the L7/L12 stalk has been found to be an indicator of tRNA and factor binding; see Agrawal et al., 1998; Malhotra et al., 1998). Thus, the observation of two copies of L1 in that reconstruction could be explained by assuming a dramatic response of the L1-bearing RNA strand to the presence of these ligands in a ribosome subpopulation. Although in both studies the ribosomes investigated are not in defined functional states, the dependency of the L1 position on the preparation conditions probably means that the L1 protuberance can undergo a considerable back and forth movement during protein synthesis. We note that the L1 protuberance in the position observed in this study closes the intersubunit space from one end and probably blocks the exit of the tRNA. It is tempting to speculate that EF3, which is implicated in an ATPase-dependent release of the tRNA from the E-site (Triana-Alonso et al., 1995), might be involved in the control of the L1 movement. Furthermore, the stalk of the yeast 80S ribosome is mobile and seen to move by ∼15 Å towards the central protuberance upon EF2 binding (Figure 5). No such drastic movement in response to factor binding has been observed in the E.coli 70S ribosome (Agrawal et al., 1998, 1999).
Interactions between EF2 and the 60S subunit
Several ribosomal components have been implicated in the interaction with EF-G/EF2. It is well established that the ribosomal stalk plays an important role in the interaction with elongation factors (e.g. Moazed et al., 1988; Uchiumi et al., 1990). In prokaryotes, the stalk is built from a pentameric L8 complex with the composition L10⋅(L7/L12)4, protein L11 and a part of domain II of 23S rRNA. L11 and L10⋅(L7/L12)4 bind to 23S rRNA in the region between positions 1028 and 1124, which includes the binding site for thiostrepton to what is called the GTPase-associated center of the rRNA, an extremely well conserved RNA fold, which is exchangeable between prokaryotes and eukaryotes (Musters et al., 1991; Thompson et al., 1993). EF-G has been found to protect from modification this rRNA region plus the so-called α-sarcin–ricin loop around position 2660 (see Wilson and Noller, 1998 and references therein). Ricin and α-sarcin cleave prokaryotic 23S and eukaryotic 28S rRNA in that loop and abolish elongation factor-dependent ribosomal functions (Hausner et al., 1987; Wool et al., 1992). Recently, the α-sarcin–ricin loop could be placed in cryo-EM maps of the 70S E.coli ribosome (15 Å map: Agrawal et al., 2000; 11.5 Å map: Gabashvili et al., 2000) and in a 5 Å resolution X-ray map of the 50S subunit from Haloarcula marismortui (Ban et al., 1999), and it was proposed that this RNA interacts directly with the G domain of the elongation factors.
In contrast to the rRNA regions implicated in the interaction with the elongation factor, the protein components of the eukaryotic stalk have no significant sequence homology to their prokaryotic counterparts. The yeast stalk is composed of four acidic proteins (rpP1α, rpP1β, rpP2α and rpP2β), proteins rpP0 and rpL12 (formerly called L15), and the segment of 26S rRNA forming the GTPase-associated center (Briones et al., 1998). Acidic proteins in yeast are dispensable for growth, unlike the L7/L12 counterpart in E.coli, which is essential. Of all the P-proteins, only rpP0 is firmly bound to the particle and essential for growth (Santos and Ballesta, 1994). The acidic proteins exist as a cytoplasmic pool apparently freely exchangeable with ribosome-bound forms, change their association with the ribosome depending on the growth phase (Saenz-Robles et al., 1990), and nevertheless strongly affect the pattern of proteins being translated (Remacha et al., 1995).
