Abstract
Viroids replicate through a rolling-circle mechanism in which the infecting circular RNA and its complementary (–) strand are transcribed. The precise site at which transcription starts was investigated for the avocado sunblotch viroid (ASBVd), the type species of the family of viroids with hammerhead ribozymes. Linear ASBVd (+) and (–) RNAs begin with a UAAAA sequence that maps to similar A+U-rich terminal loops in their predicted quasi-rod-like secondary structures. The sequences around the initiation sites of ASBVd, which replicates and accumulates in the chloroplast, are similar to the promoters of a nuclear-encoded chloroplastic RNA polymerase (NEP), supporting the involvement of an NEP-like activity in ASBVd replication. Since RNA folding appears to be kinetically determined, the specific location of both ASBVd initiation sites provides a mechanistic insight into how the nascent ASBVd strands may fold in vivo. The approach used here, in vitro capping and RNase protection assays, may be useful for investigating the initiation sites of other small circular RNA replicons.
Keywords: avocado sunblotch viroid/catalytic RNAs/rolling-circle replication
Introduction
Viroids are the simplest well characterized pathogens. They are molecular parasites of plants composed solely of a small circular RNA of 246–399 nucleotides (nt) with a high secondary structure content and without any apparent coding capacity (for reviews, see Diener, 1991; Flores et al., 1997, 2000; Symons, 1997). There is convincing evidence that viroid replication occurs only through RNA intermediates (Grill and Semancik, 1978), by a rolling-circle mechanism with two possible pathways (Branch and Robertson, 1984). Avocado sunblotch viroid (ASBVd) (Symons, 1981), the type species of the family Avsunviroidae (Flores et al., 2000), follows the symmetrical pathway (Hutchins et al., 1985; Daròs et al., 1994). In brief, the infecting monomeric circular RNA, to which the (+) polarity is arbitrarily assigned, is transcribed by an RNA polymerase to yield oligomeric (–) strands that are then processed to unit length and ligated to the monomeric (–) circular RNA, via an unconventional processing activity and an RNA ligase, respectively. This RNA species serves as the template for the second half of the cycle, which is a symmetrical version of the first. In contrast, replication of potato spindle tuber viroid (PSTVd) (Diener, 1971b; Gross et al., 1978), the type species of the other family, Pospiviroidae, follows the asymmetrical pathway in which the oligomeric (–) strands, resulting from the first transcription step, serve directly as the template for synthesis of the complementary RNAs, which, after cleavage and ligation, lead to the final product of the cycle, the monomeric (+) circular RNA (Branch and Robertson, 1981; Owens and Diener, 1982; Branch et al., 1988; Feldstein et al., 1998). Another demarcating difference between the type species of these two families is that replication and accumulation of PSTVd occur in the nucleus (Diener, 1971a; Spiesmacher et al., 1983; Harders et al., 1989), while in ASBVd these two processes take place in the chloroplast (Bonfiglioli et al., 1994; Lima et al., 1994; Navarro et al., 1999).
Some aspects of the rolling-circle mechanism are known, particularly the nature of the RNA polymerases presumably involved in the first step (Mühlbach and Sänger, 1979; Flores and Semancik, 1982; Schindler and Mühlbach, 1992; Navarro et al., 2000), and of the cleavage activity catalyzing the second step, which, in the three members of the family Avsunviroidae, is a hammerhead ribozyme embedded in the strands of both polarities (Hutchins et al., 1986; Hernández and Flores, 1992; Navarro and Flores, 1997). However, one intriguing question on viroid replication that remains unanswered is whether transcription of viroid strands is initiated at a defined position within the molecule, or at random, a feasible alternative from a theoretical perspective considering that the circular nature of the template enables its complete transcription irrespective of where transcription is initiated. In the first case, the sequences adjacent to the initiation site should provide an insight into the structure of conserved regions functionally important for promoting transcription. Moreover, the initiation site of the nascent viroid strands greatly influences RNA folding in vivo, which may differ considerably from the thermodynamically most stable secondary structure of the full-length RNA. Metastable conformations have been proposed to play an important role in viroid replication (Qu et al., 1993).
Results
The termini of naturally occurring monomeric linear ASBVd (+) molecules
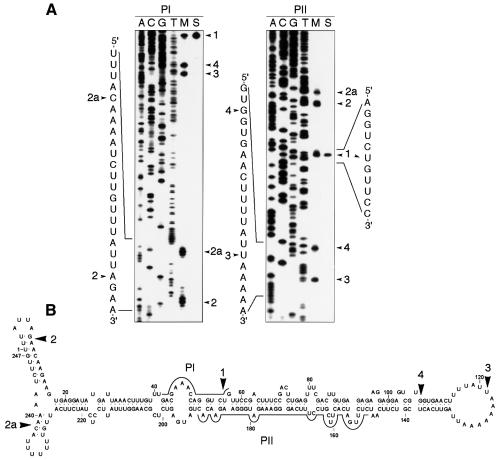
Figure 1 shows the ASBVd-specific RNAs isolated from infected tissue as revealed by non-denaturing PAGE and northern blot hybridization, prominent among which are the monomeric linear and circular ASBVd forms that comigrate in band m (Daròs et al., 1994; Navarro et al., 1999). Primer extension experiments using the ASBVd linear forms, obtained from band m by denaturing PAGE, and the ASBVd (–) primers PI or PII, led consistently to five cDNAs in approximately similar ratios (Figure 2A, lane M). In line with previous results (Marcos and Flores, 1993), four of the corresponding 5′ termini mapped at residues U56, A9, U121 and G108 of the ASBVd (+) RNA and are referred to as termini 1, 2, 3 and 4, respectively; the fifth reverse transcription stop was found at residue A239 of the ASBVd (+) RNA and is named terminus 2a (Figure 2A). Control primer extension experiments with circular viroid monomers generated unit-length and longer transcripts (Marcos and Flores, 1993; data not shown), revealing that the transcription stops did not result from premature termination caused by regions of the template rich in secondary structure. Parallel extension controls using the linear monomeric ASBVd (+) RNA resulting from self-cleavage during in vitro transcription of the linearized pAS19d plasmid, which contains a dimeric insert of the viroid (Navarro et al., 1999), identified exclusively terminus 1 (Figure 2A, lane S). Therefore, only the functional significance for this terminus—a consequence of autocatalytic processing in vitro (Hutchins et al., 1986) and in vivo (Marcos and Flores, 1993; Daròs et al., 1994), mediated by a hammerhead ribozyme—could be established. The remaining 5′ termini could reflect: (i) additional processing sites; (ii) other sites particularly sensitive to degradation either in vivo or during the in vitro manipulation of the RNA; and (iii) the precise site at which transcription of ASBVd (+) RNAs is initiated.
