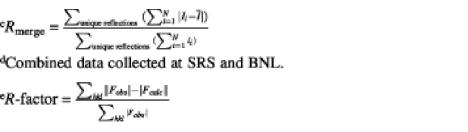Abstract
Chloramphenicol (Cm), produced by the soil bacterium Streptomyces venezuelae, is an inhibitor of bacterial ribosomal peptidyltransferase activity. The Cm-producing streptomycete modifies the primary (C-3) hydroxyl of the antibiotic by a novel Cm-inactivating enzyme, chloramphenicol 3-O-phosphotransferase (CPT). Here we describe the crystal structures of CPT in the absence and presence of bound substrates. The enzyme is dimeric in a sulfate-free solution and tetramerization is induced by ammonium sulfate, the crystallization precipitant. The tetrameric quaternary structure exhibits crystallographic 222 symmetry and has ATP binding pockets located at a crystallographic 2-fold axis. Steric hindrance allows only one ATP to bind per dimer within the tetramer. In addition to active site binding by Cm, an electron-dense feature resembling the enzyme’s product is found at the other subunit interface. The structures of CPT suggest that an aspartate acts as a general base to accept a proton from the 3-hydroxyl of Cm, concurrent with nucleophilic attack of the resulting oxyanion on the γ-phosphate of ATP. Comparison between liganded and substrate-free CPT structures highlights side chain movements of the active site’s Arg136 guanidinium group of >9 Å upon substrate binding.
Keywords: antibiotic inactivation/chloramphenicol/kinase/phosphotransferase/resistance
Introduction
Chloramphenicol (Cm) was first discovered in 1947 as an antibiotic in cultures of the producer Streptomyces venezuelae and is now synthesized chemically. This important antibiotic inhibits protein biosynthesis by binding reversibly to the 50S prokaryotic ribosomal subunits (Vining and Westlake, 1984) at a site that blocks the peptidyl transferase step in protein biosynthesis. Resistance to Cm due to enzymic inactivation of the antibiotic is normally because of chloramphenicol acetyltransferase (CAT) activity (Shaw and Leslie, 1991). CAT modifies Cm by acetylation, yielding 3-O-acetyl-Cm, which is very weakly bound by ribosomes and lacks antibiotic activity. Although the CAT mechanism for resistance to Cm is widespread in bacteria, including some actinomycetes, it is not used by the producing organism to protect itself against its own toxic products (Shaw and Hopwood, 1976). The 3-phosphoester of Cm has been identified in S.venezuelae (Mosher et al., 1995), suggesting that the producing organism has a mechanism of Cm resistance that has not been encountered in other streptomycetes or microbial systems. The enzyme responsible for this novel inactivation of Cm by phosphorylation, chloramphenicol 3-O-phosphotransferase (CPT), has a polypeptide chain of 178 residues (mol. wt 19 kDa; Mosher et al., 1995). The protein is active either as a homodimer in the absence of sulfate or as a tetramer in the presence of ammonium sulfate, the precipitant used for crystallization. The closely related enzymes shikimate kinase (Krell et al., 1998) and adenylate kinase (Dreusicke et al., 1988) are both monomeric enzymes. CPT uses ATP as phosphoryl donor to transfer the γ-phosphate to the primary (C-3) hydroxyl of Cm. The 3-phosphoryl-Cm product is biologically inactive as an antibiotic. Phosphorylation, unlike acetylation, is easily reversible through removal of the protecting group by an extracellular phosphatase that arms the antibiotic to carry out its defensive role in the environment.
In more than four decades, during which Cm has been used to treat infections in humans and animals, phosphorylation has not been reported as a mechanism for inactivating this antibiotic. Phosphorylation has been observed as a common means to obtain resistance to other antibiotics (Cundliffe, 1992); for example, the aminoglycoside kinases catalyse the phosphorylation of a wide range of important aminoglycoside antibiotics including neomycin B and kanamycin (Shaw et al., 1993). However, the similarity between these aminoglycoside phosphotransferases and CPT is limited to their functional roles within the bacterial cells, since these proteins show no significant sequence similarity to CPT. In contrast, CPT bears the most significant sequence similarity (43%) to the enzyme shikimate kinase type II of Escherichia coli, with a sequence identity of 16% across its entire 178 residues.
The CPT open reading frame has been identified (Mosher et al., 1995) and a protein with CPT activity has been overexpressed, purified and crystallized. The three-dimensional structure of CPT could not be determined by molecular replacement (Ellis et al., 1999) using the shikimate kinase crystal structure (Krell et al., 1998) as a search model. To address this novel resistance mechanism, we have crystallized the selenomethionyl isoform of the enzyme to obtain ab initio phases. Here we report the three-dimensional structures of: (i) the substrate-free CPT at 2.7 Å resolution; (ii) its ternary complex with Cm and a non-hydrolysable ATP analogue (ATPγS) at 2.6 Å resolution; (iii) the binary CPT–ATP complex at 2.5 Å resolution; and (iv) the binary CPT–Cm complex at 2.8 Å resolution.
Results
The enzyme fold
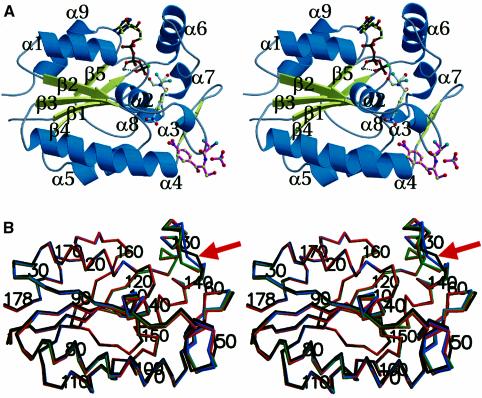
CPT consists of a central five-stranded parallel β-sheet flanked on either side by two α-helices (Figure 1). The core of the CPT structure forms a classical mononucleotide-binding fold. The four CPT structures (CPT, CPT–Cm, CPT–ATP and CPT–ATPγS–Cm), shown superimposed in Figure 1B, are very similar. The overall root mean square deviation (r.m.s.d.) for the Cα positions of residues 1–178 for any liganded CPT structure aligned with the substrate-free CPT is 0.5 Å. The 178 Cα atoms of either the complexed CPT–Cm or CPT–ATPγS–Cm structure align to the CPT–ATP structure with an overall r.m.s.d. of 0.28 and 0.22 Å, respectively, whereas the best superimposition is seen when comparing the CPT–Cm and ternary CPT structures (0.16 Å). The largest discrepancies in Cα positions occur for residues 135–137, where the Cα positions of the substrate-free structure have shifted >3.7 Å compared with the three other bound CPT structures (Figure 1B).
