Abstract
Transcription factor GATA-1 is essential for normal erythropoiesis. GATA-binding sites are consistently found in promoters or enhancers of genes expressed selectively in erythroid cells. To discover novel GATA-1-regulated genes, we searched for GATA-1-activated transcripts in G1E cells, an erythroid line derived from GATA-1– embryonic stem cells. By subtractive analysis, we identified a new ATP-binding cassette (ABC) transporter that is strongly and rapidly induced by GATA-1. This protein, named ABC-me (for ABC-mitochondrial erythroid), localizes to the mitochondrial inner membrane and is expressed at particularly high levels in erythroid tissues of embryos and adults. ABC-me is induced during erythroid maturation in cell lines and primary hematopoietic cells, and its overexpression enhances hemoglobin synthesis in erythroleukemia cells. The ABC proteins participate in diverse physiological processes by coupling ATP hydrolysis to the transport of a variety of substrates across cell membranes. We speculate that ABC-me, a newly identified erythroid-expressed ABC superfamily member, may mediate critical mitochondrial transport functions related to heme biosynthesis.
Keywords: ABC transporter/erythropoiesis/GATA-1/hemoglobin/mitochondria
Introduction
Studies of globin gene regulation and red blood cell development have provided valuable insights into the mechanisms of eukaryotic gene expression and tissue formation. Erythroid differentiation involves a coordinated series of events ultimately directed toward maximizing hemoglobin synthesis for oxygen transport and delivery by mature red blood cells. The generation of hemoglobin requires acquisition of iron, synthesis of heme and expression of the α- and β-globin proteins. These processes are tightly coupled so as to prevent accumulation of toxic metabolites. Remarkably, many of these aspects of terminal erythroid maturation appear to be regulated in large part by the essential nuclear protein GATA-1.
GATA-1 is the founding member of a family of transcription factors distinguished by a zinc finger domain that recognizes the DNA consensus motif (A/T)GATA(A/G) (Evans and Felsenfeld, 1989; Tsai et al., 1989; Orkin, 1992; Weiss and Orkin, 1995a). Functionally important GATA motifs have been identified in the regulatory regions of virtually all erythroid-expressed genes, including globins, heme biosynthetic enzymes, erythroid transcription factors and erythroid membrane proteins. Gene targeting studies in mice and embryonic stem (ES) cells demonstrate that GATA-1 is required for normal red blood cell development (Pevny et al., 1991; Fujiwara et al., 1996). GATA-1– embryos die from severe anemia between days 10.5 and 11.5 of gestation (E10.5–E11.5). In the absence of GATA-1, both yolk sac (i.e. primitive) and adult-type (i.e. fetal liver/bone marrow or definitive) erythroid precursors undergo developmental arrest and apoptosis at the proerythroblast stage (Weiss et al., 1994; Weiss and Orkin, 1995b; Fujiwara et al., 1996). Thus, in addition to controlling erythroid-specific aspects of development, GATA-1 also modulates cell survival, in part through interaction with cytokine and death receptor signaling pathways, and by regulating expression of the anti-apoptosis gene bcl-x (De Maria et al., 1999; Gregory et al., 1999).
Given the pleiotropic functions of GATA-1-regulated genes, it is likely that the discovery and analysis of additional target genes should provide valuable new insights into erythroid biology. Of particular interest are genes that control or augment the erythroid differentiation process. Previously, we reported the characterization of G1E cells, a murine GATA-1– erythroid line derived from gene-targeted ES cells (Weiss et al., 1997). G1E cells proliferate as developmentally arrested proerythroblasts and undergo terminal maturation after restoration of GATA-1 function. Expression of a conditional, estrogen-activatable form of GATA-1 renders G1E cells estrogen dependent for erythroid maturation (Tsang et al., 1997; Gregory et al., 1999). This experimental system allows the elucidation of the pivotal role of GATA-1 in erythroid differentiation and provides a convenient tool to search for new GATA-1 target genes in subtraction-based screens. Using this approach, we isolated and characterized a new member of the ABC (ATP-binding cassette) superfamily, a large group of proteins that transport a variety of compounds across cell membranes (Higgins, 1992; Croop, 1998). This protein, which we named ABC-me (for ABC-mitochondrial erythroid), is expressed at particularly high levels in mitochondria of erythrocyte precursors. The importance of ABC transporters is underscored by their functional diversity and involvement in numerous acquired and inherited diseases (Croop, 1998). ABC-me represents a newly identified family member of particular relevance to erythroid biology. Its tissue expression pattern and subcellular localization, together with functional data provided by forced overexpression in erythroleukemia cells, suggest an important role for ABC-me in hemoglobin synthesis.
Results
Identification of ABC-me, a GATA-1-induced ABC transporter
G1E cells are a murine GATA-1– erythroblast cell line that undergoes terminal erythroid maturation after GATA-1 function is restored (Weiss et al., 1997). G1E-ER2 cells are a derivative of G1E cells engineered to express stably GATA-1–ER [full-length GATA-1 fused to the ligand-binding domain of the estrogen receptor (Gregory et al., 1999)]. Treatment with β-estradiol induces synchronous erythroid maturation of G1E-ER2 cells.
A PCR-based subtraction analysis was performed to identify transcripts that are induced in G1E-ER2 cells 8 h after β-estradiol treatment. At this early time point, expression of erythroid genes (such as globins and band 3) has increased in these cells, but they have not yet undergone growth arrest or acquired the morphological features associated with terminal maturation.
Two thousand and forty-seven clones were analyzed from the subtracted pool. Clones were immobilized on nitrocellulose filters and hybridized separately to either the subtracted pool or a reverse-subtracted cDNA pool in which tester and driver populations were switched. Two hundred and twelve clones hybridized preferentially to the subtracted probe. Sequencing and blot hybridization analysis demonstrated that 132 of these clones (62%) encoded α- or β-globin, erythroid differentiation markers that are strongly induced by GATA-1 in G1E-ER2 cells (Gregory et al., 1999). The remaining 80 clones were analyzed on northern blots containing RNA from tester and driver sources; 43 of these transcripts were observed to be induced during G1E-ER2 differentiation. The NCBI BLAST program was used to identify these clones (Table I).
Table I. Genes induced during the differentiation of G1E-ER2 cells.
| No. of clones | GenBank accession No. | Sequence ID |
|---|---|---|
| Known murine genes | ||
| 132 | NM_008218.1 or NM_008219.1 | α- or β-globin (determined by sequencing and/or hybridization) |
| 6 | NM_010240.1 | ferritin light chain 1 (Ftl1) |
| 2 | S56599.1 | AP56 (acetaminophen-binding protein) |
| 2 | U78030 | Bcl-x |
| 2 | NM_011029.1 | P40-8, functional (P40-8) |
| 1 | M30779 | 78 kDa glucose-regulated protein |
| 1 | NM_007438.1 | aldolase 1, A isoform (Aldo1) |
| 1 | AF149782 | β-minor globin |
| 1 | AF183960.1 | carbon catabolite repression 4 protein |
| 1 | M95513 | interleukin 3 (IL-3), IL-5 and granulocyte–macrophage colony-stimulating factor (GM-CSF) receptor B-subunit (AIC2B) |
| 1 | M29395 | orotidine-5′-monophosphate decarboxylase |
| 1 | M28664.1 | porphobilinogen deaminase (PBG deaminase) |
| 1 | AF151711 | protein kinase LKB1 |
| 1 | NM_009150.1 | selenium-binding protein 1 (Selenbp1) |
| 1 | (M61895 M58017) | similar to human endothelial leukocyte adhesion molecule I (ELAM1) |
| 1 | (L12016) | similar to rat tricarboxylate transport protein precursor |
| 1 | NM_009121 | spermidine/spermine N1-acetyltransferase (Sat) |
| 1 | NM_009415 | triosephosphate isomerase (TPI) |
| 1 | AF096288.1 | uncoupling protein-2 (Ucp2) gene |
| 1 | AF176520 | WD repeat-containing F-box protein FBW5 |
| 1 | X53824 | X16 |
| Mouse ESTs | ||
| 3 | AI120230 | mouse EST |
| 2 | AW323886 | mouse EST |
| 1 | AA413926 | mouse EST |
| 1 | AI840582 | mouse EST |
| 1 | AV074492 | mouse EST |
| 1 | AA259278 | mouse EST |
| No match | ||
| 5 | no match | |
| 1 | ABC-me |
A PCR-based subtraction protocol was used to find transcripts induced during the differentiation of G1E-ER2 cells following estradiol activation of a GATA-1–ER fusion protein. One hundred and thirty-two clones were identified as adult globins (α or β) by hybridization and/or sequencing. Forty-three other induced clones, individually sequenced and confirmed by northern blotting as transcripts induced during G1E-ER2 differentiation, are listed above with their corresponding identities determined by the NCBI BLAST program. Accession numbers in parentheses pertain to non-murine correspondences.