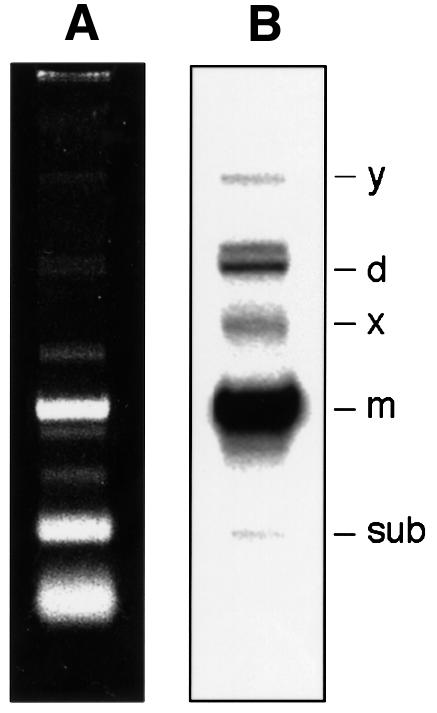
Fig. 1. Analysis of RNAs from ASBVd-infected leaves by non-denaturing PAGE and ethidium bromide staining (A) and northern blot hybridization with an RNA probe to detect ASBVd (+) strands (B). Bands d, m and sub correspond to the dimeric, monomeric and subgenomic ASBVd RNAs, respectively, and bands y and x to two ASBVd-specific complexes (Daròs et al., 1994; Navarro et al., 1999).
Fig. 2. (A) Primer extension analysis with oligonucleotides PI (left) and PII (right) using as template the monomeric linear ASBVd RNAs eluted from band m of a non-denaturing gel (lane M), or the ASBVd (+) monomeric linear RNA generated by self-cleavage during in vitro transcription of the linearized pAS19d plasmid containing a dimeric ASBVd insert (lane S). Sequencing ladders were obtained with the same oligonucleotides and recombinant plasmids containing a monomeric ASBVd insert. Arrowheads indicate positions of the extension products in the 6% sequencing gel and of their corresponding 5′ termini in the written sequences in the margins. (B) Location on the secondary structure predicted for the ASBVd (+) circular monomeric RNA of the 5′ termini (marked by arrowheads) mapped by primer extension. Continuous lines denote positions to which PI and PII are complementary.
The initiation site of ASBVd (+) strands
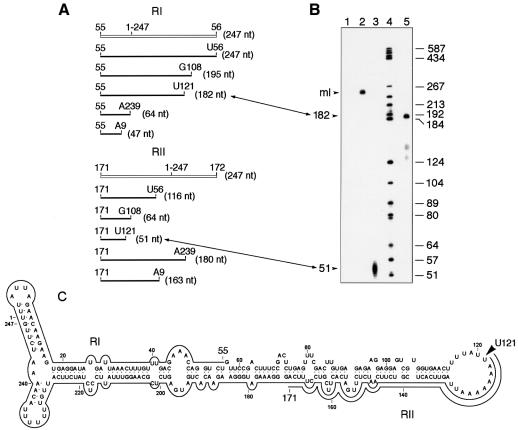
In chloroplasts, the 5′ termini of primary transcripts, but not those resulting from their processing, have a free triphosphate group that can be specifically capped in vitro with [α-32P]GTP and guanylyltransferase (Auchincloss and Brown, 1989). This labeling, combined with RNase protection assays (RPA), has been used to map the transcription start sites of chloroplast genes (Vera and Sugiura, 1992). Application of this approach to the linear ASBVd RNAs isolated from band m showed that a fraction was susceptible to in vitro capping (Figure 3B, lane 2), indicating the presence of one or more 5′ termini with a triphosphate group. Control experiments on in vitro capping of the ASBVd (+) and (–) RNAs resulting from self-cleavage during in vitro transcription of either strand of the linearized pAS19d plasmid revealed that only those RNAs expected to have a triphosphate group at their 5′ end were effectively labeled (data not shown), confirming the specificity of the guanylyltransferase reaction. The in vitro capped linear ASBVd RNAs from band m were then analyzed by RPA. They were hybridized with the unlabeled ASBVd (–) riboprobes RI or RII and digested with a mixture of RNases A and T1 in a high ionic strength buffer, and the protected fragments were identified by denaturing PAGE. Only one prominent protected fragment migrating between the DNA markers of 184–192 and 51–57 nt was detected with riboprobes RI and RII, respectively (Figure 3B, lanes 3 and 5), whereas no protected products were obtained when the riboprobes were not included in the hybridization mixtures (Figure 3B, lane 1). When estimating the exact size of the protected RNA fragments, it must be taken into account that they are expected to move slightly more slowly than DNA markers of the same size (in nucleotides). Comparisons with the predicted sizes of the protected fragments if all 5′ termini mapped by primer extension were in vitro capped (Figure 3A) enabled sizes of 182 and 51 nt to be assigned to the fragments protected with riboprobes RI and RII, respectively. Results obtained with both riboprobes were consistent with each other and showed a single initiation site of ASBVd (+) strands at residue U121, which is located in the A+U-rich right terminal loop of the secondary structure proposed for the ASBVd (+) RNA (Figure 3C).
Fig. 3. In vitro capping and ribonuclease protection assay (RPA). Monomeric linear ASBVd RNAs eluted from band m of a non-denaturing gel were capped in vitro, annealed to non-radioactive ASBVd (–) riboprobes RI or RII and treated with a mixture of RNases, and the protected fragments were analyzed in a 6% sequencing gel. (A) Schematic representation of riboprobes RI or RII (white lines) and of the expected fragments protected by them if all 5′ termini obtained by primer extension were capped (black lines). (B) Analysis by PAGE in a 6% sequencing gel of protected fragments. Lanes 1 and 2, product of the in vitro capping reaction with and without RNase treatment, respectively, in the absence of any complementary riboprobe. Lanes 3 and 5, RNase-protected fragments by riboprobes II and I, respectively, with their sizes in nucleotides indicated on the left. Lane 4, labeled DNA markers with their sizes in nucleotides indicated on the right. The position of linear monomeric ASBVd RNAs is indicated as ml. Correspondence between fragments determined by primer extension and by RPA is indicated by lines with double arrowheads connecting (A) and (B). (C) Location on the secondary structure predicted for the ASBVd (+) circular monomeric RNA of the residue U121, marked by an arrowhead, at which synthesis of ASBVd (+) strands starts. Continuous lines denote fragments protected by riboprobes RI and RII.
