Abstract
We analyzed the targeting of histone acetyltransferase (HAT) complexes by DNA-binding activators during transcriptional activation and the resulting distribution of acetylated histones. An in vitro competition assay was developed to acetylate and transcribe a nucleosomal array template in the presence of excess non-specific chromatin, which mimics in vivo conditions. Stimulation of transcription from the nucleosomal array template under competitive conditions by the SAGA and NuA4 HAT complexes depended on the presence of the Gal4-VP16 activator, which recognizes sites in the promoter and directly interacts with these HATs. Importantly, the stimulation of transcription by SAGA and NuA4 depended on the presence of Gal4-VP16 during histone acetylation, and Gal4-VP16-bound nucleosomal templates were acetylated preferentially by SAGA and NuA4 relative to the competitor chromatin. While targeting of the SAGA complex led to H3 acetylation of promoter-proximal nucleosomes, targeting of the NuA4 complex led to a broader domain of H4 acetylation of >3 kbp. Thus, either promoter-proximal H3 acetylation by SAGA or broadly distributed acetylation of H4 by NuA4 activated transcription from chromatin templates.
Keywords: chromatin/HATs/targeted acetylation/transcriptional regulation/yeast
Introduction
The level of acetylation of the N-terminal tails of the core histones results from a balance between the opposing actions of histone acetyltransferases (HATs) and histone deacetylases (HDACs) (reviewed in Kuo and Allis, 1998; Grant and Berger, 1999). Several nuclear HAT complexes with nucleosomal HAT activity (hence, likely to be involved in transcriptional regulation) have been identified in Saccharomyces cerevisiae, including the SAGA, NuA4, ADA and NuA3 complexes. While SAGA, ADA and NuA3 preferentially acetylate nucleosomal histone H3, NuA4 shows a preference for histone H4 (Eberharter et al., 1999; Allard et al., 1999; Grant and Berger, 1999; John et al., 2000). The cellular functions of these HAT complexes are not yet fully understood; however, it appears that each individual HAT is required for the regulation of only a subset of yeast genes. For example, genome-wide expression analysis indicated that Gcn5p, the catalytic subunit of the SAGA and ADA complexes, is absolutely required for the expression of only 5% of yeast genes (Holstege et al., 1998). Gcn5p has been shown to participate in the regulation of genes involved in amino acid biosynthesis (via the Gcn4 activator) and respiration (via the HAP complex) (Georgakopoulos and Thireos, 1992). It also participates in mating type switching and sucrose and phosphate metabolism (reviewed in Belotserkovskaya and Berger, 1999). Esa1, the catalytic subunit of yeast NuA4, has been less studied but also shows gene-specific effects (Galarneau et al., 2000). Thus, an important issue regarding the mechanism of action of HATs and other chromatin-modifying complexes is how they recognize their target genes in the context of the entire genome. The prevailing hypothesis is that HATs, as well as other chromatin-modifying complexes, are ‘recruited’ specifically to certain genes due to their ability to interact with DNA-binding proteins that regulate transcription (Struhl, 1998). Recent in vivo and in vitro experiments have shown that indeed the targeting of the yeast SWI/SNF complex can occur by interactions with acidic transcription activation domains (Natarajan et al., 1999; Neely et al., 1999; Yudkovsky et al., 1999; Wallberg et al., 2000). Moreover, this interaction is required for in vitro stimulation of transcription from chromatin templates in the presence of competitor chromatin (Neely et al., 1999; Wallberg et al., 2000). HDAC complexes have also been shown to repress the transcription of specific genes based on their ability to associate with co-repressors and locally deacetylate histones (Kadosh and Struhl, 1998a,b; Rundlett et al., 1998; reviewed in Struhl, 1998). Along the same lines, a number of studies support the hypothesis that the regulation of gene expression by some HAT complexes is mediated by their interaction with DNA-binding proteins and subsequent histone acetylation. For example, several human co-activator proteins that interact with nuclear receptors possess intrinsic HAT activity (reviewed in Janknecht and Hunter, 1996; Xu et al., 1999), and the acetylation of nucleosomes associated with promoter elements in vivo correlates with transcriptional activation (Kuo et al., 1998; Parekh and Maniatis, 1999). Also, the yeast SAGA and NuA4 complexes have been shown to interact directly with the acidic activation domain and selectively stimulate transcription driven by these activators (Utley et al., 1998; Ikeda et al., 1999; Wallberg et al., 1999) (see below). However, in addition to histones, there is evidence that transcription factors and other non-histone proteins might be relevant targets for HAT complexes (reviewed in Brown et al., 2000). Moreover, acetyl-CoA has recently been shown to increase in vitro transcription in the absence of histone proteins (Galasinski et al., 2000). In light of these studies, it is increasingly important to identify the relevant targets of the HAT complexes in these assays in order to understand their function in transcription activation.
Another important aspect of the function of the HAT complexes in transcription is the distribution of acetylated histones resulting from the recruitment of HATs to specific promoters. Studies that measure the in vivo distribution of acetylated histones relative to a gene sequence reveal the consequence of the combined action of all HATs and HDACs that might act on a gene. The data obtained from these studies have resulted in two models of acetylated histone distribution. One model proposes that a broad domain of several kilobase pairs surrounding a gene poised for transcription is associated with hyperacetylated histones. It is supported by studies of the chicken β-globin gene cluster, which detected a higher level of acetylated histones within a broad DNase I-sensitive domain of 33 kb that encompassed transcriptionally active globin genes (Hebbes et al., 1992, 1994). The antibodies used in these studies bind acetylated lysines, and preferentially recognize acetylated forms of H3 and H4 (Hebbes et al., 1988; Vettese-Dadey et al., 1996). In contrast, a second model proposes that the distribution of acetylated histones is restricted to a promoter-proximal region. For example, a 600 bp region of the yeast HIS3 promoter (including the UAS and TATA boxes) was found to be associated with hyperacetylated H3 in a process that depended on the HAT activity of Gcn5 (Kuo et al., 1998). Moreover, the Gcn5-dependent acetylation of H3 in a UASgal–CYC1–lacZ construct was localized to the promoter and did not extend 1.5 kb downstream into the coding region (Kuo et al., 1998). A similar pattern of promoter-localized acetylation was observed for the human interferon-β (IFN-β) promoter. For this gene, the domain of H3 and H4 hyperacetylation was restricted to 600 bp around the start site of transcription and thus spanned between two and three nucleosomes. The data suggest that the localized histone acetylation required the targeting of p300/CBP by the IRF-3 activator (Parekh and Maniatis, 1999). Slightly different results were obtained from the study of the yeast HO promoter, where the authors observed a Gcn5p-dependent domain of acetylation of H3 (and H4) that extended throughout a 1 kb region upstream of the promoter, i.e. spanning 6–7 nucleosomes. This region includes transcription factor-binding sites and the TATA box, but does not extend into the coding region of the gene (Krebs et al., 1999). Interestingly, the association of Gcn5 with this promoter depends on the recruitment of the SWI/SNF remodeling complex by Swi5p (Cosma et al., 1999).
In order to investigate further the function of the activation domain–HAT complex interactions in transcription activation by SAGA and NuA4, we have employed a competitive assay that analyzes the ability of yeast HATs to acetylate a nucleosomal template and stimulate transcription from its promoter in the presence of an excess of non-template chromatin, as would occur in vivo. These studies revealed that transcriptional stimulation by SAGA and NuA4 requires Gal4-VP16-mediated targeting of histone acetylation by these complexes to the template nucleosomal array. While acetylation either of H4 by NuA4 or of H3 by SAGA is sufficient for transcription stimulation, the distribution of acetylated histones resulting from targeting of these two complexes differs. Gal4-VP16 targeting of the SAGA complex primarily results in the acetylation of the histone H3 contained in the first few nucleosomes surrounding the promoter. In contrast, targeting of the NuA4 complex leads to a broader domain of histone H4 acetylation that spreads across the 3 kb nucleosomal array.
Results
Recruitment of SAGA and NuA4 by Gal4-VP16 in vitro is required for enhanced transcription in the presence of competitor chromatin
To address whether HAT complexes can enhance expression specifically from particular promoters upon recruitment, we have investigated the ability of SAGA and NuA4 to regulate Gal4-VP16-driven transcription in an in vitro system containing a large excess of competitor cellular chromatin. In particular, we analyzed the ability of the acidic activator Gal4-VP16 to direct the activity of these complexes to a specific promoter under these competitive conditions, and we determined the effect of this specific, activator-targeted acetylation on transcription. As a negative control, we used the NuA3 HAT complex, which does not interact with acidic activators (Utley et al., 1998).
