Abstract
A key stage during homologous recombination is the processing of the Holliday junction, which determines the outcome of the recombination reaction. To dissect the pathways of Holliday junction processing in a eukaryote, we have targeted an Escherichia coli Holliday junction resolvase to the nuclei of fission yeast recombination-deficient mutants and analysed their phenotypes. The resolvase partially complements the UV and hydroxyurea hypersensitivity and associated aberrant mitoses of an rqh1– mutant. Rqh1 is a member of the RecQ subfamily of DNA helicases that control recombination particularly during S-phase. Significantly, overexpression of the resolvase in wild-type cells partly mimics the loss of viability, hyper-recombination and ‘cut’ phenotype of an rqh1– mutant. These results indicate that Holliday junctions form in wild-type cells that are normally removed in a non-recombinogenic way, possibly by Rqh1 catalysing their reverse branch migration. We propose that in the absence of Rqh1, replication fork arrest results in the accumulation of Holliday junctions, which can either impede sister chromatid segregation or lead to the formation of recombinants through Holliday junction resolution.
Keywords: helicase/Holliday junction/recombination/RecQ/resolvase
Introduction
Homologous recombination fulfils a number of beneficial functions within the cell, including repairing double strand breaks (DSBs), directing the segregation of homologous chromosomes at meiosis I and reforming collapsed replication forks. However, if left unchecked, recombination can have deleterious consequences, for example recombination between repetitive elements can lead to chromosome rearrangements (e.g. deletions, duplications, translocations, etc.), and allelic recombination in somatic cells can result in loss of heterozygosity and so contribute to cellular carcinogenesis. Control of recombination is particularly pertinent during DNA replication where stalling or collapse of the replication fork, caused either by collisions with DNA lesions and nucleoprotein complexes, or limiting concentrations of nucleotides, exposes both single-stranded DNA (ssDNA) and DNA strand breaks to the cell’s recombination machinery.
Holliday junctions (HJs) are key intermediates of both homologous and site-specific recombination reactions. In homologous recombination, HJs are formed by strand invasion followed by strand exchange between recombining DNA molecules. This reaction is catalysed by the RecA/Rad51 family of proteins (for a review, see Baumann and West, 1998). Recently, it has been suggested that HJs may also form by the regression of stalled replication forks that involves the annealing of the two newly synthesized strands (Seigneur et al., 1998; Viguera et al., 2000). This reaction does not require a strand exchange protein like RecA/Rad51 and may occur spontaneously to relieve the superhelical tension that accumulates ahead of the replication fork.
Once formed, an HJ must be removed from DNA in order for the two recombining molecules to segregate successfully at cell division. HJs can be removed by targeted endonucleolytic cleavage, by branch migration to strand breaks and potentially by the action of topoisomerases. During homologous recombination, endonucleolytic cleavage resolves the HJ into classical crossover or non-crossover recombinant products. Branch migration to strand breaks can similarly generate recombinant DNAs, but may also abort the recombination reaction if the HJ is driven to breaks that were used to actuate strand exchange. This latter reaction is often called reverse branch migration. For HJs formed by replication fork regression, cleavage results in the collapse of the fork and the generation of a recombinogenic DNA end (Seigneur et al., 1998). Branch migration may also collapse a regressed fork if appropriate strand breaks are present, but equally may re-establish the fork by driving the reannealing of nascent with parental DNA strands. Such reverse branch migration may be an important mechanism for limiting unnecessary recombination.
Enzymes that process HJs in Escherichia coli have been identified and characterized in some detail (for a review, see Sharples et al., 1999). RuvAB and RecG are specialist DNA helicases that catalyse HJ branch migration, and RuvC and RusA are endonucleases that resolve HJs into recombinant products. Equivalent enzymes that function in the nuclei of eukaryotes have yet to be identified. For more than a decade, a number of laboratories have attempted to identify junction-processing enzymes in eukaryotes by looking for activities in fractionated cell-free extracts capable of branch migrating or resolving model HJs. These studies have detected resolvase activity from a number of sources including mammalian cells (Hyde et al., 1994), but as yet no nuclear acting enzyme has been formally identified.
To provide an alternative way of identifying pathways of recombination intermediate processing in eukaryotes, we have constructed a recombinant HJ resolvase that is targeted to the nuclei of eukaryotic cells. Using this resolvase, we have screened a range of Schizosaccharomyces pombe recombination/repair mutants for complementation or alteration of their mutant phenotypes. Here we report that the UV and hydroxyurea (HU) hypersensitivities of an rqh1– mutant are partially complemented by our recombinant HJ resolvase.
Rqh1 (also called Rad12 and Hus2) is a member of the RecQ subfamily of DNA helicases (Murray et al., 1997; Stewart et al., 1997; Davey et al., 1998). Named after the recQ gene in E.coli, this family is widespread and includes Sgs1 from Saccharomyces cerevisiae and BLM, WRN, RECQL, RECQL4 and RECQL5 from human (for a review, see Chakraverty and Hickson, 1999). Each family member consists of a common central domain of ∼600 amino acids that includes the seven conserved motifs of the DExH-box helicases, and, where determined, has a 3′–5′ polarity of DNA unwinding. ‘RecQ’ helicases also interact with topoisomerases in ways that are yet to be fully understood (Wu et al., 1999). The importance of the ‘RecQ’ helicases is exemplified by their involvement with human disease. Bloom’s (BS), Werner’s (WS) and Rothmund–Thomson syndromes, which are characterized variously by abnormal growth, immunodeficiency, cancer predisposition and premature ageing, are caused by defects in BLM, WRN and RECQL4, respectively (Ellis et al., 1995; Yu et al., 1996; Kitao et al., 1999).
Defects in ‘RecQ’ helicases typically result in elevated levels of recombination, and problems with DNA replication and chromosome segregation (Chakraverty and Hickson, 1999). In the case of rqh1– mutants, hyper-recombination is particularly marked following exposure to UV and depletion of deoxynucleotides by HU, which both perturb replication fork progression and result in S-phase arrest. rqh1– cells appear to recover normally from S-phase arrest and continue to complete bulk DNA synthesis. Despite this, a large percentage of cells are unable to segregate their chromosomes properly at division, resulting in unequal chromosome segregation and cells with the ‘cut’ (cell untimely torn) phenotype where chromosomes are bisected by the septum (Stewart et al., 1997).
Here we show that the partial complementation of the UV and HU hypersensitivities of an rqh1– mutant by our recombinant HJ resolvase correlates with a reduction in cells with the ‘cut’ phenotype. This indicates that HJs accumulate in rqh1– cells that, either directly or after further processing, impede the segregation of sister chromatids. High-level expression of the resolvase in wild-type S.pombe cells partly mimics the ‘cut’ and hyper-recombination phenotypes of an rqh1– mutant, indicating that HJs are formed during vegetative growth that would normally be removed in a non-recombinogenic way. These findings are rationalized by a model in which HJs, formed as a consequence of replication fork blockage, are removed by an Rqh1-dependent pathway. In the absence of Rqh1, HJs either remain as a physical link between sister chromatids, preventing their segregation, or are resolved to generate a DSB that is repaired by recombination. The applicability of this model to other ‘RecQ’ DNA helicases is discussed.
Results
A new system for probing pathways of recombination intermediate processing in eukaryotes
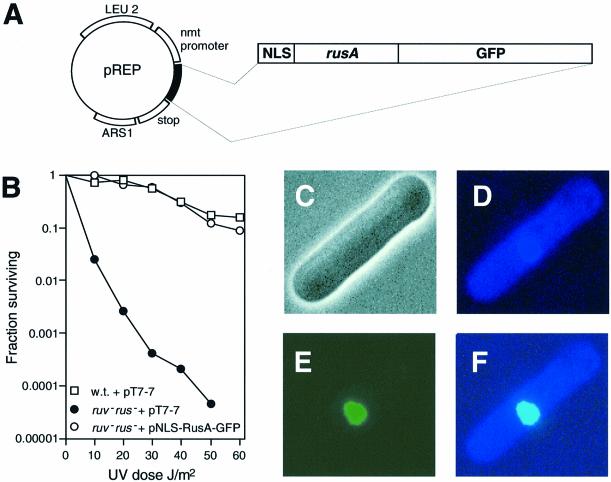
To establish a new way of identifying pathways of recombination intermediate processing in eukaryotes, we have developed a recombinant HJ resolvase that is targeted to the nuclei of eukaryotic cells. This allows us to detect genes involved in HJ processing by complementation or alteration of their mutant phenotypes. The E.coli RusA protein forms the basis of our recombinant resolvase. RusA is highly selective for HJs and cleaves them by introducing symmetrically related nicks into two strands of the same polarity immediately 5′ of CC dinucleotides at the junction crossover (Chan et al., 1997). To target RusA to eukaryotic nuclei, the sequence of the SV40 T-antigen nuclear localization signal (NLS) was fused immediately 5′ to the rusA initiation codon (Figure 1A). To be able to detect expression of NLS–RusA in vivo, the gene for green fluorescent protein (GFP) was fused in-frame to the 3′ end of rusA (Figure 1A). The resultant chimeric gene, when expressed at low levels in E.coli, fully complements the UV sensitivity of a resolvase-deficient strain, indicating that NLS–RusA–GFP is able to resolve HJs (Figure 1B).