Fig. 1. (A) Stereo cartoon drawing (Bacon and Anderson, 1988; Kraulis, 1991; Merritt and Murphy, 1994) of the CPT–ATPγS–Cm protomer; α-helices are depicted by blue helical ribbons and β-strands by yellow arrows. One ATPγS, two ligands of Cm and one sulfate anion bound to the enzyme are shown in ball-and-stick representation. Oxygen atoms are coloured in red, nitrogen in blue, sulfur in green, carbon in yellow, phosphate in black, magnesium in grey and chlorine in light blue. This colour coding is used in all figures. Helices α3 and α8 are short 310 helices. For clarity, strands β2′ and β2′′ following helix α3 are not labelled. Members of the nucleoside monophosphate (NMP) kinase family characteristically undergo large conformational changes during catalysis. One flexible region responsible for this movement is the NMP-binding site that is formed by a series of α-helices located after β-strand β2. The second flexible region is the so-called lid domain, a region of varied size and structure following β-strand β4 (Müller et al., 1996). For example, in adenylate kinase, the NMP binding domain undergoes a rigid-body rotation of ∼40°, covering a movement of 8 Å, while the lid domain moves ∼30 Å, thereby undergoing an almost 90° hinge bending rotation (Schulz et al., 1990); both movements are enormous for such a small protein. An analogous closure upon substrate binding is likely to be found in CPT. (B) Stereo Cα-trace superimposition of substrate-free CPT (black), CPT–ATP (light blue), CPT–Cm (orange) and CPT–ATPγS–Cm (green) shown in the same orientation as in (A). Every tenth Cα atom is labelled. The largest r.m.s.d. between the Cα positions among the four CPT structures is found for the following residues: Ala50 (1.5 Å), Glu51 (1.5 Å), Ala58 (1.5 Å), Gly134 (1.6 Å), Asp135 (2.8 Å) and Arg136 (3.7 Å). The red arrow indicates the location of Arg136, a crucial residue involved in stabilizing the developing negative charge in the phosphoryl transfer reaction.
Quaternary structure
Sedimentation ultracentrifugation experiments suggest the enzyme to be dimeric in a sulfate-free solution and tetrameric in the presence of 320 mM ammonium sulfate, the precipitant used for crystallization. However, sedimentation ultracentrifugation studies of CPT under more physiological conditions (5–10 mM phosphate) are consistent with a dimeric oligomer (data not shown).
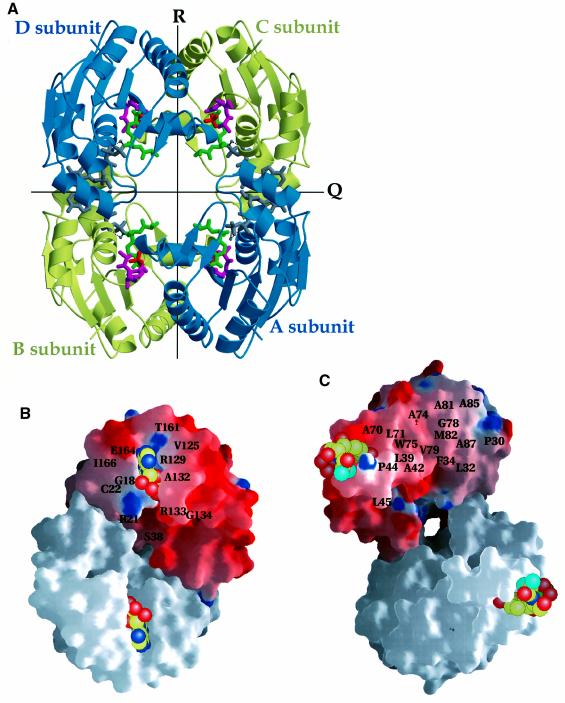
Remarkably, CPT crystallizes with one polypeptide chain per asymmetric unit, which results in a large solvent content of 0.86 and a packing density, Vm (Matthews, 1968), of 8.8 Å3/Da. The self-rotation function and the native Patterson did not reveal any significant peaks above the noise level other than the origin peak. Application of the crystallographic symmetry operators results in a tetramer with point group 222 (Figure 2A). The mutually perpendicular molecular dyads are defined as a right-handed set of axes P, Q, R. The crystallographic tetramer packs within the unit cell such as to make very loose 3-fold related contacts, burying an insignificant solvent-accessible surface area of 30 Å2. The N-terminal residues of the protein and residues located on α-helix α6 are responsible for these mainly water-mediated tetramer–tetramer interactions. Somewhat more extensive tetramer–tetramer contacts are found between 4-fold related oligomers with a buried solvent-accessible surface area of ∼50 Å2. Finally, extensive tetramer–tetramer contacts are observed between 2-fold related tetramers. Less than 700 Å2 are buried involving residues located on α-helix α9.
Fig. 2. (A) Cartoon drawing of the crystallographic CPT homotetramer with point group 222. The three different molecular dyads comprise a right-handed orthogonal set of axes P, Q, R as originally defined for the three 2-fold axes of lactate dehydrogenase (Rossmann et al., 1973). Two of the dyads (Q and R) are indicated by solid lines and the tetramer is shown along its P dyad. The four subunits are labelled A–D. Ligands are shown: ATPγS, grey; Cm, green; Cm–SO42–, magenta and red. (B) View onto crystallographic Q dyad interface showing R-axis-related subunits A and B (and C and D). The P-axis is horizontal, the Q-axis vertical and the R-axis perpendicular to the plane of the drawing. Some residues making inter-subunit contacts are labelled. The electrostatic surface potential (by using program GRASP; Nicholls et al., 1991) is shown for the A subunit (red, negative; blue, positive; white, uncharged), while a white surface is shown for the crystallographically 2-fold (R)-related B subunit. The adenylates of the bound ATPs are clearly visible, whereas the three phosphates are buried and not visible in this figure. The two interfaces are complementary by a 180° rotation around the horizontal P-axis. (C) View onto the crystallographic R dyad interface showing Q-axis-related subunits A and C (and B and D). The P-axis is horizontal, the R-axis vertical and the Q-axis perpendicular to the plane of the drawing. The electrostatic surface potential is shown for the A subunit, while a white surface is shown for the crystallographically 2-fold (Q)-related C subunit. The secondary Cm–SO42– is shown in both subunits (see Discussion). Several hydrophobic side chains participate in the inter-subunit interactions; these include residues Pro30, Trp31, Leu32, Ala33, Phe34, Leu39, Ala42, Met43, Pro44, Met47, Leu71, Ala74, Trp75, Gly78, Val79, Ala81, Met82, Ala85 and Ala87.
The inter-subunit interactions across the Q dyad involve residues located on α-helices α1, α6 and α9. The interface interactions across the Q dyad interface implicate six residues (Gly18, Cys22, Val125, Ala132, Gly134 and Ile166) that participate in hydrophobic interactions (Figure 2B), six residues (Arg21, Arg38, Ala132, Arg133, Thr161 and Arg164) that promote inter-subunit hydrogen bonds, and one salt link between residues Glu164 and Arg129. In addition, six water molecules are found at this interface bridging the Q-axis-related subunits. Here, dimerization results in a buried surface area of ∼1000 Å2 per subunit (Figure 2B).