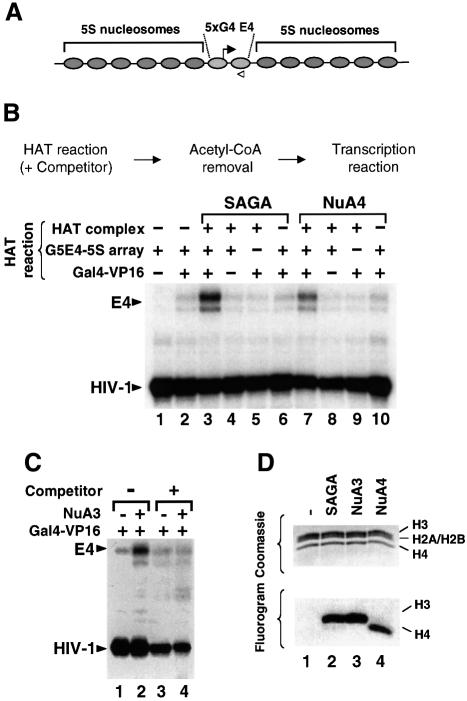
The G5E4-5S nucleosomal array, which contains the minimal E4 promoter and five Gal4-binding sites assembled into an evenly spaced array of nucleosomes (Figure 1A; Utley et al., 1998; Ikeda et al., 1999), was used to test the importance of targeting histone acetylation for transcriptional activation mediated by the SAGA and NuA4 complexes. The assay used separates the acetylation of histones by these HATs from the process of transcription by means of the removal of acetyl-CoA after the HAT reaction (Figure 1B). In brief, the G5E4-5S nucleosomal array was treated with SAGA or NuA4 in the presence or absence of Gal4-VP16. Importantly, competitor chromatin was included in all reactions to challenge the ability of the HAT complexes to acetylate the G5E4-5S template in the absence of targeting. Acetyl-CoA was removed using spin columns to terminate the histone acetylation reaction, and column eluates were analyzed for their transcriptional potential by adding HeLa nuclear extract and rNTPs. When Gal4-VP16 was omitted during the acetylation reaction, it was added to spin column eluates so that it was present in all transcription reactions. Thus, in this assay, Gal4-VP16-targeted acetylation was restricted to the period preceding the removal of acetyl-CoA, while HAT-independent transcriptional activation by Gal4-VP16 was kept constant in all samples. As a consequence of this, transcriptional differences generated from HAT reactions performed in either the presence or absence of Gal4-VP16 should be attributable to activator–HAT interactions that confer directed histone acetylation. Therefore, this assay distinguishes between the ability of Gal4-VP16 to activate transcription in a HAT-independent manner by interacting with components of the general transcription machinery and its potential role in facilitating transcription by directing histone acetylation to particular nucleosomes.
Fig. 1. Targeted histone acetylation by SAGA and NuA4 is required for stimulated transcription in vitro under competitive conditions. (A) Construct used in the transcription experiments, showing the position of the Gal4 DNA-binding sites and the 5S rDNA repeats. The chromatin template was generated as described in Materials and methods. The filled arrow signals the initiation site and the direction of transcription, while the open arrowhead shows the position and orientation of the oligonucleotide used for RNA analysis by primer extension. (B) Transcription assay examining the influence of targeted histone acetylation on transcription. The G5E4-5S nucleosomal array was transcribed following HAT reactions including (+) or omitting (–) Gal4-VP16, the G5E4-5S array and Superose 6-purified HAT complexes as indicated. All HAT reactions contained acetyl-CoA, a 50-fold molar excess of purified chromatin relative to the G5E4-5S array and an HIV-1 plasmid as an internal control for recovery. As indicated in the top diagram, acetyl-CoA was removed from the reaction after the acetylation step and before the transcription step. In lanes 3–10, spin column eluates lacking either Gal4-VP16 (lanes 4 and 8), the G5E4-5S array (lanes 5 and 9) or HAT complexes (lanes 6 and 10) were supplemented with the omitted component, so that transcription was performed under constant conditions. (C) The G5E4-5S nucleosomal array was transcribed following HAT reactions in the presence (+) or absence (–) of competitor chromatin and the NuA3 complex as indicated. All lanes contained Gal4-VP16. As in (B), the HIV-1 plasmid was included in the reaction as a recovery control. (D) The Superose 6 fractions of partially purified SAGA (lane 2), NuA3 (lane 3) and NuA4 (lane 4) were tested for their ability to acetylate nucleosomal histones. The samples were separated by SDS–PAGE, and the gel was stained with Coomassie Blue to determine the position of the core histones (top panel) and treated for fluorography to reveal the acetylation pattern (lower panel).
The influence of targeted acetylation by SAGA and NuA4 on transcription is examined in lanes 3–10 of Figure 1B. The addition of Gal4-VP16 to the HAT reaction promoted transcription stimulation by the SAGA or NuA4 complexes (compare lane 3 or 7 with lane 2). In contrast, when Gal4-VP16 was omitted from the HAT reaction and added only after acetyl-CoA depletion, transcriptional stimulation by SAGA and NuA4 was not observed (compare lane 4 or 8 with lane 2). Thus, in the presence of competitor chromatin, Gal4-VP16 was required to mediate increased transcription by SAGA and NuA4. The HAT reactions contained only DNA, histones, Gal4-VP16 and purified HAT complex. To determine whether the observed stimulation might result from the acetylation of Gal4-VP16 itself, acetyl-CoA was removed from a reaction containing only activator and HAT complexes, prior to the addition of the template. Transcription levels from these reactions were very similar to that generated with the unacetylated template (compare lane 5 or 9 with lane 2), indicating that the potential acetylation of Gal4-VP16 is not responsible for facilitating transcription in this assay. Similarly, low levels of transcription were observed when the HAT complexes were added after acetyl-CoA removal (lanes 6 and 10). As a control, we examined whether NuA3, which does not interact directly with Gal4-VP16, was able to enhance transcription from the G5E4-5S nucleosomal template under competitive conditions. In this experiment, all reactions contained Gal4-VP16, but differed with respect to the presence of competitor chromatin and NuA3. As shown in Figure 1C, NuA3 was capable of mediating increased transcription from the G5E4 promoter in the absence of competitor chromatin, where it could modify the template (compare lanes 1 and 2). In contrast, in the presence of competitor chromatin, NuA3 did not increase transcription beyond background levels in spite of the pre-incubation of the template with Gal4-VP16 (compare lane 4 with lane 3). In conclusion, the experiments presented in Figure 1 reveal that only those HAT complexes that can physically associate with Gal4-VP16 are capable of facilitating transcription under competitive conditions. Furthermore, these data show that transcriptional stimulation by the SAGA and NuA4 complexes depends on their ability to acetylate histones on the G5E4-5S nucleosomal array.
Gal4-VP16 directs the acetylase activity of SAGA and NuA4 to activator-bound nucleosomal arrays in the presence of competitor chromatin
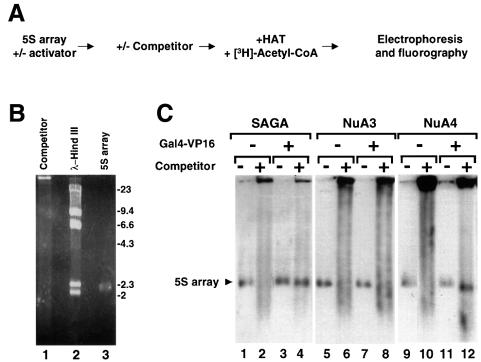
To visualize directly the effect of Gal4-VP16 on acetylation of the G5E4-5S nucleosomal array, we developed an assay that distinguishes between the acetylation of histones contained in the transcription template and those present in the competitor chromatin. Briefly, a 10- to 30-fold mass excess of competitor chromatin was added to the nucleosomal array pre-bound with Gal4-VP16. Purified SAGA, NuA3 or NuA4, normalized by their nucleosome HAT activity (Figure 1D), were then added. HAT reactions were carried out in the presence of [3H]acetyl-CoA, so that the histones that became acetylated were labeled with tritiated acetyl groups. Gal4-VP16 was subsequently competed from the template by incubation with a large molar excess of double-stranded oligonucleotide containing a consensus Gal4 DNA- binding site. Finally, the samples were separated by agarose electrophoresis and visualized by fluorography (Figure 2A).
Fig. 2. Gal4-VP16 targets the HAT activity of SAGA and NuA4, but not that of NuA3, to the G5E4-5S array in the presence of competitor chromatin. (A) Diagram of the experimental protocol. (B) Ethidium bromide-stained agarose gel showing the migration profiles of the chromatin used as competitor (lane 1) and the reconstituted G5E4-5S array (lane 3) under the electrophoretic conditions used in (C). Lane 2 is HindIII-digested λ DNA. (C) The reconstituted array was incubated in the absence (lanes 1, 2, 5, 6, 9 and 10) or presence (3, 4, 7, 8, 11 and 12) of Gal4-VP16, and competitor chromatin was added to even-numbered lanes as indicated. Next, the reactions were incubated with SAGA, NuA3 or NuA4 in the presence of [3H]acetyl-CoA, and Gal4-VP16 was competed off by incubation with an oligonucleotide corresponding to the consensus Gal4-binding site. The samples were then separated by agarose gel electrophoresis. Finally, the gels were treated for fluorography and exposed. The arrowhead indicates the position of the 5S array (also compare with lane 3 of B).