Fig. 1. Construction of an HJ resolvase that is targeted to the eukaryotic nucleus. (A) Schematic of pREP-rus plasmid. (B) Effect of pNLS-RusA-GFP on survival of a ruv– rus– E.coli strain following UV irradiation. Survival is compared with wild-type and ruv– rus– strains transformed with the empty expression vector pT7-7. (C–F) Phase contrast and fluorescence microscopy images of a wild-type S.pombe cell transformed with pREP1-rus, grown in the absence of thiamine, and stained with DAPI. (C) Phase contrast image. (D) DAPI fluorescent image. (E) GFP image. (F) Merged image of (D) and (E).
For the studies presented here, the NLS–RusA–GFP gene has been cloned into S.pombe expression plasmids that utilize the thiamine-repressible nmt1 promoter. pREP1 contains a full-strength nmt1 promoter, whereas pREP41 and pREP81 contain attenuated versions. As an approximate guide, the derepressed activity of the pREP1 promoter is 100-fold more than that of its counterpart in pREP41 and 1000-fold more active than that in pREP81 (Forsburg, 1993). In each case, thiamine repression is not absolute. The pREP-NLS-RusA-GFP constructs are referred to as pREP(1, 41 or 81)-rus throughout. Schizosaccharomyces pombe cells transformed with pREP-rus, and grown in the absence of thiamine, exhibit subcellular green fluorescence that co-localizes perfectly with 4′,6-diamidino-2-phenylindole (DAPI)-stained nuclear DNA (Figure 1D–F). This shows that NLS–RusA–GFP is targeted efficiently to the nucleus of fission yeast.
NLS–RusA–GFP partially complements the HU hypersensitivity of an rqh1– mutant
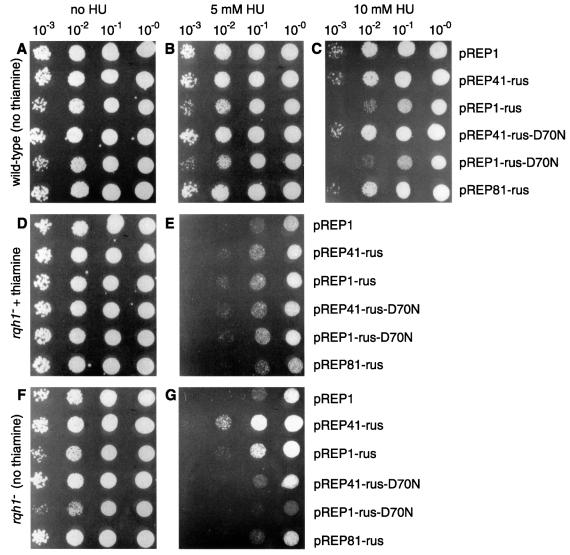
Using the pREP-rus constructs, we have begun to screen known S.pombe recombination/repair mutants systematically for complementation or alteration of their phenotypes. From this screen, partial complementation of the HU hypersensitivity of an rqh1– mutant was detected. To show this clearly, wild-type and rqh1 deletion (rqh1Δ) strains were transformed with the pREP-rus constructs and the empty pREP1 vector. The sensitivities of the transformants to HU were then compared by spotting serial dilutions of cells onto nutrient agar plates containing 0, 5 or 10 mM HU (Figure 2). As expected, the wild-type transformants show little sensitivity to 5 mM HU (Figure 2B), whereas the rqh1Δ transformants are hypersensitive (Figure 2E). However, when NLS–RusA–GFP is expressed, by the removal of thiamine from the medium, both the pREP1-rus and pREP41-rus rqh1Δ transformants show a marked increase in resistance (Figure 2G). This is not strain specific since similar results were obtained for the rqh1.r12 (rad12.502) strain that contains a point mutation in Rqh1’s putative ATP-binding site (data not shown).
Fig. 2. Effect of NLS–RusA–GFP on the HU sensitivity of an S.pombe rqh1– strain. (A–G) Wild-type (A–C) and rqh1– (D–G) strains, transformed with plasmids as indicated, were cultured in the presence (D and E) or absence (A–C and F–G) of thiamine, serially diluted and spotted onto appropriately supplemented EMM plates containing the indicated amounts of HU. In each case, the neat spot (10–0) represents 105 cells plated. The strains used were MCW45 and MCW7.
Complementation of the rqh1Δ strain’s sensitivity to HU is dependent on the level of NLS–RusA–GFP expression. The pREP41-rus level of NLS–RusA–GFP produces the greatest improvement in survival (>10-fold compared with the pREP1 control) (Figure 2G). In contrast, the pREP81-rus level produces no noticeable effect, and the pREP1-rus level has a general negative effect on viability (Figure 2A–C and F–G), which presumably offsets some of its benefits to the rqh1– mutant in the presence of HU. From these observations, we note that more complete complementation may be possible with NLS–RusA–GFP levels that are intermediate between those of pREP41-rus and pREP1-rus.
To establish whether NLS–RusA–GFP’s ability to complement rqh1– is dependent on its ability to resolve HJs, we altered codon 70 of the rusA component of our pREP-rus constructs from GAC (aspartate) to AAC (asparagine). This D70N mutation has previously been shown to abolish RusA’s ability to cleave HJs, but does not reduce its junction binding (Bolt et al., 1999). Both pREP41-rus-D70N and pREP1-rus-D70N fail to complement the HU hypersensitivity of the rqh1– mutant (Figure 2G). From these data, we conclude that NLS–RusA–GFP’s ability to complement rqh1– is dependent on its ability to resolve HJs.
NLS–RusA–GFP partially complements the UV hypersensitivity of an rqh1– mutant
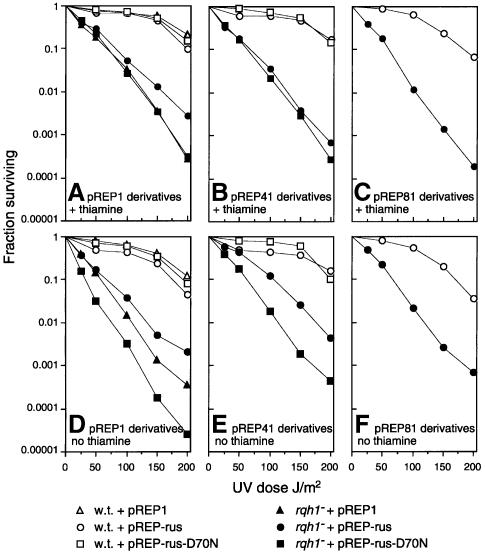
In addition to HU, rqh1– mutants are also hypersensitive to DNA-damaging agents such as UV light. To see whether an HJ resolvase can complement the defects that cause hypersensitivity to UV, we examined the effect that NLS–RusA–GFP has on the UV sensitivity of an rqh1Δ strain (Figure 3). Under repressed conditions, the pREP-rus plasmids have little effect on the sensitivity of rqh1– cells (Figure 3A–C), apart from pREP1-rus, which slightly improves survival, particularly at high UV doses (Figure 3A). This effect of pREP1-rus under repressed conditions is consistent with complementation by relatively low levels of NLS–RusA–GFP (∼20% of the pREP41 derepressed level). Under derepressed conditions, both pREP1-rus and pREP41-rus, but not pREP81-rus, give improvements in survival (Figure 3D–F). As with HU, the greatest benefit is seen with pREP41-rus, which reduces cell death by ∼10-fold. Again, the ability to complement depends on the catalytic competence of NLS–RusA–GFP since neither pREP1-rus-D70N nor pREP41-rus-D70N give any improvement in survival (Figure 3D and E). In fact, pREP1-rus-D70N produces quite a marked negative effect on the viability of rqh1Δ, especially upon irradiation (Figure 3D; data not shown). These results are similar to those obtained with HU and suggest that the underlying defect in an rqh1– mutant that causes sensitivity to UV is the same as that which causes sensitivity to HU.
Fig. 3. Effect of NLS–RusA–GFP on UV-irradiated wild-type and rqh1– cells. Wild-type and rqh1– cells, transformed with plasmids and cultured in the presence or absence of thiamine as indicated, were assayed for colony formation before and after UV irradiation. The fraction surviving is expressed relative to unirradiated cells.