To confirm the reliability of this approach, two additional controls were taken. First, the linear ASBVd RNAs eluted from band m and resuspended in water were boiled for 3 min and rapidly cooled on ice to facilitate access of the guanylyltransferase to other 5′ ends bearing a triphosphate that could be embedded within the secondary structure of the RNA; RPA failed to reveal capping of any additional 5′ end apart from U121 (data not shown). Secondly, three monomeric-length ASBVd DNAs were obtained by PCR amplification with three pairs of specific primers. In each pair, (+) and (–) primers were adjacent, with those of the (+) polarity starting at positions G100, G108 (corresponding to terminus 4 identified by primer extension, see Figure 2B) and G181, and preceded by the T7 RNA polymerase promoter. Although in vitro transcriptions from these templates are expected to produce full-length ASBVd (+) RNAs with 5′ triphosphate ends within double-stranded regions (Figure 2B), this did not preclude their in vitro capping catalyzed by the guanylyltransferase (data not shown).
Synthesis of ASBVd (–) strands starts in a region structurally similar to that containing the initiation site of (+) strands
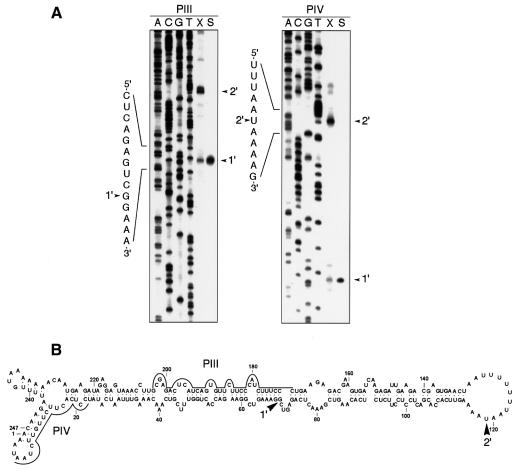
Preliminary attempts to investigate the initiation site of ASBVd (–) strands by RPA, using the in vitro capped linear ASBVd RNAs isolated from band m and the unlabeled ASBVd (+) riboprobes RIII and RIV, did not reveal any consistently protected fragment (data not shown). This was not surprising considering it has been shown (Daròs et al., 1994; Navarro et al., 1999) that band m contains very little, if any, of the monomeric (–) linear ASBVd RNA; this RNA is accompanied by a vast excess of the linear and circular forms of the complementary polarity, which could outcompete most of the riboprobes used in RPA. To circumvent this problem, the linear ASBVd RNAs were isolated from band x (Figure 1), generated from a double-stranded RNA and therefore containing equivalent amounts of the two complementary strands (Daròs et al., 1994; Navarro et al., 1999). Primer extension experiments with ASBVd (+) primers PIII and PIV revealed two major 5′ termini, one mapping to residue G69 and the other, more abundant one mapping to residue U119 of the ASBVd (–) RNA (the same numbers are used in both polarities), referred to as termini 1′ and 2′, respectively (Figure 4, lane X). Since control extensions with the same primers of the linear monomeric ASBVd (–) RNA resulting from self-cleavage during in vitro transcription of the linearized pAS19d plasmid identified G69 as the self-cleavage site of ASBVd (–) strands (Figure 4, lane S) (Hutchins et al., 1986), U119 was the only remaining candidate to be the initiation point of the synthesis of ASBVd (–) strands.
Fig. 4. (A) Primer extension analysis with oligonucleotides PIII (left) and PIV (right) using as template the monomeric linear ASBVd RNAs obtained from band x (lane X), or the ASBVd (–) linear RNA generated by self-cleavage during in vitro transcription of the linearized pAS19d plasmid containing a dimeric ASBVd insert (lane S). Sequencing ladders were obtained with the same oligonucleotides and recombinant plasmids containing a monomeric ASBVd insert. Arrowheads indicate the extension product and the nucleotide position at their 5′ end in the written sequence. (B) Location on the secondary structure predicted for the ASBVd (–) circular monomeric RNA of the 5′ termini (marked by arrowheads) mapped by primer extension. Continuous lines denote positions to which PIII and PIV are complementary. Numbering is the same as in the (+) polarity.
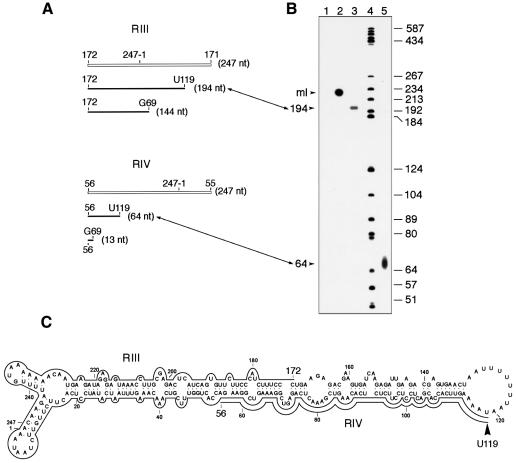
RPA with in vitro capped monomeric linear ASBVd RNAs isolated from band x and the ASBVd (+) riboprobes RIII or RIV, resulted in one protected fragment for each riboprobe of a slightly lower mobility than the 192 and 64 nt DNA markers, respectively (Figure 5B, lanes 3 and 5). In comparison with the predicted sizes of the protected fragments, if each of the two 5′ termini mapped by primer extension were capped in vitro (Figure 5A), sizes of 194 and 64 nt were assigned to the fragments protected by riboprobes RIII and RIV, respectively. Results obtained with the two riboprobes were consistent with each other and showed that U119 is the single initiation site of synthesis of the ASBVd (–) polarity strand. Interestingly, this site is located in an A+U-rich terminal loop (Figure 5C) structurally similar to that containing the initiation site of the ASBVd (+) strand. In fact, in the (+) and (–) polarities, the two initiation sites are only 2 nt apart (Figures 3C and 5C). This is an economical solution for an RNA as small as ASBVd, which within 247 nt must contain the signals for initiation, processing and ligation of both polarity strands, as well as for targeting it to the chloroplast and for inducing a pathogenic alteration. The sequences forming the central region of the quasi-rod-like secondary structures proposed for the ASBVd (+) and (–) RNAs have a clear functional role in the self-cleavage of both polarity strands during replication (Hutchins et al., 1986; Daròs et al., 1994). The results reported here allow a second ASBVd structural domain, the A+U-rich terminal loops, to be associated with a defined function: determining where synthesis of viroid strands starts.
