Abstract
Translating ribosomes bypass a 50 nt coding gap in order to fuse the information found in the two open reading frames (ORFs) of bacteriophage T4 gene 60. This study investigates the underlying mechanism by focusing on the competition between initiation of bypassing and termination at the end of the first ORF. While nearly all ribosomes initiate bypassing, no more than 50% resume translation in the second ORF. Two previously described cis-acting stimulatory signals are critical for favoring initiation of bypassing over termination. Genetic analysis of these signals supports a working model in which the first (a stem–loop structure at the junction between the first ORF and the coding gap) interferes with decoding in the A-site, and the second (a stretch of amino acids in the nascent peptide encoded by the first ORF) destabilizes peptidyl-tRNA–mRNA pairing.
Keywords: gene 60/hopping/L9/RF1/tRNA2Gly
Introduction
One of three stop codons (UAG, UGA or UAA) defines the 3′ boundaries of all open reading frames (ORFs). Ribosomes terminate translation at these triplets through the help of release factor (RF) proteins that ‘decode’ the stop codon and trigger hydrolysis of the peptidyl-tRNA bond, releasing the new polypeptide (Buckingham et al., 1997; Nakamura and Ito, 1998). The efficiency of termination appears to be determined by the competition between RF binding and alternative translational events that allow ribosomes to continue decoding. These events include read-through, frameshifting and bypassing. In read-through, a tRNA decodes the stop codon and translation continues in the 0 frame until the next terminator. In frameshifting, the linear readout of the code is disrupted and the ribosome switches to one of the alternative frames. In some cases, an incoming tRNA ignores the first nucleotide of the stop codon in the A-site and reads the +1 codon (Farabaugh et al., 1993). In other cases, the peptidyl-tRNA detaches from the mRNA and pairs with a suitable overlapping codon, resetting the reading frame (Craigen and Caskey, 1986; Weiss et al., 1987). In bypassing, this peptidyl-tRNA slippage is more extensive and the tRNA re-pairs with the mRNA at a non-overlapping codon (Weiss et al., 1987; O’Connor et al., 1989; for a mammalian example, see Chittum et al., 1998). Bypassing and frameshifting also occur at rare sense codons or codons under aminoacyl-tRNA limitation, suggesting that ribosome pausing contributes to initiation of peptidyl-tRNA slippage (Gallant and Foley, 1980; Weiss and Gallant, 1983; Belcourt and Farabaugh, 1990; Kane et al., 1992; Gallant and Lindsley, 1998).
Some genes with two ORFs require efficient stop codon read-through, frameshifting or bypassing to be translated (Farabaugh, 1996; Gesteland and Atkins, 1996; Atkins et al., 1999). The coupling of these ORFs is specified by the local mRNA sequence context, aided in most cases by discrete cis-acting signals that re-program ribosomes.
Bacteriophage T4 gene 60, an extreme example of this class of genes, represents an ideal model system to study how cis-acting signals re-program ribosomes. Fifty non-coding nucleotides (coding gap) separate the two ORFs and must be bypassed in order to synthesize the full-length protein, a topoisomerase subunit (Huang et al., 1988). Efficient bypassing of the gene 60 coding gap in Escherichia coli requires two matching GGA codons flanking the coding gap, which serve as the sites of peptidyl-tRNA detachment and re-pairing (referred to as take-off and landing sites). Additional required elements include a UAG stop codon that defines the 3′ end of the first ORF, a stem–loop structure that contains the stop codon and take-off site GGA within its stem, and a cis-acting signal in the nascent peptide consisting of a stretch of charged and hydrophobic amino acids specified by codons preceding the gap (Weiss et al., 1990) (Figure 1).
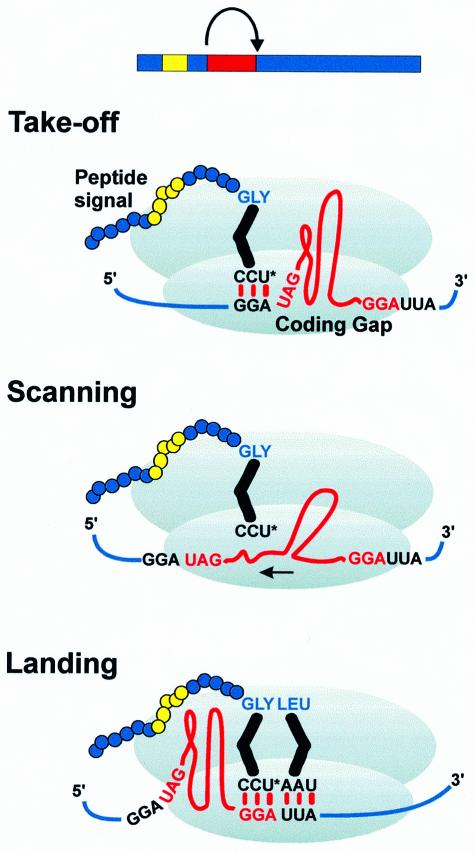
Fig. 1. T4 gene 60 bypassing requires matching GGA codons flanking an optimally sized 50 nt coding gap, a stop codon, a stem–loop structure and a nascent peptide signal. In the current model, peptidyl-tRNA2Gly detaches from the take-off site GGA, scans the mRNA as it slides through the P-site, and re-pairs with the landing-site GGA.
The proposed mechanism of bypassing depends on the pairing properties of peptidyl-tRNA2Gly (the cognate tRNA for GGA) and the movement of mRNA through the ribosome (Herr et al., 1999). In the first stage (take-off), the peptidyl-tRNA–mRNA complex dissociates and an initial movement of the mRNA occurs that prevents peptidyl-tRNA2Gly re-pairing with the take-off site GGA. During the second stage (scanning), peptidyl-tRNA2Gly probes the mRNA for a landing site as the mRNA continues to slide through the decoding center of the ribosome. In the third stage (landing), peptidyl-tRNA2Gly pairs with the landing site GGA, resetting the reading frame. Standard translation resumes with decoding of the new codon in the A-site (UUA).
In this minimal model, the matching GGA codons act as road signs, specifying where bypassing begins and ends. Less obvious are the roles of the stop codon, the stem–loop and the nascent peptide signal. The stop codon may either pause the ribosome, allowing time for peptidyl-tRNA slippage to occur, or interact with specific translational components that actively stimulate take-off. Of all the signals, the nascent peptide signal and the stem–loop seem the most likely to re-program ribosomes actively into the scanning mode. These signals might stimulate take-off by promoting peptidyl-tRNA slippage or limiting recognition of the UAG by RF1 (the cognate RF for UAG). Alternatively, they may facilitate mRNA movement through the ribosome during scanning. Additional insight into the mechanism of bypassing can be gained by understanding the role of ribosomal protein L9 in translation. Mutations in the gene for L9 (rplI) enhance simple bypassing of stop codons and partially suppress mutations in the gene 60 stem–loop (Herbst et al., 1994; Adamski et al., 1996), suggesting that L9 normally serves to limit bypassing.