As expected, the induced clones reflect many processes known to be important for definitive erythroid maturation. In addition to α- and β-globins, genes encoding proteins involved in heme synthesis (PBG deaminase), iron transport (ferritin light chain), apoptosis regulation (bcl-x) and glycolysis (triosephosphate isomerase and aldolase) were identified in our subtractive screen.
Six GATA-1-induced transcripts isolated showed little or no homology to DDBJ/EMBL/GenBank sequences, and nine corresponded to expressed sequenced tags (ESTs). Among the former group, we discovered a 173 bp fragment encoding a polypeptide with homology to the ABC superfamily of transporters (Allikmets et al., 1995). In northern blot analysis, this probe hybridizes to a single mRNA species of ∼4.1 kb, which is strongly up-regulated in G1E-ER2 cells following β-estradiol-induced activation of GATA-1 and during dimethylsulfoxide (DMSO)-induced maturation of murine erythroleukemia (MEL) cells (Figure 1). This gene was named ABC-me.
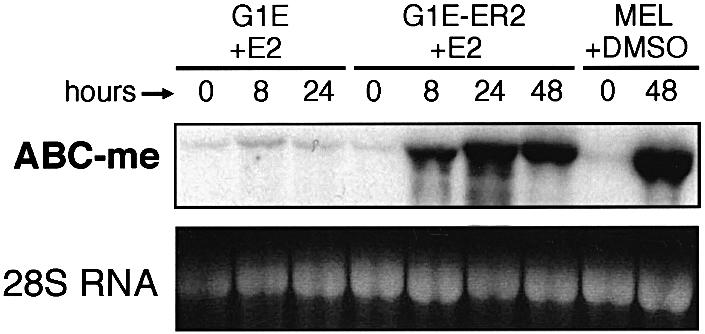
Fig. 1. ABC-me RNA is induced during terminal erythroid maturation. Northern blot analysis. G1E-ER2 cells were stimulated to differentiate by treatment with β-estradiol (E2). G1E cells are the parental line and do not express the β-estradiol-inducible GATA-1–ER fusion protein. MEL cells were induced to differentiate by treatment with DMSO for 48 h. Each lane contains 15 µg of total cellular RNA.
Expression pattern of ABC-me
The original 173 bp ABC-me fragment was used to screen a MEL cell cDNA library, and probes derived from longer clones were used to analyze the tissue expression pattern of ABC-me. Northern blot analysis using total RNA reveals that ABC-me transcripts are most abundant in sites of definitive hematopoiesis, including adult bone marrow and fetal liver (Figure 2A). Finer analysis using poly(A)+-selected RNA revealed expression in several other adult tissues such as liver and kidney, and lower expression in heart, brain and spleen (Figure 2B).
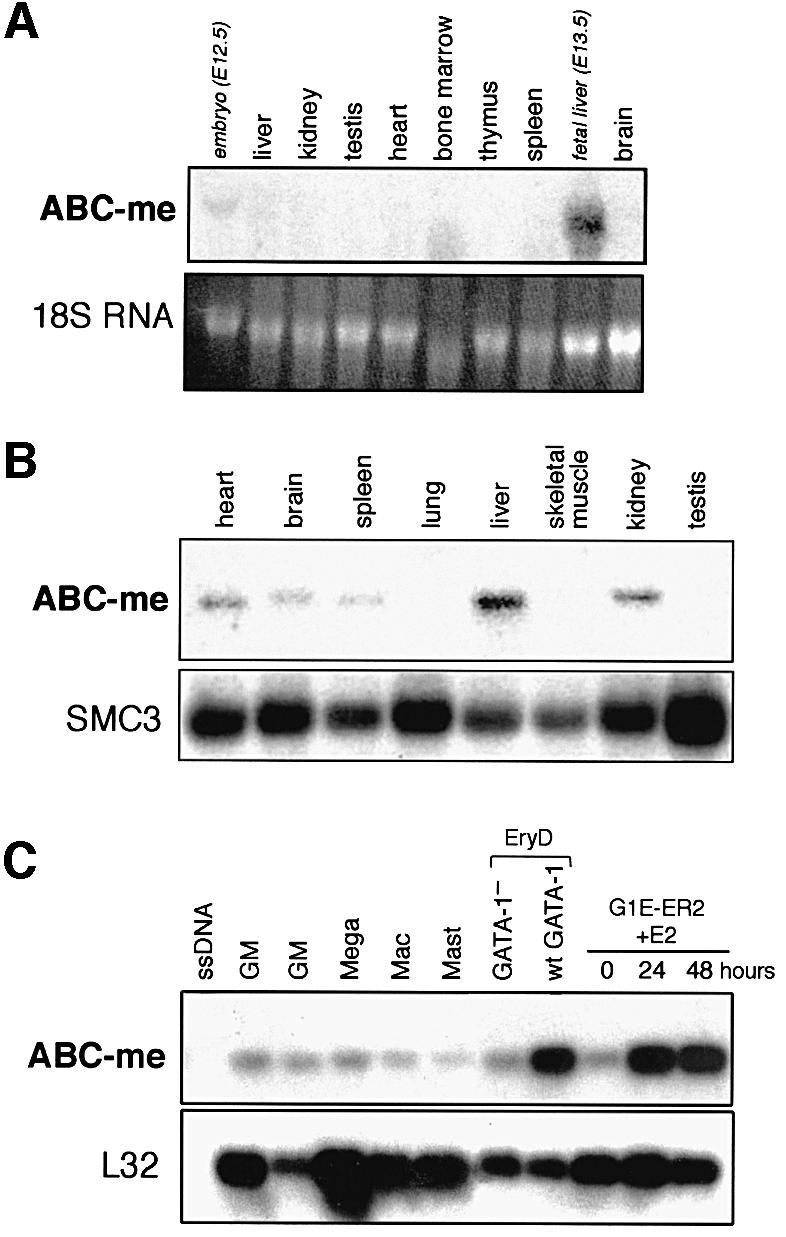
Fig. 2. Tissue expression pattern of ABC-me. (A) Northern blot analysis using total cellular RNA from various mouse tissues. The ABC-me signal is highest in fetal liver and adult bone marrow. Each lane contains 15 µg of total cellular RNA. Italicized labels denote embryonic tissues; all others are adult. (B) Northern blot analysis using poly(A)+-selected RNA from murine adult tissues. As an RNA loading control, the hybridization signal from SMC3, a ubiquitously expressed gene (unpublished data), is shown in the lower panel. Based on RNA size markers (not shown), the major ABC-me transcript is ∼4.1 kb. (C) Expression of ABC-me in primary hematopoietic colonies derived from in vitro differentiation of murine ES cells. Total cellular RNA from individual colonies was amplified by reverse-transcribed PCR and analyzed for ABC-me expression by blot hybridization using a radiolabeled probe from the 3′ end of the cDNA. Expression of the ubiquitous ribosomal transcript L32 is shown in the lower panel. EryD, definitive erythroid; GM, granulocyte–macrophage; Mac; macrophage; Mast, mast cell; Mega, megakaryocyte; ssDNA, salmon sperm DNA used as a negative control for hybridization. The GATA-1– EryD colony was derived from in vitro differentiation of ES cells containing a targeted disruption of the GATA-1 gene.