NLS–RusA–GFP reduces the proportion of rqh1– cells that ‘cut’ due to exposure to HU
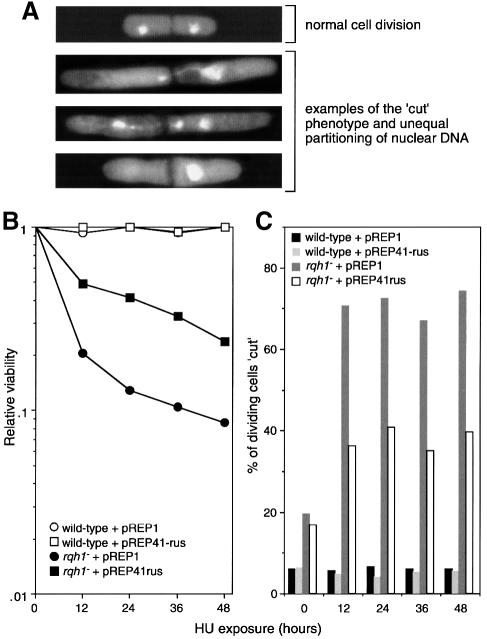
Exposure to HU, which perturbs DNA replication by depleting deoxynucleotides, or UV irradiation, which generates bulky lesions in DNA that impede the progress of the replication fork, cause S-phase arrest in S.pombe. This involves the cessation of DNA synthesis and the inhibition of mitosis by negative regulation of the cyclin-dependent kinase Cdc2 (Caspari and Carr, 1999). During the arrest, cells continue to grow and therefore display an elongated morphology. rqh1– cells exposed to either UV or HU undergo a seemingly normal S-phase arrest, re-enter the cell cycle at the same time as wild-type cells and proceed to complete bulk DNA synthesis (Stewart et al., 1997). Despite this, a large proportion of rqh1– cells undergo a subsequent aberrant mitosis where they fail to segregate their sister chromosomes properly. Such cells are described as ‘cut’ and are distinguished by their septa, which often bisect the single nucleus, or by a division that creates one anucleate daughter (Figure 4A). These defects correlate with the hypersensitivity of rqh1– mutants to HU and UV (Stewart et al., 1997).
Fig. 4. Effect of NLS–RusA–GFP on the ‘cut’ phenotype of rqh1– cells. (A) Examples of rqh1– cells undergoing normal cell division (in the absence of HU) and aberrant divisions (after exposure to HU). Images are of DAPI-stained cells viewed by fluorescence microscopy. DAPI stains nuclear DNA brightly. It also stains the rest of the cytoplasm weakly, allowing visualization of septa as dark lines across the cells. (B) Effect of NLS–RusA–GFP on the HU sensitivity of an rqh1– mutant. The strains were MCW45 and MCW7 transformed with plasmids as indicated. Relative viability was calculated by dividing the number of viable cells after the addition of HU by the number that were viable before its addition. (C) Effect of NLS–RusA–GFP on the proportion of dividing cells that display the ‘cut’ phenotype. Cells from the cultures described in (B) were scored for the ‘cut’ phenotype. See Materials and methods for further details.
To see whether NLS–RusA–GFP’s ability to complement the HU hypersensitivity of rqh1– correlates with a reduction in aberrant mitoses, wild-type and rqh1Δ cells, transformed with pREP1 and pREP41-rus, were incubated in liquid media containing HU and analysed at intervals for viability and the percentage of dividing cells showing the ‘cut’ phenotype (Figure 4). Wild-type cells were resistant to HU and showed no decline in survival over a 48 h incubation (Figure 4B). In accord with this, very few dividing wild-type cells exhibit the ‘cut’ phenotype (Figure 4C). In contrast, rqh1Δ cells show a marked decline in survival when incubated in HU, which correlates with an increase in the percentage of dividing cells that ‘cut’ (Figure 4B and C). Interestingly, even in the absence of HU, quite a large percentage of dividing rqh1Δ cells ‘cut’, and this correlates with a general reduction in viability (Figure 4C; data not shown). As in the spot tests in Figure 2, the expression of NLS–RusA–GFP from pREP41-rus results in a significant improvement in survival of rqh1Δ cells exposed to HU (Figure 4B). This improvement correlates well with the reduction in the percentage of these cells that ‘cut’ upon division (Figure 4C) and was not observed with pREP41-rus-D70N (data not shown). A similar correlation between improved UV resistance and a reduction in the number of ‘cut’ cells with pREP41-rus in rqh1– was also found (data not shown). From these data, we conclude that the HU and UV hypersensitivity of rqh1– cells relates directly to a reduced ability to segregate sister chromatids properly. Furthermore, the ability to complement these defects by expression of a resolvase indicates that the sister chromatids are unable to segregate because they are linked via HJs.
pREP41-rus does not suppress the hyper-recombination of rqh1– cells
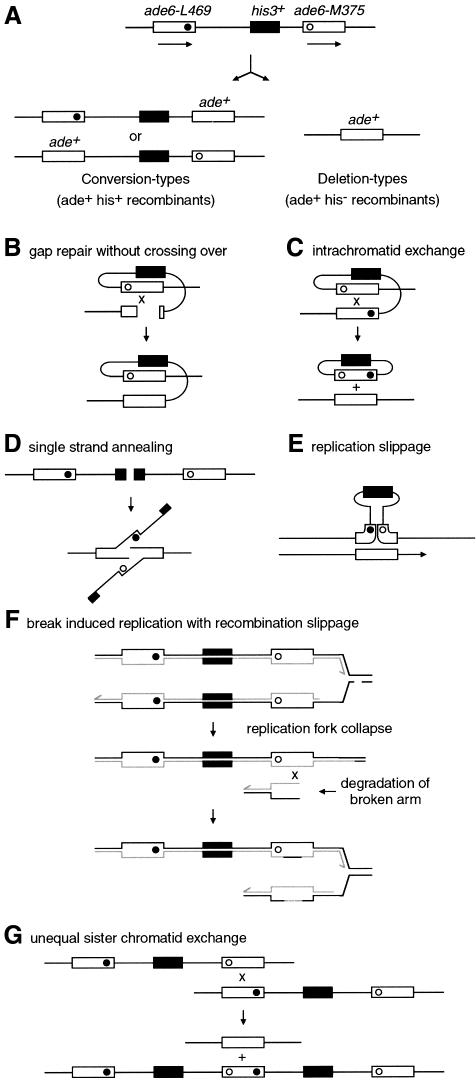
Like other members of the ‘recQ’ family, rqh1– cells display a hyper-recombination phenotype (Stewart et al., 1997). Hyper-recombination may be explained by the resolution of some of the HJs that evidently accumulate in rqh1– cells following S-phase arrest. If this is true, then the addition of an HJ resolvase should not affect the frequency of recombinant formation per viable cell in an rqh1– strain. To test this, and to see what other effects a resolvase might have on mitotic recombination, we used strains containing a non-tandem direct repeat of ade6– heteroalleles to measure Ade+ recombinant frequencies (Figure 5). Such intrachromosomal direct repeats can recombine to generate recombinant products by multiple, and sometimes overlapping, pathways (Klein, 1995; Paques and Haber, 1999). In our case, a functional his3+ gene placed between the repeats enables us to distinguish Ade+ recombinants that are either His– (deletion types) or His+ (conversion types), which arise predominantly via different pathways (Figure 5).
Fig. 5. A non-tandem direct repeat of heteroalleles for measuring recombination. (A) Schematic of intrachromosomal recombination substrate and recombinant products. Solid and open circles represent the ade6-L469 and ade6-M375 mutations, respectively. (B–G) Some of the possible mechanisms of deletion- and conversion-type recombinant formation. (B) DSB/gap repair involving intrachromatid recombination. If the recombination intermediate is resolved to give a non-crossover product, then this can generate an ade+ his+ conversion type. (C) Intrachromatid exchange between the direct repeats generates a single copy of the gene in the chromosome and an excised circle bearing the second copy together with the his+ gene. (D) Single strand annealing (SSA) can repair a DSB between two direct repeats by resection of the broken ends to expose two complementary single strands that anneal. Repair involves removal of the non-homologous 3′ ends. (E) Replication slippage involving detachment of a nascent strand and mispairing within repeats upon reattachment. (F) Break-induced replication (BIR). This may occur when a replication fork collapses after encountering a single strand break in the template DNA. Recombination can restore the fork by reattaching the broken end. If the broken arm of the fork is degraded, then recombination may occur between non-equivalent repeats, resulting in the formation of deletion-type recombinants. In this version of the BIR model, the reformation of the replication fork is accompanied by the formation and resolution of an HJ (not shown). In other versions, replication is envisaged to proceed via a migrating bubble or D-loop (Paques and Haber, 1999). (G) Unequal sister chromatid exchange can result in a deletion on one chromatid and a triplication on the other chromatid. Such unequal or slipped recombination can generate conversion-type recombinants if recombination intermediates are processed without crossing over.