More extensive and predominantly hydrophobic interactions are formed between R-axis-related subunits A and B (and C and D; Figure 2C). Several secondary structural elements contribute up to 19 residues that participate in extensive inter-subunit van der Waals contacts. These involve residues located on the C-terminal end of α-helix α1 and its following loop, helix α3, and α-helices α2 and α4 and their following loops. The inter-subunit interactions here implicate 14 inter-subunit hydrogen bonds, and four water molecules are found bridging this interface. This interface buries about twice as large a surface area (2060 Å2 per subunit) upon dimerization.
Nucleotide binding
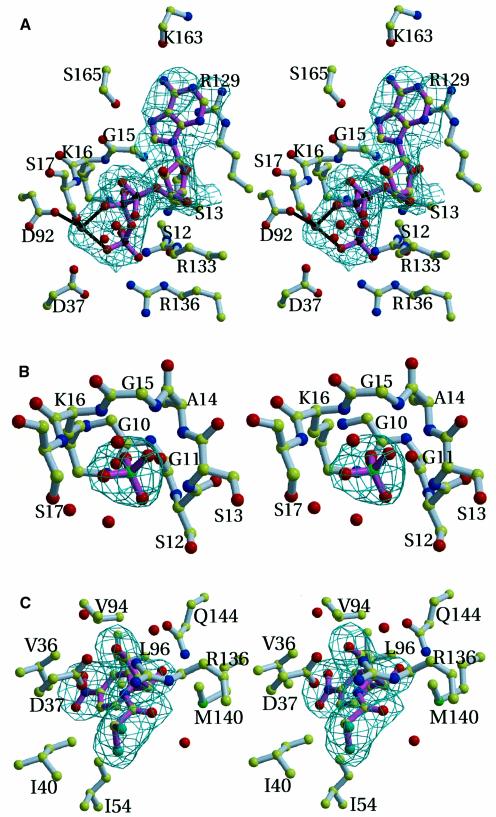
Many of the interactions made by ATP occur with α-helix α1 and its preceding loop (Figure 1). The γ-phosphate of ATP is within hydrogen bonding distance of the hydroxyl of Ser12, while the side chains and the amide nitrogens of Lys16 and Ser17, as well as the nitrogen of Ser13, interact with the β-phosphate (Figure 3A). Hydrogen bonds are formed between the α-phosphate of ATP and the oxygen bridging the α- and β-phosphorus with the nitrogen amide of Gly15. Additionally, the adenylate is in van der Waals contact with Arg129, and N7 of the adenylate is hydrogen bonded to the side chains of Ser165 (3 Å), while N6 interacts with the carbonyl of Lys163 (2.8 Å). The side chain of Arg133 is in hydrophobic contact with carbon atom C5 bridging the ribose with the α-phosphate as well as in ionic interactions with the oxygen bridging the β- and γ-phosphorus. Binding of ATPγS in the CPT–ATPγS–Cm ternary complex is very similar to that of ATP in the CPT–ATP binary complex. The main chain of Arg136 has moved 1.6 Å towards the nucleotide in the CPT–ATPγS–Cm ternary structure compared with the CPT–ATP structure. The guanidinium group of Arg136 is almost within hydrogen bonding distance (3.2 Å) of the γ-phosphate of the nucleotide in both structures. Although the electron density is clear for the entire residue 136 in the CPT–ATP structure, its temperature factors refined to high values.
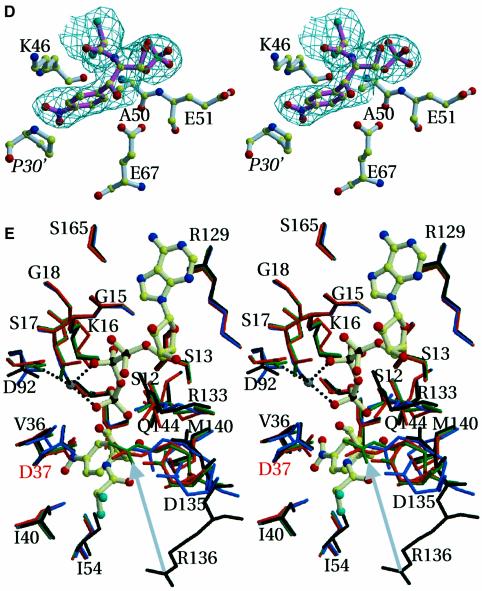
Fig. 3. Stereo views of ligands binding to CPT. Final σA weighted Fo – Fc omit electron density map for ligands bound to the enzyme. The bonds of the ligands are drawn in pink, while the bonds of the enzyme are shown in white. For clarity, water molecules (drawn as red spheres) are not labelled. (A), (B), (C) and (E) are in the same orientation. (A) Residues in contact with ATP. The catalytic magnesium site is shown and possible protein ligands coordinating the Mg2+ are indicated. The two water molecules shown interact with the α- and γ-phosphates, respectively. The contour level of the electron density map is 2.7σ and the resolution is 2.5 Å. (B) Sulfate binding to the substrate-free enzyme. This anion coincides with the β-phosphate position of ATP and ATPγS in the CPT–ATP and CPT–ATPγS–Cm structures, respectively. This sulfate is also found in the CPT–Cm crystal structure. The Nζ of Lys16 engages in electrostatic interactions with the sulfate ion in the substrate-free enzyme (2.8 Å) and is further away in the CPT–Cm crystal structure (4.1 Å). Arg133 forms further electrostatic interactions with the anion (not shown), as seen in the binding of the β-phosphate of ATP (A). Additionally, the two water molecules shown are within hydrogen bonding distance of the anion. The contour level of the electron density map is 3σ and the resolution is 2.7 Å. (C) Residues binding Cm as seen in the CPT–Cm binary complex. Arg136 is slightly further away in the ternary CPT structure (4 Å) and refined to a much larger temperature factor (∼63 Å2, compared with 35 Å2 in the CPT–Cm structure). The two water molecules shown interact with the primary hydroxyl of the antibiotic, while a further water molecule is in hydrogen bonding contact with Cm’s carbonyl. The contour level of the electron density map is 3.5σ and the resolution is 2.8 Å. (D) The hydrophobic pocket binding the distinct secondary Cm, which is hydrogen-bonded to a sulfate anion, as observed in the CPT–Cm binary complex. The ion is positioned relative to Cm such that Cm–SO42– resembles the product 3-phosphoryl-Cm. The contour level of the electron density map is 2.8σ and the resolution is 2.8 Å. Pro30′ belongs to an R-axis-related protomer. (E) Superimposition of the substrate-free CPT (black) onto CPT in complex with ligands (CPT–ATP, light blue; CPT–Cm, orange; CPT–ATPγS–Cm, green). Residues are labelled at positions corresponding to the substrate-free (black) CPT structure. Asp37, the proposed catalytic base that deprotonates the hydroxyl group on Cm, is labelled in red. Its side chain points towards Cm in CPT–Cm (orange), while in CPT–ATP it points towards the γ-phosphate (blue). It assumes positions between these in the substrate-free protein (black) and in the ternary complex (green). Asp135 points towards the antibiotic binding site in the substrate-free CPT structure, while its side chain is directed away from Cm in the ternary complex. The movement of the proposed transition state stabilizing side chain (Arg136) is indicated by a grey arrow. The three substrate-bound CPT structures were solved by using the substrate-free structure. All three ligand bound structures have been refined independently to give similar positions for Asp135 and Arg136. In the ternary CPT–ATPγS–Cm structure, the distance between the oxygen of the reactive hydroxyl of Cm and the γ-phosphorous atom is 3.1 Å. Nucleophilic addition to Pγ is assisted by charge dispersal by the metal ion and by the bond between Arg136 and the γ-phosphate. The geometry of their interaction supports a penta-coordinate geometry at the phosphorus.