To examine ABC-me expression in erythroid and other hematopoietic cell types, we analyzed mRNA levels in primary hematopoietic colonies derived from in vitro differentiation of normal ES cells (Figure 2C). We also examined ABC-me expression in primary GATA-1– erythroid colonies derived from ES cells containing a targeted disruption in the GATA-1 locus. Cells within these mutant colonies are developmentally arrested at the proerythroblast stage and eventually undergo apoptosis (Weiss et al., 1994), although in this experiment mutant colonies were harvested before the onset of cell death.
Individual hematopoietic colonies in methylcellulose cultures were identified by morphology, aspirated with a micropipet and analyzed for ABC-me transcripts by poly(A)+ PCR analysis (Brady and Iscove, 1993). As shown in Figure 2C, ABC-me expression was highest in wild-type erythroid colonies, similar to levels observed in G1E-ER2 cells following activation of GATA-1. ABC-me was expressed at lower levels in various non-erythroid colonies. In addition, GATA-1– erythroid colonies were deficient in ABC-me expression compared with age-matched wild-type colonies, consistent with a role for GATA-1 in ABC-me expression during normal erythropoiesis.
Thus, ABC-me is expressed predominantly in erythroid cells, especially during later stages of maturation when GATA-1 function is essential (Weiss et al., 1994). ABC-me RNA was induced within 3 h after estradiol-induced activation of GATA-1 in G1E-ER2 cells and within 12 h after DMSO treatment to induce MEL cell maturation (not shown). The rapid onset of ABC-me induction in G1E-ER2 cells, and the presence of GATA-binding motifs in the gene promoter region (not shown), suggest that ABC-me expression may be regulated directly by GATA-1.
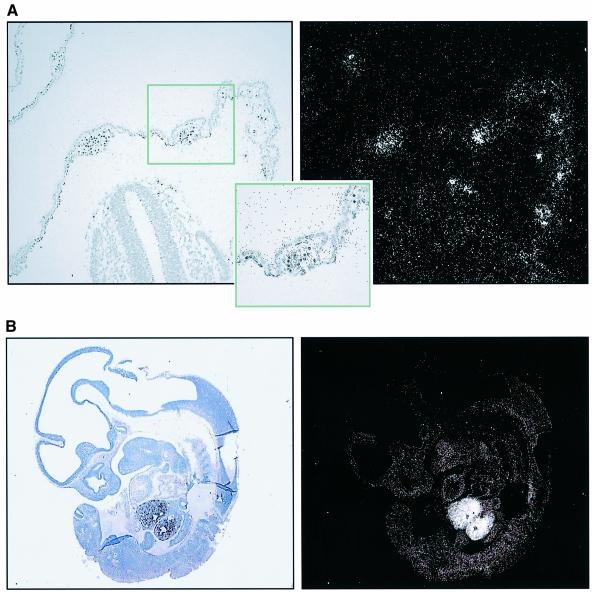
The expression pattern of ABC-me during embryonic development was analyzed by RNA in situ hybridization. Analysis of primitive hematopoiesis was performed with E12 rat embryos, whose larger size relative to stage-equivalent E10 mouse embryos allows for more convenient analysis. At this stage, ABC-me transcripts are abundant in hematopoietic cells in the yolk sac blood islands (Figure 3A). Analysis in later stage embryos (E13 mouse) revealed intense expression in fetal liver, the predominant site of definitive hematopoiesis at that stage. Higher magnification views of E13 mouse embryos demonstrated low levels of ABC-me transcripts in all non-hematopoietic tissues (not shown). Thus, ABC-me mRNA is highly expressed in embryonic sites of primitive and definitive hematopoiesis, and at lower levels in other tissues. That the majority of hematopoietic cells in yolk sac blood islands and fetal liver are erythroid is consistent with our findings that ABC-me was strongly expressed in primary erythroid colonies and during maturation of MEL and G1E-ER2 cells.
Fig. 3. ABC-me is expressed in hematopoietic tissues of the embryo. (A) In situ hybridization to E12 rat embryo, which is stage equivalent to E10 mouse. Staining for tissue histology is shown on the left and hybridization signal on the right. A yolk sac blood island is magnified in the insert. (B) In situ hybridization to E13 mouse embryo showing abundant ABC-me expression in fetal liver.
Structure of ABC-me
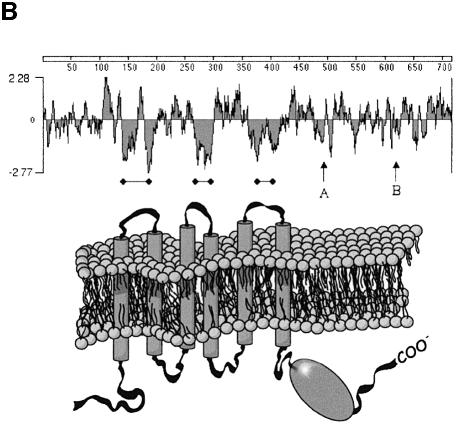
The longest ABC-me cDNA isolated is 4080 bp, close to full length as judged by the size of the single RNA band seen on northern blots (see Figure 2). A poly(A)+ stretch is present at the 3′ end, and the 5′ end extends to a major transcription start site determined by primer extension and rapid amplification of cDNA ends (RACE) analyses (not shown). An open reading frame of 716 amino acids begins at nucleotide 40. An in-frame stop codon located 27 bp upstream of the presumed ATG initiation codon is present in cDNA and genomic DNA clones (not shown). The predicted polypeptide (Figure 4A) has an expected mass of 77.2 kDa and contains three hydrophobic membrane-spanning loops, followed by the configuration of ATP-binding motifs that distinguish the ABC family, including Walker A (GX7GX4GKST), Walker B (RX8ILLLDE) and SGGQ signature (Walker et al., 1982; Higgins, 1992) (Figure 4A and B).
Fig. 4. Primary amino acid structure of ABC-me protein. (A) Amino acid sequence alignment of murine ABC-me with related proteins human TAP-2 and ABC-7. Amino acids that are identical to ABC-me are indicated by black boxes. Conserved elements that comprise the ATP-binding pocket, including the Walker A and B motifs and the SGGQ signature, are overlined. (B) A Kyte–Doolittle hydrophilicity plot of ABC-me with the predicted membrane-spanning regions indicated by black bars. The locations of Walker A and B sites are indicated by arrows. A cartoon depicting the protein arrangement within the membrane is shown below. For the topology prediction, a window of 17 amino acids and a critical hydropathy threshold index of 1.58 were used.
Within the ATP-binding pocket, ABC-me is highly related to all other family members, especially the multidrug resistance (MDR) proteins (Figure 4A; data not shown). Within the domain containing the membrane-spanning loops, which typically encodes substrate specificity, ABC-me shows relatively strong similarity to the endoplasmic reticulum peptide transporter TAP-2 (Bahram et al., 1991) (24% identity) (Figure 4A), the mitochondrial transporter M-ABC-1 (Hogue et al., 1999) (23% identity) and several less well characterized proteins in yeast, Aspergillus and Drosophila (not shown). In contrast, the membrane-spanning domains of ABC-me are more distantly related to human ABC7 (ATM1p homolog), a mitochondrial exporter of iron–sulfur clusters (14% identity) (Leighton and Schatz, 1995; Allikmets et al., 1999; Kispal et al., 1999). ABC-me is 78% identical to a partial protein sequence encompassing an ABC domain encoded by a human EST (EST 20237, NCBI) (Allikmets et al., 1995).
The functional unit of ABC transporters, comprised of one or more polypeptides that assemble in the membrane, is defined by two ATP-binding domains and two hydrophobic regions, each containing three transmembrane loops (Higgins, 1992; Croop, 1998). ABC-me resembles a ‘half-transporter’ (Figure 4B) and is therefore predicted to function as a homo- or heterodimer (Boscoboinik et al., 1990; Dean and Allikmets, 1995).