Wild-type and rqh1Δ strains, containing the ade6– heteroallelic repeat, were transformed with the pREP- rus plasmids, and spontaneous and UV-induced Ade+ recombinant frequencies were determined under repressed and derepressed conditions (Table I). Wild-type cells containing pREP1, in both the presence and absence of thiamine, yield spontaneous Ade+ recombinants at a frequency of ∼2–3 × 10–4 per viable cell (Table I). Approximately 30% of these recombinants are conversion types and 70% are deletion types (Figure 6B). UV irradiation, at 50 J/m2, produces no significant increase in the total frequency of Ade+ recombinants, but does abolish the bias in favour of deletion types such that ∼50% of recombinants are now conversion types (Figure 6B; Table I). At higher UV doses there is a marked increase in the total number of recombinants, with the majority of extra recombinants being conversion types (data not shown).
Table I. Spontaneous and UV-induced mitotic recombination frequencies.
| Thiamine | UVa | Mean frequency of Ade+ recombinantsper 104 viable cells |
|||
|---|---|---|---|---|---|
| Deletiontype (His–) | Conversiontype (His+) | Total(His– + His+) | |||
| Wild type | |||||
| pREP1 | – | – | 1.97 | 0.72 | 2.69 |
| + | – | 1.73 | 0.75 | 2.48 | |
| – | + | 1.12c | 1.09b | 2.21 | |
| + | + | 1.52 | 1.71b | 3.23 | |
| pREP41-rus | – | – | 2.13 | 0.74 | 2.87 |
| + | – | 1.44 | 0.76 | 2.20 | |
| – | + | 1.33c | 1.12b | 2.45 | |
| + | + | 0.87c | 1.53b | 2.40 | |
| pREP1-rus | – | – | 4.63d | 1.40 | 6.03d |
| + | – | 2.43 | 0.76 | 3.19 | |
| – | + | 5.60e | 1.57 | 7.17e | |
| + | + | 2.78 | 1.43b | 4.21 | |
| pREP1-rus-D70N | – | – | 2.45 | 0.90 | 3.35 |
| + | – | 1.60 | 0.63 | 2.23 | |
| – | + | 4.76f | 5.81g | 10.57e | |
| + | + | 1.26 | 3.52b | 4.78 | |
| rqh1– | |||||
| pREP1 | – | – | 5.24h | 0.91 | 6.15h |
| + | – | 4.25h | 1.66h | 5.91h | |
| – | + | 23.24h | 9.75h | 32.99h | |
| + | + | 19.84h | 11.35h | 31.19h | |
| pREP41-rus | – | – | 6.45 | 1.50 | 7.95 |
| + | – | 5.43 | 0.99 | 6.42 | |
| – | + | 24.96 | 10.40 | 35.36 | |
| + | + | 26.29 | 11.77 | 38.06 | |
| pREP1-rus | – | – | 12.50i | 1.48 | 13.98i |
| + | – | 7.36 | 2.01 | 9.37 | |
| – | + | 44.50e | 8.85 | 53.35e | |
| + | + | 26.32 | 10.32 | 36.64 | |
| pREP1-rus-D70N | – | – | 7.03 | 1.57 | 8.60 |
| + | – | 5.72 | 1.11 | 6.83 | |
| – | + | 53.73e | 17.84j | 71.57e | |
| + | + | 20.15 | 8.27 | 28.42 | |
a50 J/m2.
bIndicated value significantly (p <0.05) greater than –UV equivalent.
cIndicated value significantly (p <0.01) less than –UV equivalent.
dIndicated value significantly (p <0.0005) greater than pREP1, pREP1-resolvaseD70N and + thiamine equivalents.
eIndicated value significantly (p <0.0005) greater than pREP1 and + thiamine equivalents.
fIndicated value significantly (p <0.02) greater than pREP1 and + thiamine equivalents.
gIndicated value significantly (p <0.01) greater than all other wild-type conversion-type values.
hIndicated value significantly (p <0.0005) greater than wild-type equivalent.
iIndicated value significantly (p <0.03) greater than pREP1, pREP1-resolvaseD70N and + thiamine equivalents.
jIndicated value significantly (p <0.04) greater than all other rqh1– conversion-type values.
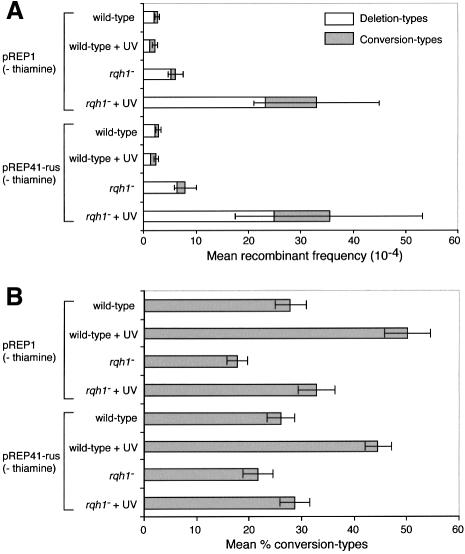
Fig. 6. Spontaneous and UV-induced recombination in wild-type and rqh1– cells, and the effect of pREP41-rus. (A) Mean frequency of deletion- and conversion-type recombinants. (B) The percentage of total recombinants that are conversion types. Error bars represent the 95% confidence limits.
By measuring the formation of Ade+ recombinants from diploid strains that were heterozygous at the ade6 locus, Stewart et al. (1997) previously showed that rqh1– cells have a hyper-recombination phenotype after exposure to HU. Using our recombination substrate, we have found that rqh1Δ cells also exhibit significantly higher frequencies of both spontaneous (∼2-fold higher) and UV-induced (∼10-fold higher) recombination than wild type (Figure 6A; Table I). Moreover, the majority of the extra recombinants are deletion types such that, even upon UV induction, only ∼30% of the total recombinants are conversion types (Figure 6B). These data indicate that Rqh1 is required both under normal growth conditions and following DNA damage to suppress recombination, particularly that which generates deletion-type recombinants.
The addition of pREP41-rus has no significant effect on the recombination frequencies of wild-type or rqh1Δ cells (Figure 6; Table I). This is consistent with the prediction that complementation of the UV sensitivity and ‘cutting’ of rqh1– cells does not involve suppression of recombination.
Effects of high-level expression of NLS–RusA–GFP on wild-type and rqh1– cells
During the course of these studies, we noted that both pREP1-rus and pREP1-rus-D70N have a negative effect on the viability of wild-type and rqh1– strains that is exacerbated further by UV or HU. Quantitation of this effect shows that under normal growth conditions both plasmids reduce viability by ∼30–40% in either wild-type or rqh1– strains when compared with pREP1, pREP41-rus or pREP41-rus-D70N equivalent cultures (data not shown). Reduced viability correlates with an increase in the frequency of cells with the ‘cut’ phenotype, e.g. ∼50% of wild-type cells containing either pREP1-rus or pREP1-rus-D70N in the absence of thiamine show an aberrant mitosis compared with <10% for those containing pREP1 (data not shown).
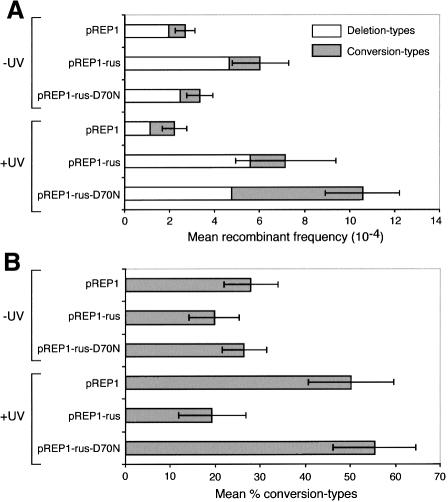
The reduced viability and associated increased ‘cut’ phenotype mediated by high-level expression of catalytically active and inactive NLS–RusA–GFP are reminiscent of the rqh1– phenotype. To see whether, like rqh1–, NLS–RusA–GFP also affected recombination, Ade+ recombinant frequencies in wild-type and rqh1– cells, containing the ade6– heteroallelic repeat, and pREP1, pREP1-rus and pREP1-rus-D70N, were compared (Table I; Figure 7). In the absence of UV, pREP1-rus stimulates the formation of recombinants by up to 2-fold in both wild-type and rqh1– cells, with the majority of extra recombinants being deletion types (Figure 7; Table I). Much of this stimulation of recombination is dependent on junction resolution since a statistically significant increase in recombination is not observed with pREP1-rus-D70N (Figure 7; Table I). In contrast, both pREP1-rus and pREP1-rus-D70N stimulate, by 2- to 5-fold, the formation of deletion-type recombinants in wild-type and rqh1– cells after exposure to UV (Figure 7; Table I). However, whereas pREP1-rus has no effect on the level of conversion-type recombinants under these conditions, pREP1-rus-D70N increases conversion types by up to 5-fold over the pREP1 level (Figure 7; Table I).