Nearly half of all ribosomes translating gene 60 bypass the coding gap successfully (productive bypassing) (Maldonado and Herr, 1998). The fate of those ribosomes that fail to bypass is unclear, although there are several possibilities (collectively called unproductive bypassing). The most obvious explanation for unproductive bypassing is that ribosomes terminate translation at the UAG in an RF1-dependent manner, and the bypassing efficiency simply reflects the competition between take-off and termination. Alternatively, an abortive termination event may be required for take-off, and bypassing efficiency may reflect the rate at which termination is subverted in favor of take-off. Other possible explanations account for unproductive bypassing without invoking RF1. Perhaps all ribosomes take-off from the first GGA, and competing events during scanning prevent 50% of them from landing at the downstream GGA.
Here we investigate the mechanism of gene 60 bypassing by focusing on how take-off is enhanced at the expense of termination. We assess the importance of the local sequence context of the UAG at the take-off site, whether specific bypassing signals interfere with decoding of the stop codon by RF1, and whether known mutants in translational components that alter bypassing efficiency influence termination efficiency.
Results
Previously, we developed an in vivo assay for bypassing efficiency based on a glutathione S-transferase (GST)–gene 60 fusion. This fusion allows detection of proteins due to both productive and unproductive bypassing (Maldonado and Herr, 1998). Expression levels of both products are monitored by pulse–chase labeling of total protein with [35S]methionine followed by separation on SDS–PAGE and quantitation of the appropriate radioactive products. Each estimate of bypassing efficiency represents the level of protein derived from productive bypassing divided by the total protein derived from both productive and unproductive bypassing (Maldonado and Herr, 1998; Herr et al., 1999).
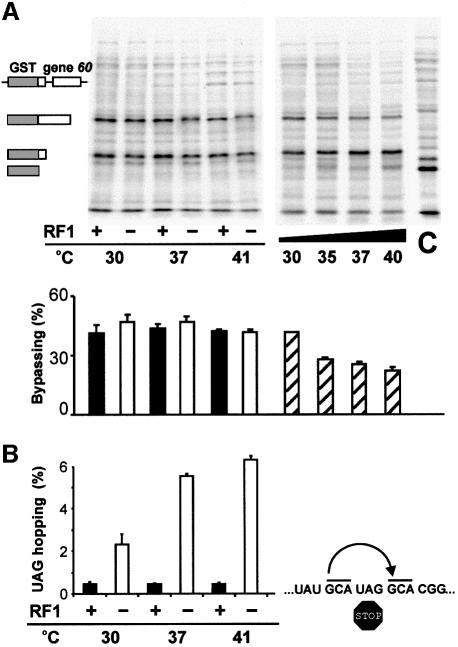
In order to test whether bypassing competes directly with termination, bypassing efficiency was measured under conditions in which RF1 was either defective or overexpressed (Figure 2). To determine the effects of inactivating RF1, isogenic strains were constructed that differed only in the presence or absence of a temperature-sensitive (ts) allele of prfA, the gene for RF1. The resulting mutant protein binds the ribosome less efficiently at the non-permissive temperature (Zhang et al., 1994). Inactivation of RF1 by growth at 41°C elevates simple bypassing of a stop codon (stop-hopping), flanked by matching GCA codons, to levels that are 10 times that of a wild-type (WT) control (Figure 2B). Despite this reduction in RF1 activity, gene 60 bypassing efficiency under the same growth conditions is essentially the same as the WT control (Figure 2A); ∼50% of ribosomes still fail to bypass the coding gap. In these experiments, a slight decrease in the mobility of the product due to productive bypassing is apparent in the sample from the strain expressing the ts RF1. The decreased mobility is consistent with read-through of the UAG stop codon at the end of the GST–gene 60 fusion and addition of nine amino acids to the C-terminus. This provides an internal control for RF1 inactivation under these growth conditions.
Fig. 2. The competition between bypassing and termination. (A) Pulse–chase analysis of isogenic strains expressing a GST–gene 60 fusion (pGS1): WT, black bars; a strain expressing a ts RF1, white bars; a strain overexpressing RF1 from a ts promoter, striped bars. The GST–gene 60 fusion and products due to productive (45.1 kDa) or unproductive bypassing (31.7 kDa) are represented schematically to the left of the autoradiograph. The control (C) is an isogenic strain expressing just GST (26.3 kDa). (B) β-galactosidase assay measurements of +6 bypassing (UAG hopping; see bypassing window to the right) demonstrating inactivation of the ts RF1 with increasing temperature.
To discern whether termination competes with take-off when RF1 levels are elevated, bypassing efficiency was measured in a strain with a plasmid overexpressing prfA from the λ pL promoter. This plasmid carries the ts λ repressor allele, cI857, so that prfA expression is induced at elevated temperature. Bypassing efficiency decreases as the promoter is induced, consistent with RF1 overexpression causing increased termination at the take-off site UAG (Figure 2).
These observations suggest that under normal conditions the efficiency of bypassing is not defined by the competition between termination and take-off, and yet RF1 can recognize the stop codon and compete with bypassing when intracellular levels are elevated. This seems to eliminate models in which RF1 stimulates take-off. Under normal conditions, either RF1 recognition of the stop codon is highly inefficient or initiation of bypassing occurs much faster than formation of a stable termination complex. What elements of gene 60 bias the competition between termination and take-off so heavily in favor of take-off?
The UAG at the take-off site lies in one of the weakest possible contexts for RF1 recognition. Two different groups have characterized the importance of the nucleotide just 3′ of UAG on general RF1 decoding (Pedersen and Curran, 1991; Poole et al., 1995). Although the studies disagree on the rank order of nucleotides preferred by RF1 (U>G>C>A versus G>U>A>C), both suggest that RF1 recognizes the UAGC quadruplet at the take-off site poorly. Another group, investigating the influence of the codon immediately 5′ of the stop codon, found that termination efficiency is lowest (by a factor of two) when GGA or GGG immediately precedes UAG (Zhang et al., 1996). When all GGA codons in the cell are decoded by a mutant tRNA2Gly with a 3′CCU5′ anticodon, instead of tRNA2Gly, termination efficiency improves to levels seen with other codons in the 5′ position (Zhang et al., 1998). A final study found that acidic residues in the –2 amino acid position (relative to the tRNA in the P-site) also diminish RF1 recognition of UAG codons (Mottagui-Tabar et al., 1994). In gene 60, the –2 amino acid at take-off is aspartic acid. Together, these studies raise the possibility that the local sequence context of the UAG is the key feature that limits the influence of RF1 on bypassing. To explore this possibility, we assessed the individual contributions of the above elements to bypassing efficiency.