Fig. 5. In vitro capping and ribonuclease protection assay (RPA). ASBVd RNAs from band x of a non-denaturing gel were capped in vitro, annealed to non-radioactive ASBVd (+) riboprobes RIII or RIV and treated with a mixture of RNases and the protected fragments were analyzed in a 6% sequencing gel. (A) Schematic representation of riboprobes RIII or RIV (white lines) and of the expected fragments protected by them if the two 5′ termini obtained by primer extension were capped (black lines). (B) Analysis by PAGE in a 6% sequencing gel of protected fragments. Lanes 1 and 2, product of the in vitro capping reaction with and without RNase treatment, respectively, in the absence of any complementary riboprobe. Lanes 3 and 5, RNase-protected fragments by riboprobes III and IV, respectively, with their sizes in nucleotides indicated on the left. Lane 4, labeled DNA markers with their sizes in nucleotides indicated on the right. The position of linear monomeric ASBVd RNAs is marked as ml. Correspondence between fragments determined by primer extension and by RPA is indicated by lines with double arrowheads connecting (A) and (B). (C) Location on the secondary structure predicted for the ASBVd (–) circular monomeric RNA of the residue U119, marked by an arrowhead, at which synthesis of ASBVd (–) strands starts. Continuous lines denote fragments protected by riboprobes RIII and RIV. Numbering is the same as in the (+) polarity.
On the other hand, the specific location of both ASBVd initiation sites implies that synthesis of the lower strands of the most stable structures predicted for both ASBVd RNAs (Figures 3C and 5C) is completed before synthesis of their quasi-complementary upper strands begins. Because RNA folding mechanisms can be complex, involving parallel pathways and kinetic traps (Thirumalai, 1998), the growing ASBVd strands may well adopt metastable conformations potentially relevant in vivo. Indeed, computer-assisted analysis showed that the upper and lower strands of the ASBVd RNAs can both form such alternative structures (data not shown).
Conserved sequences flanking the initiation sites of ASBVd (+) and (–) strands support the involvement of a common RNA polymerase in their synthesis
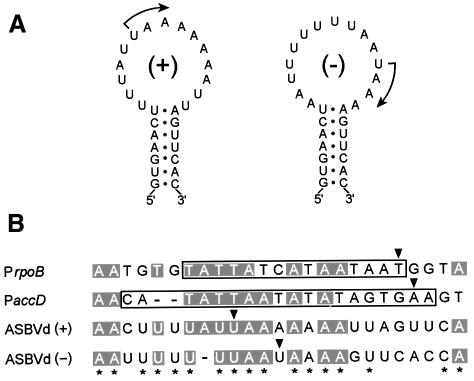
There is a striking similarity between the sequences near the two ASBVd initiation sites (Figure 6A). In particular, ASBVd (+) and (–) RNAs start with the same sequence, UAAAA (Figures 3 and 5). This strongly suggests that viroid promoters are formed, at least in part, by the sequences flanking the two initiation sites, which are presumably recognized by the same transcription factor(s) that then recruits the RNA polymerase. As already indicated, ASBVd replicates in the chloroplast, where two RNA polymerases have been characterized: a plastid-encoded polymerase (PEP) with a multisubunit structure similar to the Escherichia coli enzyme (Sugiura, 1992), and a single-unit nuclear-encoded polymerase (NEP) resembling phage RNA polymerases (Allison et al., 1996). PEP appears to be sensitive to tagetitoxin (Mathews and Durbin, 1990), whereas the inhibitor has no effect on NEP-transcribed genes (Liere and Maliga, 1999) or on ASBVd (Navarro et al., 2000), suggesting that polymerization of viroid strands is mediated by an NEP-like activity. Interestingly, the two best characterized NEP promoters (Liere and Maliga, 1999) have extensive similarity with the sequences around ASBVd initiation sites (Figure 6B), reinforcing the involvement of an NEP-like activity in transcription of both polarity strands of ASBVd.
Fig. 6. (A) Initiation sites, marked with arrows, in the A+U-rich terminal loops of ASBVd (+) and (–) RNAs. (B) Alignment of the sequences around both ASBVd initiation sites with the two best characterized promoters, and their flanking sequences, of the NEP involved in transcription of some chloroplast genes. rpoB and accD refer to genes coding for the β subunit of the PEP and for a subunit of the acetyl-CoA carboxylase, respectively. The sequences of their corresponding promoters PrpoB (15 nt) and PaccD (19 nt) are boxed. Conserved residues in at least three of the four sequences are in a gray background and conserved residues between both polarity strands of ASBVd are marked by asterisks. Arrowheads indicate initiation sites and dashes denote gaps.
Discussion
By in vitro capping of the linear monomeric ASBVd RNAs isolated from infected tissue and RPA, single initiation sites of both polarity strands of this viroid have been mapped, providing evidence that transcription is promoter-driven rather than being initiated randomly. Primer extension analysis of the same templates cannot provide an unequivocal answer because all 5′ termini are determined irrespective of whether they are produced by specific initiation, termination or processing. However, a combination of the two analytical approaches, primer extension and in vitro capping plus RPA, has independently confirmed the precise initiation sites and helped to delimit them at the nucleotide level. Both initiation sites have been found embedded in A+U-rich regions located in terminal hairpin loops of the quasi-rod-like structures predicted for the ASBVd (+) and (–) monomeric circular RNAs (Figures 3C and 5C), enabling the allocation of a defined functional role to these structural domains. Since the ASBVd (+) and (–) monomeric circular RNAs are the initial templates of the two symmetrical halves of the rolling-circle replication mechanism proposed for this viroid, the structural similarity between the regions surrounding the two initiation sites suggests that a similar mechanism operates in assembling the replication complexes that initiate and elongate the ASBVd RNAs of both polarities. ASBVd (+) RNAs accumulate in infected tissue at concentrations considerably higher than their (–) counterparts (Hutchins et al., 1985; Daròs et al., 1994), maybe because of minor sequence differences around both initiation sites that would affect the affinity for RNA polymerase-associated transcription factors; alternatively, this may reflect the distinct in vivo stability of both ASBVd strands. It is also worth noting in this context that sequence heterogeneity in ASBVd variants has been found predominantly in both terminal loops (Pallás et al., 1988; Rakowski and Symons, 1989; Semancik and Szychowski, 1994). However, mutations in the right terminal loop are U or A insertions, and A→U or U→A substitutions, which maintain its A+U-rich character.