Fig. 7. The effect of high-level expression of NLS–RusA–GFP on spontaneous and UV-induced recombination in wild-type cells. (A) Mean frequency of deletion- and conversion-type recombinants. (B) The percentage of total recombinants that are conversion types. Error bars represent the 95% confidence limits.
To interpret these results, it must be remembered that RusA is highly specific for HJs, but is unable to resolve every junction that it binds to due to its dependence on specific sequences at the junction crossover for cleavage (Chan et al., 1997). Furthermore, once bound, it can prevent other proteins from processing the junction by physical exclusion or by holding the junction in a conformation that is not recognized (Bolt et al., 1999). With this in mind, the key conclusion from the above data is that HJs are formed, both during normal growth and following exposure to UV or HU, that would normally be removed by a non-recombinogenic mechanism. We suspect that NLS–RusA–GFP competes with this pathway such that many HJs are bound and remain unprocessed, thereby preventing sister chromatid segregation. However, some of the HJs bound by the catalytically active NLS–RusA–GFP may be resolved, which would promote recombinant formation either directly or by collapsing replication forks. The stimulation of UV-induced recombination by the D70N mutant is harder to explain. Holding on to the HJ and delaying its normal processing could provide more time for heteroduplex DNA to be utilized for the formation of conversion-type recombinants. The greater stability of the RusA D70N–HJ complex compared with that formed by wild-type RusA (Bolt et al., 1999) may explain its greater ability to promote this type of recombinant. Holding on to the HJ may also promote the formation of deletion-type recombinants either by enabling an endogenous nuclease to resolve the HJ or by causing an aberrant mitosis that could expose DNA fragments for recombination in surviving daughter cells.
Discussion
We present a novel system for dissecting pathways of recombination intermediate processing in eukaryotes. It makes use of a recombinant HJ resolvase that is targeted to the eukaryotic nucleus. This resolvase is a potent reagent for the identification of genes that are required for processing recombination intermediates in mitotic and meiotic cells by complementation or modification of their mutant phenotypes. In this study, the system is used to reveal that Rqh1, a member of the RecQ subfamily of DNA helicases, is required for processing HJs in mitotic S.pombe cells. This not only demonstrates the utility of the system, but is also an important advance in our understanding of the biological roles of a helicase whose human homologues are associated with disease.
The partial complementation of rqh1– mutant phenotypes by an HJ resolvase
Rqh1, like other members of the RecQ subfamily of DNA helicases, plays an important role in controlling recombination. Various laboratories have shown that rqh1– mutants are hypersensitive to UV and HU (Murray et al., 1997; Stewart et al., 1997; Davey et al., 1998). This sensitivity correlates with an increased frequency of recombinant formation and a tendency for cells to ‘cut’ during mitotic growth (Stewart et al., 1997). We have observed that rqh1– cells also exhibit reduced viability during normal growth that is associated with increased ‘cutting’ and a slight hyper-recombination phenotype. However, our main finding is that the expression of a catalytically active HJ resolvase partially complements both the UV and HU hypersensitivity of rqh1– cells. Moreover, complementation correlates with a reduction in the frequency of cells that ‘cut’ upon division, but hyper-recombination is not suppressed.
The most economical explanation for these results is that UV and HU, by perturbing DNA replication, stimulate recombination. Rqh1 acts as an anti-recombinase that controls much of this attempted recombination. Without Rqh1, recombination is allowed to proceed, resulting in increased recombinant formation. However, many of these recombination events fail to be processed through to viable recombinant products and get stuck at an intermediate stage where DNA molecules remain linked via HJs. In the case of intersister chromatid recombination, unresolved HJs prevent the sister chromatids from segregating properly during mitosis, resulting in reduced viability. The addition of an HJ resolvase overcomes this problem by unlinking the sister chromatids. So, improved viability comes not from suppressing recombination, but by converting more recombination intermediates into viable recombinant products.
Why only partial complementation?
If the UV and HU sensitivity of rqh1– cells solely reflects a problem with segregating sister chromatids that are linked via HJs, then it might be expected that an HJ resolvase would fully complement these phenotypes, yet only partial complementation is observed. The reason for this is uncertain, but the following are four possible explanations. (i) Plasmid copy number is highly variable in S.pombe such that the level of expression of NLS–RusA–GFP is different between cells. Therefore, since the level of resolvase is critical for complementation, the degree of complementation represents the average for a given population of cells. (ii) NLS–RusA–GFP is unable to resolve a subset of HJs due to its requirement for specific sequences at the junction crossover to catalyse strand cleavage (Chan et al., 1997). (iii) Not all sister chromatids are linked via HJs; other recombination intermediates such as D-loops could also prevent proper segregation. (iv) Rqh1 is required for other functions in addition to preventing the accumulation of HJs, e.g. Rqh1, by exposing regions of ssDNA to a strand exchange protein (e.g. Rhp51), could promote certain recombination reactions required for the tolerance or repair of DNA damage. Such a role is envisaged for RecQ in E.coli, which is believed to function with RecJ, a 5′–3′ single-stranded exonuclease, to expose ssDNA onto which the strand exchange protein RecA can nucleate (Harmon and Kowalczykowski, 1998; Courcelle and Hanawalt, 1999). Rqh1 may also function with topoisomerase III (Topo III) in promoting both the disruption of recombination intermediates and decatenation of late stage replicons (Goodwin et al., 1999).
How does Rqh1 limit the accumulation of HJs?
The data presented here indicate that HJs accumulate in rqh1– cells. Rqh1 could limit this accumulation by: (i) preventing recombination from initiating; (ii) aborting recombination once it has initiated; or (iii) resolving HJs to recombinant products. Since rqh1– mutants display hyper-recombination, we can probably discount the latter possibility. Our further speculations rely heavily on the characterization of other ‘RecQ’ helicases. Sgs1 has been shown to interact both genetically and physically with Topo III and, since top3– mutants display hyper-recombination, it has been speculated that the Sgs1–Topo III complex acts as a ‘eukaryotic reverse gyrase’ that suppresses recombination by a positive supercoiling activity (Gangloff et al., 1994). Such a model may also apply to Rqh1 since it too has a genetic interaction with Topo III (Goodwin et al., 1999). Another idea for how ‘RecQ’ helicases could prevent recombination initiation comes from the observation in E.coli that RecQ together with RecJ selectively degrades the nascent lagging strand at stalled replication forks (Courcelle and Hanawalt, 1999). Courcelle and Hanawalt suggest that this promotes RecA binding, which in turn protects the fork from further processing. Alternatively, it could limit the possibility that replication fork regression would generate an HJ by the annealing of nascent strands (see below). Recent studies of BLM, Sgs1 and RecQ indicate that this family of helicases will unwind a wide range of DNA substrates, including three- and four-way junctions (Harmon and Kowalczykowski, 1998; Bennett et al., 1999; Chakraverty and Hickson, 1999). Furthermore, E.coli RecQ has been shown to disrupt recombination intermediates formed by RecA in reconstituted strand exchange reactions (Harmon and Kowalczykowski, 1998), and BLM has been shown to exhibit some specificity for the HJ and to catalyse branch migration of HJs formed by RecA in vitro (Karow et al., 2000). These results suggest that Rqh1 could abort recombination by unwinding the invading DNA strand (D-loop) and/or by catalysing the reverse branch migration of an HJ. Alternatively, a propensity for unwinding DNA junctions may only reflect a role for Rqh1 in removing secondary structures from DNA to aid replication fork progression (Cromie et al., 2000).
Evidence for other pathways of non-recombinogenic HJ processing
A key result in this study is that high-level expression of NLS–RusA–GFP in wild-type cells partly mimics the reduced viability, ‘cut’ phenotype and hyper-recombination of an rqh1– mutant. This dominant-negative effect indicates that HJs are formed, even in the presence of Rqh1, that are normally removed in a non-recombinogenic way. Clearly, Rqh1 is a prime candidate for catalysing the non-recombinogenic removal of HJs by catalysing their reverse branch migration. However, the observation that NLS–RusA–GFP also stimulates recombination in an rqh1– mutant indicates that there must be other pathways for the non-recombinogenic processing of HJs, although these are clearly insufficient for dealing with the extra accumulation of HJs in rqh1– cells.