The G5E4-5S nucleosomal array migrates as a discrete band upon gel electrophoresis (Figure 2B, lane 3), while the competitor chromatin migrates as a smear due to its heterogeneous length (lane 1). As shown in Figure 2C, all of the HATs tested in this assay were capable of acetylating the G5E4-5S nucleosomal array efficiently in the absence of competitor chromatin (lanes 1, 5 and 9). Upon the addition of competitor chromatin (lanes 2, 6 and 10), instead of the discrete signal corresponding to the acetylation of the 5S array, a smear corresponding to the acetylation of the competitor chromatin was observed. This indicates that the excess chromatin successfully competed the HAT activities away from the 5S nucleosomal array. However, when the template was pre-bound with Gal4-VP16, both SAGA and NuA4 were able to acetylate the nucleosomes contained in the array in the presence of the competitor chromatin (lanes 4 and 12). In contrast, the addition of Gal4-VP16 did not rescue the inability of NuA3 to acetylate the array histones in the presence of competitor chromatin (compare lane 6 with lane 8). Thus, the specific acetylation of the G5E4-5S array by either SAGA or NuA4 under competitive conditions was observed only in the presence of Gal4-VP16. This observation correlates both with the ability of the HAT complexes to interact with Gal4-VP16 and with their ability to increase transcription in the presence of competitor chromatin. Taken together, these data provide direct evidence that both SAGA and NuA4 can facilitate transcription by directing histone acetylation to activator-bound promoters as a result of their physical interaction with DNA-binding transcriptional activators.
Establishment of an in vitro system to analyze the distribution of histones acetylated by the HAT complexes
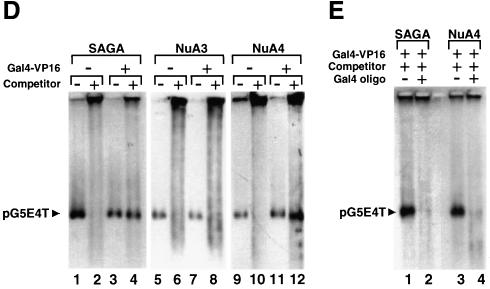
In vivo experimental data suggest that transcription-related acetylation can be localized to a small number of nucleosomes within the promoter region of some genes. For other genes, however, the domain of acetylated histones extends over the whole coding region (see above). In light of this, we were interested in determining the extent of the domain of acetylated chromatin generated by the SAGA and NuA4 HAT complexes upon recruitment by Gal4-VP16. Because the use of the G5E4-5S array to address this issue would be hampered by the repetitive 5S sequences in the template, we used the pG5E4T plasmid as the source of the nucleosomal template (Lin et al., 1988). pG5E4T is the parental plasmid used to generate the G5E4-5S construct (Utley et al., 1998; Ikeda et al., 1999) and, therefore, contains the same promoter fragment (i.e. five Gal4 DNA-binding sites upstream of the adenovirus 2 E4 minimal promoter) but lacks the 5S rDNA repeats (Figure 3A).
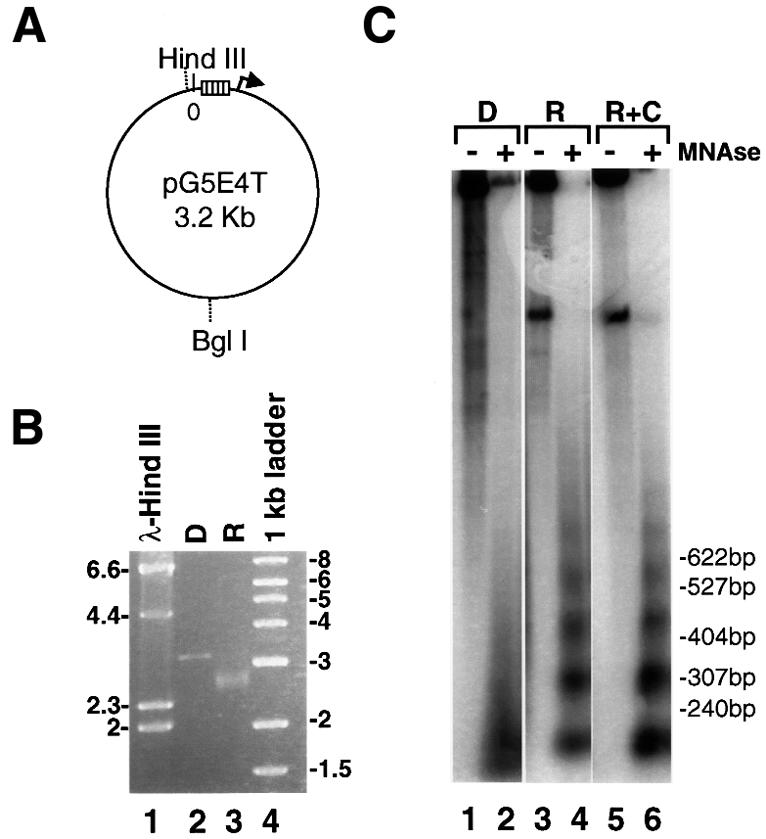
Fig. 3. The HAT activity of SAGA and NuA4, but not that of NuA3, is recruited to the pG5E4T nucleosomal array by Gal4-VP16. (A) Diagram showing the location of the Gal4-binding sites (open rectangles) and the adenovirus 2 E4 minimal promoter (arrow) in the pG5E4T plasmid, as well as selected restriction enzyme sites. (B) Agarose electrophoresis of the in vitro reconstituted pG5E4T template (R) and of non-reconstituted DNA (D). Lane 1 corresponds to HindIII-digested λ DNA and lane 4 is the 1 kb ladder from NEB. (C) The in vitro reconstitution of pG5E4T generates a regular nucleosomal ladder upon MNase digestion. Naked DNA (D) and reconstituted array +/– competitor chromatin (R and R+C, respectively) were either mock digested (–) or digested with appropriate amounts of MNase (+). DNA was extracted from these reactions, separated by electrophoresis using 32P-labeled pBR322/MspI as the molecular weight marker, and subjected to Southern blotting using hexanucleotide-labeled pG5E4T DNA as a probe. (D) Fluorography of agarose gels showing the targeting of the different HAT activities to the reconstituted pG5E4T array. The experiment was performed as described in Figure 2C. (E) The recruitment of SAGA and NuA4 to the pG5E4T template depends on the ability of Gal4-VP16 to bind DNA. The experiment was performed as in (D), but all reactions contain Gal4-VP16 and competitor chromatin, as indicated. A large molar excess of an oligonucleotide corresponding to the consensus Gal4-binding site (Gal4 oligo) was added either after the HAT reaction (lanes 1 and 3, also labeled –) or together with Gal4-VP16 (lanes 2 and 4, labeled +).
The pG5E4T plasmid was linearized by restriction enzyme digestion and assembled into chromatin in vitro by the salt dilution transfer method (Steger et al., 1998). G5E4T does not possess known octamer-positioning properties and it was assembled into chromatin in the absence of nucleosome-spacing factors. As a result of this, we characterized the structure of the reconstituted nucleosomal template in some detail. As expected, the mobility of the reconstituted template was different from that of the histone-free DNA upon agarose electrophoresis (Figure 3B). Furthermore, the apparent absence of free DNA indicated that the G5E4T DNA was reconstituted efficiently into nucleosomes. Micrococcal nuclease (MNase) digestion (Figure 3C) produced the characteristic nucleosomal ladder for the nucleosomal template (R) but not for free DNA (D). The nucleosome repeat was visible up to six or seven nucleosomes. The addition of a 10- to 30-fold mass excess of competitor chromatin had no visible effect on the organization of the nucleosomal template as assayed by this technique (R+C). Finally, when short probes complementary to different regions of the template DNA (see Figure 5A) were used in Southern blots of the MNase digests, the MNase pattern observed was indistinguishable from that obtained with the full-length probe (data not shown). This result suggested that octamers were distributed along the entire template DNA.