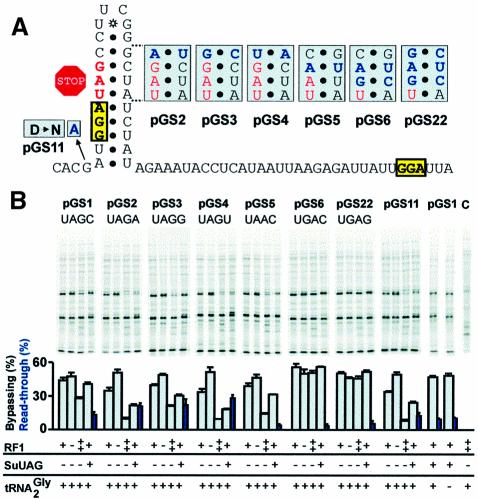
Changing the nucleotide 3′ of the UAG stop codon to A, G or U (along with the appropriate compensatory mutations in order to maintain base-pairing in the stem) lowers the efficiency of bypassing in a WT strain from 44% to 34, 39 and 33%, respectively (pGS2-4; Figure 3). Inactivation of RF1 restores bypassing efficiency to each mutant and each shows an increased sensitivity to elevated RF1 levels (Figure 3). This suggests that the mutations reduce bypassing by increasing RF1 recognition of the UAG. Interestingly, the relative decreases in efficiency do not follow either of the RF1 preference orders described above. If the 3′ context were the sole reason for the reduction in bypassing in these constructs, then mutation of the 3′ position to G or A should have a more pronounced effect than U. A more likely explanation for these results is that stem–loop stability governs termination efficiency. The variants with A⋅U or U⋅A base pairs in this position of the stem are predicted to be less stable than those with G⋅C or C⋅G.
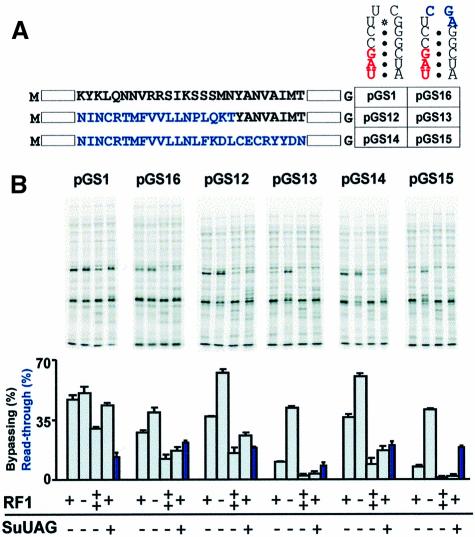
Fig. 3. Analysis of importance of stop codon context for bypassing efficiency. (A) Gene 60 coding gap and flanking sequence showing mutations (in blue). (B) Pulse–chase analysis of isogenic strains, which differ as indicated, expressing mutant GST–gene 60 constructs. pGS1 is the WT. C (far right) is a strain over-expressing RF1 in which the GST–gene 60 fusion has not been induced. Read-through levels in the strain expressing an amber suppressor tRNA are indicated by blue bars and correspond to the level of single read-through product (see the text). The read-through product for pGS16 is detectable upon more extensive electrophoresis.
Expression of these constructs in a strain with an amber suppressor tRNA provides a second indication of whether the 3′ nucleotide influences decoding of the UAG during take-off. Like termination, amber suppression is sensitive to the identity of the nucleotide 3′ of the UAG stop codon (A = G>C>U) (Bossi and Roth, 1980; Miller and Albertini, 1983; Pedersen and Curran, 1991). Purines favor suppression, presumably by stabilizing codon–anticodon pairing through stacking interactions. Read-through of the UAG at the take-off site extends the protein encoded by the first ORF by six amino acids (single read-through product) giving rise to a product that migrates on an SDS–PAGE gel just above the product due to unproductive bypassing. Because the next two stop codons in the 0 frame are also UAG codons (UAGA and UAGG), the potential exists for double and triple read-through products that add 11 and 41 amino acids to the C-terminus with decreasing efficiency. Given the difficulty in quantitating all products resulting from decoding the take-off site UAG, a relative estimate was determined by dividing the level of single read-through product by the combined total protein produced by single read-through, productive bypassing and unproductive bypassing.
In the context of gene 60, amber suppression increases 1.5- to 2-fold when the C just 3′ of the UAG is mutated to A, G or U (Figure 3B). The suppressor tRNA [a derivative of tRNA2Gly (anticodon = 3′AUC5′) (Herr et al., 1999)] decodes UAGU, UAGG and UAGA with nearly equal efficiencies. Unexpectedly, the presence of an amber suppressor tRNA decreases the amount of productive bypassing product relative to unproductive bypassing product (reported as bypassing efficiency to allow comparison with other bypassing efficiency estimates) in a manner reminiscent of RF1 overproduction. This is unexpected because according to a simple competition model, amber suppression should divert ribosomes equally from take-off and termination. The reduction in the ratio of these two products appears dependent on the stability of the stem–loop (variants with C⋅G or G⋅C pairs in this position are less sensitive than those with A⋅U or U⋅A pairs), suggesting that stem stability influences accessibility of the stop codon to tRNA as well as RF1.
To test whether aspartic acid in the –2 amino acid position reduces termination efficiency, this position was mutated to asparagine (Figure 3; pGS11). The observed reduction in bypassing (from 44 to 34%) is reversed by inactivating RF1 and enhanced by overexpressing RF1. The ratio of the products due to productive and unproductive bypassing also decreases in the presence of an amber suppressor tRNA. Together, these results suggest that aspartic acid in the –2 amino acid position influences the competition between decoding in the A-site and take-off.
Previous experiments have hinted that the identity of the tRNA of peptidyl-tRNA may be important for bypassing. Changing the take-off and landing site codons from GGA to GCA lowers bypassing efficiency ∼40% (Weiss et al., 1990). To test whether maximum bypassing efficiency requires peptidyl-tRNA2Gly, we used a strain in which all GGA codons are decoded by a mutant form of tRNA2Gly with a 3′CCU5′ anticodon (Table I; MRA291). Bypassing efficiency in both this strain and the appropriate control strain (Table I; MRA289; contains both the mutant tRNA2Gly and WT tRNA2Gly) is indistinguishable from WT, suggesting that peptidyl-tRNA2Gly does not limit RF1 recognition of the UAG at the take-off site (Figure 3).
Table I. Bacterial strains.