A hypothetical model for Rqh1 controlling recombination resulting from problems during DNA replication
Rqh1 appears to control recombination that arises predominantly from problems during DNA replication. A connection with DNA replication is indicated by several observations: (i) rqh1– mutants are hypersensitive to HU; (ii) rqh1– mutants are only more sensitive to UV than are wild type during S-phase; (iii) rqh1– mutants show synthetic lethality with mutants that are defective in components of the replicative apparatus that are involved in chain elongation; and (iv) Rqh1 is required for the Cds1-dependent pathway for recovery from DNA damage that is specific to S-phase (Murray et al., 1997; Stewart et al., 1997). Furthermore, it has been shown that, at least in S.cerevisiae, UV and HU depend on DNA replication to stimulate recombination (Galli and Schiestl, 1995, 1996).
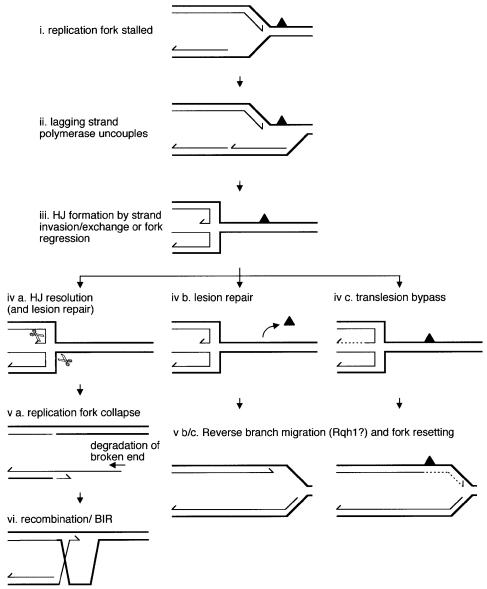
Growing evidence indicates that it is a common occurrence for a replication fork to stall or collapse during DNA synthesis (Kuzminov, 1995; Rothstein et al., 2000). Such events are precipitated by collision with other nucleoprotein complexes (e.g. transcription and repair complexes), encounter with lesions, breaks or secondary structure in the helix, and exhaustion of the raw materials for new synthesis. Of these events, only encounter with a strand break will cause the immediate collapse of a replication fork, the detached arm, with its exposed end, being highly recombinogenic. Salvage of this situation would depend on recombination ‘reattaching’ the broken arm of the fork. A stalled replication fork may also suffer collapse. This may occur by the resolution of an HJ formed either from a strand invasion reaction catalysed by a RecA/Rad51-like protein or by the regression of the replication fork such that the complementary nascent strands anneal (Figure 8) (Seigneur et al., 1998). At least in bacteria, which depend on bidirectional replication from a single origin, it may be necessary to collapse a stalled fork in order to complete synthesis of the chromosome. This is because the resultant recombination would involve strand exchange intermediates (D-loops) that can act as sites for the reassembly of replisomes (Liu and Marians, 1999). However, in eukaryotes, replication occurs from multiple origins, with forks converging on each other. Therefore, it may not be so critical to reassemble replisomes at stalled forks since any shortfall in replication could be compensated by the opposing fork. In fact, it may be quite hazardous for eukaryotes to provoke break-induced replication (BIR) unnecessarily since its associated recombination runs the risk of being illegitimate, causing chromosome deletions, duplications or other potentially harmful rearrangements. This is especially true in eukaryotes whose genomes contain more repetitive elements than those of bacteria and are therefore more prone to illegitimate recombination. Regression of the replication fork, however, may be desirable in order to provide room for the repair of blocking lesions or to enable translesion synthesis by template switching (Figure 8). Rqh1 may control this necessary event by catalysing the reverse branch migration of HJs to prevent their resolution and consequent risk of illegitimate recombination. Such reverse branch migration would also reset the replication fork, which could either act as a termination point for the opposing fork, or continue synthesis if the replisome can reassemble.
Fig. 8. Model for Rqh1 controlling recombination at stalled replication forks. In this version of the model, the replication fork is blocked by a UV photoproduct (solid triangle) in the leading strand template. The lagging strand polymerase uncouples from the leading strand polymerase and continues synthesis past the lesion site (Cordeiro-Stone et al., 1997). An HJ is formed either by a strand invasion reaction catalysed by a Rad51-like protein bound to the single-stranded region exposed on the leading strand template or by regression of the fork. Fork regression or strand exchange may provide room for repair enzymes to operate. Alternatively, the leading strand may bypass the lesion by using the extended lagging strand as a template (dashed line) (Higgins et al., 1976). Reverse branch migration of the HJ would re-establish the replication fork, which could continue synthesis unimpeded. Resolution of the HJ collapses the replication fork and promotes BIR.
In this model, the extra deletion-type recombinants that we observe in rqh1– cells would most probably derive from recombination or replication slippage during BIR, whereas the extra conversion-type recombinants could arise from increased strand exchange or fork regression, both involving slippage, at stalled replication forks. The fact that an HJ resolvase promotes predominantly deletion-type recombinants is also consistent with it stimulating BIR by collapsing replication forks, as shown in Figure 5F. This is similar to what happens in E.coli where DSBs, formed at stalled replication forks by RuvABC, can result in deletions (Bierne et al., 1997; Michel et al., 1997; Seigneur et al., 1998). Of course, if two converging forks collapse close to each other within the heteroallelic direct repeat substrate, then both conversion- and deletion-type recombinants may arise during the repair of the resultant DSB via a variety of different intra- or interchromatid processes (Figure 5).
Increasing the frequency of replication fork collapse is not necessarily the only way that recombination could be stimulated in rqh1– cells. Lesion-containing single strand gaps, generated by the replisome avoiding lesions in the lagging strand template by skipping onto the next Okazaki fragment, are recombinogenic especially if converted into a DSB (West et al., 1982). Furthermore, ends created by the fragmentation of DNA during aberrant mitoses may promote recombination in surviving daughter cells. This may also explain why pREP1-rus-D70N promotes the formation of deletion-type recombinants in UV-irradiated wild-type and rqh1– cells (Table I).
HJ processing by other ‘RecQ’ helicases
The model we propose above for Rqh1 may be generally applicable. Like rqh1– cells, recQ–, sgs1–, BS and certain WRN cells display hyper-recombination and, at least in the case of BS cells, this includes increased sister chromatid exchange, particularly during S-phase when the cells are exposed to bromodeoxyuridine (Kuhn and Therman, 1986; Cheng et al., 1990; Watt et al., 1996; Hanada et al., 1997). Furthermore, the hyper-recombination of sgs1– cells is largely suppressed by expression of either BLM or WRN, suggesting that the mechanisms for limiting recombination are conserved (Yamagata et al., 1998). The link with DNA replication is also a conserved feature of ‘RecQ’ family members. Phenotypes such as retarded replication fork progression, extended S-phase, abnormal replication intermediates and sensitivity to S-phase-specific agents, that are variously associated with different ‘recQ’ mutants, may, at least in some cases, be explained by difficulties in recovering from replication fork stalling or collapse (Chakraverty and Hickson, 1999). Problems with chromosome segregation are also seen in some ‘recQ’ family members, which may be a consequence of HJ accumulation as in rqh1– cells, e.g. sgs1– cells display mitotic/meiotic chromosome non-disjunction (Watt et al., 1995).
Perhaps the clearest evidence in eukaryotes that HJ accumulation and replication fork stalling and collapse are associated comes from studies of the rDNA repeat array in S.cerevisiae. Here Fob1 creates a unidirectional block to replication fork progression that induces recombination (Kobayashi et al., 1998; Defossez et al., 1999). These events correlate with the direct detection of HJs in the rDNA array that reach their highest level during S-phase (Zou and Rothstein, 1997). Interestingly, HJ formation depends very much on Rad52, but not on the strand exchange protein Rad51 (Zou and Rothstein, 1997). This suggests that replication forks that are blocked by Fob1 regress to form HJs. The ability of Rad52 to promote the annealing of complementary strands may aid this reaction (Mortensen et al., 1996). Furthermore, Rad52’s ability to bind and protect DNA ends may also prevent the newly annealed nascent strands from being degraded (Van Dyck et al., 1999). Resolution of the HJ would collapse the fork, creating a recombinogenic end. One consequence of recombination in the rDNA array is the formation of extachromosomal rDNA circles (ERCs). The accumulation of ERCs, at least in S.cerevisiae, may promote ageing by titrating essential replication and transcription factors (Sinclair and Guarente, 1997). Interestingly, sgs1– mutants accumulate ERCs more rapidly than wild-type cells and have shorter life spans (Sinclair and Guarente, 1997; Sinclair et al., 1997). By catalysing the reverse branch migration of HJs, Sgs1 may limit recombination at stalled replication forks and thereby prolong life.