Fig. 5. Gal4-VP16 directs the HAT activity of the SAGA complex to promoter-proximal nucleosomes. The experimental set-up for these ‘scanning ChIPS’ is similar to that used in Figure 4: the template was incubated in the absence (–) or presence (+) of Gal4-VP16, and competitor chromatin was added where indicated. Next, the reactions were incubated with SAGA and acetyl-CoA (or mock acetylated in the absence of HAT complex), MNase digested and immunoprecipitated with anti-acetylated H3 antibody. DNA was extracted from the bound and unbound fractions and slot-blotted. The membranes were hybridized successively with a series of short probes (between 250 and 300 bp) that scan the length of the template, generated by PCR and labeled by primer extension from random hexanucleotides (Boehringer Mannheim). (A) Diagram showing the localization of the different probes when the plasmid is digested with BglI. (B) Average values and standard deviation of normalized data from three repeats of the experiment. The background signal (–HAT) was subtracted from the values obtained in the presence of SAGA for each condition. The numbers under the graph show the ratio of proximal (average of +A and –A) versus distal (average of +C and –C) signal for each condition tested. ND, not determined. (C) The reconstituted array was pre-incubated with Gal4-VP16 and competitor chromatin was added. Then, the template was acetylated by SAGA in the presence of acetyl-CoA, reactions were digested with MNase (+) or mock-digested (–), and the immunoprecipitation and slot blot were carried out as described above.
Consistent with the results shown in Figure 2C, efficient acetylation of the pG5E4T nucleosomal template by either SAGA or NuA4 under competitive conditions was observed only in the presence of Gal4-VP16 (Figure 3D). As in the case of the G5E4-5S nucleosomal array template, NuA3 was not able to acetylate the pG5E4T template under competitive conditions, regardless of the presence of Gal4-VP16. This result demonstrates that the VP16-mediated recruitment of the HAT complexes to this nucleosomal template is identical to their recruitment to the G5E4-5S array. Next, we tested whether the ability of SAGA and NuA4 to acetylate the nucleosomal template could be prevented if the activator could no longer bind the nucleosomal array. To this end, we added a large molar excess of a double-stranded oligonucleotide containing a Gal4-binding site simultaneously with the activator. In these conditions, the specific acetylation of the array by both SAGA and NuA4 was prevented (Figure 3E, lanes 2 and 4). In contrast, the addition of oligonucleotide after the acetylation reaction had no effect (Figure 3E, lanes 1 and 3, and also all lanes in Figures 2C and 3D). This result shows that, under competitive conditions, the binding of Gal4-VP16 to cognate sites in the promoter is required for the targeting of the acetyltransferase activity of the SAGA and NuA4 complexes to the nucleosomal array.
In vitro chromatin immunoprecipitation assays illustrate the specific targeting of the HAT activity of SAGA and NuA4 to the Gal4 template under competitive conditions
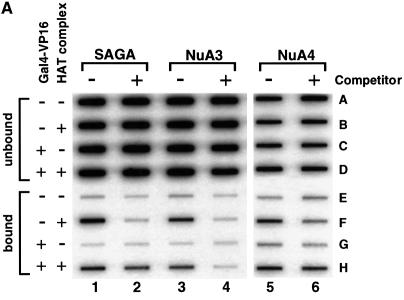
We measured chromatin acetylation generated by SAGA and NuA4 upon targeting using the chromatin immunoprecipitation (ChIP) technique, which has previously been used to study chromatin acetylation in vivo (Kuo and Allis, 1999). The initial set-up of these experiments was very similar to that used in the fluorography experiments presented in Figures 2 and 3. The reconstituted pG5E4T array was incubated with Gal4-VP16 in the presence of a large mass excess of competitor chromatin. The templates were then acetylated in the presence of unlabeled acetyl-CoA by each of the three HAT complexes tested (SAGA, NuA3 and NuA4). Next, the reactions were immunoprecipitated with antibodies specific for the acetylated forms of the core histones (Upstate Biotechnology). The antibody used for the SAGA and NuA3 experiments recognizes acetylated lysines 9 and 14 in the N-terminal tail of H3 and thus overlaps with the preferential sites of acetylation of these HATs (Grant et al., 1999; S.John and J.L.Workman, unpublished data). The antibody used for the NuA4 experiments was generated against a histone H4 N-terminal tail peptide acetylated in four lysines (amino acids 5, 8, 12 and 16). After the immunoprecipitation, DNA was extracted from the unbound and bound fractions, slot blotted and then hybridized with radiolabeled pG5E4T DNA.
The result of one of these experiments is shown in Figure 4A. In the absence of competitor chromatin, the addition of SAGA, NuA3 or NuA4 to the acetylation reaction increased the amount of DNA immunoprecipitated by the antibodies specific for the acetylated forms of H3 (SAGA and NuA3) or H4 (NuA4). Moreover, this effect was independent of the pre-incubation of the nucleosomal array with Gal4-VP16 (compare rows E with F and G with H of odd-numbered columns). This result is consistent with the fluorography data presented in Figures 2 and 3, and with previous data showing that none of these yeast HATs requires an activator to acetylate nucleosomal templates when these are the only substrate available in the reaction (e.g. see Figure 1D). The picture is quite different under competitive conditions. As before, NuA3 was unable to acetylate the array in the presence of competitor chromatin (see rows F and H of column 4). However, for both SAGA and NuA4, the increased amount of DNA immunoprecipitated under competitive conditions was dependent on the pre-incubation of the array with Gal4-VP16 (compare row H with F for columns 2 and 6). This result is in agreement with the fluorography data presented in Figures 2 and 3. Figure 4B–D shows the quantitation of 3–4 independent experiments performed with SAGA (Figure 4B), NuA3 (Figure 4C) or NuA4 (Figure 4D). The negative control columns, where no HAT was added (–HAT), show the basal level of template immunoprecipitated by each antibody under the different conditions (+/– Gal4-VP16, +/– competitor). This background includes any endogenous acetylation of the histones used to reconstitute the DNA template, and any material immunoprecipitated non-specifically by the antibodies. For all the HATs, the columns where the complexes were added (+) in the absence of competitor chromatin (–) show an increase in the amount of material immunoprecipitated. Interestingly, we noticed that the activity of NuA4 seems to be slightly diminished by the presence of Gal4-VP16, while those of SAGA and NuA3 seem to be unaffected (compare columns +/– Gal4-VP16 in the absence of competitor, for the different HATs). The possible implications of this observation will be discussed later. The fold stimulation elicited by each HAT in the different conditions tested is summarized in Figure 4E. These values were calculated by dividing the amount of material immunoprecipitated by the antibody after acetylation by the HAT complexes by the amount immunoprecipitated in the absence of the complexes. The graph clearly shows that all the HATs functioned equally well in the absence of competitor chromatin, independently of the presence of the activator. However, while SAGA and NuA4 retained the ability to acetylate the template nucleosomal array in the presence of competitor chromatin in an activator-dependent manner, NuA3 did not.
Fig. 4. In vitro ChIP assays confirm the targeting of the HAT activity of SAGA and NuA4 to the pG5E4T array in the presence of competitor chromatin. (A) The reconstituted array was incubated in the absence (rows A, B, E and F) or presence (rows C, D, G and H) of Gal4-VP16 as in Figure 3. Competitor chromatin was added as indicated (columns 2, 4 and 6). After acetylation by SAGA (columns 1 and 2), NuA3 (3 and 4) or NuA4 (5 and 6), the substrates were immunoprecipitated with antibodies directed against acetylated H3 (SAGA and NuA3) or H4 (NuA4). DNA was extracted from the unbound (rows A–D) and bound (rows E–H) fractions and applied to a Zeta-Probe membrane by slot blot. The membrane was hybridized with hexanucleotide-labeled pG5E4T DNA. (B–D) Graphic representation of the data shown in (A). The membranes were exposed to a PhosphorImager and quantitated. The graphs show the average and standard deviation of 3–4 repeats of each experiment with the SAGA (B), NuA3 (C) or NuA4 (D) complexes. (E) Fold stimulation by each HAT under the different conditions. The y-axis corresponds to the ratio of the material immunoprecipitated in the presence of each HAT complex (% IP with HAT) divided by the amount precipitated in the absence of the complex (% IP without HAT).
In summary, the experiments presented thus far show that, under competitive conditions, the in vitro acetylation of a nucleosomal template that contains Gal4-binding sites depends on the ability of the different HAT complexes to interact with Gal4-VP16. From a more general point of view, the data suggest that the specific interaction of HATs with DNA-binding activators can target the HAT activity of these complexes to particular promoters in chromatin.