Subcellular localization
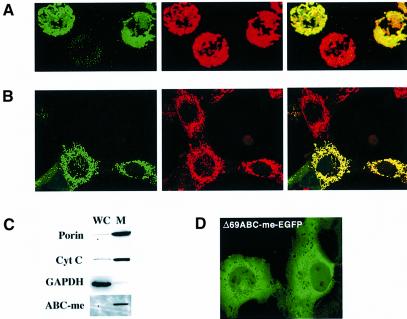
To understand the function of ABC-me better, we determined its subcellular localization. Enhanced green fluorescent protein (EGFP) was fused in-frame to the C-terminus of ABC-me and introduced into a mammalian expression vector. Following transfection into various cell lines, distribution of the chimeric protein was examined by confocal fluorescence microscopy. ABC-me–EGFP co-localized with the mitochondria-specific dyes MitoTracker Red or tetramethylrhodamine ethyl ester (TMRE) in all cells tested, including NIH 3T3 and MEL (Figure 5A and B), as well as COS, L, 293 and G1E (not shown). ABC-me–EGFP remained in the mitochondria during induced erythroid maturation of MEL and G1E cells (Figure 5A; data not shown). Thus, ABC-me localizes to mitochondria in many different cellular contexts and is expected to be a mitochondrial protein in both erythroid and non-erythroid tissues in vivo. Biochemical fractionation followed by western blotting also demonstrated that ABC-me–EGFP is concentrated in mitochondria (Figure 5C).
Fig. 5. ABC-me is a mitochondrial protein. (A) Expression of EGFP-tagged ABC-me in MEL cells. Cells that express ABC-me-EGFP emit a green signal (left panel) that co-localizes with the red signal emitted by the mitochondria-specific dye TMRE (middle panel). The red and green images are superimposed in the right panel. The images are projections of three-dimensional reconstructions of serial confocal microscopy images. (B) Expression of ABC-me–EGFP in NIH 3T3 fibroblasts. Cells were transfected with an ABC-me–EGFP expression plasmid and analyzed 48 h later. The ABC-me–EGFP signal is shown in green (left panel). Mitochondria were stained red using the dye CMXRos (MitoTracker Red, middle panel). Images are superimposed in the right panel. (C) Western blot analysis of MEL cells expressing the ABC-me–EGFP fusion protein. Eighty micrograms of protein from whole cells (WC) and purified mitochondria (M) were analyzed using antibodies against EGFP, the cytosolic protein GAPDH, and the mitochondrial proteins porin and cytochrome c (Cyt C). ABC-me–EGFP is ∼80-fold enriched in the mitochondrial fraction, as determined by densitometry (not shown). (D) Mitochondrial targeting is encoded within the N-terminus of ABC-me. In COS cells the deletion mutant Δ69ABC-me–EGFP (green signal) localizes predominantly to the cytoplasm and is excluded from mitochondria.
Mitochondrial targeting is often mediated through peptide sequences at the N-terminus (Neupert, 1997). To investigate this possibility in ABC-me, amino acids 2–69 were removed and the truncated protein was expressed in COS cells. As shown in Figure 5D, Δ69ABC-me–EGFP distributed to the cytosol, indicating that the N-terminus of ABC-me is indeed necessary for mitochondrial localization.
The results of these subcellular localization studies and the primary amino acid sequence of ABC-me suggest that it is a mitochondrial membrane protein. To verify this possibility, we analyzed intact mitochondria and purified mitochondrial membranes from 293T cells expressing a V5-epitope-tagged version of ABC-me (Figure 6A). Western blot analysis showed that V5–ABC-me and the mitochondrial membrane protein porin co-purified in the membrane fraction. In contrast, citrate synthase, a soluble mitochondrial matrix protein, was detected in whole mitochondria but was excluded from the membrane fraction. Localization of ABC-me to the membrane fraction of mitochondria is consistent with its predicted role as a transporter protein.
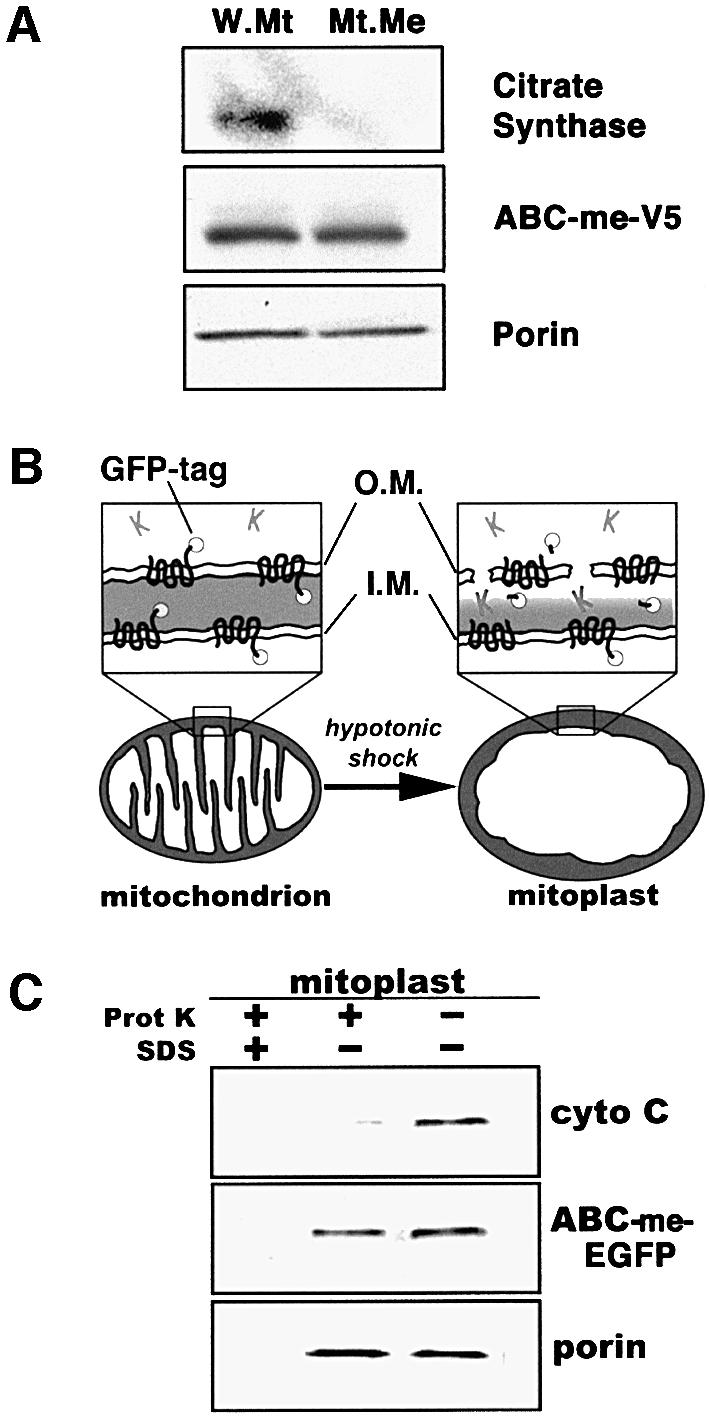
Fig. 6. ABC-me localizes to the inner mitochondrial membrane. (A) Western blot analysis of ABC-me in whole mitochondria (W.Mt) and mitochondrial membrane fraction (Mt.Me). Whole mitochondria were isolated from 293T cells expressing a V5 epitope-tagged version of ABC-me and subjected to membrane extraction followed by western blotting. ABC-me, as detected by anti-V5 antibody, is present in both whole mitochondria and purified mitochondrial membranes. As controls for efficiency of membrane fractionation, samples were analyzed with antibodies against the mitochondrial membrane protein porin and the soluble matrix enzyme citrate synthase. (B) Submitochondrial localization of ABC-me through analysis of mitoplasts. Four different mitochondrial membrane configurations are possible for ABC-me–EGFP. Disruption of the outer membrane by hypotonic shock to form mitoplasts renders protein domains within the cytosol and intermembrane space susceptible to proteinase K activity (shown as ‘K’ in the cartoon), while moieties within the mitochondrial matrix or membrane bilayer are relatively proteinase resistant. Solubilization of the membranes with detergent renders all proteins sensitive to proteinase treatment. (C) Mitoplasts derived from ABC-me–EGFP-expressing MEL cells were treated with proteinase K with or without SDS as indicated, and analyzed by western blotting. ABC-me–GFP was detected using an anti-GFP antibody. As controls, samples were analyzed for the integral membrane protein porin, which is known to be proteinase resistant in non-detergent-treated mitoplasts, and cytochrome c, a soluble protein that resides in the intermembrane space. Proteinase K resistance of ABC-me–EGFP in mitoplasts indicates that the protein resides in the inner membrane with the hydrophilic C-terminus directed into the mitochondrial matrix. OM, outer mitochondrial membrane; IM, inner mitochondrial membrane.