On the basis of sensitivity to tagetitoxin, an NEP-like activity has been proposed as the chloroplast RNA polymerase involved in the synthesis of ASBVd strands (Navarro et al., 2000). The two NEP promoters that have so far been subjected to a more detailed functional analysis, corresponding to rpoB and accD genes coding for the β subunit of the PEP and for a subunit of the acetyl-CoA carboxylase, respectively, are exclusively composed of a short sequence of 15–19 nt placed immediately upstream of the transcription start sites (Liere and Maliga, 1999). The similarity between these promoters and the sequences around the ASBVd initiation sites (Figure 6B) suggests that these sequences constitute or form part of the promoters of ASBVd, which within 247 nt has to accommodate other functional determinants. However, the parallels between transcription of the rpoB and accD genes, and of ASBVd strands, must be regarded with care considering the different nature (DNA versus RNA) and structure (double-stranded versus partially double-stranded) of the corresponding templates. It is possible that the partially double-stranded character of the ASBVd terminal hairpins containing the initiation sites, which have stems with identical sequences (Figure 6A), may serve as a recognition signal for the proposed NEP-like activity and favor its template switching from DNA to RNA. Previous reports have shown that the DNA-dependent T7 RNA polymerase, which is structurally similar to NEP, can recognize and replicate two small single-stranded RNAs that, like ASBVd, have an A+U-rich composition and compact folding, and initiate replication at unpaired residues close to a double-stranded region (Konarska and Sharp, 1989, 1990). On the other hand, the location of both ASBVd initiation sites in terminal hairpin loops parallels the situation found in the promoters of viral and subviral RNAs of some positive-stranded plant RNA viruses in which terminal hairpin loops are critical for promoter activity (Lauber et al., 1997; Carpenter and Simon, 1998).
Little is known about the nature of the initiation sites in other viroid and viroid-like RNAs. Using a different experimental approach, based on in vitro transcription studies with a nuclear extract from potato cells supplemented with PSTVd monomeric (+) circular RNA, initiation of PSTVd (–) strands has been proposed to occur at two different positions, neither of which is located at terminal loops (Riesner et al., 1999). In the case of the viroid-like RNA of human hepatitis delta virus (HDV), which has a circular structure, a predicted rod-like conformation and a symmetric rolling-circle mode of replication, there is an interesting parallel with ASBVd. A subgenomic polyadenylated complementary RNA, the presumed mRNA for the HDV delta antigen, has been detected in HDV-infected tissues (Hsieh et al., 1990). Rapid amplification of cDNA ends has shown that the 5′ terminus of the subgenomic RNA is near the so-called top terminal loop of the rod-like secondary structure proposed for HDV RNA (Gudima et al., 1999). The finding of this 5′ terminus has led to the hypothesis that it is the initiation site of all complementary HDV RNAs and, as a corollary, that close to this site there is a region in HDV genomic RNA that acts as a promoter. Regarded as support for this view, a 29 nt fragment close to the top terminal loop of the rod-like structure of HDV RNA showed promoter activity when expressed as a cDNA, and a slightly larger fragment was also shown to act as a promoter for transcription in the other direction (Macnaughton et al., 1993). Moreover, an HDV RNA of genomic polarity corresponding to this region was able to direct synthesis of complementary RNA in a nuclear extract transcription assay (Beard et al., 1996), and an HDV RNA of antigenomic polarity, corresponding to the other terminal hairpin, served as a template for efficient and specific synthesis of genomic RNA in an alternative in vitro transcription system based on HeLa cell nuclear extracts (Filipovska and Konarska, 2000). Therefore, the available evidence, although not yet conclusive, suggests that the initiation sites of HDV RNAs, and one or both RNA promoters, are also located close to the terminal hairpin loops of the proposed rod-like structures.
It has long been accepted that nascent RNA can begin to adopt local sequential folding in the course of transcription (Boyle et al., 1980; Kramer and Mills, 1981). Simulations, using in vitro transcription of a PSTVd cDNA template with T7 RNA polymerase, have revealed the generation of metastable structures that probably play an important role in viroid replication (Repsilber et al., 1999). Furthermore, computer simulations of RNA folding pathways with algorithms that approximate the kinetic features of the RNA folding process, including folding during transcription, have also been used to predict functional metastable structures (Schmitz and Steger, 1996; Gultyaev et al., 1998). Critical parameters in these simulations are the elongation rate of the RNA polymerase, which can only be guessed, taking as a reference the rates estimated for polymerases transcribing their standard templates, and the site where transcription is initiated in vivo. Therefore, results reported here on mapping the initiation sites of ASBVd (+) and (–) RNAs are particularly relevant in this context and should help to improve simulations of their corresponding folding pathways.
The experimental approach used in the present work to identify the intiation sites of ASBVd is based on the characterization of a chemical group (a 5′ triphosphate) that specifically marks where transcription starts in vivo. Therefore, it is different from the in vitro transcription assays applied in the cases of PSTVd and HDV RNAs, and has the additional advantage that it allows mapping of the initiation sites of both polarity strands. The application of this approach, or modifications thereof, to other viroid and viroid-like RNAs may help to determine where synthesis of these RNAs starts, as well as to provide information about the possible nature of the promoters directing this synthesis and about RNA folding pathways.