Profiles of targeted acetylation by SAGA and NuA4
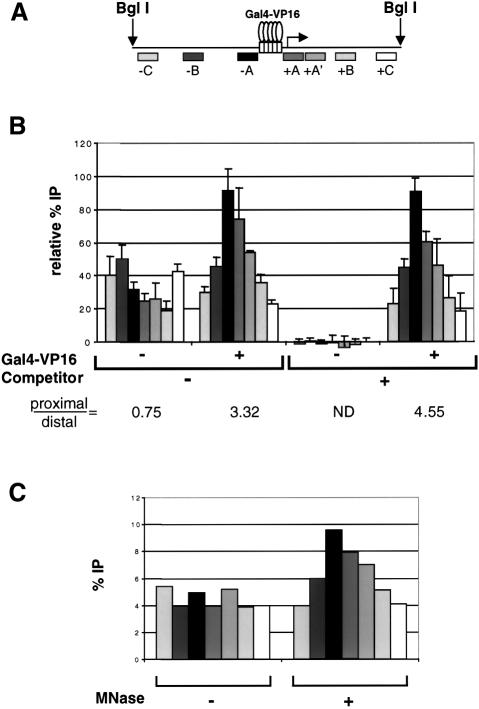
In order to define the nucleosomal domain that was acetylated by the HAT complexes upon recruitment, we performed ‘scanning ChIP’ assays. After the acetylation step, the pG5E4T nucleosomal array template was digested with MNase. This material was then immunoprecipitated with antibodies specific for the acetylated forms of the histones (see above). DNA was extracted from the bound and unbound fractions, and slot-blotted. To control for the immunoprecipitation conditions, the membranes were initially hybridized with the full-length probe used in Figure 4. Next, the membranes were stripped and hybridized successively with a series of probes that span the length of the array. The intensity of the signal obtained with each probe reveals the degree of association of that particular fragment of the template DNA with acetylated histones.
All the probes used in these ‘scanning ChIP’ assays are between 250 and 300 bp long (Figure 5A). Those probes that hybridize in the downstream half of the BglI-linearized array (oriented in the direction of transcription from the E4 promoter) are indicated with plus signs, while those that hybridize upstream of the Gal4 DNA-binding sites have minus signs. The first set of symmetrical probes, which we called +/–A, hybridize immediately adjacent to either side of the Gal4-binding sites. The second set of probes (+/–B) are complementary to sequences 800 bp to 1 kb away from the sites. The third set (+/– C) hybridize to either end of the BglI-linearized fragment, ∼1.5 kb away from the Gal4 sites. Additionally, we used probe +A′, which hybridizes immediately downstream of probe +A, in the direction of transcription and, therefore, is centered ∼500 bp downstream of the Gal4 sites.
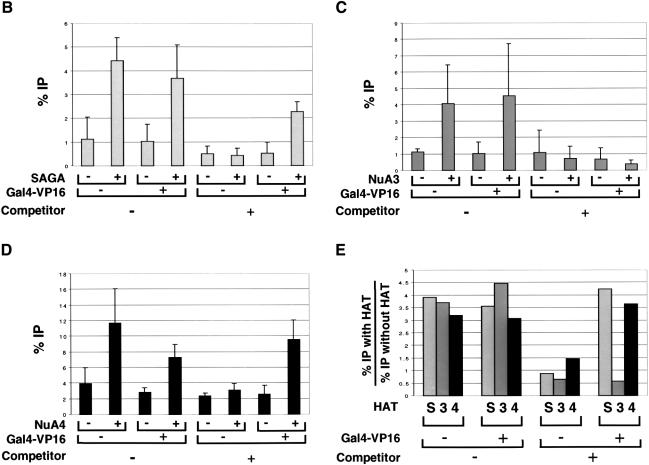
Figure 5B shows the pattern obtained upon immunoprecipitation of the BglI-linearized pG5E4T nucleosomal template acetylated by SAGA in the presence or absence of Gal4-VP16 and competitor chromatin. When pG5E4T was the only template present in the reaction (– Gal4-VP16, – competitor), the immunoprecipitation of the different fragments was distributed quite evenly across the length of the template. This suggests that the complex had no strong preference for nucleosomes placed in any particular position. In fact, the signal seems to be higher with the probes that hybridize furthest away from the Gal4 sites. This is confirmed by the fact that the ratio of proximal to distal signals (average of +/–A divided by the average of +/– C, values shown under the graph) is <1. Thus, under these conditions, it is likely that SAGA has a slight preference for nucleosomes localized close to the ends of the linearized template. However, when the array was pre-incubated with Gal4-VP16 (+Gal4-VP16, – competitor), the SAGA-mediated acetylation peaked sharply around the Gal4 sites, and progressively decreased towards both ends of the array. This observation illustrates that the activator directs the activity of the HAT complex to those nucleosomes that are adjacent to the Gal4 sites. When competitor chromatin was added (–Gal4-VP16, +competitor), the signal obtained with all the probes drops to background levels in the absence of Gal4-VP16. This result agrees with our observation that SAGA is unable to acetylate the array under competitive conditions in the absence of the activator. However, when Gal4-VP16 was included (+Gal4-VP16, +competitor), the acetylation was rescued and once again peaked sharply around the Gal4 sites in a pattern very similar to that observed in the absence of competitor chromatin (compare the proximal/distal ratios). Therefore, the activator specifically directs the acetyltransferase activity of the SAGA complex to the promoter region in both the absence and presence of competitor chromatin. As a control, we show that detection of this peak of acetylation required digestion of the array with MNase. Figure 5C shows that a peak of acetylation flanking the Gal4 sites was observed when the array was digested with MNase, while all probes yield approximately the same signal in the absence of MNase digestion.
The profiles of histone acetylation generated by the NuA4 complex are presented in Figure 6. In the absence of competitor and activator (–Gal4-VP16, – competitor), NuA4, like SAGA, appeared to have a slight preference for the nucleosomes located toward the ends of the reconstituted array, as indicated by a proximal/distal ratio of <1. In agreement with the result presented in Figure 4 for the undigested template, inclusion of Gal4-VP16 seemed to decrease the total level of acetylation of the template by NuA4 (+Gal4-VP16, – competitor). One possible explanation for this effect is that interactions of NuA4 with the activator sequestered the complex, reducing its turnover to acetylate additional template arrays. In contrast to the profile obtained from the SAGA experiments under these conditions, the peak of promoter-proximal acetylation was much less pronounced for NuA4. In other words, the NuA4 complex acetylated nucleosomes over a much broader range of the template than the SAGA complex. Nevertheless, the addition of competitor chromatin in the absence of Gal4-VP16 abolished the acetylation of the Gal4 template, and inclusion of Gal4-VP16 restored the profile of acetylation observed in the absence of competitor chromatin (compare columns +/–Gal4-VP16, +competitor). Thus, while the NuA4 complex was clearly targeted to the template arrays by Gal4-VP16, it acetylated a broader distribution of nucleosomes across the array than did the targeted SAGA complex. As for SAGA, we show that the peak of acetylation is dependent on MNase digestion of the template (Figure 6B).
Fig. 6. Scanning ChIPs for the NuA4 complex. Experiments identical to those described in detail in the legend for Figure 5 were carried out for NuA4, except that anti-acetylated H4 antibody was used for the immunoprecipitation step. (A) Average and standard deviation of normalized data from 3–4 repeats of the scanning ChIP performed on BglI-linearized reconstituted template. (B) Control for MNase digestion (see Figure 5C).
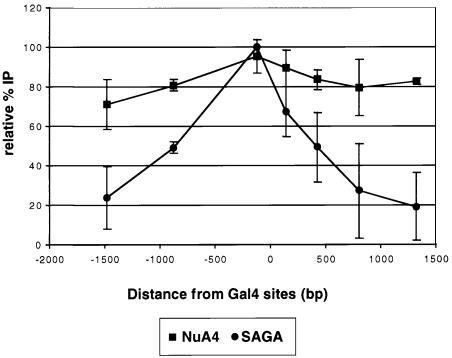
The difference in the acetylation profiles generated upon Gal4-VP16-mediated targeting of the SAGA and NuA4 complexes in the presence of competitor chromatin is illustrated further in the graph presented in Figure 7. Upon Gal4-VP16 targeting, acetylation by SAGA strongly peaked adjacent to the Gal4 sites and dropped to ∼20% of the peak at 1500 bp away (i.e. 7–8 nucleosome lengths). In contrast, we did not observe a strong peak of targeted NuA4 acetylation adjacent to the Gal4 sites, and acetylation 1500 bp away was 70–80% of that adjacent to the bound activator.
Fig. 7. The domain of acetylation generated by NuA4 upon Gal4-VP16 targeting is broader than that observed for SAGA. BglI-linearized pG5E4T nucleosomal arrays were pre-incubated with Gal4-VP16 and competitor chromatin was added. Scanning ChIPs were performed as described for previous figures. The data were normalized to the highest peak for comparison purposes. The x-axis shows the distance from the Gal4 sites in base pairs.
Discussion
We have analyzed the mechanism by which yeast HAT complexes might be targeted to specific promoters in the context of cellular chromatin to regulate the expression of particular genes, using a defined in vitro system with purified components. Our data show that, under competitive conditions that resemble the in vivo situation more closely than previous experiments, activator-mediated targeted acetylation of template nucleosomes is required for the enhanced transcription effected by the SAGA and NuA4 complexes. We also show that the activator-dependent acetylation of a template can occur in a step independent of the initiation of transcription. Furthermore, we demonstrate that the interaction of SAGA and NuA4 with Gal4-VP16, a DNA-binding, chimeric acidic activator (Sadowski et al., 1988), is sufficient to direct the HAT activity of these complexes to specific nucleosomal templates under competitive conditions. This targeting results in a sharp, localized region of acetylation by the SAGA complex, with maximal acetylation of those octamers that are immediately adjacent to the Gal4-binding sites. In contrast, targeted acetylation of the template nucleosomal array by the NuA4 complex is more widely spread and generates a broader domain of acetylation.