To localize ABC-me more precisely, we isolated mitochondria from MEL cells expressing the ABC-me–EGFP fusion protein and disrupted their outer membrane by hypotonic shock to create mitoplasts (Glick, 1995; Leighton, 1998) (Figure 6B). This procedure renders proteins and protein domains within the intermembrane space susceptible to digestion by proteinase K. Protein moieties within the inner and outer membrane bilayers and mitochondrial matrix are relatively inaccessible to added proteinase. Thus, in the absence of detergent, the membrane-spanning regions of ABC-me–EGFP should be proteinase resistant. The hydrophilic EGFP moiety, which is fused to the ATP-binding cassette at the C-terminus of ABC-me, will be proteinase resistant only if it resides in the mitochondrial matrix.
Western blot analysis (Figure 6C) showed that, in mitoplasts, cytochrome c (which resides in the intermembrane space) was almost entirely degraded by proteinase K, while ABC-me–EGFP remained intact. As expected, porin, which resides almost entirely within the outer membrane bilayer, is also proteinase resistant (Glick, 1995). Both ABC-me–EGFP and porin were degraded by proteinase K after solubilization of the membrane with detergent. These findings indicate that ABC-me resides in the inner mitochondrial membrane with the hydrophilic C-terminus containing the ATP-binding cassette directed into the mitochondrial matrix. This configuration suggests that ABC-me transports substrate(s) from the matrix into the intermembrane space (Higgins, 1992). The band recognized by anti-GFP was not reduced in size in proteinase-treated mitoplasts, suggesting that the intermembrane space loops of ABC-me are relatively inaccessible. This observation is consistent with the short intermembrane loops predicted by hydrophilicity plots (Figure 4B).
Forced overexpression of ABC-me enhances hemoglobin biosynthesis
The mitochondrial localization and high-level erythroid expression of ABC-me suggest a potential role in heme biosynthesis, iron metabolism or both. If this were the case, overexpression of ABC-me during erythroid maturation might be expected to enhance the rate of hemoglobin accumulation but not affect the timing of its onset. We tested this possibility by examining the effects of forced overexpression in MEL cells (Figure 7). ABC-me cDNA was inserted into a mammalian expression vector and transfected into MEL cells to generate stably expressing clones. In each of the three ABC-me-transfected clones analyzed, the expression level of the stable transgene in uninduced cells was comparable to that of the endogenous gene after 48 h of DMSO-induced maturation, as determined by northern blot analysis (not shown). Flow cytometry of isolated mitochondria showed that overexpression of ABC-me in MEL cells does not alter mitochondrial size distribution. In addition, there were no obvious alterations in the shape or number of mitochondria per cell, as determined by three-dimensional confocal microscopy (not shown).
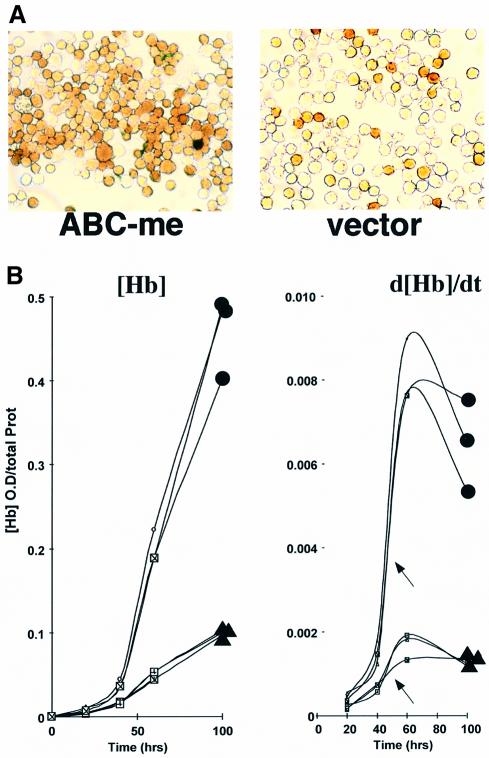
Fig. 7. Forced overexpression of ABC-me enhances hemoglobin production during MEL cell differentiation. (A) MEL cells stably transfected with either an ABC-me expression plasmid or vector only were treated with DMSO and stained for hemoglobin with benzidine after 55 h. One representative clone of each is shown. (B) Kinetics of hemoglobin production during DMSO-induced differentiation in MEL cell clones stably transfected with either an ABC-me expression plasmid (filled circles) or vector only (filled triangles). Hemoglobin production is plotted against time in the graph on the left, and the rate of hemoglobin production, d[Hb]/dt, is plotted on the right. Three separate vector-expressing clones and three ABC-me-expressing clones were tested. The onset of accelerated hemoglobin synthesis in ABC-me- and vector-expressing cells was similar, as indicated by arrows.
Hemoglobin synthesis in ABC-me-transfected and control MEL cells was analyzed by two methods: (i) the proportion of hemoglobin-positive cells was assessed by staining with the benzidine reagent (Orkin et al., 1975); (ii) the hemoglobin content of total cell lysates was quantified by direct spectroscopic measurement (Cioe et al., 1981). Together, these methods allowed us to assess both the rate of hemoglobin synthesis in the population and the extent of hemoglobin production per cell. During DMSO-induced differentiation of polyclonal populations and three clonal lines, overexpression of ABC-me enhanced both the number of benzidine-positive cells and the intensity of benzidine staining (Figure 7A; data not shown). In addition, total hemoglobin content was consistently 2- to 4-fold higher than that in cells expressing vector alone (Figure 7B). In both assays, overexpression of ABC-me had no effect on the spontaneous differentiation rate of MEL cells in the absence of DMSO (Figure 7B; data not shown).
During DMSO-induced differentiation of control MEL cells, the increase in rate of hemoglobin production was non-linear, with a sharp increase between 40 and 60 h (shown by arrows in Figure 7B, right panel). This period, which presumably represents maturation of the hemoglobin synthesis pathway, occurred at a similar time in MEL cells overexpressing ABC-me. Overexpressed ABC-me alone does not initiate differentiation or hemoglobin synthesis; rather, it augments hemoglobin production during DMSO-induced cell maturation.
Regulation of ABC-me transcript level by heme
Numerous components of the heme biosynthesis pathway are regulated by substrates, intermediaries and end metabolites, such as iron and heme (Ponka, 1997, 1999). To test whether ABC-me is subject to similar feedback regulation, we examined mRNA levels during MEL cell differentiation in the presence of various heme biosynthesis metabolites or inhibitors (Figure 8). Steady-state ABC-me transcripts are significantly reduced by addition of heme at a physiological concentration (5 µM; Veltman and Maines, 1986). This reduction is probably due to the combined effects of exogenously supplied heme and endogenous heme production, since it can be alleviated by the addition of the heme biosynthesis inhibitor succinyl acetone (SA). SA decreases heme biosynthesis intermediates by inhibiting delta-aminolevulinic acid dehydratase (ALA-D), the second enzyme in the pathway. Therefore, since ABC-me levels are unchanged by added SA alone, it is less likely that heme affects ABC-me through the regulation of another heme-dependent intermediate of the pathway. Finally, the effect of added heme on steady-state ABC-me mRNA level cannot be attributed to increased intracellular iron released by heme metabolism, as iron alone has no effect on ABC-me expression. Consistent with this observation, an iron response element (IRE) consensus sequence, which would be expected to destabilize the mRNA in an iron-dependent fashion, is not present in the 3′ untranslated region of the mRNA.