Materials and methods
RNA extraction and purification
Nucleic acids from leaves of ASBVd-infected avocado plants (Persea americana Mill.) were extracted (Dellaporta et al., 1983) and fractionated on non-ionic cellulose (CF11; Whatman) with STE (50 mM Tris–HCl pH 7.2, 100 mM NaCl, 1 mM EDTA) containing 35% ethanol (Navarro et al., 1999). Polysaccharides were removed with methoxyethanol (Bellamy and Ralph, 1968). RNAs were partitioned with 2 M LiCl overnight at 4°C and the soluble fraction recovered. ASBVd RNAs were separated by non-denaturing PAGE and some of the RNAs were eluted and separated again by denaturing PAGE to obtain the ASBVd monomeric circular and linear forms (Navarro et al., 1999).
Plasmid construction
ASBVd DNAs with the complete viroid sequence were obtained by RT–PCR using two pairs of adjacent primers of opposite polarities. First-strand cDNA was synthesized on purified ASBVd monomeric circular forms with primer PI (5′-CAGACCTGGTTTCGTC-3′) or PV (5′-TGAAGAGACGAAGTGATC-3′) and avian myeloblastosis virus (AMV) reverse transcriptase. For synthesis of second-strand cDNA, aliquots (1/20) of these preparations were PCR-amplified with PI and PVI (5′-TTCCGACTTTCCGACTCTGAG-3′), or PV and PIII (5′-GGGAAA GATGGGAAGAACACTGATGAGTCTCGC-3′), and 2.5 U of cloned Pfu DNA polymerase using the buffer recommended by the manufacturer (Stratagene). The PCR cycling profile (30 cycles) was 95°C for 40 s, 60°C for 30 s and 72°C for 2 min, with a final extension step at 72°C for 15 min. DNA products of the expected length were separated by non-denaturing PAGE, eluted and cloned into the EcoRV site of pBluescript II KS+ (Stratagene), giving rise to recombinant plasmids pAS9 and pAS14 with inserts obtained with PI and PV, and PVI and PIII, respectively.
Primer extension analysis
Labeling of primers at their 5′ ends with T4 polynucleotide kinase and [γ-32P]ATP (3000 Ci/mmol; Amersham) was according to standard procedures (Sambrook et al., 1989). Each labeled primer (100 ng) was allowed to anneal to 100 ng of purified ASBVd RNAs in sterile water at 80°C for 3 min and snap-cooled on ice. Primer extension reactions (20 µl final volume) were carried out in 50 mM Tris–HCl pH 8.3, containing 75 mM KCl, 3 mM MgCl2, 10 mM dithiothreitol (DTT), 1 mM each of dATP, dCTP, dGTP and dTTP, 125 µg/ml actinomycin D and 200 U of reverse transcriptase (SuperScript II RNase H–; Life Technologies). After incubation at 42°C for 1 h, the reaction mixture was heated at 70°C for 15 min and the cDNAs were analyzed by PAGE on 6% sequencing gels. The exact size of the extension products was determined by running in parallel sequence ladders obtained with the same primer and a recombinant plasmid with a complete ASBVd insert. ASBVd-specific oligonucleotides used in primer extensions were PI, PII (5′-GTGTTCTTCCCATCTTTCCCTGAAGAGACGAAGTG-3′), PIII and PIV (5′-TTAGAACAAGAAGTGAGAGG-3′).
In vitro capping
Monomeric linear ASBVd RNAs obtained from band m of non-denaturing gels (Figure 1) were capped in vitro in a reaction mixture (20 µl final volume) containing 50 mM Tris–HCl pH 7.9, 1.25 mM MgCl2, 6 mM KCl, 2.5 mM DTT, 100 U of human placental RNase inhibitor (Amersham), 50 µCi [α-32P]GTP (3000 Ci/mmol; Amersham) and 10 U of guanylyltransferase (Life Technologies). Following incubation at 37°C for 90 min, labeled RNAs were extracted with phenol–chloroform, recovered by ethanol precipitation and passed through a Sephadex G-50 spun column (1 ml) to remove the unincorporated labeled precursor. In vitro capping of linear ASBVd RNAs from band x of non-denaturing gels (Figure 1) was as above except that the amount of [α-32P]GTP was doubled and fresh guanylyltransferase (10 U) was added to the reaction mixture after 45 min with the incubation extended for another 45 min.
Ribonuclease protection assay
An RPA II kit was used according to the instruction manual provided by the supplier (Ambion). Briefly, in vitro capped ASBVd RNAs were hybridized overnight at 65°C in 20 µl of hybridization buffer (80% deionized formamide, 100 mM sodium citrate pH 6.4, 300 mM sodium acetate pH 6.4 and 1 mM EDTA) with a molar excess of non-radioactive ASBVd-specific riboprobes, and then digested with a mixture of RNase A (10 U/ml) and T1 (400 U/ml). Protected fragments were identified by PAGE in 6% sequencing gels. In protection assays using linear monomeric ASBVd RNAs obtained from band x of non-denaturing gels (Figure 1), the in vitro capped RNAs were denatured with 10 mM methylmercuric hydroxide for 10 min at room temperature in the presence of the non-radioactive riboprobe, and the hybridization buffer (20 µl) contained 80% deionized formamide, 40 mM PIPES–NaOH pH 6.4, 40 mM NaCl and 5 mM EDTA. Unlabeled ASBVd (–) riboprobes RI and RII were transcribed with T7 RNA polymerase from XbaI-linearized plasmids pAS14 and pAS9, respectively. Unlabeled ASBVd (+) riboprobes RIII and RIV were transcribed with T3 RNA polymerase from XhoI-linearized plasmids pAS9 and pAS14, respectively.
Acknowledgments
Acknowledgements
We thank A.Ahuir for excellent technical assistance, Drs A.Vera, C.Hernández, J.A.Daròs, V.Pallás and J.Taylor for critical reading of the manuscript and/or suggestions, and Barraclough-Donnellan for English revision. R.F. was supported by grant PB98-0500 from the Comisión Interministerial de Ciencia y Tecnología de España. J.-A.N. was the recipient of a predoctoral fellowship from the Generalidad Valenciana.