In the CPT–ATP and CPT–ATPγS–Cm tetrameric crystal structures, the nucleotide binds at its dimer A–C (and B–D) subunit interface with its O3 oxygen of the ribose ring only 1.8 Å away from the O3 oxygen of a Q-axis-related ligand (Figure 2A). Temperature factors only refined to comparable values of the protein structure if an occupancy of one half was used for all nucleotide atoms except the β-phosphate. The steric hindrance together with the refined temperature factors suggest half-occupancy of the nucleotide whereby binding of one nucleotide blocks binding of its 2-fold related counterpart. The lower temperature factors and stronger electron density of the β-phosphate suggest that this site is fully occupied in both 2-fold related protomers. We conclude that one subunit binds the nucleotide, while the other binds a sulfate anion at the β-phosphate position of ATP.
The crystal structures of the nucleotide-free protein, CPT and CPT–Cm, show a bound sulfate anion coinciding with the β-phosphate location of ATP or ATPγS in the CPT–ATP or CPT–ATPγS–Cm crystal structure, respectively (Figure 3B). The sulfate anion participates in hydrogen bonds with several backbone amides residing on residues 10–17, which wrap around the sulfate anion.
Magnesium in the active site
CPT activity is reduced to 48% when the catalytic magnesium ion is replaced by a larger divalent cation such as Mn2+ (data not shown). During refinement, Fo – Fc electron density maps were calculated from models comprising all atoms except the subsequently identified magnesium cation. The strongest peak in the difference electron density maps calculated from either these CPT–ATP or CPT–ATPγS–Cm models showed a peak height of >12 times the noise level. This peak found in both CPT–ATP and CPT–ATPγS–Cm structures represents the best candidate for the binding site of the catalytic magnesium ion (Figure 3A). Possible ligands include one oxygen atom each from the β- (2.6 Å) and γ-phosphates (2.3 Å) of ATP, and the side chains of Ser17 (2.5 Å) and Asp92 (2.5 Å).
Cm binding
The crystal structures of CPT–Cm and CPT–ATPγS–Cm show that Cm binds the active site in a tunnel within the CPT subunit. The tunnel is lined by many hydrophobic residues that interact with Cm (for example, Val36, Ile54, Val94, Leu96 and Met140; Figure 3C), and is also lined by Phe56 in the CPT–ATPγS–Cm structure. The side chain of Asp37 is hydrogen bonded (2.6 Å) to the reactive hydroxyl group of Cm. Furthermore, the Nε of Gln144 is positioned 2.9 Å away from the carbonyl of Cm. The guanidinium group of Arg136 is almost within hydrogen bonding distance of the Cm’s reactive hydroxyl (3.2 Å). Superimposition of the adenylate kinase structure (Abele and Schulz, 1995) onto the CPT–Cm or CPT–ATPγS–Cm structure shows that the Cm binding site coincides with the AMP binding pocket.
Surprisingly, an electron dense feature was found in the CPT–Cm and CPT–ATPγS–Cm structures, located in a hydrophobic pocket lined by residues of helix α3. This electron density resembles that observed for Cm in the active site (Figure 3D). Moreover, all crystals were grown from the same batch of protein, with the exception of the selenomethionyl isoform of the protein, and the only experimental difference between co-crystallizing the CPT–Cm and CPT–ATPγS–Cm crystals compared with the CPT and CPT–ATP crystals was the inclusion of 2 mM Cm in the crystallization solution. We therefore modelled a second molecule of Cm into this electron density.
The benzyl ring of this secondary Cm makes van der Waals contacts with residues Lys46, Ala50 and Glu67, as well as with Pro30 located on the loop α1–β2 of the R-axis-related protomer (Figure 3D). Hydrogen bonding interactions include the side chain of Glu67 with the antibiotic’s carbonyl group. Differences between the CPT–Cm and CPT–ATPγS–Cm structures are significant only in the orientations of side chains Glu67 and Glu51, and their refined high temperature factors (74–104 Å2) indicate high mobility. A sulfate anion is within hydrogen bonding distance of the C-3 hydroxyl of Cm (3 Å), its phosphorylation site. This sulfate anion is further hydrogen bonded to the amide nitrogen of Cm (3 Å), and the ion’s only contact with CPT is one hydrogen bond to the amide nitrogen of Glu51. It is therefore not surprising that this anion is found only when Cm is present, in the CPT–Cm and CPT–ATPγS–Cm structures but not in the Cm-free structures CPT and CPT–ATP. The Cm–SO42– complex closely resembles the structure of the product of CPT, 3-phosphoryl-Cm. The Cm–SO42– resides 13 Å away from the Cm bound to the active site (Figure 1A).
Although the electron density for the secondary Cm binding site is strong and unambiguous, the temperature factors refined only to values similar to those of other ligands when occupancy of one half was used. We conclude that binding of Cm at this secondary Cm binding site is much weaker than at its catalytic site.
Discussion
Overall structure of the mononucleotide-binding fold
The precise ordering of the β-strands 23145 in the parallel β-sheet classifies CPT as belonging to the same structural family as the nucleoside monophosphate (NMP) kinases. This family includes adenylate kinase (Dreusicke et al., 1988) and shikimate kinase (Krell et al., 1998). Although CPT shows greatest sequence similarity to shikimate kinase (43%), superimposition of 131 Cα positions results in a large overall r.m.s.d. of 8.3 Å. The highest ranking Cα coordinate match using the DALI server (Holm and Sander, 1997) is found for adenylate kinase from Bacillus stearothermophilus (Berry and Phillips, 1998) with an r.m.s.d. of 5.5 Å for 157 Cα positions.