| Strain | Genotype | Reference |
|---|---|---|
| CSH142 | ara Δ(gpt-lac)5 | Miller (1992) |
| CSH142bypG19U | ara Δ(gpt-lac)5 glyT(G19U) | Herr (1999) |
| CSH142bypC40G | ara Δ(gpt-lac)5 glyT(C40G) | Herr (1999) |
| MRA289 | rph Δ(lac-proAB) supD(serU) glyV55(SuGGA/G) | Zhang (1998) |
| MRA291 | rph Δ(lac-proAB) supD(serU) glyV55(SuGGA/G)glyT(SuUGA/G) | Zhang (1998) |
| MRA7 | rph prfA1 zcg::Tn10 | Zhang (1994) |
| AH133 | ara Δ(gpt-lac)5 prfA1 zcg::Tn10 | this study |
| AH133bypG19U | ara Δ(gpt-lac)5 prfA1 glyT(G19U) zcg::Tn10 | this study |
| AH133bypC40G | ara Δ(gpt-lac)5 prfA1 glyT(C40G) zcg::Tn10 | this study |
| AH156 | ara Δ(gpt-lac)5 ΔrplI | this study |
| AH156bypG19U | ara Δ(gpt-lac)5 ΔrplI glyT(G19U) | this study |
| AH156bypC40G | ara Δ(gpt-lac)5 ΔrplI glyT(C40G) | this study |
| AH157 | ara Δ(gpt-lac)5 ΔrplI prfA1 zcg::Tn10 | this study |
| AH157bypG19U | ara Δ(gpt-lac)5 ΔrplI prfA1 glyT(G19U) zcg::Tn10 | this study |
| AH156bypC40G | ara Δ(gpt-lac)5 ΔrplI prfA1 glyT(C40G) zcg::Tn10 | this study |
In addition to examining the influence of the local sequence context, we tested whether the stop codon must be UAG (Figure 3; pGS5, pGS6). In E.coli, RF1 decodes UAG and UAA and RF2 decodes UGA and UAA. Changing the stop codon to UAA incurs a slight reduction in bypassing (44 to 39%), which is suppressed by inactivation of RF1 and enhanced by RF1 overproduction. This suggests that RF1 competes more effectively than RF2 for the UAA in this context. When the stop codon is UGA, bypassing efficiency increases from 44 to 55%. As expected, given that UGA is decoded solely by RF2, the UGA mutant is insensitive to RF1 inactivation and overproduction. This confirms that with the other mutants, RF1 competes with bypassing by decoding the take-off site UAG and not by derailing scanning ribosomes. Like RF1, RF2 shows a distinct preference order for the nucleotide 3′ of the stop codon (U>G>A>C) (Poole et al., 1995). Changing the 3′ nucleotide downstream of UGA from C to G causes only a slight reduction in bypassing efficiency, which suggests that RF2 competes poorly with bypassing when present at normal levels regardless of the 3′ nucleotide. Surprisingly, the presence of the amber suppressor tRNA reduces bypassing in the UAA mutant without causing read-through. Because no reduction in bypassing is seen when the stop codon is UGA, the effect must be specific for a stop codon with UA in the first two positions. This raises the interesting possibility that increased residence time in the A-site by a near-cognate tRNA (without peptidyl transfer) may interfere with take-off. Perhaps a similar mechanism accounts for the other reductions in take-off efficiency caused by an amber suppressor tRNA.
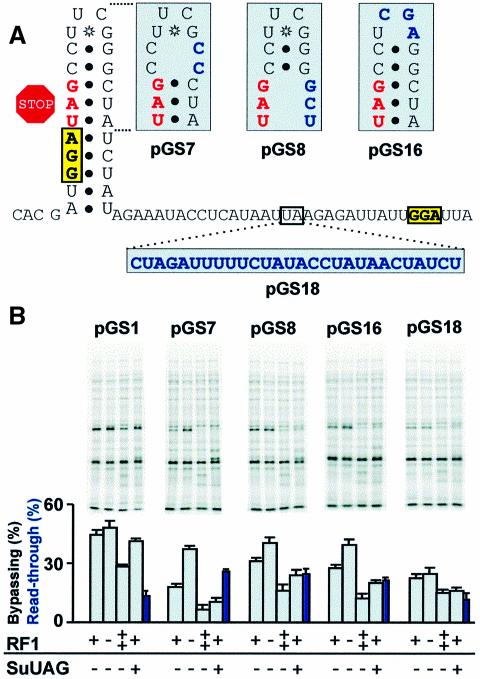
Our analysis hints that the stability of the stem–loop may be more important for limiting decoding of the UAG than the immediate context of the stop codon. Mutations that destabilize the stem or extend its length have previously been shown to reduce bypassing efficiency (Weiss et al., 1990). To test whether the stem–loop influences the competition between take-off and termination, we disrupted the top two C⋅G pairs of the stem (pGS7), the potential base pairs involving the stop codon (pGS8) and the stable UUCG tetraloop at the top of the stem (pGS16) (Figure 4A). These mutations reduce bypassing efficiency 60, 30 and 30%, respectively (Figure 4B). Inactivation of RF1 suppresses each mutation, indicating that they reduce bypassing by allowing increased termination. Consistent with increased recognition of the stop codon, the stem mutants are more sensitive than WT to elevated levels of RF1 or an amber suppressor tRNA. This provides strong evidence that the stem–loop stimulates take-off by influencing the competition between general decoding of the UAG and peptidyl-tRNA slippage. We tested whether any of the other bypassing signals acted in a similar manner.
Fig. 4. Stimulation of take-off by the stem–loop. (A) Stem–loop and coding gap mutations (in blue). (B) Pulse–chase analysis of isogenic strains, which differ as indicated, expressing mutant GST–gene 60 constructs. Read-through levels in the strain expressing an amber suppressor tRNA (blue bars). Read-through product for pGS16 migrates more quickly than other read-through products, but is visible upon more extensive electrophoresis (not shown).
Insertions in the coding gap have previously been shown to reduce bypassing (Weiss et al., 1990). In the GST–gene 60 fusion, an insertion of 26 nucleotides between the stem–loop and the GAG sequence reduces bypassing efficiency 2-fold (Figure 4; pGS18). Inactivation of RF1 does not suppress the defects associated with this mutant nor is the mutant more susceptible than WT to elevated RF1 levels. This suggests that ribosomes fail to bypass the enlarged coding gap for a reason other than increased termination.
A role for the nascent peptide signal in stimulating bypassing was previously established using lacZ reporter constructs (Weiss et al., 1990). Changing the reading frame of codons 17–34 reduces bypassing ∼140-fold. Extending the change in reading frame through codon 42 reduces bypassing 1000-fold. For cloning purposes, these constructs carried a mutation in the tetraloop (creating a XhoI site) that in the presence of a WT nascent peptide reduced bypassing <2-fold (Weiss et al., 1990) (see pGS16 for mutation). The mutation altering codons 17–34, in the presence of a WT stem–loop, reduces bypassing only 9-fold, indicative of synergy between the stem–loop and the nascent peptide signal (A.J.Herr, C.C.Nelson, R.F.Gesteland and J.F.Atkins, in preparation).
To test whether the nascent peptide signal influences the competition between take-off and termination, four GST–gene 60 constructs were made in which the changes in reading frame between codons 17–34 and 17–42 were introduced alone or in tandem with the tetraloop mutation. Surprisingly, the nascent peptide mutations by themselves (Figure 5A; pGS12, pGS14) reduce bypassing efficiency modestly (to 36%) (Figure 5B). In contrast, combining the nascent peptide and tetraloop mutations (pGS13, pGS15) reduces bypassing ∼80% relative to WT and >60% relative to the tetraloop mutation alone (pGS16). These results indicate that in our assay, the WT stem–loop largely masks the effects of the nascent peptide mutations in WT cells.