Conclusion
The spectrum of mutant phenotypes, interactions with other proteins and in vitro versatility of ‘RecQ’ family members are symptomatic of multiple roles in vivo. We have shown that, for at least one ‘recQ’ family member, mutant phenotypes can be partly explained by the accumulation of HJs that interfere with chromosome segregation. It will be interesting to use our resolvase in other ‘recQ’ mutant cells to establish whether this is a common problem. Such insight will aid dissection of the multifarious phenotypes of ‘recQ’ mutants, which in turn should increase our understanding of the defects that underlie the genetic instability of diseases such as Bloom’s, Werner’s and Rothmund–Thomson’s syndromes.
Materials and methods
Strains
Escherichia coli K-12 strains AB1157 (wild-type) and AM888 (ΔruvAC65 ΔrusA::kan) are described in Whitby and Dixon (1997). For testing HU and UV sensitivity and assessing ‘cut’ phenotypes, the S.pombe strains used were MCW45 (wild type; h+ leu1-32 his3-D1), MCW7 (rqh1Δ::ura4+ h+ ura4-D18 leu1-32) and MCW2 (rad12-502 h+ ura4D-18 leu1-32 his3-D1). The strains designated MCW2 and MCW7 were a gift from A.Carr (MRC Cell Mutation Unit, University of Sussex, UK). The strains used to measure recombination frequencies were FO163 (wild type; h– ura4D-18 leu1-32 his3-D1 ade6-M375 int::pUC8/his3+/ade6-L469) and FO506 (rqh1Δ::ura4+ derivative of FO163).
Plasmids
To construct the NLS–RusA–GFP chimeric gene, a PCR-based strategy was used to clone rusA with the SV40 T-antigen NLS sequence fused in-frame to its 5′ end. The GFP gene was subsequently subcloned into the NLS-rusA plasmids. The D70N mutation was introduced by site-directed mutagenesis of the pREP-rus constructs using a ‘QuikChange’ site-directed mutagenesis kit (Stratagene). Further details of plasmid constructions are available on request.
Media
LB broth and agar supplemented with 125 µg/ml ampicillin were used for bacterial culture. Media for S.pombe have been described (Gutz et al., 1974). The complete medium was yeast extract medium (YES) and the minimal medium was Edinburgh minimal medium (EMM), each supplemented with appropriate amino acids. Thiamine was added to media at 1.35 µg/ml where appropriate. The medium used to select ade+ recombinants was YES lacking adenine and supplemented with 200 µg/ml guanine to prevent adenine uptake. HU was added to media as indicated.
Spot assays
Cell cultures growing exponentially in selective minimal medium were adjusted to a density of 1 × 107 cells/ml and then serially diluted as indicated. Aliquots (10 µl) of each dilution were spotted onto selective minimal medium containing HU as indicated. Plates were typically incubated for 4 days at 30°C before being photographed. All spot tests were repeated at least three times in independent experiments to ensure reproducibility.
HU sensitivity and ‘cut’ phenotype
Strains were grown at 30°C in selective medium lacking thiamine until early log phase. A known number of cells was then assessed for viability before and at time points after the addition of 10 mM HU by plating onto selective minimal medium. Cells were also fixed, stained with DAPI and analysed for the number of dividing cells showing the ‘cut’ phenotype as classified in Figure 3A. Only cells clearly displaying a septum were counted as dividing, and at least 200 of these were assessed randomly for each time point. All data points represent the mean of at least two independent experiments.
UV sensitivity
Sensitivity of E.coli strains to UV was measured as described previously (Whitby and Dixon, 1997). For S.pombe, a known number of cells were plated from exponentially growing cultures and UV irradiated. All data points represent the mean of at least two independent experiments.
Recombination assay
Mitotic recombination was assayed by the recovery of Ade+ recombinants from strains containing the intrachromosomal recombination substrate shown in Figure 5A. Strains were grown on selective minimal medium with and without thiamine for 4–5 days at 30°C. For each assay, five single colonies of each strain were plated at a density of between 104 and 106 cells per plate onto medium selective for Ade+. Plates were unirradiated or UV irradiated to select for spontaneous and UV-induced recombinants, respectively. Appropriately diluted cells were also plated onto complete medium and treated as above to determine the relevant cell titre. After 5 days incubation at 30°C, the number of recombinants and cell titre were determined. The Ade+ recombinants on the selective plates were then replicated onto appropriate minimal medium to determine the proportion of conversion-type (Ade+ His+) and deletion-type (Ade+ His–) recombinants. Each recombination frequency in Table I represents the mean value from 3–9 independent assays (i.e. 15–45 separate colonies). Two-sample t-tests were used to analyse the recombinant frequencies for all individual colonies and so assess the statistical significance of differences in recombination frequencies between given strains.
Acknowledgments
Acknowledgements
We thank David Sherratt for his advice and support. This work was supported by a project grant from the Medical Research Council and latterly by a Wellcome Trust Senior Research Fellowship awarded to M.C.W.
References
- Baumann P. and West,S.C. (1998) Role of the human RAD51 protein in homologous recombination and double-stranded-break repair. Trends Biochem. Sci., 23, 247–251. [DOI] [PubMed] [Google Scholar]
- Bennett R.J., Keck,J.L. and Wang,J.C. (1999) Binding specificity determines polarity of DNA unwinding by the Sgs1 protein of S.cerevisiae.J. Mol. Biol., 289, 235–248. [DOI] [PubMed] [Google Scholar]
- Bierne H., Ehrlich,S.D. and Michel,B. (1997) Deletions at stalled replication forks occur by two different pathways. EMBO J., 16, 3332–3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolt E.L., Sharples,G.J. and Lloyd,R.G. (1999) Identification of three aspartic acid residues essential for catalysis by the RusA Holliday junction resolvase. J. Mol. Biol., 286, 403–415. [DOI] [PubMed] [Google Scholar]
- Caspari T. and Carr,A.M. (1999) DNA structure checkpoint pathways in Schizosaccharomyces pombe.Biochimie, 81, 173–181. [DOI] [PubMed] [Google Scholar]
- Chakraverty R.K. and Hickson,I.D. (1999) Defending genome integrity during DNA replication: a proposed role for RecQ family helicases. BioEssays, 21, 286–294. [DOI] [PubMed] [Google Scholar]
- Chan S.N., Harris,L., Bolt,E.L., Whitby,M.C. and Lloyd,R.G. (1997) Sequence specificity and biochemical characterization of the RusA Holliday junction resolvase of Escherichia coli.J. Biol. Chem., 272, 14873–14882. [DOI] [PubMed] [Google Scholar]
- Cheng R.Z., Murano,S., Kurz,B. and Shmookler Reis,R.J. (1990) Homologous recombination is elevated in some Werner-like syndromes but not during normal in vitro or in vivo senescence of mammalian cells. Mutat. Res., 237, 259–269. [DOI] [PubMed] [Google Scholar]
- Cordeiro-Stone M., Zaritskaya,L.S., Price,L.K. and Kaufmann,W.K. (1997) Replication fork bypass of a pyrimidine dimer blocking leading strand DNA synthesis. J. Biol. Chem., 272, 13945–13954. [DOI] [PubMed] [Google Scholar]
- Courcelle J. and Hanawalt,P.C. (1999) RecQ and RecJ process blocked replication forks prior to the resumption of replication in UV-irradiated Escherichia coli.Mol. Gen. Genet., 262, 543–551. [DOI] [PubMed] [Google Scholar]
- Cromie G.A., Millar,C.B., Schmidt,K.H. and Leach,D.R. (2000) Palindromes as substrates for multiple pathways of recombination in Escherichia coli.Genetics, 154, 513–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey S., Han,C.S., Ramer,S.A., Klassen,J.C., Jacobson,A., Eisenberger,A., Hopkins,K.M., Lieberman,H.B. and Freyer,G.A. (1998) Fission yeast rad12+ regulates cell cycle checkpoint control and is homologous to the Bloom’s syndrome disease gene. Mol. Cell. Biol., 18, 2721–2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Defossez P.A., Prusty,R., Kaeberlein,M., Lin,S.J., Ferrigno,P., Silver,P.A., Keil,R.L. and Guarente,L. (1999) Elimination of replication block protein Fob1 extends the life span of yeast mother cells. Mol. Cell, 3, 447–455. [DOI] [PubMed] [Google Scholar]