CPT and other nucleotide-binding enzymes contain the Walker A-motif (Walker et al., 1982), a short conserved stretch of sequence G-XXXX-GKT/S (10-G-GSSA-GKS-17 in CPT), near the N-terminus on the loop following β-strand β1 (Figure 1). This motif is often considered to be the signature of a phosphate-binding loop (P-loop), a giant anion hole that accommodates the β-phosphate of ATP by donating hydrogen bonds from several backbone amides (Saraste et al., 1990; Smith and Rayment, 1996). In all four CPT structures presented here, either a sulfate anion or the β-phosphate of the nucleotide binds the P-loop in a similar fashion (Figure 3A and B). In many P-loop proteins, the binding site for the magnesium cation essential for enzyme activity is associated with the P-loop. This cation binding involves hexa-coordination of the Mg2+ by one oxygen atom from each of the β- and γ-phosphates of the bound nucleotide, two water molecules and two protein ligands. One protein ligand is invariably the hydroxyl side chain of the Thr/Ser at the last position of the Walker A-motif. Our identified magnesium site is located analogously as reported for binding of the catalytic metal ion to P-loop proteins. It should be noted that the distances between the magnesium and its ligands in CPT (that were not restrained during refinement) are slightly larger (2.3–2.6 Å) than those seen in most P-loop proteins, which range from 2.0 to 2.3 Å. However, the coordinate errors as estimated from the Luzzati plot (Luzzati, 1952) or σA are 0.33 and 0.31 Å, respectively, for the CPT–ATP model at 2.5 Å resolution. Moreover, a crucial ligand for the metal, the γ-phosphate of ATP, is only present with half an occupancy while the cation has a full occupancy of one. Taking these factors into account, we conclude that magnesium is indeed bound at the identified location.
Secondary product binding site
Streptomyces venezuelae, like certain other antibiotic producers, may have an additional mechanism for self-protection, namely antibiotic export by an efflux protein (Davies, 1994). An open reading frame (ORF1) that has been identified upstream of the cpt gene encodes an efflux protein for 3-phosphoryl-Cm, rather than for Cm itself (Mosher et al., 1995). The efflux protein is thought to function in the presence of CPT to facilitate the export of inactivated antibiotic. The observed alternative product binding site could play a possible role in the resistance phenotype, either regulatory or as a ‘carrier’ to the efflux protein. However, solution binding studies have been unable to characterize a secondary product binding site, and partial occupancy of this site at the 2 mM Cm concentrations used in our crystallization experiments would indicate low occupancy at in vivo levels of ∼0.5 mM Cm (Shapiro and Vining, 1983).
The CPT oligomer
The CPT tetramer of point group 222 has two distinct crystallographically related 2-fold interfaces (Figure 2A). First, the ATP binding pockets are located between Q-axis-related subunits A and C (and B and D) and steric hindrance allows only one nucleotide to bind per Q-axis-related dimer within the crystallographic tetramer. However, there is no kinetic evidence to support half-site reactivity. Unfortunately, the high binding constant of ATP (Km of 2 mM in 50 mM HEPES pH 7) rules out UV optical techniques to provide independent measures of binding and stoichiometry due to the absorbance of the nucleotide. Secondly, the additional product binding site is located at the R-axis-generated crystallographic interface, although most contacts with the Cm ligand are made by one subunit (B and C; Figures 2A and 3D). The greater hydrophobicity, the significantly larger buried solvent-accessible surface area, and the ability for ligands to bind without steric hindrance would argue in favour of the A–B (and C–D) dimer being the solution dimer.
Does CPT undergo induced fit movements?
Identification of the sulfate anion bound to the nucleotide-free enzyme (CPT and CPT–Cm crystal structures) at the enzyme’s ATP-binding site raises the question of whether the substrate-free crystal structure represents the functional apo-form of the enzyme. The circular dichroism (CD) spectra of CPT indicate that large changes in secondary structure occur in the presence of either Mg2+-ATP or sulfate, but not in the presence of Cm (data not shown). In fact, the CD spectra of the CPT–ATP binary complex and CPT in the presence of 400 mM ammonium sulfate are superimposable, implying that similar changes in the enzyme’s secondary structure are induced by either ATP or sulfate. It is possible that these changes occur because of a domain closure upon ATP or sulfate binding. ATP buries 76% of its solvent-accessible surface area upon binding, while 68% of the antibiotic solvent-accessible surface area is occluded upon binding and consistent with the need to shield the catalytic centre against omnipresent water to avoid becoming ATPases (Koshland, 1958; Jencks, 1975). The active site region, in particular the 3-hydroxyl of Cm and the γ-phosphate of ATP, is completely buried and not solvent accessible. Because Cm is found buried in a tunnel within the protein, ATP binding would not be inconsistent with a domain closure fashioning the Cm-binding site. The substrate-free structure of CPT crystallized in the presence of the sulfate anion, however, does not indicate a domain movement upon ATP binding as is observed in the other members of the nucleotide-binding superfamily including hexokinase (Bennett and Steitz, 1980) and adenylate kinase (Müller et al., 1996), or phosphoglycerate kinase (Bernstein et al., 1997). It is possible, however, that the sulfate ion binding has provoked domain closure. The observed decrease in the dissociation constant (Kd) for Cm from 425 to 195 µM in the presence of 32 mM ammonium sulfate would argue in favour of such a possibility. The lesser effect on Km values of the ternary complexes than on Km values of the sulfate-free enzyme would be consistent with a role for ATP in promoting domain closure to facilitate ternary complex formation. However, the conformational movement is not rate determining in Cm phosphorylation, as the presence of sulfate does not affect enzymatic turnover (kcat; data not shown).
Proposed reaction mechanism
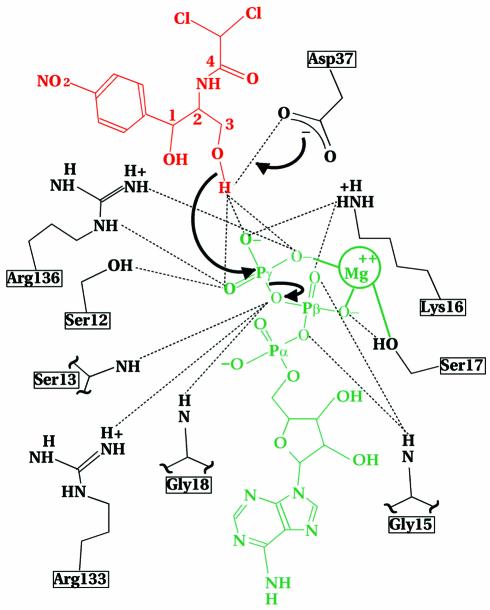
Steady-state kinetic data indicate substrate phosphorylation requiring prior ternary complex formation, with no preferred order for substrate binding (data not shown). An in-line mechanism for phosphoryl transfer would require the presence of a catalytic base to abstract a proton from the attacking hydroxyl group. Consideration of the CPT–ATPγS–Cm structure suggests the reaction mechanism shown in Figure 4, whereby CPT undergoes a simple SN2-type phosphate transfer. The side chain carboxyl of Asp37 is the only enzyme functional group within hydrogen bonding distance of the primary C-3 alcohol of the antibiotic and therefore capable of participating directly as a general base to deprotonate the C-3 hydroxyl.
Fig. 4. Structure of Cm and interaction of nucleophile, base and phosphate group that might occur in a general base-catalysed mechanism during the early steps in the CPT reaction. Cm is shown in red, Mg2+-ATP in green and CPT in black. The hydrogen bonding network is indicated as found in the ternary crystal structure. The carboxylate of Asp37 serves as a general base to deprotonate the primary (C-3) alcohol of the antibiotic. The resulting oxyanion of Cm then attacks the γ-phosphate of ATP. Collapse of the pentacovalent transition state yields the products 3-phosphoryl-Cm and ADP. Specific interactions proposed for stabilization of the penta-covalent transition state by Arg136 are shown.