References
- Allison L.A., Simon,L.D. and Maliga,P. (1996) Deletion of rpoB reveals a second distinct transcription system in plastids of higher plants. EMBO J., 15, 2802–2809. [PMC free article] [PubMed] [Google Scholar]
- Auchincloss A.H. and Brown,G.G. (1989) Soybean mitochondrial transcripts capped in vitro with guanylyltransferase. Biochem. Cell Biol., 67, 315–319. [Google Scholar]
- Beard M.R., Macnaughton,T.B. and Gowans,E.J. (1996) Identification and characterization of a hepatitis delta virus RNA transcriptional promoter. J. Virol., 70, 4986–4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellamy A.R. and Ralph,R.K. (1968) Recovery and purification of nucleic acids by means of cetyltrimethylammonium bromide. Methods Enzymol., 12, 156–160. [Google Scholar]
- Bonfiglioli R.G., McFadden,G.I. and Symons,R.H. (1994) In situ hybridization localizes avocado sunblotch viroid on chloroplast thylakoid membranes and coconut cadang cadang viroid in the nucleus. Plant J., 6, 99–103. [Google Scholar]
- Boyle J., Robillard,G.T. and Kim,S.H. (1980) Sequential folding of transfer RNA: a nuclear magnetic resonance study of successively longer tRNA fragments with a common 5′ end. J. Mol. Biol., 139, 601–625. [DOI] [PubMed] [Google Scholar]
- Branch A.D. and Robertson,H.D. (1981) Longer-than-unit-length viroid minus strands are present in RNA from infected plants. Proc. Natl Acad. Sci. USA, 78, 6381–6385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branch A.D. and Robertson,H.D. (1984) A replication cycle for viroids and other small infectious RNAs. Science, 223, 450–454. [DOI] [PubMed] [Google Scholar]
- Branch A.D., Benenfeld,B.J. and Robertson,H.D. (1988) Evidence for a single rolling circle in the replication of potato spindle tuber viroid. Proc. Natl Acad. Sci. USA, 85, 9128–9132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter C.D. and Simon,A.E. (1998) Analysis of sequence and predicted structures required for viral satellite RNA accumulation by in vivo genetic selection. Nucleic Acids Res., 26, 2426–2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daròs J.A., Marcos,J.F., Hernández,C. and Flores,R. (1994) Replication of avocado sunblotch viroid: evidence for a symmetric pathway with two rolling circles and hammerhead ribozyme processing. Proc. Natl Acad. Sci. USA, 91, 12813–12817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellaporta S.L., Wood,J. and Hicks,J.B. (1983) A plant DNA minipreparation: version II. Plant Mol. Biol. Rep., 1, 19–21. [Google Scholar]
- Diener T.O. (1991) Subviral pathogens of plants: viroids and viroidlike satellite RNAs. FASEB J., 5, 2808–2813. [DOI] [PubMed] [Google Scholar]
- Diener T.O. (1971a) Potato spindle tuber ‘virus’: a plant virus with properties of a free nucleic acid. III. Subcellular location of PSTV-RNA and the question of whether virions exist in extracts or in situ.Virology, 43, 75–89. [DOI] [PubMed] [Google Scholar]
- Diener T.O. (1971b) Potato spindle tuber ‘virus’. IV. A replicating low molecular weight RNA. Virology, 45, 411–428. [DOI] [PubMed] [Google Scholar]
- Feldstein P.A., Hu,Y. and Owens,R.A. (1998) Precisely full length, circularizable, complementary RNA: an infectious form of potato spindle tuber viroid. Proc. Natl Acad. Sci. USA, 95, 6560–6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipovska J. and Konarska,M.M. (2000) Specific HDV RNA-templated transcription by pol II in vitro.RNA, 6, 41–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores R. and Semancik,J.S. (1982) Properties of a cell-free system for synthesis of citrus exocortis viroid. Proc. Natl Acad. Sci. USA, 79, 6285–6288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores R., Di Serio,F. and Hernández,C. (1997) Viroids: the non-coding genomes. Semin. Virol., 8, 65–73. [Google Scholar]
- Flores R., Randles,J.W., Bar-Joseph,M. and Diener,T.O. (2000) Viroids. In van Regenmortel,M.H.V. et al. (eds), Virus Taxonomy. Seventh Report of the International Committee on Taxonomy of Viruses. Academic Press, San Diego, CA, pp. 1009–1024. [Google Scholar]
- Grill L.K. and Semancik,J.S. (1978) RNA sequences complementary to citrus exocortis viroid in nucleic acid preparations from infected Gynura aurantiaca. Proc. Natl Acad. Sci. USA, 75, 896–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross H.J., Domdey,H., Lossow,C., Jank,P., Raba,M., Alberty,H. and Sänger,H.L. (1978) Nucleotide sequence and secondary structure of potato spindle tuber viroid. Nature, 273, 203–208. [DOI] [PubMed] [Google Scholar]
- Gudima S., Dingle,K., Wu,T.T., Moraleda,G. and Taylor,J. (1999) Characterization of the 5′ ends for polyadenylated RNAs synthesized during replication of hepatitis delta virus. J. Virol., 73, 6533–6539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gultyaev A.P., van Batenburg,F.H.D. and Pleij,C.W.A. (1998) Dynamic competition between alternative structures in viroid RNAs simulated by an RNA folding algorithm. J. Mol. Biol., 276, 43–55. [DOI] [PubMed] [Google Scholar]
- Harders J., Lukacs,N., Robert-Nicoud,M., Jovin,J.M. and Riesner,D. (1989) Imaging of viroids in nuclei from tomato leaf tissue by in situ hybridization and confocal laser scanning microscopy. EMBO J., 8, 3941–3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández C. and Flores,R. (1992) Plus and minus RNAs of peach latent mosaic viroid self cleave in vitro via hammerhead structures. Proc. Natl Acad. Sci. USA, 89, 3711–3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh S.-Y., Chao,M., Coates,L. and Taylor,J. (1990) Hepatitis delta virus genome replication: a polyadenylated mRNA for delta antigen. J. Virol., 64, 3192–3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchins C.J., Keese,P., Visvader,J.E., Rathjen,P.D., McInnes,J.L. and Symons,R.H. (1985) Comparison of multimeric plus and minus forms of viroids and virusoids. Plant Mol. Biol., 4, 293–304. [DOI] [PubMed] [Google Scholar]