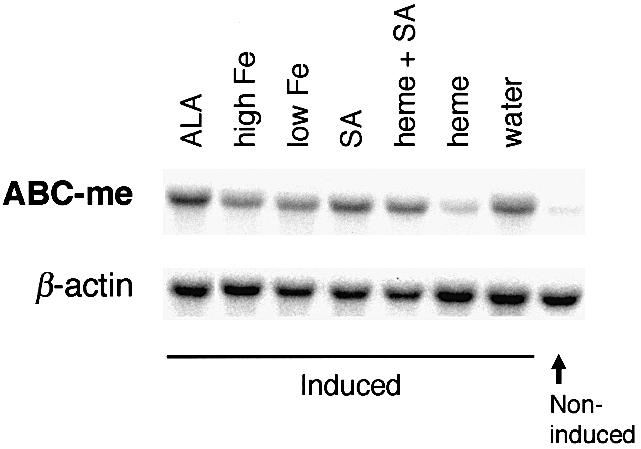
Fig. 8. Regulation of ABC-me transcript levels in MEL cells. MEL cells were induced to differentiate with DMSO in the presence of the indicated compounds and RNA was collected for northern blot analysis after 72 h. Northern blot analysis was performed using ABC-me and β-actin probes. The results shown here were reproduced in three separate experiments. Each lane represents 9 µg of total cellular RNA. ALA, aminolevulinic acid; Fe, iron added as holotransferrin (high, 100 µg/ml; low, 5 µg/ml); SA, succinyl acetone, 0.5 mM; heme, added as hemin, 5 µm.
Discussion
Erythroid differentiation is characterized by the de novo construction or activation of numerous enzymatic and transport pathways, which are responsible for the functional and structural changes seen in the maturing erythroid cell. With the hypothesis that key proteins within these pathways may be downstream of an essential transcriptional regulator of erythroid differentiation, we performed a subtraction screen for genes induced during GATA-1-dependent differentiation of an erythroid cell line. As expected, our screen revealed numerous well-characterized genes that participate in specialized erythroid function. Among the GATA-1-induced genes, we identified ABC-me, a novel mitochondrial membrane transport protein that is expressed at high levels in erythrocyte precursors and enhances hemoglobin production in differentiating MEL cells.
Eukaryotic ABC transporters perform diverse functions within various organelles including the plasma membrane, peroxisome, endoplasmic reticulum and mitochondrion. Members of this large superfamily have been implicated in numerous human diseases, including cystic fibrosis (CFTR) (Riordan et al., 1989), adrenoleukodystrophy (ALDP) (Mosser et al., 1993), Zellweger’s syndrome (PMP70) (Gartner et al., 1992), progressive familial intrahepatic cholestasis (SPGP) (Strautnieks et al., 1998) and Stargardt macular dystrophy (ABCR) (Zhang et al., 1999). ABC-me is a novel member of the ABC superfamily with a potentially important role in erythroid development.
The induction of ABC-me expression during MEL and G1E cell maturation parallels that of other molecules involved in terminal erythropoiesis, most of which are believed to be regulated by GATA-1 (Orkin, 1992; Weiss and Orkin, 1995a). The relative deficiency of ABC-me transcripts in GATA-1– erythroid colonies is consistent with a regulatory hierarchy in which ABC-me is positioned downstream of GATA-1 during erythroid maturation. Whether the ABC-me gene is a direct target for GATA-1 action remains an open question. In favor of this possibility, ABC-me mRNA is induced within 3 h following GATA-1 activation in G1E cells, and several GATA-consensus motifs are present in the promoter region of the ABC-me gene (not shown).
As a ‘half-ABC transporter’ comprising one ATP-binding domain and three transmembrane loops, ABC-me is predicted to function as a homo- or heterodimer (Dean and Allikmets, 1995). Two previously identified ABC transporters also localize to mitochondria and so could be heterodimerization partners for ABC-me. M-ABC1, a mitochondrial transporter of unknown function, is expressed ubiquitously (Hogue et al., 1999). ABC7, the human homolog of the yeast mitochondrial iron–sulfur cluster transporter ATM1p (Kispal et al., 1999), is expressed in many tissues, especially heart, skeletal muscle and pancreas (Shimada et al., 1998; Allikmets et al., 1999). ABC-me is more closely related to M-ABC1 than ABC7 within the transmembrane domains that encode the predicted heterodimerization surfaces and substrate-binding motifs, suggesting a greater degree of functional similarity and potential for heterodimerization. In contrast to ABC-me, there is no evidence to suggest that ABC7 or M-ABC1 is expressed at particularly high levels during erythroid maturation.
Mitochondria participate in many essential and specialized functions during erythroid differentiation, including hemoglobin synthesis, energy production, integration of apoptotic signals and gradual self-elimination. While ABC-me could act in any of these processes, effects of its expression in MEL cells favor a role in hemoglobin production. ABC-me does not induce spontaneous MEL cell differentiation; rather, it appears to enhance the function of hemoglobin synthesis machinery that is coordinately induced during erythroid maturation triggered by DMSO. Hence, under some circumstances, ABC-me may be rate limiting for hemoglobin synthesis during erythropoiesis. The effects of ABC-me contrast with those of other proteins reported to enhance MEL cell maturation, such as the basic helix–loop–helix protein SCL/tal-1 (Aplan et al., 1992) and hematopoietic ring finger protein-1 (HERF-1) (Harada et al., 1999), each of which can trigger differentiation independent of DMSO.
The predicted structure, expression pattern, submitochondrial localization and overexpression effects of ABC-me suggest that it may have a role in heme biosynthesis, which occurs at especially high rates in erythroid cells. The first and the three last steps of heme biosynthesis occur in the mitochondrion; the steps in between are cytosolic. While the enzymes involved in heme synthesis are well characterized, little is known about how biosynthetic intermediates are shuttled across mitochondrial membranes. We speculate that ABC-me may be involved in one or more of these transport processes. Consistent with non-erythroid requirements for heme, lower levels of ABC-me expression are seen in many tissues, most notably liver, where the rate of heme synthesis for cytochrome production is relatively high. Our observation that added heme represses ABC-me mRNA expression is also compatible with a role in heme biosynthesis, which is subject to end-product inhibition at several steps in the metabolic pathway (Ponka, 1997, 1999).
The importance of mitochondria in erythroid cell physiology is illustrated by the sideroblastic anemias, a heterogeneous group of acquired and inherited disorders characterized by ineffective erythropoiesis with failure to incorporate iron into heme, and accumulation of iron complexes in the mitochondria of erythroid precursors. The best characterized inherited form is caused by mutations in the X-linked gene encoding the erythroid mitochondrial heme biosynthetic enzyme aminolevulinic acid synthetase. Another inherited form, X-linked sideroblastic anemia with cerebellar ataxia (XLSA/A), is caused by mutations in the mitochondrial transporter ABC7 (Allikmets et al., 1999), which is believed to export iron–sulfur clusters from the mitochondrion to the cytosol (Kispal et al., 1999). As an erythroid mitochondrial transporter implicated in hemoglobin production, ABC-me is thus a strong candidate gene for autosomally inherited forms of sideroblastic anemia (Kasturi et al., 1982; Jardine et al., 1994).
Materials and methods
Cell lines
G1E-ER2 cells were cultured as described (Gregory et al., 1999). Upon activation of GATA-1 by β-estradiol, G1E-ER2 cells undergo synchronous cell cycle arrest followed by terminal erythroid maturation.
MEL cells were grown in Dulbecco’s modified Eagle’s medium (Gibco-BRL) supplemented with 10% fetal calf serum (Gibco-BRL). To induce erythroid differentiation, cells at an initial density of 105 cells/ml were cultured in medium containing 1.9% DMSO (Sigma). Cells were stained with benzidine to detect hemoglobin production (Orkin et al., 1975) or subjected to spectroscopic determination of hemoglobin as described (Cioe et al., 1981).
Subtractive analysis
Total cellular RNA was isolated using Trizol (Gibco-BRL) and enriched for poly(A)+ RNA using the FastTrack 2.0 kit (Invitrogen). The PCR-Select cDNA Subtraction Kit (Clontech) was used to find transcripts induced during the differentiation of G1E-ER2 cells following estradiol activation of the GATA-1–ER fusion protein. Tester cDNA was created from poly(A)+ RNA prepared from G1E-ER2 cells treated with β-estradiol for 8 h. Driver cDNA was similarly prepared from cells treated with ethanol vehicle alone. Subtraction was performed according to the manufacturer’s instructions, with 12 cycles of PCR amplification. The pool of subtracted cDNAs was cloned into pT-Adv (Clontech).