The interactions between EF2 and the ribosomal P-protein stalk observed in this work must be the morphological correlate of their known biochemical and functional interactions. It has been observed, for example, that EF2 protects the GTPase-associated center in 28S rRNA to which the acidic proteins bind (Holmberg and Nygard, 1994; Uchiumi and Kominami, 1994). The factor has been chemically cross-linked to mammalian P0 and P2 proteins (Uchiumi et al., 1986) and anti-P-protein antibodies are known to block EF2 binding (Uchiumi et al., 1990). Most recently, it has been found that exchange of the E.coli L10⋅(L7/L12)4 complex by eukaryotic P-proteins allows interaction of the changed E.coli ribosome with the eukaryotic EF2 (Uchiumi et al., 1999). Also of interest is the observation that resistance mutations to sordarin, the antibiotic used in this study, can be found in both EF2 and rpP0 (Capa et al., 1998; Gomez-Lorenzo and Garcia-Bustos, 1998; Justice et al., 1998, 1999). Ribosomal stalk proteins rpP0, rpP1α and rpP2β influence the sensitivity to sordarin derivative GM193663 in different ways and, at least in the case of rpP0 mutants, without significantly affecting the binding of the inhibitor to the ribosome⋅EF2⋅complex (Gomez-Lorenzo and Garcia-Bustos, 1998; Justice et al., 1999). It would appear that a modified ribosomal stalk can, by an unknown mechanism, allow EF2 to function even when bound to a sordarin molecule. All these observations strongly indicate that EF2 and stalk components interact physically and functionally to bring about translation elongation, correlating nicely with the observed contacts in the cryo-EM images.
Domain IV of EF2 and its role in translocation
In E.coli a rearrangement of domains III, IV and V relative to domains G and II was found to be necessary to fit the EF-G X-ray structure into the cryo-EM maps of the E.coli 70S ribosome in complex with EF-G, which led to the proposal that a conformational change takes place in EF-G upon binding (Agrawal et al., 1998, 1999). The density attributed to EF2 appears to be more similar to the ribosome-bound conformation of EF-G than to the X-ray structure of EF-G (Ævarsson et al., 1994; Czworkowski et al., 1994). This indicates that a similar conformational change upon binding takes place in EF2 as well. This conformational change might therefore be a universal feature of protein synthesis.
Domain IV of EF-G/EF2 has been strongly implicated in the function of the elongation factor by several lines of evidence. The structure of this domain (Ævarsson et al., 1994; Czworkowski et al., 1994) mimics the acceptor stem of the tRNA within the ternary complex EF-Tu⋅tRNA⋅GTP (Nissen et al., 1995). In contrast to EF-G, EF2 contains a post-translationally modified histidine named diphthamide (Foley et al., 1995 and references therein). Sequence alignments have placed this amino acid at the tip of domain IV, roughly corresponding to the position of the anticodon within tRNA. ADP ribosylation of the diphthamide residue by bacterial toxins like diphtheria toxin render EF2 inactive (Oppenheimer and Bodley, 1981). Two studies on the activity of bacterial EF-G with domain IV deleted have yielded results that were partially contradictory (Rodnina et al., 1997; Martemyanov and Gudkov, 1999). Nevertheless, in both studies it was shown that a complete EF-G cycle is not possible without domain IV.
In our cryo-EM study we have located the distal end of domain IV of EF2 near the cleft of the 40S subunit, at the presumed binding site of aa-tRNA. This is in agreement with the reported protection by EF2 of 18S rRNA segments forming part of the decoding center and of 28S rRNA segments in the peptidyltransferase ring (Holmberg and Nygard, 1994). This position is very similar to that of the corresponding domain of E.coli EF-G (Agrawal et al., 1998, 1999). However, the end of domain IV of S.cerevisiae EF2 is broadened due to the presence of extra mass, which makes intensive contacts with the 80S ribosome.