Fig. 5. Stimulation of take-off by the nascent peptide signal. (A) The amino acid sequences of the nascent peptide signal and nascent peptide signal mutants (blue) were placed in combination with a WT stem or a stem carrying a mutation in the tetraloop. (B) Pulse–chase analysis of isogenic strains, which differ as indicated, expressing mutant GST–gene 60 constructs. Read-through levels in the presence of amber suppressor tRNA are indicated by blue bars. Multiple read-through products are visible for nascent peptide mutants expressed in the amber suppressor strain. These read-through products are also present when the nascent peptide is WT (visible on more extensive electrophoretic analysis), but more apparent in the mutants because the mutation increases the mobility of all GST–gene 60 products.
Inactivation of RF1 reveals that the nascent peptide mutations enable termination to compete with take-off. Whereas WT bypassing efficiency barely increases with RF1 inactivation (from 44 to 48%), the efficiency in nascent peptide mutants increases nearly 2-fold (from 36 to 63%) (Figure 5). The observation that inactivation of RF1 causes the nascent peptide mutants to bypass more efficiently than WT may indicate that the nascent peptide signal contributes to the process of RF1-independent unproductive bypassing. RF1 inactivation strongly stimulates bypassing in the nascent peptide/tetraloop double mutants (a 4-fold increase), implying that the synergistic effect of combining the mutations affecting the nascent peptide and the tetraloop is largely due to increased termination. All nascent peptide mutant variants show a marked decrease in bypassing efficiency in response to both elevated levels of RF1 and the presence of an amber suppressor tRNA, suggesting that the nascent peptide signal enhances take-off at the expense of general decoding in the A-site.
An important mechanistic question is raised by the observation that both the nascent peptide signal and the stem–loop influence the competition between take-off and general decoding in the A-site. Do these signals target the same process or do they have distinct functions? One way to address this question is to test whether extragenic mutations have differential effects on mutations in these signals. Mutations in the gene for ribosomal protein L9 (rplI) suppress both stem–loop mutants and nascent peptide/tetraloop double mutants (Herbst et al., 1994), which suggests that the signals may have overlapping functions. But no experiments have addressed whether an L9 deficiency suppresses a nascent peptide mutation in an otherwise WT context.
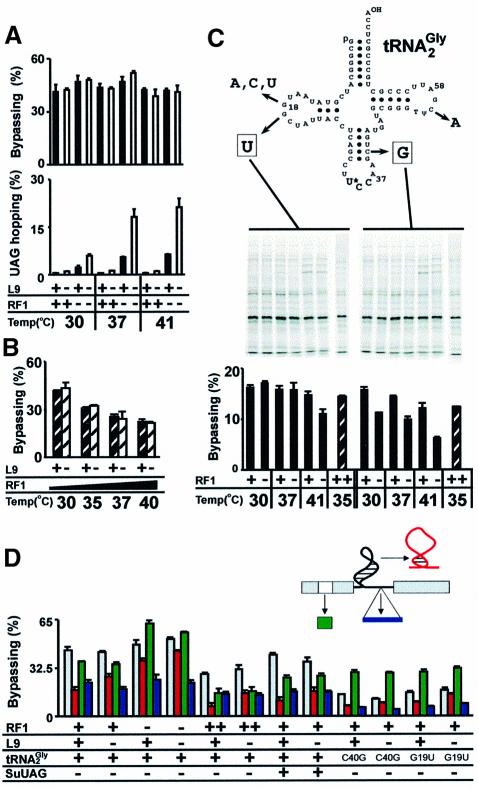
L9 clearly influences the competition between take-off and termination. In the absence of any stimulatory signals, L9 deficient ribosomes bypass a stop codon (stop-hopping, see Figure 2) with a 2-fold increased efficiency (1%) (Figure 6A) (Herbst et al., 1994). The combination of RF1 and L9 deficiencies is synergistic, elevating stop-hopping to ∼20%. This is 40-fold higher than when both factors are active and 3-fold higher than when only RF1 is defective. The intact bypassing signals of gene 60 neutralize the activities of both RF1 and L9 in WT cells. Bypassing in strains deficient in one or both of the factors is essentially indistinguishable from WT (Figure 6A), and overexpression of RF1 lowers bypassing equally in strains with, and without, L9 (Figure 6B).
Fig. 6. (A) Bypassing and UAG hopping (see Figure 2) when RF1 and L9 are inactivated. WT strain, black columns; L9 deficient strain, white columns. (B) Overexpression of RF1 in WT and L9 deficient strains. WT, black striped bars; L9 deficient strain, white striped bars. (C) Bypassing in tRNA2Gly mutants when RF1 is mutant or overexpressed. Secondary structure prediction of tRNA2Gly shows positions of mutations that reduce bypassing efficiency. Autoradiograph shows pulse–chase analysis of tRNA2Gly mutant strains expressing WT gene 60 (pGS1) under conditions in which RF1 is inactive or overexpressed. (D) Influence of ribosomal protein L9 and tRNA2Gly on bypassing efficiency with mutant gene 60 variants. WT gene 60 (pGS1; gray bars), a stem mutant (pGS7; red bars), a nascent peptide mutant (pGS12; green bars) and a mutant with an insertion in the coding gap (pGS18; blue bars).
To determine whether the stem–loop or a combination of signals counters the activity of L9, a stem–loop mutant (pGS7) and a nascent peptide mutant (pGS12) were assayed in strains with or without L9 under conditions in which RF1 was defective or overexpressed. The influence of L9 on the bypassing defect associated with insertions in the coding gap was also tested. As shown previously, the L9 deficiency partially suppresses the defect in the stem–loop, but not the coding gap (Figure 6D) (Herbst et al., 1994). Importantly, an L9 deficiency also fails to reverse the take-off defect found in the nascent peptide mutant. Combining the RF1 and L9 deficiencies enhances suppression of the stem mutant in an additive fashion, and an L9 deficiency diminishes the effects of RF1 overexpression or the presence of an amber suppressor tRNA. These observations suggest that the stem–loop neutralizes L9 during the competition between take-off and termination and that its activity is distinct from the nascent peptide signal.
L9 is not the only component of the translational apparatus implicated in bypassing. Mutations in the gene for tRNA2Gly (glyT) reduce WT bypassing 3- to 5-fold (Figure 6C) (Herr et al., 1999). One mutation (C40G) destabilizes the anticodon stem. The rest destabilize the elbow region formed by the interaction between the dihydrouridine (D) loop and ribothymidine (T) loop. Because glyT is an essential gene (Murgola and Pagel, 1980) and overexpression of the mutant variants in WT cells lowers bypassing significantly, these mutant tRNA2Gly variants likely derail bypassing after decoding the take-off site GGA (Herr et al., 1999). The decoding efficiencies of the mutant tRNA variants are 10- to 20-fold lower than WT tRNA2Gly (Herr et al., 1999), suggesting that the mutants may be defective in re-pairing with the landing site GGA.
The effect of mutant tRNA2Gly variants on the competition between take-off and termination was tested (Figure 6C). The mutants are not suppressed by inactivation of RF1 and appear even less sensitive than WT to increased RF1 levels, suggesting that they initiate bypassing readily. Curiously, the bypassing defect of the C40G mutant is enhanced by the presence of the ts RF1, even at the permissive temperature, as if WT RF1 is required for bypassing in this mutant. This seems to run counter to the observation that termination competes with take-off when RF1 levels are elevated. One possible explanation is that the C40G variant in the P-site enhances recognition of the take-off site UAG by the ts RF1.