Upon deprotonation of the C-3 hydroxyl of Cm, the resultant nucleophile could then attack the γ-phosphate of ATP to yield a pentacovalent transition state through an in-line mechanism (Figure 3E), with nucleophilic group and leaving group in the two apical positions of a trigonal bipyramid. Arg136 could hydrogen bond to the γ-phosphate group of ATP, thereby stabilizing the pentacovalent transition state. Mg2+ favours formation of product through neutralization of the developing negative charge. The side chains of Ser12, Lys16, Ser17 and Arg133 further stabilize the transferred phosphoryl group. Ser17 coordinates the catalytic cation. Furthermore, this side chain neutralizes the developing negative charge on the γ-phosphate by participating in a hydrogen bond. Collapse of the pentacovalent transition state yields the final product, 3-phosphoryl-Cm. Superimposition of the complexed and substrate-free CPT crystal structures reveals a significant movement of Asp135 and Arg136 residing on loop α6–α7 of the lid domain (Figure 3E). Upon substrate binding, the main chain shifts as much as 3.7 Å towards the active site. In both CPT and CPT–ATP crystal structures, these residues 135 and 136 refined to high temperature factors. However, the backbone and the Cβ positions show continuous and unambiguous electron density. Because of ATP’s half occupancy, the movement of residues 135–137 associated with nucleotide binding is also expected to occur only half of the time. However, we were unable to identify electron density corresponding to a second conformation for these residues for the ATP unbound state of the enzyme. The reorientation of Arg136 induced by substrate binding provides compelling evidence for its role during catalysis. The guanidinium group of Arg136 moves >9 Å into the active site, allowing formation of a hydrogen bond with the oxygens of the γ-phosphate of ATP (Figure 3E). For these reasons, we propose that Arg136 promotes bond cleavage through its role of stabilizing the developing negative charge on the equatorial oxygen atom of the γ-phosphate in the transition state.
Two aspartate residues are amongst the strictly invariant amino acid residues found not only in all protein kinases (reviewed in Cox et al., 1994; Goldsmith and Cobb, 1994; Bossemeyer, 1995) but also in bacterial phosphotransferases responsible for antibiotic resistance, suggesting a general role for binding ATP and phosphorylation of substrates (Brenner, 1987). For example, in the cAMP dependent protein kinase (cAPK), the invariant Asp166 is thought to be the catalytic residue (Bossemeyer et al., 1993). In CPT, Asp37 is oriented similarly towards the substrate’s hydroxyl to fulfil the same catalytic function. Furthermore, the invariant Asp184 in cAPK coordinates the high affinity metal cation (Bossemeyer et al., 1993) and Asp92 in CPT plays its role. Moreover, Arg136 in CPT is oriented towards the γ-phosphate of ATP as seen for Lys168 binding to the nucleotide in cAPK (Bossemeyer et al., 1993). Thus, the proposed CPT catalytic mechanism involves similar functional groups as identified in protein kinases.
Materials and methods
Preparation of selenomethionyl CPT
The seleno-l-methionine (SeMet) isoform of CPT was produced in Escherichia coli HMS174 (DE3), freshly transformed with the expression vector pET:cpt (Ellis et al., 1999). The cells were grown in M9 minimal media supplemented with 4 g/l glucose; 10 mg/l thiamine and biotin; 50 mg/l isoleucine, leucine and valine; 100 mg/l lysine, phenylalanine and threonine; and 60 mg/l SeMet. The six amino acids inhibit methionine biosynthesis in a non-auxotrophic prokaryotic strain. The cells were allowed to grow overnight in a shaking incubator at 37°C, then were diluted with freshly supplemented media 15 min prior to induction with isopropyl-β-d-thiogalactopyranoside (IPTG). After a further 7 h of incubation, the cells were harvested and CPT was purified by ammonium sulfate precipitation followed by hydrophobic interaction chromatography, as described previously (Ellis et al., 1999). Electrospray mass spectrometry analysis of the SeMet CPT (K.Lilley, unpublished results) indicated substitution of all six methionine residues.
Crystallization and data collection
Crystals of SeMet CPT were obtained as described for the native enzyme (Ellis et al., 1999). These crystals also belong to space group I4132 (a = 200.0 Å), with one polypeptide chain in the asymmetric unit, a solvent content of 0.86 and a volume to protein mass ratio (Vm) of 8.8 Å3/Da. Co-crystals of CPT in complex with ATP, Cm and both ATPγS and Cm were grown under conditions similar to those used for native CPT (Ellis et al., 1999) except that saturating concentrations of Cm (2 mM) and/or ATP/ATPγS (10 mM) and magnesium chloride (10 mM) were included in the crystallization drop. Crystals were cryoprotected by including 35% glycerol in the mother liquor. A CCD detector was used to collect data from a single, flash-frozen crystal of selenomethionyl protein at three wavelengths at beam line BM14 at ESRF, Grenoble. An X-ray fluorescence spectrum was recorded and used to select the wavelength optima for subsequent MAD data collection. Data were collected at 0.9785 Å (the inflection point of the fluorescence spectrum, f′ minimum), 0.9783 Å (f′′ maximum) and 0.8855 Å (remote high-energy wavelength). All three data sets were collected from the same crystal at a crystal-to-detector distance of 240 mm and using 0.5° oscillations per image. The data were collected by using inverse beam geometry with each set of data measured in a single pass. Data (2.7 Å resolution) at the three wavelengths were processed independently using the programs DENZO and SCALEPACK (Otwinowski and Minor, 1997). Data statistics are given in Table I. CPT–ATP, CPT–Cm and CPT–ATPγS–Cm X-ray data were collected at 100 K on a CCD detector on beamline 9.6 at SRS, Daresbury, with the wavelength set to 0.87 Å and processed with DENZO and SCALEPACK (Otwinowski and Minor, 1997; Table I). A further data set of CPT–Cm was collected at a wavelength of 1.1 Å at beamline X12-C of the National Synchrotron Light Source, Brookhaven National Laboratory (BNL) on a Brandeis CCD detector and the data were merged with the CPT–Cm data set collected at SRS.