For many years it was generally accepted that EF-G (as well as EF2) follows the classical scheme of G-proteins. However, this view has recently been challenged and various models have been proposed according to which the chemical energy stored in the GTP molecule is directly used for the movement of the tRNA⋅mRNA complex (Abel and Jurnak, 1996; Rodnina et al., 1997). Domain IV of EF-G/EF2, a key player in these models, is thought to achieve the tRNA⋅mRNA movement either by directly pushing the A-site-bound tRNA or by exerting the motor function on 16S rRNA and thereby inducing the translocation (Rodnina et al., 1999). Whether either of these active push models is correct, or domain IV has a more passive role as a ‘door-stop’ in occupying the A-site region and preventing a reversed translocation when the bound factor allows movement of the tRNAs (Wilson and Noller, 1998), remains an open question. In any case, domain IV of EF2 seems to be more efficiently constructed to fulfill tasks related to this particular function than domain IV of EF-G as it reaches deeper into the ribosome. The fact that the broadened end of domain IV is shaped to follow the topology of the anticodon stem–loop of the P-site-bound tRNA might suggest a direct interaction during translocation, in line with a mechanism whereby a conformational change of EF2 directly drives the tRNA relocation. Moreover, the additional interactions of domain IV with the head of 40S and the bridge B2a might facilitate a conformational rearrangement of the 18S rRNA and a transmission of this signal to the 60S subunit.
With the caveat that the inhibitors used to stabilize the prokaryotic ribosome⋅EF-G (fusidic acid) and the eukaryotic ribosome⋅EF2 (sordarin GM193663) complexes could introduce subtly different distortions of the system, the topological differences found between prokaryotes and eukaryotes in the functionally significant region are striking. Protein synthesis in bacteria is a fast and highly efficient process. That a eukaryotic elongation factor might be more efficiently constructed to fulfill a particular step of the elongation cycle does not necessarily mean that protein biosynthesis as a whole is more efficient in eukaryotes than in prokaryotes. On the contrary, prokaryotic ribosomal machines are likely to be adapted to fast and accurate protein synthesis requiring less cooperation with external elongation factors.
That much larger displacements of parts of the ribosomal machine occur for the eukaryotic 80S ribosome from S.cerevisiae than for the bacterial 70S ribosome from E.coli, as observed in the cases of the L1 protuberance and the P-protein stalk, could mean that generally much larger movements take place in eukaryotic ribosomes. However, an alternative explanation is that the movements in bacterial ribosomes are in fact on the same scale, but take place only in short-lived transition states of the elongation cycle so that they have thus far escaped observation. Both explanations would imply that more force is needed to shift gears for the eukaryotic translational machinery. Eukaryotic ribosomes might be less streamlined with respect to translational efficiency because they are involved in more complex tasks within the cell, such as co-translational protein transport, than their bacterial counterparts and are subjected to a more complicated translational control of gene expression (see Hershey et al., 1996).
Materials and methods
Ribosomes were isolated from S.cerevisiae strain W303 cells grown in YPD medium to an OD600 of 1. Washed cells were broken in lysis buffer [30 mM HEPES–KOH, 100 mM potassium acetate, 12.5 mM magnesium acetate, 2 mM dithiothreitol (DTT), 1 mM phenylmethylsulfonyl fluoride, 8.5% d-mannitol, pH 7.4] by vortexing with glass beads for 10 1-min periods, with intervening 1 min cooling in ice. Unbroken cells and debris were pelleted at 4000 g for 10 min, and the supernatant was then centrifuged at 50 000 g for 30 min. This S-50 supernatant was then centrifuged for 4 h at 100 000 g. The ribosomal pellet was resuspended in lysis buffer without d-mannitol, and placed on top of a sucrose gradient made of solutions of 40 and 5% sucrose in lysis buffer without d-mannitol. It was then centrifuged for 14 h at 20 000 r.p.m. on an SW28 rotor. The gradient was fractionated and only the 80S ribosomes were taken and pelleted again.
EF2 was purified from the post-ribosomal supernatant by ion-exchange chromatography as described previously (Dasmahapatra and Chakraburtty, 1981). The binding reaction was prepared in a total volume of 50 µl, using 3 pmol of 80S ribosomes, 50 pmol of EF2 and 2.5 nmol of GTP, in 80 mM HEPES–KOH, 10 mM magnesium acetate, 20 mM potassium acetate and 1 mM DTT buffer. All components were mixed in ice and then incubated for 10 min at 30°C. Then 1 nmol of the sordarin derivative GM193663 was added to the mixture and incubated for another 15 min at 30°C. The complex was immediately applied to the electron microscope grid and prepared for cryo-microscopy according to standard methods (Wagenknecht et al., 1988).