- Hutchins C.J., Rathjen,P.D., Forster,A.C. and Symons,R.H. (1986) Self-cleavage of plus and minus RNA transcripts of avocado sunblotch viroid. Nucleic Acids Res., 14, 3627–3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konarska M.M. and Sharp,P.A. (1989) Replication of RNA by the DNA-dependent RNA polymerase of phage T7. Cell, 57, 423–431. [DOI] [PubMed] [Google Scholar]
- Konarska M.M. and Sharp,P.A. (1990) Structure of RNAs replicated by the DNA-dependent T7 RNA polymerase. Cell, 63, 609–618. [DOI] [PubMed] [Google Scholar]
- Kramer F.R. and Mills,D.R. (1981) Secondary structure formation during RNA synthesis. Nucleic Acids Res., 9, 5109–5124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauber E., Guilley,H., Richards,K., Jonard,G. and Gilmer,D. (1997) Conformation of the 3′-end of beet necrotic yellow vein benevirus RNA 3 analysed by chemical and enzymatic probing and mutagenesis. Nucleic Acids Res., 25, 4723–4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liere K. and Maliga,P. (1999) In vitro characterization of the tobacco rpoB promoter reveals a core sequence motif conserved between phage-type plastid and plant mitochondrial promoters. EMBO J., 18, 249–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima M.I., Fonseca,M.E.N., Flores,R. and Kitajima,E.W. (1994) Detection of avocado sunblotch viroid in chloroplasts of avocado leaves by in situ hybridization. Arch. Virol., 138, 385–390. [DOI] [PubMed] [Google Scholar]
- Macnaughton T.B., Beard,M.R., Chao,M., Gowans,E.J. and Lai,M.M.C. (1993) Endogenous promoters can direct the transcription of hepatitis delta virus RNA from a recircularized cDNA template. Virology, 196, 629–636. [DOI] [PubMed] [Google Scholar]
- Marcos J.F. and Flores,R. (1993) The 5′ end generated in the in vitro self-cleavage reaction of avocado sunblotch viroid RNA is present in naturally occurring linear viroid molecules. J. Gen. Virol., 74, 907–910. [DOI] [PubMed] [Google Scholar]
- Mathews D.E. and Durbin,R.D. (1990) Tagetitoxin inhibits RNA synthesis directed by RNA polymerases from chloroplasts and Escherichia coli. J. Biol. Chem., 265, 493–498. [PubMed] [Google Scholar]
- Mühlbach H.P. and Sänger,H.L. (1979) Viroid replication is inhibited by α-amanitin. Nature, 278, 185–188. [DOI] [PubMed] [Google Scholar]
- Navarro B. and Flores,R. (1997) Chrysanthemum chlorotic mottle viroid: unusual structural properties of a subgroup of viroids with hammerhead ribozymes. Proc. Natl Acad. Sci. USA, 94, 11262–11267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro J.A., Daròs,J.A. and Flores,R. (1999) Complexes containing both polarity strands of avocado sunblotch viroid: identification in chloroplasts and characterization. Virology, 253, 77–85. [DOI] [PubMed] [Google Scholar]
- Navarro J.A., Vera,A. and Flores,R. (2000) A chloroplastic RNA polymerase resistant to tagetitoxin is involved in replication of avocado sunblotch viroid. Virology, 268, 218–225. [DOI] [PubMed] [Google Scholar]
- Owens R.A. and Diener,T.O. (1982) RNA intermediates in potato spindle tuber viroid replication. Proc. Natl Acad. Sci. USA, 79, 113–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallás V., García-Luque,I., Domingo,E. and Flores,R. (1988) Sequence variability in avocado sunblotch viroid (ASBV). Nucleic Acids Res., 16, 9864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu F., Heinrich,C., Loss,P., Steger,G., Tien,P. and Riesner,D. (1993) Multiple pathways of reversion in viroid conservation of structural domains. EMBO J., 12, 2129–2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakowski A.G. and Symons,R.H. (1989) Comparative sequence studies of variants of avocado sunblotch viroid. Virology, 173, 352–356. [DOI] [PubMed] [Google Scholar]
- Repsilber D., Wiese,S., Rachen,M., Schröder,A.W., Riesner,D. and Steger,G. (1999) Formation of metastable RNA structures by sequential folding during transcription: time-resolved structural analysis of potato spindle tuber viroid (–)-stranded RNA by temperature-gradient gel electrophoresis. RNA, 5, 574–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riesner D., Fels,A., Repsilber,D., Schmitz,A., Schrader,O., Schröder,A. and Steger,G. (1999) Structural motifs involved in replication and pathogenicity of potato spindle tuber viroid. In XI International Congress of Virology, Sydney, Australia, p. 29. [Google Scholar]
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Schindler I.M. and Mühlbach,H.P. (1992) Involvement of nuclear DNA-dependent RNA polymerases in potato spindle tuber viroid replication: a reevaluation. Plant Sci., 84, 221–229. [Google Scholar]
- Schmitz M. and Steger,G. (1996) Description of RNA folding by ‘simulated annealing’. J. Mol. Biol., 255, 254–266. [DOI] [PubMed] [Google Scholar]
- Semancik J.S. and Szychowski,J.A. (1994) Avocado sunblotch disease: a persistent viroid infection in which variants are associated with differential symptoms. J. Gen. Virol., 75, 1543–1549. [DOI] [PubMed] [Google Scholar]
- Spiesmacher E., Mühlbach,H.P., Schnölzer,M., Haas,B. and Sänger,H.L. (1983) Oligomeric forms of potato spindle tuber viroid (PSTV) and of its complementary RNA are present in nuclei isolated from viroid-infected potato cells. Biosci. Rep., 3, 767–774. [DOI] [PubMed] [Google Scholar]
- Sugiura M. (1992) The chloroplast genome. Plant Mol. Biol., 19, 149–168. [DOI] [PubMed] [Google Scholar]
- Symons R.H. (1981) Avocado sunblotch viroid: primary sequence and proposed secondary structure. Nucleic Acids Res., 9, 6527–6537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symons R.H. (1997) Plant pathogenic RNAs and RNA catalysis. Nucleic Acids Res., 25, 2683–2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thirumalai D. (1998) Native secondary structure formation in RNA may be a slave to tertiary folding. Proc. Natl Acad. Sci. USA, 95, 11506–11508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vera A. and Sugiura,M. (1992) Combination of in vitro capping and ribonuclease protection improves the detection of transcription start sites in chloroplasts. Plant Mol. Biol., 19, 309–311. [DOI] [PubMed] [Google Scholar]