Potentiation of transcription upon targeting
Different subunits of SAGA can interact with TATA box-binding protein (Spt3 and Spt8) and with acidic activators (Adas) (reviewed in Winston and Sudarsanam, 1998). SAGA also contains TAF proteins previously thought to be exclusive components of TFIID (Grant et al., 1998a), a discovery that further strengthened the role of this complex in transcriptional regulation. In addition, both the NuA4 and SAGA complexes contain Tra1 as their largest subunit (Grant et al., 1998b; Saleh et al., 1998; Allard et al., 1999). Tra1 is the yeast homolog of the human TRRAP protein implicated as a co-activator of transcription factors c-Myc and E2F (McMahon et al., 1998, 2000). Thus, the subunit composition of the SAGA and NuA4 complexes is consistent with the concept that they could posses a co-activator function in transcription. Indeed, SAGA components have been shown to facilitate TBP binding to the GAL1 promoter in vivo in a step subsequent to activator binding (Dudley et al., 1999).
In addition to potential direct roles for the SAGA and NuA4 complexes as classic ‘co-activators’ (i.e. bridging factors between activators and basal transcription factors), they also contain HAT subunits, Gcn5 and Esa1, respectively (Grant et al., 1997; Allard et al., 1999). Our in vitro studies illustrate the importance of targeting the histone acetylation activity of SAGA and NuA4 in the activation of transcription from chromatin templates. In fact, the data support the notion that an important function of both SAGA and NuA4 in promoting transcription stimulation from chromatin templates depends on their ability to acetylate template nucleosomes. These assays have been conducted in the presence of a large excess of competitor chromatin. Under these conditions, the ability of the HAT complexes to acetylate the nucleosome array template and enhance transcription depended strictly on their competence to interact with the Gal4-VP16 activator.
In addition to histones, many HATs can also acetylate non-histone proteins (reviewed in Brown et al., 2000). Such an activity does not appear to contribute to the transcriptional stimulation observed here. The only proteins present in the targeted acetylation reactions were HAT complexes, histones and Gal4-VP16. When we excluded the nucleosome template from the acetylation reaction, and added it only after acetyl-CoA had been removed, we observed no increase in transcription. This result excludes the possibility that the transcriptional effect we observed was due to the acetylation of the activator by the HATs or by the HAT complexes acetylating one of their own subunits. Similarly, as the basal transcription factors were added after the acetyl-CoA had been removed, the effect we observed could not result from the acetylation of basal transcription factors by the HATs. Thus, these results demonstrate that the main mechanism by which HATs potentiate transcription in our in vitro system is through acetylation of the histone octamers associated with the template.
Targeting of histone acetylation by the SAGA and NuA4 complexes
An important aspect of the function of HAT complexes is how they locate their target genes among other genes in the genome. For several HAT complexes, this targeting is mediated through DNA-binding transcription activators. The interaction of particular HAT complexes with transcription factors has been demonstrated both in yeast (Berger et al., 1992; Marcus et al., 1994; Utley et al., 1998) and in humans (reviewed in Xu et al., 1999). Moreover, in vivo evidence suggests that some HATs might act upon specific genes in an activator-dependent manner (see, for example, Korzus et al., 1998; Krebs et al., 1999; Parekh and Maniatis, 1999). However, in vivo studies are complicated by the many components present, and thus have the caveat that the effects observed might be indirect. In this study, we have taken advantage of a biochemical assay with purified components to investigate the targeting of the enzymatic activity of native HAT complexes. Using two novel approaches, fluorography of acetylated products in agarose gels and in vitro ChIP assays, we have shown targeted histone acetylation by the two yeast HAT complexes that interact with acidic activators. SAGA and NuA4 were able to acetylate nucleosomes assembled onto specific templates in the presence of an excess of competitor chromatin in a manner dependent on the presence and DNA-binding ability of Gal4-VP16. In contrast, NuA3, which is unable to interact with acidic activators, correspondingly was unable to acetylate this template under competitive conditions regardless of the presence of the activator. Thus, these studies provide biochemical evidence in a purified system for targeting of histone acetylation to chromatin templates as a result of direct interactions of DNA-binding activators with HAT complexes.
The first links between acetylation of histones and transcription came from immunoprecipitation studies using antibodies generated against acetylated lysines. These studies revealed that transcriptionally active (or potentially active) genomic regions were associated with hyperacetylated histones (Hebbes et al., 1988, 1992, 1994) and, in contrast, that transcriptionally repressed regions were associated with hypoacetylated histone H4 (O’Neill and Turner, 1995) or histones H3 and H4 (Braunstein et al., 1996). Other experiments, however, have suggested that HATs generate localized domains of acetylated histones in the promoter region of target genes. Data from the Allis group suggest that Gcn5-dependent histone H3 acetylation of promoters might span only 2–3 nucleosomes (Kuo et al., 1998). A more detailed study of the IFN-β promoter in human cells showed an infection-dependent domain of H3 and H4 acetylation that also extends for 2–3 nucleosomes (Parekh and Maniatis, 1999). In contrast, the study of the HO promoter in yeast suggested a more extended domain of Gcn5-dependent H3 (and H4) acetylation, corresponding to 6–7 nucleosomes over the regulatory region of the gene, which did not extend into the coding region (Krebs et al., 1999).
Interestingly, our in vitro study shows a different profile of acetylation for SAGA and NuA4 relative to the promoter. While both HAT complexes are targeted to the same promoter via interaction with Gal4-VP16, the domain of acetylation generated by SAGA is more restricted than the one generated by NuA4. In vivo experiments have shown that LexA-tethered Gcn5 was able to increase transcription of reporter genes (Candau et al., 1997). This result suggests that the localized acetylation of nucleosomes by Gcn5-dependent HAT complexes might be sufficient to increase transcription. Although NuA4 seems to generate a more extended domain of acetylation upon targeting, our current data do not allow us to determine whether this is required for the transcriptional enhancement mediated by NuA4. It is entirely possible that the acetylation of the promoter-proximal nucleosomes by NuA4 is sufficient to increase initiation of transcription. In this view, the extended domain of acetylation might have other functions, perhaps related to elongation of transcription or chromatin fiber decondensation, which are not detected in the assay used here. We are currently carrying out additional experiments to explore whether these HAT complexes truly function in mechanistically different processes. This difference would be particularly interesting in view of the different substrate specificities of the SAGA and NuA4 complexes, as it might reveal that H3 and H4 acetylation could have diverse roles in the different aspects of transcriptional regulation.
Materials and methods
Plasmids and templates
pG5E4T, a kind gift from M.Carey (Lin et al., 1988), contains five consensus Gal4 DNA-binding sites upstream of the adenovirus 2 E4 minimal promoter, and was linearized with BglI so that the Gal4 sites are central to the DNA fragment (New England Biolabs). The template used in the transcription experiments was obtained as described (Ikeda et al., 1999) by restriction enzyme digestion of p2085S-G5E4 and contains the same enhancer and promoter as pG5E4T, cloned into an array of rRNA 5S sequences in order to force the organization of the central region into nucleosomes. In all cases, the DNA fragments were gel purified after restriction enzyme digestion and quantitated by absorbance at λ = 260 nm. Then, the purified DNA was mixed in 2 M NaCl with a 1:1 molar ratio of core histones purified from HeLa cells, and reconstituted in vitro by step dilution as described (Steger et al., 1998). The efficiency of the reconstitution was determined by comparing the electrophoretic mobility of the nucleosomal array with that of naked DNA on 1.2% agarose, 1× TAE gels run for 240 V/h and stained with ethidium bromide a posteriori. The reconstitution was also tested by MNase digestion followed by Southern blotting. Naked DNA and reconstituted array +/– competitor chromatin purified from HeLa cells were digested with varying amounts of Sigma MNase for 5 min at room temperature. After the reactions were stopped by incubation in 20 mM Tris pH 7.5, 50 mM EDTA, 1% SDS, 250 ng of tRNA and 200 ng of proteinase K for >1 h at 50°C, the DNA was purified by phenol–chloroform extraction and ethanol precipitation. Then, the samples were resuspended in gel loading dye and separated on a 1.6% agarose, 1× TAE gel together with 32P-labeled MspI-digested pBR322 as molecular weight marker, and transferred to a Zeta-Probe (Bio-Rad) membrane by Southern blotting. Finally, the membrane was hybridized with probes generated by random hexanucleotide primer extension against full-length plasmid DNA or against different parts of the template DNA (see below), exposed to a Phosphor Screen and quantitated using the ImageQuant software from Molecular Dynamics.