To calculate ribosome occupancy, the binding reaction was performed under the same conditions but in the presence of a 3H-labeled sordarin derivative. A PD10 column (Amersham Pharmacia Biotech) was used to remove the unbound sordarin. Background controls were performed by omitting either EF2 or 80S ribosomes from the reaction mixture. The occupancy with sordarin was ∼60%. Since only sordarin bound to EF2/80S ribosome complexes can be detected, the EF2 occupancy of 80S ribosomes was at least 60%.
Micrographs were taken at defoci between 1.0 and 2.0 µm by using the low-dose protocol (10 e–/Å2) on a Philips EM420 (FEI/Philips, Eindhoven), at a magnification of 52 200 (± 2%). Micrographs were checked for the presence and size of Thon rings, drift and astigmatism by optical diffraction, and scanned on a Hi-Scan drum scanner (Eurocore/Saint-Denis) with an accuracy of 1018 d.p.i., corresponding to a pixel size of 4.78 Å on the object scale.
For the 3D reconstruction of 80S⋅EF2⋅sordarin complexes, a total of 17 716 particles from 39 micrographs were used. They were distributed in 10 defocus groups, as determined by positions of Thon rings. In the case of the empty ribosome, 13 372 particles from 24 micrographs, distributed over eight defocus groups, were used. Ribosomes were selected by a semi-automated selection procedure, whereby the particle candidates are directly compared with the reference set of 83 quasi-evenly spaced projections (Penczek et al., 1994) of an existing best-resolution reconstruction (Beckmann et al., 1997). Image processing was conducted as described previously (Malhotra et al., 1998).
Resolution values given are based on a 0.5 cut-off for the Fourier shell correlation curve (Malhotra et al., 1998), yielding 17.5 Å for the 80S⋅EF2⋅sordarin complex and 18.9 Å for the vacant ribosome.
The difference map was calculated after adjustment of the standard deviation of densities in both reconstructions to the same level. The vacant ribosome was used as a reference structure and it was subtracted from the 80S⋅EF2⋅sordarin complex. To isolate distinct fragments of the ribosome, connected clusters of voxels that are above a predefined threshold were identified. The latter method, preferable due to relatively smaller errors, was used to find the boundaries of EF2.
Supplementary data
Supplementary data to this paper are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We would like to thank Amy Heagle for the preparation of illustrations, and the members of the New Antifungal Targets Group and Biochemistry Group at Glaxo Wellcome for their valuable suggestions. This work was supported by grant DGICYT/PB94/0032, the Foundation Ramón Areces, and NIH grant R37 GM29169 (to J.F.).