Further evidence for a distinction between the nascent peptide signal and the stem–loop comes from analysis of the bypassing signal mutants in glyT mutant strains. Strikingly, bypassing in these mutants is twice as efficient with a mutant nascent peptide than with a WT nascent peptide, regardless of whether L9 is present (Figure 6D). In contrast, bypassing decreases 2-fold with the stem–loop mutation. Inactivation of L9 suppresses this defect, suggesting that the stem–loop is still required for bypassing in the glyT mutants. Given the evidence suggesting that the defect of these tRNA2Gly mutants is in landing, these results suggest that the nascent peptide signal may remain active following take-off and provide additional evidence that the role of the nascent peptide signal is distinct from the stem–loop.
Discussion
Nearly all ribosomes translating the first ORF of gene 60 appear to avoid termination by initiating bypassing. Only half of these ribosomes resume translation in the second ORF. How are ribosomes so efficiently re-programmed to take-off rather than terminate? And what defines the efficiency of landing? We begin by examining what influences take-off efficiency and then address the roles of the stop codon, the stem–loop and the nascent peptide signal in the mechanism of bypassing.
In the simplest view of bypassing, take-off efficiency reflects the competition between decoding in the A-site and slippage of the peptidyl-tRNA. Two events must occur for slippage: dissociation of peptidyl-tRNA–mRNA pairing and movement of the mRNA so that the peptidyl-tRNA cannot re-pair with the take-off codon. Recent models of 70S ribosome functional complexes, based on X-ray crystallography, reveal six ‘fingers’ of electron density from the 30S subunit that likely stabilize peptidyl-tRNA–mRNA pairing (Cate et al., 1999). This tight grip on the peptidyl-tRNA–mRNA complex in the P-site contrasts with the limited contacts made to the aminoacyl- tRNA–mRNA complex in the A-site. By holding peptidyl-tRNA–mRNA pairing tightly and aminoacyl-tRNA–mRNA pairing loosely, the ribosome selects for stable aminoacyl-tRNA–mRNA interactions without losing its position on the mRNA.
Cognate interactions, once made in the A-site, presumably limit mRNA movement should dissociation of P-site pairing occur. However, the possibility of take-off is not eliminated until peptidyl transfer/hydrolysis of the peptidyl-tRNA bond. During termination, the interval between ‘decoding’ and hydrolysis of the peptidyl-tRNA is much slower (2 s for UAG) than the corresponding interval in chain elongation (30 ms) (Freistroffer et al., 1997). The observation that even without special signals, a low level of peptidyl-tRNA slippage occurs at stop codons or rare codons suggests that the fingers of the 30S subunit occasionally ‘lose their grip’. It may be possible for slippage to occur at overlapping codons without complete dissociation of the mRNA–ribosome contacts, but during bypassing, these contacts must be broken in order for the mRNA to slide through the P-site. Thus, for high efficiency take-off, peptidyl-tRNA–mRNA and ribosome–mRNA contacts must be actively displaced and/or the rate of A-site occupancy must be reduced.
The ability of stop codons to stimulate bypassing is well known (Weiss et al., 1987). This has generally been attributed to slow decoding by RFs. Bypassing also occurs at rare codons in highly expressed genes (Kane et al., 1992) or just before codons under aminoacyl-tRNA limitation (Gallant and Lindsley, 1998), suggesting that specific recognition of the stop codon is not required for take-off. In keeping with this view, our results suggest that the role of the stop codon in gene 60 is simply to pause the ribosome. The best evidence for this is the decrease in bypassing efficiency caused by elevated levels of RF1. This indicates that low A-site occupancy is important for gene 60 bypassing and eliminates a model in which an abortive termination event triggers take-off. Consistent with a pausing model, the local sequence context surrounding the UAG appears to make a minor, but significant contribution to efficient bypassing.
There may still be something distinctive about having a stop codon in this position. As expected, replacing this codon with common sense codons reduces bypassing 10- to 50-fold (Weiss et al., 1990). Yet even a rare codon (AGA), which should be read slowly, reduces take-off substantially (A.J.Herr, C.C.Nelson, R.F.Gesteland and J.F.Atkins, in preparation). One, perhaps unlikely, explanation is that the high expression level of the GST–gene 60 fusion reduces the effective concentration of RF1 below even the rarest aminoacyl-tRNAs. Alternatively, the gene 60 bypassing signals may be more effective at preventing stable RF–mRNA interactions than codon–anticodon pairing. Another possibility is that initiation of bypassing occurs during the interval between decoding and peptidyl transfer/hydrolysis in concert with ejection of the RF or tRNA. Because peptidyl transfer occurs so much more quickly than hydrolysis, sense codons may favor elongation rather than take-off. By any of these models, a highly efficient amber suppressor tRNA should lower take-off as effectively as a sense codon, but this is not the case. Take-off appears to outstrip UAG read-through by a significant margin, despite the fact that the amber suppressor tRNA expression system used here achieves nearly 100% suppression of a UAGG stop codon on a different mRNA (data not shown). The explanation for this discrepancy may lie with subtle differences in recognition rates or stability of different cognate codon–anticodon interactions. The observation that the presence of an amber suppressor tRNA can reduce bypassing efficiency in a construct with a UAA stop codon, rather than UAG, suggests that a tRNA need not contribute to the growing peptide chain to interfere with take-off. Thus, the degree of A-site occupancy may be critical for efficient take-off. Whether RF-independent recognition of the stop codon by the ribosome plays a role in the mechanism can not yet be ruled out.
Genetic data indicate that the nascent peptide signal and the stem–loop stimulate take-off by separate mechanisms. Only stem–loop mutants are suppressed by the lack of ribosomal protein L9 and only nascent peptide mutants suppress the bypassing defects associated with mutant peptidyl-tRNA2Gly. Given what must be overcome for efficient bypassing, this suggests that one signal destabilizes the peptidyl-tRNA–mRNA–ribosome complex and the other limits decoding in the A-site. Either signal could indirectly trigger take-off by altering some component of the ribosome that normally limits bypassing. Alternatively, the signals may act directly. Cross-linking experiments show that nascent peptides of gene 60 and ompA exhibit the necessary flexibility to contact the decoding center directly (Choi and Brimacombe, 1998; Choi et al., 1998). Likewise, the proximity of the stem–loop to the site of peptidyl-tRNA slippage places it in position to influence the decoding process or the stability of peptidyl-tRNA–mRNA pairing. Assigning the functions of the nascent peptide signal and the stem–loop is difficult given the interconnected nature of pausing and slippage, but some inferences can be drawn by the analysis of the mutations that effect L9 and tRNA2Gly.