Detection of HAT targeting by fluorography
Approximately 100 ng of the nucleosomal array template were pre-incubated with Gal4-VP16 (Utley et al., 1998) in binding/HAT reaction buffer (Steger et al., 1998) for 10 min at room temperature. Sheared chromatin purified from HeLa cells (or 0.6 M NaCl buffer; Côté et al., 1995) was added in a 10- to 30-fold mass excess where indicated. Next, the reactions were incubated with partially purified HAT complexes (Superose 6 fractions; Eberharter et al., 1998) and [3H]acetyl-CoA for 30 min at 30°C. A large mass excess of a double-stranded oligonucleotide containing a Gal4 consensus site was added at this point, and the reactions were incubated further for 1 h at 37°C. For the experiment presented in Figure 3E, the oligonucleotide was added concurrently with Gal4-VP16. The samples were then subjected to electrophoresis on 0.5 cm thick, 1.2% agarose gels, run in 1× TAE for 280 V/h together with molecular weight markers. Gels were fixed by agitation for 1 h in 10% acetic acid, 10% methanol, EN3HANCEd for 3 h (NEN Life Science Products, Inc.), rinsed for 1 h in ddH2O, dried and exposed to Fuji Film.
HAT assays
Superose 6 fractions of all the HATs used in the experiments were purified and assayed for their ability to acetylate nucleosomal substrates as described (Grant et al., 1997; Eberharter et al., 1998).
ChIP assays
The fluorography reactions were scaled up for the ChIP assays. After the acetylation step, commercially available kits (Upstate Biotechnology) were used, following the standard protocol (Kuo and Allis, 1999) modified for in vitro conditions. DNA was extracted from the unbound and bound material by proteinase K digestion followed by phenol–chloroform extraction and ethanol precipitation. Denatured samples were applied to Zeta-Probe membranes using a Bio-Rad Bio-Dot microfiltration apparatus. The membranes were then hybridized as per the manufacturer’s directions with a full-length probe corresponding to the fragment reconstituted into chromatin. For the data presented in Figures 4–6, the samples were incubated with MNase and 3 mM CaCl2 after the acetylation reaction. The digestion was stopped with the addition of a 2-fold excess of EGTA, and the samples were immunoprecipitated as before. After DNA purification and slot blot, the membranes were hybridized successively with probes of ∼250–300 bp generated by PCR amplification from the pG5E4T template. Considering that the HindIII restriction enzyme site is located at bp 7 and that the Gal4-binding sites extend from bp 25 to 128, the probes used were: +A (bp 129–418), +A′ (430–680), +B (801–1066), +C (1327–1582), –A (3025–25), –B (2252–2535) and – C (1665–1920) (Figure 5A). All the probes were labeled with [α-32P]dCTP by random hexanucleotide primer extension (Boehringer Mannheim) using Klenow polymerase (New England Biolabs), and purified from the unincorporated nucleotides by means of QIAQUICK PCR purification kit (Qiagen).
In vitro transcription
Transcription was performed as described elsewhere (Steger et al., 1998). To remove acetyl-CoA from HAT reactions, the samples were placed over MicroSpin S-300 HR columns (Pharmacia) equilibrated in HAT reaction buffer without acetyl-CoA. Gels were quantitated after exposure to Phosphor Screens.
Acknowledgments
Acknowledgements
We are grateful to Marilyn Pray-Grant, Patrick Grant and Anton Eberharter for purifying the HAT fractions used in these assays. We thank Michael Carey for providing us with the pG5E4T plasmid, and Keiko Ikeda for pG5E4-5S. In addition, we thank Jacques Côté for communicating data before publication and for many useful discussions, and other members of the Workman laboratory and the Penn State Gene Regulation Group for critical comments. This work was supported by a grant from NIGMS to J.L.W. D.J.S. was an HHMI Associate and J.L.W. is an HHMI Associate Investigator.
References
- Allard S., Utley,R.T., Savard,J., Clarke,A., Grant,P.A., Brandl,C.J., Pillus,L., Workman,J.L. and Côté,J. (1999) NuA4, an essential transcription adaptor/histone H4 acetyltransferase complex containing Esa1p and the ATM-related cofactor Tra1p. EMBO J., 18, 5108–5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belotserkovskaya R. and Berger,S.L. (1999) Interplay between chromatin modifying and remodeling complexes in transcriptional regulation. Crit. Rev. Eukaryot. Gene Express., 9, 221–230. [DOI] [PubMed] [Google Scholar]
- Berger S.L., Piña,B., Silverman,N., Marcus,G.A., Agapite,J., Regier,J.L., Triezenberg,S.J. and Guarente,L. (1992) Genetic isolation of ADA2: a potential transcriptional adaptor required for function of certain acidic activation domains. Cell, 70, 251–265. [DOI] [PubMed] [Google Scholar]
- Braunstein M., Sobel,R.E., Allis,C.D., Turner,B.M. and Broach,J.R. (1996) Efficient transcriptional silencing in Saccharomyces cerevisiae requires a heterochromatin histone acetylation pattern. Mol. Cell. Biol., 16, 4349–4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C.E., Lechner,T., Howe,L. and Workman,J.L. (2000) The many HATs of transcription coactivators. Trends Biochem. Sci., 25, 15–19. [DOI] [PubMed] [Google Scholar]
- Candau R., Zhou,J.X., Allis,C.D. and Berger,S.L. (1997) Histone acetyltransferase activity and interaction with ADA2 are critical for GCN5 function in vivo.EMBO J., 16, 555–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosma M.P., Tanaka,T. and Nasmyth,K. (1999) Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell, 97, 299–311. [DOI] [PubMed] [Google Scholar]
- Côté J., Utley,R.T. and Workman,J.L. (1995). Basic analysis of transcription factor binding to nucleosomes. In Adolph,K.W. (ed.), Methods in Molecular Genetics, Vol. 6, Microbial Gene Techniques. Academic Press, Inc., San Diego, CA, pp. 108–128. [Google Scholar]
- Dudley A.M., Rougeulle,C. and Winston,F. (1999) The Spt components of SAGA facilitate TBP binding to a promoter at a post-activator-binding step in vivo.Genes Dev., 13, 2940–2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberharter A., John,S., Grant,P.A., Utley,R.T. and Workman,J.L. (1998) Identification and analysis of yeast nucleosomal histone acetyltransferase complexes. Methods, 15, 315–321. [DOI] [PubMed] [Google Scholar]
- Eberharter A., Sterner,D.E., Schieltz,D., Hassan,A.H., Yates,J.R.,III, Berger,S.L. and Workman,J.L. (1999) The ADA complex is a distinct histone acetyltransferase complex in Saccharomyces cerevisiae. Mol. Cell. Biol., 19, 6621–6631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galarneau L. et al. (2000) Multiple links between the NuA4 histone acetyltransferase complex and epigenetic control of transcription. Mol. Cell, in press. [DOI] [PubMed] [Google Scholar]
- Galasinski S.K., Lively,T.N., Grebe De Barron,A. and Goodrich,J.A. (2000) Acetyl coenzyme A stimulates RNA polymerase II transcription and promoter binding by transcription factor IID in the absence of histones. Mol. Cell. Biol., 20, 1923–1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgakopoulos T. and Thireos,G. (1992) Two distinct yeast transcriptional activators require the function of the GCN5 protein to promote normal levels of transcription. EMBO J., 11, 4145–4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant P.A. and Berger,S.L. (1999) Histone acetyltransferase complexes. Semin. Cell Dev. Biol., 10, 169–177. [DOI] [PubMed] [Google Scholar]
- Grant P.A. et al. (1997) Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev., 11, 1640–1650. [DOI] [PubMed] [Google Scholar]
- Grant P.A., Schieltz,D., Pray-Grant,M.G., Steger,D.J., Reese,J.C., Yates,J.R.,III and Workman,J.L. (1998a) A subset of TAF(II)s are integral components of the SAGA complex required for nucleosome acetylation and transcriptional stimulation. Cell, 94, 45–53. [DOI] [PubMed] [Google Scholar]
- Grant P.A., Schieltz,D., Pray-Grant,M.G., Yates,J.R.,III and Workman,J.L. (1998b) The ATM-related cofactor Tra1 is a component of the purified SAGA complex. Mol. Cell, 2, 863–867. [DOI] [PubMed] [Google Scholar]