References
- Abel K. and Jurnak,F. (1996) A complex profile of protein elongation: translating chemical energy into molecular movement. Structure, 4, 229–238. [DOI] [PubMed] [Google Scholar]
- Ævarsson A., Brazhnikov,E., Garber,M., Zheltonosova,J., Chirgadze,Y., Al-Karadaghi,S., Svensson,L.A. and Liljas,A. (1994) Three-dimensional structure of the ribosomal translocase: elongation factor G from Thermus thermophilus. EMBO J., 13, 3669–3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal R.K., Penczek,P., Grassucci,R.A. and Frank,J. (1998) Visualization of elongation factor G on the Escherichia coli 70S ribosome: the mechanism of translocation. Proc. Natl Acad. Sci. USA, 95, 6134–6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal R.K., Heagle,A.B., Penczek,P., Grassucci,R.A. and Frank,J. (1999) EF-G-dependent GTP hydrolysis induces translocation accompanied by large conformational changes in the 70S ribosome. Nature Struct. Biol., 6, 643–647. [DOI] [PubMed] [Google Scholar]
- Agrawal R.K., Heagle,A.B. and Frank,J. (2000) Studies of elongation factor G-dependent tRNA translocation by three-dimensional cryo-electron microscopy. In Garrett,R.A., Douthwaite,S.R., Liljas,A., Matheson,A.T., Moore,P.B. and Noller,H.F. (eds), The Ribosome: Structure, Function, Antibiotics and Cellular Interactions. ASM Press, Washington, DC, pp. 53–62. [Google Scholar]
- Ban N., Nissen,P., Hansen,J.C., Capel,M., Moore,P.B. and Steitz,T.A. (1999) Placement of protein and RNA structures into a 5 Å-resolution map of the 50S ribosomal subunit. Nature, 400, 841–847. [DOI] [PubMed] [Google Scholar]
- Beckmann R., Bubeck,D., Grassucci,R., Penczek,P., Verschoor,A., Blobel,G. and Frank,J. (1997) Alignment of conduits for the nascent polypeptide chain in the ribosome–Sec61 complex. Science, 278, 2123–2126. [DOI] [PubMed] [Google Scholar]
- Bourne H.R., Sanders,D.A. and McCormick,F. (1991) The GTPase superfamily: conserved structure and molecular mechanism. Nature, 349, 117–127. [DOI] [PubMed] [Google Scholar]
- Briones E., Briones,C., Remacha,M. and Ballesta,J.P. (1998) The GTPase center protein L12 is required for correct ribosomal stalk assembly but not for Saccharomyces cerevisiae viability. J. Biol. Chem., 273, 31956–31961. [DOI] [PubMed] [Google Scholar]
- Capa L., Mendoza,A., Lavandera,J.L., Gomez de las Heras,F. and Garcia-Bustos,J.F. (1998) Translation elongation factor 2 is part of the target for a new family of antifungals. Antimicrob. Agents Chemother., 42, 2694–2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cate J.H., Yusupov,M.M., Yusupova,G.Zh., Earnest,T.N. and Noller,H.F. (1999) X-ray crystal structures of 70S ribosome functional complexes. Science, 285, 2095–2104. [DOI] [PubMed] [Google Scholar]
- Clemons W.M. Jr, May,J.L.C., Wimberly,B.T., McCutcheon,J.P., Capel,M. and Ramakrishnan,V. (1999) Structure of a bacterial 30S ribosomal subunit at 5.5 Å resolution. Nature, 400, 833–840. [DOI] [PubMed] [Google Scholar]
- Czworkowski J., Wang,J., Steitz,T.A. and Moore,P.B. (1994) The crystal structure of elongation factor G complexed with GDP, at 2.7 Å resolution. EMBO J., 13, 3661–3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasmahapatra B. and Chakraburtty,K. (1981) Protein synthesis in yeast. I. Purification and properties of elongation factor 3 from Saccharomyces cerevisiae. J. Biol. Chem., 256, 9999–10004. [PubMed] [Google Scholar]