Table I. Crystallographic data and refinement statistics.
| Data collection |
Edge |
Peak |
Remote |
CPT–Cm |
CPT–ATP |
CPT–ATPγS–Cm |
| wavelength (Å) | 0.9785 | 0.9783 | 0.88 | 0.89a, 1.1b | 0.89 | 0.89 |
| resolution (Å) | 2.7 | 2.7 | 2.7 | 2.8 | 2.5 | 2.6 |
| total data | 255 940 | 261 697 | 279 021 | 464 992 | 327 116 | 304 201 |
| unique data | 18 936 | 19 005 | 19 032 | 17 144 | 23 682 | 21 331 |
| redundancy | 13.52 | 13.77 | 11.67 | 27.12 | 13.81 | 14.26 |
| overall completeness | 0.995 | 0.996 | 1. | 1. | 0.991 | 0.997 |
| completeness (last shell) | 1. | 1. | 1. | 1. | 0.995 | 0.998 |
| F2 > 3σ (F2) (%) | 83.5 | 79.5 | 82.1 | 81.5 | 85.8 | 81.0 |
| Rmergec (overall) | 0.128 | 0.131 | 0.128 | 0.101 | 0.058 | 0.061 |
| Rmergec (last shell) | 0.368 | 0.366 | 0.417 | 0.359 | 0.177 | 0.367 |
| average F2/σ(F2) |
11.1 |
7.7 |
9.7 |
14.3 |
29.8 |
21.4 |
| Crystallographic refinement |
Se-Met CPT |
CPT–Cmd |
CPT–ATP |
CPT–ATPγS–Cm |
|
|
| no. of reflections | 18 929 | 17 118 | 23 608 | 21 191 | ||
| Final model parameters | ||||||
| no. of amino acid residues | 178 | 178 | 178 | 178 | ||
| no. of protein atoms | 1320 | 1320 | 1320 | 1320 | ||
| no. of solvent molecules | 177 | 76 | 100 | 46 | ||
| resolution range (Å) | 20–2.7 | 20–2.8 | 20–2.5 | 20–2.6 | ||
| R-factore (overall) | 0.201 | 0.226 | 0.228 | 0.233 | ||
| R-factore (last shell) | 0.240 | 0.315 | 0.305 | 0.349 | ||
| Rfreef (overall) | 0.226 | 0.238 | 0.238 | 0.247 | ||
| Rfreef (last shell) | 0.262 | 0.334 | 0.358 | 0.358 | ||
| average main chain B-factor (Å2) | 28.3 | 39.9 | 37.2 | 45.5 | ||
| average side chain B-factor (Å2) | 36.8 | 44.9 | 43.2 | 51.3 | ||
| average water molecule B-factor (Å2) | 33.8 | 36.2 | 36.9 | 38.9 | ||
| average chloramphenicol B-factor (Å2) | – | 32.7 | – | 86.6 | ||
| average Cm–SO42– B-factorg (Å2) | – | 30.6 | – | 77.6 | ||
| average nucleotide B-factorh (Å2) | – | – | 36.68 | 52.5 | ||
| R.m.s. deviations from ideal geometry | ||||||
| covalent bond lengths (Å) | 0.007 | 0.008 | 0.007 | 0.010 | ||
| bond angles (°) | 1.1 | 1.3 | 1.2 | 1.4 |
aData collected at SRS.
bData collected at BNL.
fThe free R-factor is a cross-validation residual calculated using 5% of the native data that were randomly chosen and excluded from the refinement.
gWith an occupancy of one half.
hWith an occupancy of one half for all nucleotide atoms except the β-phosphate.
Structure determination
The MAD data (Table I) were scaled together with the CCP4 (1994) program SCALEIT. Five Se sites of six expected in the asymmetric unit were determined using SOLVE (Terwilliger and Brendzen, 1996). The N-terminal SeMet was not found, probably due to disorder. Initial phases were calculated with the program MLPHARE (CCP4, 1994) and were improved by 100 cycles of solvent flattening and gradual phase extension from 3.9 to 2.7 Å resolution using the program DM (CCP4, 1994). The final R-factor for the phase extension at 2.7 Å resolution was 0.40. The Se positions allowed rapid unambiguous matching of the electron density to the sequence and construction of an atomic model by using the program O (Jones et al., 1991). The crystal structures of the enzyme in complex with ligands were solved by difference Fourier from the initial SeMet CPT model.
Crystallographic refinement
All CPT structures were refined by using the program CNS (Brünger et al., 1988, 1998) with standard protocols. The free R-value (Brünger, 1992) was monitored throughout the refinement. Table I lists the final model parameters. The data set collected at the inflection point of the fluorescence was used for refinement. The electron density was poorest for the ATP and ATPγS ligands in the CPT–ATP and ternary crystal structures, respectively. The nucleotide could be traced unambiguously into the omit electron density map but the electron density was very weak when the ligand was included in further map calculations. Temperature factors refined to ∼40–50 Å2 only with the occupancy set to 0.5. The electron density was also weak for residues 135 and 136 in the CPT and CPT–ATP crystal structures. Water molecules were initially identified in Fo – Fc maps and screened for reasonable geometry and refined thermal factor <50 Å2. Table I shows the overall crystallographic R-factor and the free R-factor for all models for all observed reflections within the indicated resolution range. A Ramachandran plot analysis of all four structures by the program PROCHECK (CCP4, 1994) indicates that 92–94% of all the residues lie in most favourable regions and 6–8% in additional allowed regions. This structure analysis also showed that all stereochemical parameters are better than expected at the given resolution.
Coordinates
The atomic coordinates have been deposited in the Protein Data Bank (PDB ID codes 1qhn, 1qhs, 1qhx and 1qhy for the SeMet CPT, CPT–Cm, CPT–ATP and CPT–ATPγS–Cm structures, respectively).
Acknowledgments
Acknowledgements
We thank Andy Thompson (ESRF) for his invaluable assistance during and after the MAD experiment, Bob Sweet (NSLS) for help during data collection at NSLS, Neil Errington (Nottingham) for performing the analytical ultracentrifugation experiment, and Sharon Naron (St Jude) for editing the manuscript. The NSLS beamline X12-C is supported by the United States Department of Energy Offices of Health and Environmental Research and of Basic Energy Sciences, and by the National Science Foundation. We are grateful to Jurgen Sygusch (Montréal) for stimulating discussions and helpful comments on the manuscript. Thanks also to our colleagues at Leicester: Bob Liddington for his continuous support, Peter Moody for fruitful discussions, and Nic Blackwell for help during data collection at ESRF. J.E. is supported by a Wellcome Trust project grant awarded to W.V.Shaw.