- Grant P.A., Eberharter,A., John,S., Cook,R.G., Turner,B.M. and Workman,J.L. (1999) Expanded lysine acetylation specificity of Gcn5 in native complexes. J. Biol. Chem., 274, 5895–5900. [DOI] [PubMed] [Google Scholar]
- Hebbes T.R., Thorne,A.W. and Crane-Robinson,C. (1988) A direct link between core histone acetylation and transcriptionally active chromatin. EMBO J., 7, 1395–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebbes T.R., Thorne,A.W., Clayton,A.L. and Crane-Robinson,C. (1992) Histone acetylation and globin gene switching. Nucleic Acids Res., 20, 1017–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebbes T.R., Clayton,A.L., Thorne,A.W. and Crane-Robinson,C. (1994) Core histone hyperacetylation co-maps with generalized DNase I sensitivity in the chicken β-globin chromosomal domain. EMBO J., 13, 1823–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holstege F.C., Jennings,E.G., Wyrick,J.J., Lee,T.I., Hengartner,C.J., Green,M.R., Golub,T.R., Lander,E.S. and Young,R.A. (1998) Dissecting the regulatory circuitry of a eukaryotic genome. Cell, 95, 717–728. [DOI] [PubMed] [Google Scholar]
- Ikeda K., Steger,D.J., Eberharter,A. and Workman,J.L. (1999) Activation domain-specific and general transcription stimulation by native histone acetyltransferase complexes. Mol. Cell. Biol., 19, 855–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janknecht R. and Hunter,T. (1996) Transcription. A growing coactivator network. Nature, 383, 22–23. [DOI] [PubMed] [Google Scholar]
- John S. Howe,L., Tafrov,S.T., Grant,P.A., Sternglanz,R. and Workman,J.L. (2000) The Something About Silencing protein, Sas3, is the catalytic subunit of NuA3, a yTAFII30-containing HAT complex that interacts with the Spt16 subunit of the yeast CP(Cdc68/Pob3)–FACT complex. Genes Dev., in press. [PMC free article] [PubMed] [Google Scholar]
- Kadosh D. and Struhl,K. (1998a) Histone deacetylase activity of Rpd3 is important for transcriptional repression in vivo.Genes Dev., 12, 797–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadosh D. and Struhl,K. (1998b) Targeted recruitment of the Sin3–Rpd3 histone deacetylase complex generates a highly localized domain of repressed chromatin in vivo.Mol. Cell. Biol., 18, 5121–5127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korzus E., Torchia,J., Rose,D.W., Xu,L., Kurokawa,R., McInerney,E.M., Mullen,T.M., Glass,C.K. and Rosenfeld,M.G. (1998) Transcription factor-specific requirements for coactivators and their acetyltransferase functions. Science, 279, 703–707. [DOI] [PubMed] [Google Scholar]
- Krebs J.E., Kuo,M.H., Allis,C.D. and Peterson,C.L. (1999) Cell cycle-regulated histone acetylation required for expression of the yeast HO gene. Genes Dev., 13, 1412–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo M.H. and Allis,C.D. (1998) Roles of histone acetyltransferases and deacetylases in gene regulation. BioEssays, 20, 615–626. [DOI] [PubMed] [Google Scholar]
- Kuo M.H. and Allis,C.D. (1999) In vivo cross-linking and immunoprecipitation for studying dynamic protein:DNA associations in a chromatin environment. Methods, 19, 425–433. [DOI] [PubMed] [Google Scholar]
- Kuo M.H., Zhou,J., Jambeck,P., Churchill,M.E. and Allis,C.D. (1998) Histone acetyltransferase activity of yeast Gcn5p is required for the activation of target genes in vivo.Genes Dev., 12, 627–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y.S., Carey,M.F., Ptashne,M. and Green,M.R. (1988) GAL4 derivatives function alone and synergistically with mammalian activators in vitro.Cell, 54, 659–664. [DOI] [PubMed] [Google Scholar]
- Marcus G.A., Silverman,N., Berger,S.L., Horiuchi,J. and Guarente,L. (1994) Functional similarity and physical association between GCN5 and ADA2: putative transcriptional adaptors. EMBO J., 13, 4807–4815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon S.B., Van Buskirk,H.A., Dugan,K.A., Copeland,T.D. and Cole,M.D. (1998) The novel ATM-related protein TRRAP is an essential cofactor for the c-Myc and E2F oncoproteins. Cell, 94, 363–374. [DOI] [PubMed] [Google Scholar]
- McMahon S.B., Wood,M.A. and Cole,M.D. (2000) The essential cofactor TRRAP recruits the histone acetyltransferase hGCN5 to c-Myc. Mol. Cell. Biol., 20, 556–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan K., Jackson,B.M., Zhou,H., Winston,F. and Hinnebusch,A.G. (1999) Transcriptional activation by Gcn4p involves independent interactions with the SWI/SNF complex and the SRB/mediator. Mol. Cell, 4, 657–664. [DOI] [PubMed] [Google Scholar]
- Neely K.E., Hassan,A.H., Wallberg,A.E., Steger,D.J., Cairns,B.R., Wright,A.P. and Workman,J.L. (1999) Activation domain-mediated targeting of the SWI/SNF complex to promoters stimulates transcription from nucleosome arrays. Mol. Cell, 4, 649–655. [DOI] [PubMed] [Google Scholar]
- O’Neill L.P. and Turner,B.M. (1995) Histone H4 acetylation distinguishes coding regions of the human genome from heterochromatin in a differentiation-dependent but transcription- independent manner. EMBO J., 14, 3946–3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parekh B.S. and Maniatis,T. (1999) Virus infection leads to localized hyperacetylation of histones H3 and H4 at the IFN-β promoter. Mol. Cell, 3, 125–129. [DOI] [PubMed] [Google Scholar]
- Rundlett S.E., Carmen,A.A., Suka,N., Turner,B.M. and Grunstein,M. (1998) Transcriptional repression by UME6 involves deacetylation of lysine 5 of histone H4 by RPD3. Nature, 392, 831–835. [DOI] [PubMed] [Google Scholar]
- Sadowski I., Ma,J., Triezenberg,S.J. and Ptashne,M. (1988) GAL4-VP16 is an unusually potent transcriptional activator. Nature, 335, 563–564. [DOI] [PubMed] [Google Scholar]
- Saleh A., Schieltz,D., Ting,N., McMahon,S.B., Litchfield,D.W., Yates,J.R.,III, Lees-Miller,S.P., Cole,M.D. and Brandl,C.J. (1998) Tra1p is a component of the yeast Ada⋅Spt transcriptional regulatory complexes. J. Biol. Chem., 273, 26559–26565. [DOI] [PubMed] [Google Scholar]
- Steger D.J., Eberharter,A., John,S., Grant,P.A. and Workman,J.L. (1998) Purified histone acetyltransferase complexes stimulate HIV-1 transcription from preassembled nucleosomal arrays. Proc. Natl Acad. Sci. USA, 95, 12924–12929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. (1998) Histone acetylation and transcriptional regulatory mechanisms. Genes Dev., 12, 599–606. [DOI] [PubMed] [Google Scholar]
- Utley R.T., Ikeda,K., Grant,P.A., Côté,J., Steger,D.J., Eberharter,A., John,S. and Workman,J.L. (1998) Transcriptional activators direct histone acetyltransferase complexes to nucleosomes. Nature, 394, 498–502. [DOI] [PubMed] [Google Scholar]
- Vettese-Dadey M., Grant,P.A., Hebbes,T.R., Crane-Robinson,C., Allis, C.D. and Workman,J.L. (1996) Acetylation of histone H4 plays a primary role in enhancing transcription factor binding to nucleosomal DNA in vitro.EMBO J., 15, 2508–2518. [PMC free article] [PubMed] [Google Scholar]
- Wallberg A.E., Neely,K.E., Gustafsson,J.A., Workman,J.L., Wright,A.P. and Grant,P.A. (1999) Histone acetyltransferase complexes can mediate transcriptional activation by the major glucocorticoid receptor activation domain. Mol. Cell. Biol., 19, 5952–5959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallberg A.E., Neely,K.E., Hassan,A.H., Gustafsson,J.A., Workman,J.L. and Wright,A.P. (2000) Recruitment of the SWI–SNF chromatin remodeling complex as a mechanism of gene activation by the glucocorticoid receptor τ1 activation domain. Mol. Cell. Biol., 20, 2004–2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston F. and Sudarsanam,P. (1998) The SAGA of Spt proteins and transcriptional analysis in yeast: past, present and future. Cold Spring Harb. Symp. Quant. Biol., 63, 553–561. [DOI] [PubMed] [Google Scholar]
- Xu L., Glass,C.K. and Rosenfeld,M.G. (1999) Coactivator and corepressor complexes in nuclear receptor function. Curr. Opin. Genet. Dev., 9, 140–147. [DOI] [PubMed] [Google Scholar]
- Yudkovsky N., Logie,C., Hahn,S. and Peterson,C.L. (1999) Recruitment of the SWI/SNF chromatin remodeling complex by transcriptional activators. Genes Dev., 13, 2369–2374. [DOI] [PMC free article] [PubMed] [Google Scholar]