- Dominguez J.M. and Martin,J.J. (1998) Identification of elongation factor 2 as the essential protein targeted by sordarins in Candida albicans. Antimicrob. Agents Chemother., 42, 2279–2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez J.M., Kelly,V.A., Kinsman,O.S., Marriott,M.S., Gomez de las Heras,F. and Martin,J.J. (1998) Sordarins: A new class of antifungals with selective inhibition of the protein synthesis elongation cycle in yeast. Antimicrob. Agents Chemother., 42, 2274–2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez J.M., Gomez-Lorenzo,M.G. and Martin,J.J. (1999) Sordarin inhibits fungal protein synthesis by blocking translocation differently to fusidic acid. J. Biol. Chem., 274, 22423–22427. [DOI] [PubMed] [Google Scholar]
- Dube P., Wieske,M., Stark,H., Schatz,M., Stahl,J., Zemlin,F., Lutsch,G. and van Heel,M. (1998a) The 80S rat liver ribosome at 25 Å resolution by electron cryomicroscopy and angular reconstruction. Structure, 6, 389–399. [DOI] [PubMed] [Google Scholar]
- Dube P., Bacher,G., Stark,H., Mueller,F., Zemlin,F., van Heel,M. and Brimacombe,R. (1998b) Correlation of the expansion segments in mammalian rRNA with the fine structure of the 80 S ribosome; a cryoelectron microscopic reconstruction of the rabbit reticulocyte ribosome at 21 Å resolution. J. Mol. Biol., 279, 403–421. [DOI] [PubMed] [Google Scholar]
- Foley B.T., Moehring,J.M. and Moehring,T.J. (1995) Mutations in the elongation factor 2 gene which confer resistance to diphtheria toxin and Pseudomonas exotoxin A. J. Biol. Chem., 270, 23218–23225. [DOI] [PubMed] [Google Scholar]
- Frank J. et al. (1995a) A model of the translational apparatus based on a three-dimensional reconstruction of the Escherichia coli ribosome. Biochem. Cell Biol., 73, 757–765. [DOI] [PubMed] [Google Scholar]
- Frank J., Zhu,J., Penczek,P., Li,Y., Srivastava,S., Verschoor,A., Radermacher,M., Grassucci,R., Lata,R.K. and Agrawal,R.K. (1995b) A model of protein synthesis based on cryo-electron microscopy of the E.coli ribosome. Nature, 376, 441–444. [DOI] [PubMed] [Google Scholar]
- Gabashvili I.S., Agrawal,R.K., Spahn,C.M.T., Grassucci,R.A., Svergun,D.I., Frank,J. and Penczek,P. (2000) Solution structure of the E.coli 70S ribosome at 11.5 Å resolution. Cell, 100, 537–549. [DOI] [PubMed] [Google Scholar]
- Gomez-Lorenzo M.G. and Garcia-Bustos,J.F. (1998) Ribosomal P-protein stalk function is targeted by sordarin antifungals. J. Biol. Chem., 273, 25041–25044. [DOI] [PubMed] [Google Scholar]
- Hausner T.P., Atmadja,J. and Nierhaus,K.H. (1987) Evidence that the G2661 region of 23S rRNA is located at the ribosomal binding sites of both elongation factors. Biochimie, 69, 911–923. [DOI] [PubMed] [Google Scholar]
- Hershey J.W.B., Mathews,M.B. and Sonenberg,N. (1996) Translational Control. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Holmberg L. and Nygard,O. (1994) Interaction sites of ribosome-bound eukaryotic elongation factor 2 in 18S and 28S rRNA. Biochemistry, 33, 15159–15167. [DOI] [PubMed] [Google Scholar]
- Justice M.C., Hsu,M.J., Tse,B., Ku,T., Balkovec,J., Schmatz,D. and Nielsen,J. (1998) Elongation factor 2 as a novel target for selective inhibition of fungal protein synthesis. J. Biol. Chem., 273, 3148–3151. [DOI] [PubMed] [Google Scholar]
- Justice M.C., Ku,T., Hsu,M.J., Carniol,K., Schmatz,D. and Nielsen,J. (1999) Mutations in ribosomal protein L10e confer resistance to the fungal-specific eukaryotic elongation factor 2 inhibitor sordarin. J. Biol. Chem., 274, 4869–4875. [DOI] [PubMed] [Google Scholar]
- Kinsman O.S. et al. (1998) Isolation and characterisation of an antifungal antibiotic (GR135402) with protein synthesis inhibition. J. Antibiot., 51, 41–49. [DOI] [PubMed] [Google Scholar]