Isolation of full-length ABC-me
A cDNA library was constructed from MEL cells, half of which were induced to differentiate by DMSO treatment for 36 h and half left untreated. The library was cloned into the plasmid vector pcDNA3.0 (Invitrogen) and screened by colony hybridization using the 173 bp ABC-me fragment described above. Plasmid DNA was purified from hybridizing clones and analyzed by restriction endonuclease digestion and DNA sequence analysis using the dideoxy-chain termination method.
Northern blot analysis
Total cellular RNA was isolated from various tissues and cell lines using Trizol, fractionated on 1.2% agarose–formaldehyde gels, transferred to Hybond C+ (Amersham Pharmacia Biotech) and hybridized with a [32P]dCTP-labeled ABC-me probe. For Figure 1, the probe was the original 173 bp cDNA fragment isolated by subtractive analysis, corresponding to nucleotides 2242–2415 of the reported cDNA. The probe used in Figure 2A and B was derived from bp 109–901 of the reported cDNA, corresponding to an EagI–KpnI fragment. This probe contains the coding region of the transmembrane domains, but not the ATP-binding cassette domain of the ABC-me protein. Blots were washed at a final stringency of 0.5× SSC at 65°C. For analysis of poly(A)+ RNA, the Mouse Multiple Tissue Northern Blot (Clontech) was hybridized to radiolabeled ABC-me probe according to the manufacturer’s instructions.
Analysis of ABC-me expression in hematopoietic colonies
Gene expression analysis in hematopoietic colonies was carried out using the poly(A)+ PCR method (Brady and Iscove, 1993). Hematopoietic colonies derived from in vitro differentiation of ES cells (Keller et al., 1993) were transferred directly into first-strand PCR buffer. Reverse transcription tailing and PCR procedures were performed as described (Brady and Iscove, 1993), with the exception that the (dT)–x oligonucleotide was shortened to 5′-GTTAACTCGAGAATTC(T)24-3′. The amplified products from the PCR reaction were separated on agarose gels, denatured and transferred to Hybond N+ membrane (Amersham). The resulting blots were hybridized with 32P-labeled random-primed cDNA fragments encoding the 3′ end of either ABC-me cDNA or the ribosomal protein L32.
In situ hybridization
In situ hybridization of ABC-me RNA to mouse and rat embryos was performed as described (Vainio et al., 1993). Probes for in situ hybridization were derived from nucleotide positions 1226–2896 of the reported cDNA.
Subcellular localization
ABC-me cDNA was fused in-frame to EGFP in the plasmid EGFP-N1 (Clontech). Cells were transfected with Fugene 6 (Roche). After 48 h, cells were incubated with 20 nM MitoTracker Red (CMXRos) (Molecular Probes) for 20 min and washed three times with phosphate-buffered saline (PBS). For TMRE staining, cells were incubated with 1 µM TMRE (Molecular Probes). A Zeiss LSM510 confocal microscope was used in multitrack mode, such that excitation and detection of EGFP and TMRE/CMXRos were performed at separate beam tracks beginning with TMRE/CMXRos, to minimize both cross-excitation and bleaching. EGFP was detected at 480 nm excitation/540 nm emission, and TMRE/CMXRos at 550 nm excitation/570 nm emission long-pass filter. Co-localization and mitochondrial structure studies in MEL cells were performed using z-slice function with 20 slices per cell (15 µm) and analyzed in a 3D-projection reconstruction mode.
Submitochondrial localization
Submitochondrial localization was performed as described (Leighton, 1998). Briefly, crude mitochondria were isolated by differential centrifugation of cell homogenate. Mitochondrial membrane fraction was purified by repeated freeze–thaw and sonication in hypotonic solution, followed by ultracentrifugation (150 000 g). For proteinase K resistance studies, mitochondria were subjected to hypotonic shock (to create mitoplasts) in 20 mM KCl in the presence or absence of 1 mg/ml proteinase K at room temperature for 30 min. Proteinase K was then inactivated by trichloroacetate. Samples were centrifuged and the mitoplast pellets were analyzed by western blotting using an Enhanced Chemiluminescence (ECL) kit (Amersham Pharmacia Biotech). ABC-me was detected by anti-EGFP antibody (Clontech) at a dilution of 1 in 100 or by anti-V5 antibody (Invitrogen) at 1 in 1000. Other antibodies used were: anti-porin (Promega), 1 in 100; anti-cytochrome c (Molecular Probes), 1 in 100; anti-human citrate synthase (Research Diagnostics), 1 in 100; and anti-GAPDH antibody (Advanced ImmunoChemicals), 1 in 500.
Overexpression of ABC-me in MEL cells
MEL cells were transfected with a DNA plasmid containing ABC-me in the mammalian expression vector pEF1α-neo (Kotkow and Orkin, 1995). Individual clones were isolated by limiting dilution in medium containing 1 mg/ml neomycin (G418; Gibco-BRL) and tested for ABC-me expression by northern blot analysis.
DDBJ/EMBL/GenBank accession number
The accession number for ABC-me is AF266284.
Acknowledgments
Acknowledgements
We thank P.Smith for help with confocal microscopy studies, I.Karavarov for help with in situ hybridization and P.Mead for help with cloning. We are grateful to Gerd Blobel, Jim Croop and Carlo Brugnara for reviewing this manuscript. O.S.S. is sponsored by the Dorot Foundation. S.H.O. is an Investigator of the Howard Hughes Medical Institute.
References
- Allikmets R., Gerrard,B., Glavac,D., Ravnik-Glavac,M., Jenkins,N.A., Gilbert,D.J., Copeland,N.G., Modi,W. and Dean,M. (1995) Characterization and mapping of three new mammalian ATP-binding transporter genes from an EST database. Mamm. Genome, 6, 114–117. [DOI] [PubMed] [Google Scholar]
- Allikmets R., Raskind,W.H., Hutchinson,A., Schueck,N.D., Dean,M. and Koeller,D.M. (1999) Mutation of a putative mitochondrial iron transporter gene (ABC7) in X-linked sideroblastic anemia and ataxia (XLSA/A). Hum. Mol. Genet., 8, 743–749. [DOI] [PubMed] [Google Scholar]
- Aplan P.D., Nakahara,K., Orkin,S.H.O. and Kirsch,I.R. (1992) The SCL gene product: a positive regulator of erythroid differentiation. EMBO J., 11, 4073–4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahram S., Arnold,D., Bresnahan,M., Strominger,J.L. and Spies,T. (1991) Two putative subunits of a peptide pump encoded in the human major histocompatibility complex class II region. Proc. Natl Acad. Sci. USA, 88, 10094–10098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boscoboinik D., Debanne,M.T., Stafford,A.R., Jung,C.Y., Gupta,R.S. and Epand,R.M. (1990) Dimerization of the P-glycoprotein in membranes. Biochim. Biophys. Acta, 1027, 225–228. [DOI] [PubMed] [Google Scholar]
- Brady G. and Iscove,N.N. (1993) Construction of cDNA libraries from single cells. Methods Enzymol., 225, 611–623. [DOI] [PubMed] [Google Scholar]
- Cioe L., McNab,A., Hubbell,H.R., Meo,P., Curtis,P. and Rovera,G. (1981) Differential expression of the globin genes in human leukemia K562(S) cells induced to differentiate by hemin or butyric acid. Cancer Res., 41, 237–243. [PubMed] [Google Scholar]
- Croop J.M. (1998) Evolutionary relationships among ABC transporters. Methods Enzymol., 292, 101–116. [DOI] [PubMed] [Google Scholar]
- Dean M. and Allikmets,R. (1995) Evolution of ATP-binding cassette transporter genes. Curr. Opin. Genet. Dev., 5, 779–785. [DOI] [PubMed] [Google Scholar]
- De Maria R. et al. (1999) Negative regulation of erythropoiesis by caspase-mediated cleavage of GATA-1. Nature, 401, 489–493. [DOI] [PubMed] [Google Scholar]