L9 has two putative RNA binding domains, separated by a long, rigid α-helix (Hoffman et al., 1994). Mutations that alter the C-terminal domain suppress stem–loop defects (Herbst et al., 1994; Adamski et al., 1996). Cross-linking experiments (Walleczek et al., 1989; Brimacombe et al., 1990) place L9 just under the L1 protuberance. Toeprinting (Adamski et al., 1996) and footprinting (Lieberman et al., 2000) data indicate that the N-terminal domain binds the three-helical junction adjacent to helix 76 of domain V of 23S rRNA. In the current model of the 50S subunit derived from cryo-electron microscopy, the N-terminal domain of L9 appears to bind just below the L1 stalk while the C-terminal domain extends toward the inter-subunit space (Matadeen et al., 1999). Upon assembly of the 70S subunit, L9 may undergo a dramatic change in position. One candidate rod-like density, some 50 Å away from the position of L9 in the 50S subunit, extends from L1 into the inter-subunit space. This raises the intriguing possibility that the C-terminal domain of L9 directly influences tRNA–mRNA–ribosome interactions.
The apparent role of L9 in standard decoding is to limit bypassing. In theory, L9 could do this by stabilizing peptidyl-tRNA–mRNA pairing, facilitating decoding in the A-site, or limiting slippage of the mRNA. The relationship between L9 and the stem–loop suggests that they both influence the same process, either A-site decoding or peptidyl-tRNA slippage. It should be emphasized that L9 could enhance A-site decoding indirectly by limiting mRNA movement through the ribosome.
If the role of L9 is to stabilize peptidyl-tRNA–mRNA pairing then it seems reasonable to expect a deficiency in L9 to enhance the phenotype of the mutant tRNA2Gly variants that reduce bypassing. These variants, which form unstable codon–anticodon interactions in the A-site, appear to retain this phenotype in the P-site. A deficiency in L9 does not alter the phenotype of these mutants, suggesting that peptidyl-tRNA–mRNA pairing stability is not influenced by L9. It could be argued that mutation of the tRNA negates any stabilization that L9 may offer, but the observation that L9 suppresses defects in the stem–loop in a strain expressing the peptidyl-tRNA2Gly variants makes this unlikely. Thus, it appears likely that the stem–loop influences A-site occupancy and the nascent peptide stimulates peptidyl-tRNA dissociation.
The relationship between the nascent peptide signal and the mutant tRNA2Gly variants supports a role for the nascent peptide signal in destabilizing peptidyl-tRNA–mRNA pairing. The defect in bypassing due to the mutant tRNA2Gly variants is incomplete, suggesting that the mutants are on the edge of being able to form a stable interaction with mRNA. The observation that mutation of the nascent peptide signal improves landing efficiency is consistent with increased stabilization of peptidyl-tRNA–mRNA pairing. Recent observations demonstrate the nascent peptide signal can stimulate peptidyl-tRNA slippage independently of the stem–loop (A.J.Herr, C.C.Nelson, R.F.Gesteland and J.F.Atkins, in preparation). Both bypassing of a stop codon and –1 frameshifting are enhanced when the nascent peptide is encoded just upstream of the slippage site.
What happens to those ribosomes that fail to bypass the coding gap? Do they dissociate spontaneously from the mRNA or are they derailed by cellular factors known to promote premature dissociation of peptidyl-tRNA (drop-off)? Ribosome recycling factor (RRF) RF3, elongation factor G (EFG), initiation factor 1 (IF1), IF2 and tmRNA have all been implicated in drop-off (Heurgue-Hamard et al., 1998; Karimi et al., 1998; Roche and Sauer, 1999). Inactivation or overexpression of a select group of these factors does not induce an overwhelming alteration in bypassing efficiency, suggesting that there may be something inherently unstable about the scanning complex that leads to spontaneous dissociation of the scanning complex (A.J.Herr, R.F.Gesteland and J.F.Atkins, submitted). Hydrolysis of the peptidyl-tRNA bond by peptidyl-tRNA hydrolase may then result in the appearance of a termination-sized product (Cuzin et al., 1967; Kossel and RajBhandary, 1968; Chapeville et al., 1969).
From these observations a working model for the initiation of bypassing emerges. Ribosomes decode the take-off site GGA and translocate the peptidyl-tRNA2Gly–GGA complex into the P-site. Following translocation, formation of the stem–loop within the A-site of the ribosome either displaces RF or inhibits recognition of the UAG. The stem may either refold in the A-site or evade the mRNA structure-destabilizing activity of the ribosome inferred from analysis of bacterial biosynthetic attenuators and translational control regions of inducible antibiotic resistance genes. Refolding or evasion could be driven in part by the stable UUCG tetraloop at the top of the stem. Formation of base-pairing interactions in the stem–loop involving the stop codon are not necessary for its activity, suggesting that steric hindrance alone may be sufficient to limit recognition of the UAG. Whatever the mechanism, repeated entry by a cognate decoding molecule (RF or tRNA) can overwhelm the stem–loop activity. Under normal conditions, limitation of decoding in the A-site complements the peptidyl-tRNA–mRNA dissociation activity of the nascent peptide signal and take-off occurs efficiently.
The proposed functions of the nascent peptide and the stem–loop may be addressed through studies examining the degree of ribosome pausing with various gene 60 mutants. Further insight into the following questions may be gained through biochemical and cryo-electron microscopic analysis of the different stages of bypassing. Does the stem–loop fold within the ribosome at take-off? Does mRNA secondary structure pass through the P-site intact? Are there conformational differences between scanning and translating ribosomes? As the details of how gene 60 re-programs translation become clear, why this mechanism is used at all and the extent of its distribution remain a mystery that may only be solved through the systematic analysis of genomes and their end products.
Materials and methods
Plasmids
The GST–gene 60 expression plasmid is a derivative of pGEX-2T and has been described elsewhere (Maldonado and Herr, 1998). For the purposes of this paper this vector is renamed pGS1. All other gene 60 variants were cloned into this vector using BamHI sites that flank gene 60. Inserts were created by a two-step PCR strategy that utilized inside primers to create the specific changes and outside primers carrying embedded BamHI sites.
To create the RF1 overexpression plasmid (pLprfA), prfA was cloned into the XbaI and SalI sites of a pACYC184 derivative, pLSuglyT, encoding chloramphenicol resistance (camr) (Herr et al., 1999). Expression from this vector is driven by the λ pL promoter (upstream of XbaI and SalI sites) and regulated by the ts λ repressor allele, cI857. We amplified the coding sequence for prfA using a forward primer containing an embedded XbaI site and a reverse primer containing an embedded SalI site.
GLZ96 contains the sequence CCCAAATATGCATAGGCACGG (amber codon in bold, matching codons underlined) cloned between the protA-Cat and lacZ ORFs of GLZ69 (Herr et al., 1999). The in-frame control carries a TAC codon instead of TAG.
The amber suppressor tRNA expression plasmid (pACYC184 derivative, camr) encodes a derivative of tRNA2Gly described previously that has a 3′AUC5′ anticodon rather than 3′CCU5′ (Herr et al., 1999). Expression of the amber suppressor tRNA is driven by a constitutive λ pL promoter.