- Malhotra A., Penczek,P., Agrawal,R.K., Gabashvili,I.S., Grassucci,R.A., Jünemann,R., Burkhardt,N., Nierhaus,K.H. and Frank,J. (1998) Escherichia coli 70S ribosome at 15 Å resolution by cryo-electron microscopy: localization of fMet-tRNAMetf and fitting of L1 protein. J. Mol. Biol., 280, 103–116. [DOI] [PubMed] [Google Scholar]
- Martemyanov K.A. and Gudkov,A.T. (1999) Domain IV of elongation factor G from Thermus thermophilus is strictly required for translocation. FEBS Lett., 452, 155–159. [DOI] [PubMed] [Google Scholar]
- Moazed D., Robertson,J.M. and Noller,H.F. (1988) Interaction of elongation factors EF-G and EF-Tu with a conserved loop in 23S RNA. Nature, 334, 362–364. [DOI] [PubMed] [Google Scholar]
- Möller W. and Maassen,J.A. (1986) On the structure, function, and dynamics of L7/L12 from Escherichia coli ribosomes. In Hardesty,B. and Kramer,G. (eds), Structure, Function and Genetics of Ribosomes. Springer-Verlag, New York, NY, pp. 309–325. [Google Scholar]
- Musters W., Gonzalves,P.M., Boon,K., Raué,H.A., Heerikhuizen,H. and Planta,R.J. (1991) The conserved GTPase center and variable V9 from Saccharomyces cerevisiae 26S rRNA can be replaced by their equivalent from other prokaryotes or eukaryotes without detectable loss of ribosomal function. Proc. Natl Acad. Sci. USA, 88, 1469–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissen P., Kjeldgaard,M., Thirup,S., Polekhina,G., Reshetnikova,L., Clark,B.F. and Nyborg,J. (1995) Crystal structure of the ternary complex of Phe-tRNAPhe, EF-Tu and a GTP analog. Science, 270, 1464–1472. [DOI] [PubMed] [Google Scholar]
- Oppenheimer N.J. and Bodley,J.W. (1981) Diphtheria toxin. Site and configuration of ADP-ribosylation of diphthamide in elongation factor 2. J. Biol. Chem., 256, 8579–8581. [PubMed] [Google Scholar]
- Orlova E.V., Dube,P., Harris,J.R., Beckman,E., Zemlin,F., Markl,J. and van Heel,M. (1997) Structure of keyhole limpet hemocyanin type 1 (KLH1) at 15 Å resolution by electron cryomicroscopy and angular reconstitution. J. Mol. Biol., 271, 417–437. [DOI] [PubMed] [Google Scholar]
- Penczek P.A., Grassucci,R.A. and Frank,J. (1994) The ribosome at improved resolution: new techniques for merging and orientation refinement in 3D cryo-electron microscopy of biological particles. Ultramicroscopy, 53, 251–270. [DOI] [PubMed] [Google Scholar]
- Remacha M., Jimenez-Diaz,A., Bermejo,B., Rodriguez-Gabriel,M.A., Guarinos,E. and Ballesta,J.P.G. (1995) Ribosomal acidic phosphoproteins P1 and P2 are not required for cell viability but regulate the pattern of protein expression in Saccharomyces cerevisiae. Mol. Cell. Biol., 15, 4754–4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodnina M.V., Savelsbergh,A., Katunin,V.I. and Wintermeyer,W. (1997) Hydrolysis of GTP by elongation factor G drives tRNA movement on the ribosome. Nature, 385, 37–41. [DOI] [PubMed] [Google Scholar]
- Rodnina M.V., Savelsbergh,A. and Wintermeyer,W. (1999) Dynamics of translation on the ribosome: molecular mechanics of translocation. FEMS Microbiol. Rev., 23, 317–333. [DOI] [PubMed] [Google Scholar]
- Saenz-Robles M.T., Remacha,M., Vilella,M.D., Zinker,S. and Ballesta,J.P.G. (1990) The acidic ribosomal proteins as regulators of the eukaryotic ribosomal activity. Biochim. Biophys. Acta, 1050, 51–55. [DOI] [PubMed] [Google Scholar]
- Santos C. and Ballesta,J.P.G. (1994) Ribosomal protein P0, contrary to phosphoproteins P1 and P2, is required for ribosome activity and Saccharomyces cerevisiae viability. J. Biol. Chem., 269, 15689–15696. [PubMed] [Google Scholar]