References
- Abele U. and Schulz,G. (1995) High-resolution structures of adenylate kinase from yeast ligated with inhibitor Ap5A, showing the pathway of phosphoryl transfer. Protein Sci., 4, 1262–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacon D.J. and Anderson,W.F. (1988) A fast algorithm for rendering space-filling molecule pictures. J. Mol. Graph., 6, 219–220. [Google Scholar]
- Bennett W.S. Jr, and Steitz,T.A. (1980) Structure of a complex between yeast hexokinase A and glucose. II. Detailed comparisons of conformation and active site configuration with the native hexokinase B monomer and dimer. J. Mol. Biol., 140, 211–230. [DOI] [PubMed] [Google Scholar]
- Bernstein B.E., Michels,P.A.M. and Hol,W.G.J. (1997) Synergistic effects of substrate-induced conformational changes in phosphoglycerate kinase activation. Nature, 385, 275–278. [DOI] [PubMed] [Google Scholar]
- Berry M.B. and Phillips,G.N.,Jr (1998) Crystal structures of Bacillus stearothermophilus adenylate kinase with bound Ap5A, Mg2+ Ap5A and Mn2+ Ap5A reveal an intermediate lid position and six coordinate octahedral geometry for bound Mg2+ and Mn2+. Proteins, 32, 276–288. [DOI] [PubMed] [Google Scholar]
- Bossemeyer D. (1995) Protein kinases—structure and function. FEBS Lett., 369, 57–61. [DOI] [PubMed] [Google Scholar]
- Bossemeyer D., Engh,R.A., Kinzel,V., Ponstingl,H. and Huber,R. (1993) Phosphotransferase and substrate binding mechanism of the cAMP-dependent protein kinase catalytic subunit from porcine heart as deduced from the 2.0 Å structure of the complex with Mn2+ adenylyl imidodiphosphate and inhibitor peptide PKI(5-24). EMBO J., 12, 849–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. (1987) Phosphotransferase sequence homology. Nature, 329, 21. [DOI] [PubMed] [Google Scholar]
- Brünger A.T. (1992) Free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature, 355, 472–475. [DOI] [PubMed] [Google Scholar]
- Brünger A.T., Kuriyan,J. and Karplus,M. (1988) Crystallographic R-factor refinement by molecular dynamics. Science, 235, 458–460. [DOI] [PubMed] [Google Scholar]
- Brünger A.T. et al. (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Cox S., Radzio-Andzelm,E. and Taylor,S.S. (1994) Domain movements in protein kinases. Curr. Opin. Struct. Biol., 4, 893–901. [DOI] [PubMed] [Google Scholar]
- Cundliffe E. (1992) Self-protection mechanisms in antibiotic producers. In Chadwick,D.J. and Whelan,J. (eds), Secondary Metabolites: Their Function and Evolution. Wiley, Chichester, UK, pp. 199–214. [DOI] [PubMed] [Google Scholar]
- Davies J. (1994) Inactivation of antibiotics and the dissemination of resistance genes. Science, 264, 375–382. [DOI] [PubMed] [Google Scholar]
- Dreusicke D., Karplus,A. and Schulz,G.E. (1988) Refined structure of porcine cytosolic adenylate kinase at 2.1 Å resolution. J. Mol. Biol., 199, 359–371. [DOI] [PubMed] [Google Scholar]
- Ellis J., Campopiano,D.J. and Izard,T. (1999) Cubic crystals of chloramphenicol phosphotransferase from Streptomyces venezuelae in complex with chloramphenicol. Acta Crystallogr. D, 55, 1086–1088. [DOI] [PubMed] [Google Scholar]
- Goldsmith E.J. and Cobb,M.H. (1994) Protein kinases. Curr. Opin. Struct. Biol., 4, 833–840. [DOI] [PubMed] [Google Scholar]
- Holm L. and Sander,C. (1997) New structure—novel fold? Structure, 5, 165–171. [DOI] [PubMed] [Google Scholar]
- Jencks W.P. (1975) Binding energy, specificity and enzyme catalysis: the circe effect. Adv. Enzymol. Relat. Areas Mol. Biol., 43, 219–410. [DOI] [PubMed] [Google Scholar]
- Jones T.A., Zou,J.-Y. and Cowan,S.W. (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Koshland D.E. Jr, (1958) Application of a theory of enzyme specificity to protein synthesis. Proc. Natl Acad. Sci. USA, 44, 98–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraulis P.J. (1991) MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr., 24, 946–950. [Google Scholar]
- Krell T., Coggins,J.R. and Lapthorn,A.J. (1998) The three-dimensional structure of shikimate kinase. J. Mol. Biol., 278, 983–997. [DOI] [PubMed] [Google Scholar]
- Luzzati V. (1952) Traitement statistique des erreurs dans la determination des structures cristallines. Acta Crystallogr., 5, 802–810. [Google Scholar]
- Matthews B.W. (1968) Solvent content of protein crystals. J. Mol. Biol., 33, 491–497. [DOI] [PubMed] [Google Scholar]
- Merritt E.A. and Murphy,M.E.P. (1994) Raster3D version 2.0—a program for photorealistic molecular graphics. Acta Crystallogr. D, 50, 869–873. [DOI] [PubMed] [Google Scholar]
- Mosher R.H., Camp,D.J., Yang,K., Brown,M.P., Shaw,W.V. and Vining,L.C. (1995) Inactivation of chloramphenicol by O-phosphorylation. J. Biol. Chem., 270, 27000–27006. [DOI] [PubMed] [Google Scholar]
- Müller C.W., Schlauderer,G.J., Reinstein,J. and Schulz,G.E. (1996) Adenylate kinase motions during catalysis: an energetic counterweight balancing substrate binding. Structure, 4, 147–156. [DOI] [PubMed] [Google Scholar]
- Nicholls A., Sharp,K. and Honig,B. (1991) Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins, 11, 281–296. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z. and Minor,W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol., 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Rossmann M.G. et al. (1973) Molecular symmetry axes and subunit interfaces in certain dehydrogenases. J. Mol. Biol., 76, 533–537. [DOI] [PubMed] [Google Scholar]
- Saraste M., Sibbald,P.R. and Wittinghofer,A. (1990) The P-loop—a common motif in ATP- and GTP-binding proteins. Trends Biochem. Sci., 15, 430–434. [DOI] [PubMed] [Google Scholar]
- Schulz G.E., Müller,C.W. and Diedrichs,K. (1990) Induced-fit movements in adenylate kinases. J. Mol. Biol., 213, 627–630. [DOI] [PubMed] [Google Scholar]
- Shapiro S. and Vining,L.C. (1983) Nitrogen metabolism and chloramphenicol production in Streptomyces venezuelae.Can. J. Microbiol., 29, 1706–1714. [DOI] [PubMed] [Google Scholar]
- Shaw K.J., Rather,P.N., Hare,R.S. and Miller,G.H. (1993) Molecular genetics of aminoglycoside resistance genes and familial relationships of the aminoglycoside-modifying enzymes. Microbiol. Rev., 57, 138–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw W.V. and Hopwood,D.A. (1976) Chloramphenicol acetylation in Streptomyces. J. Gen. Microbiol., 94, 159–166. [DOI] [PubMed] [Google Scholar]
- Shaw W.V. and Leslie,A.G.W. (1991) Chloramphenicol acetyltransferase. Annu. Rev. Biophys. Biophys. Chem., 20, 363–386. [DOI] [PubMed] [Google Scholar]
- Smith C.A. and Rayment,I. (1996) Active site comparisons highlight structural similarities between myosin and other P-loop proteins. Biophys. J., 70, 1590–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger T.C. and Brendzen,J. (1996) Bayesian weighting for macromolecular crystallographic refinement. Acta Crystallogr. D, 52, 743–748. [DOI] [PubMed] [Google Scholar]
- Vining L.C. and Westlake,D.W.S. (1984) Chloramphenicol: properties, biosynthesis and fermentation. In Vandamme,E.J. (ed.), Bio/Technology of Industrial Antibiotics. Marcel Dekker, New York, NY, pp. 387–409. [Google Scholar]
- Walker J.E., Saraste,M., Runswick,M.J. and Gay,N.J. (1982) Distantly related sequences in the α- and β-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J., 1, 945–951. [DOI] [PMC free article] [PubMed] [Google Scholar]