- Ellis N.A., Groden,J., Ye,T.Z., Straughen,J., Lennon,D.J., Ciocci,S., Proytcheva,M. and German,J. (1995) The Bloom’s syndrome gene product is homologous to RecQ helicases. Cell, 83, 655–666. [DOI] [PubMed] [Google Scholar]
- Forsburg S.L. (1993) Comparison of Schizosaccharomyces pombe expression systems. Nucleic Acids Res., 21, 2955–2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli A. and Schiestl,R.H. (1995) On the mechanism of UV and γ-ray-induced intrachromosomal recombination in yeast cells synchronized in different stages of the cell cycle. Mol. Gen. Genet., 248, 301–310. [DOI] [PubMed] [Google Scholar]
- Galli A. and Schiestl,R.H. (1996) Hydroxyurea induces recombination in dividing but not in G1 or G2 cell cycle arrested yeast cells. Mutat. Res., 354, 69–75. [DOI] [PubMed] [Google Scholar]
- Gangloff S., McDonald,J.P., Bendixen,C., Arthur,L. and Rothstein,R. (1994) The yeast type I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolog: a potential eukaryotic reverse gyrase. Mol. Cell. Biol., 14, 8391–8398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin A., Wang,S.W., Toda,T., Norbury,C. and Hickson,I.D. (1999) Topoisomerase III is essential for accurate nuclear division in Schizosaccharomyces pombe.Nucleic Acids Res., 27, 4050–4058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutz H., Heslot,H., Leopold,U. and Loprieno,N. (eds) (1974) Schizosaccharomyces pombe. Plenum Press, New York, NY. [Google Scholar]
- Hanada K., Ukita,T., Kohno,Y., Saito,K., Kato,J. and Ikeda,H. (1997) RecQ DNA helicase is a suppressor of illegitimate recombination in Escherichia coli.Proc. Natl Acad. Sci. USA, 94, 3860–3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmon F.G. and Kowalczykowski,S.C. (1998) RecQ helicase, in concert with RecA and SSB proteins, initiates and disrupts DNA recombination. Genes Dev., 12, 1134–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins N.P., Kato,K. and Strauss,B. (1976) A model for replication repair in mammalian cells. J. Mol. Biol., 101, 417–425. [DOI] [PubMed] [Google Scholar]
- Hyde H., Davies,A.A., Benson,F.E. and West,S.C. (1994) Resolution of recombination intermediates by a mammalian activity functionally analogous to Escherichia coli RuvC resolvase. J. Biol. Chem., 269, 5202–5209. [PubMed] [Google Scholar]
- Karow J.K., Constantinou,A., Li,J.-L., West,S.C. and Hickson,I.D. (2000) The Bloom’s syndrome gene product promotes branch migration of Holliday junctions. Proc. Natl Acad. Sci. USA, 97, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitao S., Shimamoto,A., Goto,M., Miller,R.W., Smithson,W.A., Lindor,N.M. and Furuichi,Y. (1999) Mutations in RECQL4 cause a subset of cases of Rothmund–Thomson syndrome. Nature Genet., 22, 82–84. [DOI] [PubMed] [Google Scholar]
- Klein H.L. (1995) Genetic control of intrachromosomal recombination. BioEssays, 17, 147–159. [DOI] [PubMed] [Google Scholar]
- Kobayashi T., Heck,D.J., Nomura,M. and Horiuchi,T. (1998) Expansion and contraction of ribosomal DNA repeats in Saccharomyces cerevisiae: requirement of replication fork blocking (Fob1) protein and the role of RNA polymerase I. Genes Dev., 12, 3821–3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn E.M. and Therman,E. (1986) Cytogenetics of Bloom’s syndrome. Cancer Genet. Cytogenet., 22, 1–18. [DOI] [PubMed] [Google Scholar]
- Kuzminov A. (1995) Collapse and repair of replication forks in Escherichia coli.Mol. Microbiol., 16, 373–384. [DOI] [PubMed] [Google Scholar]
- Liu J. and Marians,K.J. (1999) PriA-directed assembly of a primosome on D loop DNA. J. Biol. Chem., 274, 25033–25041. [DOI] [PubMed] [Google Scholar]
- Michel B., Ehrlich,S.D. and Uzest,M. (1997) DNA double-strand breaks caused by replication arrest. EMBO J., 16, 430–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortensen U.H., Bendixen,C., Sunjevaric,I. and Rothstein,R. (1996) DNA strand annealing is promoted by the yeast Rad52 protein. Proc. Natl Acad. Sci. USA, 93, 10729–10734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray J.M., Lindsay,H.D., Munday,C.A. and Carr,A.M. (1997) Role of Schizosaccharomyces pombe RecQ homolog, recombination and checkpoint genes in UV damage tolerance. Mol. Cell. Biol., 17, 6868–6875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paques F. and Haber,J.E. (1999) Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae.Microbiol. Rev., 63, 349–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein R., Michel,B. and Gangloff,S. (2000) Replication fork pausing and recombination or ‘gimme a break’. Genes Dev., 14, 1–10. [PubMed] [Google Scholar]
- Seigneur M., Bidnenko,V., Ehrlich,S.D. and Michel,B. (1998) RuvAB acts at arrested replication forks. Cell, 95, 419–430. [DOI] [PubMed] [Google Scholar]
- Sharples G.J., Ingleston,S.M. and Lloyd,R.G. (1999) Holliday junction processing in bacteria: insights from the evolutionary conservation of RuvABC, RecG and RusA. J. Bacteriol., 181, 5543–5550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair D.A. and Guarente,L. (1997) Extrachromosomal rDNA circles—a cause of aging in yeast. Cell, 91, 1033–1042. [DOI] [PubMed] [Google Scholar]
- Sinclair D.A., Mills,K. and Guarente,L. (1997) Accelerated aging and nucleolar fragmentation in yeast sgs1 mutants. Science, 277, 1313–1316. [DOI] [PubMed] [Google Scholar]
- Stewart E., Chapman,C.R., Al-Khodairy,F., Carr,A.M. and Enoch,T. (1997) rqh1+, a fission yeast gene related to the Bloom’s and Werner’s syndrome genes, is required for reversible S phase arrest. EMBO J., 16, 2682–2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dyck E., Stasiak,A.Z., Stasiak,A. and West,S.C. (1999) Binding of double-strand breaks in DNA by human Rad52 protein. Nature, 398, 728–731. [DOI] [PubMed] [Google Scholar]
- Viguera E., Hernandez,P., Krimer,D.B., Lurz,R. and Schvartzman,J.B. (2000) Visualisation of plasmid replication intermediates containing reversed forks. Nucleic Acids Res., 28, 498–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt P.M., Louis,E.J., Borts,R.H. and Hickson,I.D. (1995) Sgs1: a eukaryotic homolog of E.coli RecQ that interacts with topoisomerase II in vivo and is required for faithful chromosome segregation. Cell, 81, 253–260. [DOI] [PubMed] [Google Scholar]
- Watt P.M., Hickson,I.D., Borts,R.H. and Louis,E.J. (1996) SGS1, a homologue of the Bloom’s and Werner’s syndrome genes, is required for maintenance of genome stability in Saccharomyces cerevisiae.Genetics, 144, 935–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West S.C., Cassuto,E. and Howard-Flanders,P. (1982) Postreplication repair in E.coli: strand exchange reactions of gapped DNA by RecA protein. Mol. Gen. Genet., 187, 209–217. [DOI] [PubMed] [Google Scholar]
- Whitby M.C. and Dixon,J. (1997) A new Holliday junction resolving enzyme from Schizosaccharomyces pombe that is homologous to CCE1 from Saccharomyces cerevisiae.J. Mol. Biol., 272, 509–522. [DOI] [PubMed] [Google Scholar]
- Wu L., Karow,J.K. and Hickson,I.D. (1999) Genetic recombination: helicases and topoisomerases link up. Curr. Biol., 9, R518–R520. [DOI] [PubMed] [Google Scholar]
- Yamagata K., Kato,J., Shimamoto,A., Goto,M., Furuichi,Y. and Ikeda,H. (1998) Bloom’s and Werner’s syndrome genes suppress hyperrecombination in yeast sgs1 mutant: implication for genomic instability in human diseases. Proc. Natl Acad. Sci. USA, 95, 8733–8738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C.E. et al. (1996) Positional cloning of the Werner’s syndrome gene. Science, 272, 258–262. [DOI] [PubMed] [Google Scholar]
- Zou H. and Rothstein,R. (1997) Holliday junctions accumulate in replication mutants via a RecA homolog-independent mechanism. Cell, 90, 87–96. [DOI] [PubMed] [Google Scholar]