- Skogerson L. and Wakatama,E. (1976) A ribosome-dependent GTPase from yeast distinct from elongation factor 2. Proc. Natl Acad. Sci. USA, 73, 73–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spahn C.M.T., Grassucci,R.A., Penczek,P. and Frank,J. (1999) Direct three-dimensional localization and positive identification of RNA helices within the ribosome by means of genetic tagging and cryo-electron microscopy. Structure, 7, 1567–1573. [DOI] [PubMed] [Google Scholar]
- Stark H., Rodnina,M.V., Rinke-Appel,J., Brimacombe,R., Wintermeyer, W. and van Heel,M. (1997) Visualization of elongation factor Tu on the Escherichia coli ribosome. Nature, 389, 403–406. [DOI] [PubMed] [Google Scholar]
- Thompson J., Musters,W., Cundliffe,E. and Dahlberg,A.E. (1993) Replacement of the L11 binding region within E.coli 23S ribosomal RNA with its homologue from yeast: in vivo and in vitro analysis of hybrid ribosomes altered in the GTPase centre. EMBO J., 12, 1499–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triana-Alonso F.J., Chakraburtty,K. and Nierhaus,K.H. (1995) The elongation factor 3 unique in higher fungi and essential for protein biosynthesis is an E site factor. J. Biol. Chem., 270, 20473–20478. [DOI] [PubMed] [Google Scholar]
- Uchiumi T. and Kominami,R. (1994) A functional site of the GTPase-associated center within 28S ribosomal RNA probed with an anti-RNA autoantibody. EMBO J., 13, 3389–3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiumi T., Kikuchi,M., Terao,K., Iwasaki,K. and Ogata,K. (1986) Cross-linking of elongation factor 2 to rat-liver ribosomal proteins by 2-iminothiolane. Eur. J. Biochem., 156, 37–48. [DOI] [PubMed] [Google Scholar]
- Uchiumi T., Traut,R.R. and Kominami,R. (1990) Monoclonal antibodies against acidic phosphoproteins P0, P1 and P2 of eukaryotic ribosomes as functional probes. J. Biol. Chem., 265, 89–95. [PubMed] [Google Scholar]
- Uchiumi T., Hori,K., Nomura,T. and Hachimori,A. (1999) Replacement of L7/L12⋅L10 protein complex in Escherichia coli ribosomes with the eukaryotic counterpart changes the specificity of elongation factor binding. J. Biol. Chem., 274, 27578–27582. [DOI] [PubMed] [Google Scholar]
- van Agthoven A.J., Maassen,J.A. and Möller,W. (1977) Structure and phosphorylation of an acidic protein from 60S ribosomes and its involvement in elongation factor-2 dependent GTP hydrolysis. Biochem. Biophys. Res. Commun., 77, 989–998. [DOI] [PubMed] [Google Scholar]
- Verschoor A., Warner,J.R., Srivastava,S., Grassucci,R.A. and Frank,J. (1998) Three-dimensional structure of the yeast ribosome. Nucleic Acids Res., 26, 655–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagenknecht T., Grassucci,R. and Frank,J. (1988) Electron microscopy and computer image averaging of ice-embedded large ribosomal subunits from Escherichia coli. J. Mol. Biol., 199, 137–147. [DOI] [PubMed] [Google Scholar]
- Warner J.R. (1999) The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci., 24, 437–440. [DOI] [PubMed] [Google Scholar]
- Wilson K.S. and Noller,H.F. (1998) Mapping the position of translational elongation factor EF-G in the ribosome by directed hydroxyl radical probing. Cell, 92, 131–139. [DOI] [PubMed] [Google Scholar]
- Wool I.G., Gluck,A. and Endo,Y. (1992) Ribotoxin recognition of ribosomal RNA and a proposal for the mechanism of translocation. Trends Biochem. Sci., 17, 266–269. [DOI] [PubMed] [Google Scholar]