- Evans T. and Felsenfeld,G. (1989) The erythroid-specific transcription factor Eryf1: a new finger protein. Cell, 58, 877–885. [DOI] [PubMed] [Google Scholar]
- Fujiwara Y., Browne,C.P., Cunniff,K., Goff,S.C. and Orkin,S.H. (1996) Arrested development of embryonic red cell precursors in mouse embryos lacking transcription factor GATA-1. Proc. Natl Acad. Sci. USA, 93, 12355–12358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartner J., Moser,H. and Valle,D. (1992) Mutations in the 70K peroxisomal membrane protein gene in Zellweger syndrome. Nature Genet., 1, 16–23. [DOI] [PubMed] [Google Scholar]
- Glick B.S. (1995) Pathways and energetics of mitochondrial protein import in Saccharomyces cerevisiae.Methods Enzymol., 260, 224–231. [DOI] [PubMed] [Google Scholar]
- Gregory T., Yu,C., Ma,A., Orkin,S.H., Blobel,G.A. and Weiss,M.J. (1999) GATA-1 and erythropoietin cooperate to promote erythroid cell survival by regulating bcl-xL expression. Blood, 94, 87–96. [PubMed] [Google Scholar]
- Harada H., Harada,Y., O’Brien,D.P., Rice,D.S., Naeve,C.W. and Downing,J.R. (1999) HERF1, a novel hematopoiesis-specific RING finger protein, is required for terminal differentiation of erythroid cells. Mol. Cell. Biol., 19, 3808–3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins C.F. (1992) ABC transporters: from microorganisms to man. Annu. Rev. Cell Biol., 8, 67–113. [DOI] [PubMed] [Google Scholar]
- Hogue D.L., Liu,L. and Ling,V. (1999) Identification and characterization of a mammalian mitochondrial ATP-binding cassette membrane protein. J. Mol. Biol., 285, 379–389. [DOI] [PubMed] [Google Scholar]
- Jardine P.E., Cotter,P.D., Johnson,S.A., Fitzsimons,E.J., Tyfield,L., Lunt,P.W. and Bishop,D.F. (1994) Pyridoxine-refractory congenital sideroblastic anaemia with evidence for autosomal inheritance: exclusion of linkage to ALAS2 at Xp11.21 by polymorphism analysis. J. Med. Genet., 31, 213–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasturi J., Basha,H.M., Smeda,S.H. and Swehli,M. (1982) Hereditary sideroblastic anaemia in 4 siblings of a Libyan family—autosomal inheritance. Acta Haematol., 68, 321–324. [DOI] [PubMed] [Google Scholar]
- Keller G., Kennedy,M., Papayannopoulou,T. and Wiles,M.V. (1993) Hematopoietic differentiation during embryonic stem cell differentiation in culture. Mol. Cell. Biol., 13, 472–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kispal G., Csere,P., Prohl,C. and Lill,R. (1999) The mitochondrial proteins Atm1p and Nfs1p are essential for biogenesis of cytosolic Fe/S proteins. EMBO J., 18, 3981–3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotkow K.J. and Orkin,S.H. (1995) Dependence of globin gene expression in mouse erythroleukemia cells on the NF-E2 heterodimer. Mol. Cell. Biol., 15, 4640–4647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leighton J. (1998) Mitochondrial ABC transporters. Methods Enzymol., 292, 776–787. [DOI] [PubMed] [Google Scholar]
- Leighton J. and Schatz,G. (1995) An ABC transporter in the mitochondrial inner membrane is required for normal growth of yeast. EMBO J., 14, 188–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosser J., Douar,A.M., Sarde,C.O., Kioschis,P., Feil,R., Moser,H., Poustka,A.M., Mandel,J.L. and Aubourg,P. (1993) Putative X-linked adrenoleukodystrophy gene shares unexpected homology with ABC transporters. Nature, 361, 726–730. [DOI] [PubMed] [Google Scholar]
- Neupert W. (1997) Protein import into mitochondria. Annu. Rev. Biochem., 66, 863–917. [DOI] [PubMed] [Google Scholar]
- Orkin S.H. (1992) GATA-binding transcription factors in hematopoietic cells. Blood, 80, 575–581. [PubMed] [Google Scholar]
- Orkin S.H., Harosi,F.I. and Leder,P. (1975) Differentiation in erythroleukemic cells and their somatic hybrids. Proc. Natl Acad. Sci. USA, 72, 98–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pevny L., Simon,M.C., Robertson,E., Klein,W.H., Tsai,S.-H., D’Agati,V., Orkin,S.H. and Costantini,F. (1991) Erythroid differentiation in chimaeric mice blocked by a targeted mutation in the gene for transcription factor GATA-1. Nature, 349, 257–260. [DOI] [PubMed] [Google Scholar]
- Ponka P. (1997) Tissue-specific regulation of iron metabolism and heme synthesis: distinct control mechanisms in erythroid cells. Blood, 89, 1–25. [PubMed] [Google Scholar]
- Ponka P. (1999) Cell biology of heme. Am. J. Med. Sci., 318, 241–256. [DOI] [PubMed] [Google Scholar]
- Riordan J.R. et al. (1989) Identification of the cystic fibrosis gene: cloning and characterization of complementary DNA. Science, 245, 1066–1073. [DOI] [PubMed] [Google Scholar]
- Shimada Y. et al. (1998) Cloning and chromosomal mapping of a novel ABC transporter gene (hABC7), a candidate for X-linked sideroblastic anemia with spinocerebellar ataxia. J. Hum. Genet., 43, 115–122. [DOI] [PubMed] [Google Scholar]
- Strautnieks S.S. et al. (1998) A gene encoding a liver-specific ABC transporter is mutated in progressive familial intrahepatic cholestasis. Nature Genet., 20, 233–238. [DOI] [PubMed] [Google Scholar]
- Tsai S.F., Martin,D.I.K., Zon,L.I., D’Andrea,A.D., Wong,G.G. and Orkin,S.H. (1989) Cloning of cDNA for the major DNA-binding protein of the erythroid lineage through expression in mammalian cells. Nature, 339, 446–451. [DOI] [PubMed] [Google Scholar]
- Tsang A.P., Visvader,J.E., Turner,C.A., Fujiwara,Y., Yu,C., Weiss,M.J., Crossley,M. and Orkin,S.H. (1997) FOG, a multitype zinc finger protein, acts as a cofactor for transcription factor GATA-1 in erythroid and megakaryocytic differentiation. Cell, 90, 109–119. [DOI] [PubMed] [Google Scholar]
- Vainio S., Karavanova,I., Jowett,A. and Thesleff,I. (1993) Identification of BMP-4 as a signal mediating secondary induction between epithelial and mesenchymal tissues during early tooth development. Cell, 75, 45–58. [PubMed] [Google Scholar]
- Veltman J.C. and Maines,M.D. (1986) Alterations of heme, cytochrome P-450, and steroid metabolism by mercury in rat adrenal. Arch. Biochem. Biophys., 248, 467–478. [DOI] [PubMed] [Google Scholar]
- Walker J.E., Saraste,M., Runswick,M.J. and Gay,N.J. (1982) Distantly related sequences in the α - and β-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J., 1, 945–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss M.J. and Orkin,S.H. (1995a) GATA transcription factors: key regulators of hematopoiesis. Exp. Hematol., 23, 99–107. [PubMed] [Google Scholar]
- Weiss M.J. and Orkin,S.H. (1995b) Transcription factor GATA-1 permits survival and maturation of erythroid precursors by preventing apoptosis. Proc. Natl Acad. Sci. USA, 92, 9623–9627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss M.J., Keller,G. and Orkin,S.H. (1994) Novel insights into erythroid development revealed through in vitro differentiation of GATA-1– embryonic stem cells. Genes Dev., 8, 1184–1197. [DOI] [PubMed] [Google Scholar]
- Weiss M.J., Yu,C. and Orkin,S.H. (1997) Erythroid-cell-specific properties of transcription factor GATA-1 revealed by phenotypic rescue of a gene-targeted cell line. Mol. Cell. Biol., 17, 1642–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K., Kniazeva,M., Hutchinson,A., Han,M., Dean,M. and Allikmets,R. (1999) The ABCR gene in recessive and dominant Stargardt diseases: a genetic pathway in macular degeneration. Genomics, 60, 234–237. [DOI] [PubMed] [Google Scholar]