Bacterial strains
CSH142 has been described (Miller, 1992). AH133 was constructed as follows. A P1 lysate from MRA7 (a gift from B.Persson) was used to transduce CSH142 to tetr and prfA1. The presence of the prfA1 allele was confirmed by screening for the ts phenotype and sequencing PCR products amplified from the chromosome. AH133bypC40G and AH133bypG19U were derived in a similar fashion from CSH142bypC40G and CSH142bypG19U (Herr et al., 1999). AH156 is a CSH142 derivative containing a precise deletion/replacement of rplI (the gene for L9) with the kanamycin resistance (kanr) gene from Tn903. (Construction and characterization of this mutant will be described elsewhere.) This kanr replacement was moved by P1 transduction into AH133 to make AH157.
Bypassing efficiency assays
Bypassing efficiency estimates were made essentially as described (Herr et al., 1999) using [35S]methionine pulse–chase analysis. The only modification was the use of a 200 µl culture volume for the assays. Following pulse–chase labeling, total protein in each sample was separated on an 11% Tris–glycine SDS–polyacrylamide gel and visualized with a Molecular Dynamics PhosphorImager. Efficiency estimates represent the amount of bypassing product divided by the total of protein produced by both productive and unproductive bypassing. All efficiency estimates are corrected for background expression and for differences in methionine content between the products due to productive and unproductive bypassing.
Stop-hopping assays
To estimate the degree of RF1 inactivation, GLZ96 was assayed in the appropriate WT or prfA1 strains expressing lacIq from a pACYC184 derivative. A control (GLZ95), which lacks a stop codon between protA-Cat and lacZ, was used to estimate the levels of bypassing (or hopping) of the stop codon. β-galactosidase activity measurements were performed essentially as described (Miller, 1992).
Acknowledgments
Acknowledgements
The work was supported by grants (to J.F.A.) from NIH (GM 48152) and (to R.F.G.) from DOE (DE-FG03-99ER62732). A.J.H. was supported by an NIH training grant (#5T32GM07464-24).
References
- Adamski F.M., Atkins,J.F. and Gesteland,R.F. (1996) Ribosomal protein L9 interactions with 23S rRNA: the use of a translational bypass assay to study the effect of amino acid substitutions. J. Mol. Biol., 261, 357–371. [DOI] [PubMed] [Google Scholar]
- Atkins J.F., Böck,A., Matsufuji,S. and Gesteland,R.F. (1999) Dynamics of the genetic code. In Gesteland,R.F., Cech,T.R. and Atkins,J.F. (eds), The RNA World. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 637–673. [Google Scholar]
- Belcourt M.F. and Farabaugh,P.J. (1990) Ribosomal frameshifting in the yeast retrotransposon Ty: tRNAs induce slippage on a 7 nucleotide minimal site. Cell, 62, 339–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossi L. and Roth,J.R. (1980) The influence of codon context on genetic code translation. Nature, 286, 123–127. [DOI] [PubMed] [Google Scholar]
- Brimacombe R., Gornicki,P., Greuer,B., Mitchell,P., Osswald,M., Rinke-Appel,J., Schuler,D. and Stade,K. (1990) The three-dimensional structure and function of Escherichia coli ribosomal RNA, as studied by cross-linking techniques. Biochim. Biophys. Acta, 1050, 8–13. [DOI] [PubMed] [Google Scholar]
- Buckingham R.H., Grentzmann,G. and Kisselev,L. (1997) Polypeptide chain release factors. Mol. Microbiol., 24, 449–456. [DOI] [PubMed] [Google Scholar]
- Cate J.H., Yusupov,M.M., Yusupova,G.Z., Earnest,T.N. and Noller,H.F. (1999) X-ray crystal structures of 70S ribosome functional complexes. Science, 285, 2095–2104. [DOI] [PubMed] [Google Scholar]
- Chapeville F., Yot,P. and Paulin,D. (1969). Enzymatic hydrolysis of N-acyl-aminoacyl transfer RNAs. Cold Spring Harb. Symp. Quant. Biol., 34, 493–498. [DOI] [PubMed] [Google Scholar]
- Chittum H.S., Lane,W.S., Carlson,B.A., Roller,P.P., Lung,F.D., Lee,B.J. and Hatfield,D.L. (1998) Rabbit β-globin is extended beyond its UGA stop codon by multiple suppressions and translational reading gaps. Biochemistry, 37, 10866–10870. [DOI] [PubMed] [Google Scholar]
- Choi K.M. and Brimacombe,R. (1998) The path of the growing peptide chain through the 23S rRNA in the 50S ribosomal subunit; a comparative cross-linking study with three different peptide families. Nucleic Acids Res., 26, 887–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K.M., Atkins,J.F., Gesteland,R.F. and Brimacombe,R. (1998) Flexibility of the nascent polypeptide chain within the ribosome—contacts from the peptide N-terminus to a specific region of the 30S subunit. Eur. J. Biochem., 255, 409–413. [DOI] [PubMed] [Google Scholar]
- Craigen W.J. and Caskey,C.T. (1986) Expression of peptide chain release factor 2 requires high-efficiency frameshift. Nature, 322, 273–275. [DOI] [PubMed] [Google Scholar]
- Cuzin F., Kretchmer,N., Greenberg,R.E., Hurwitz,R. and Chapeville,F. (1967) Enzymatic hydrolysis of N-substituted aminoacyl-tRNA. Proc. Natl Acad. Sci. USA, 58, 2079–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farabaugh P.J. (1996) Programmed translational frameshifting. Annu. Rev. Genet., 30, 507–528. [DOI] [PubMed] [Google Scholar]
- Farabaugh P.J., Zhao,H. and Vimaladithan,A. (1993) A novel programmed frameshift expresses the POL3 gene of retrotransposon Ty3 of yeast: frameshifting without tRNA slippage. Cell, 74, 93–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freistroffer D.V., Pavlov,M.Y., MacDougall,J., Buckingham,R.H. and Ehrenberg,M. (1997) Release factor RF3 in E.coli accelerates the dissociation of release factors RF1 and RF2 from the ribosome in a GTP-dependent manner. EMBO J., 16, 4126–4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallant J. and Foley,D. (1980) On the causes and prevention of mistranslation. In Chambliss,G., Craven,G.R., Davies,J., Davis,K., Kahan,L. and Nomura,M. (eds), Ribosomes, Structure, Function and Genetics. University Park Press, Baltimore, MD, pp. 615–638. [Google Scholar]
- Gallant J.A. and Lindsley,D. (1998) Ribosomes can slide over and beyond ‘hungry’ codons, resuming protein chain elongation many nucleotides downstream. Proc. Natl Acad. Sci. USA, 95, 13771–13776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gesteland R.F. and Atkins,J.F. (1996) Recoding: dynamic reprogramming of translation. Annu. Rev. Biochem., 65, 741–768. [DOI] [PubMed] [Google Scholar]
- Herbst K.L., Nichols,L.M., Gesteland,R.F. and Weiss,R.B. (1994) A mutation in ribosomal protein L9 affects ribosomal hopping during translation of gene 60 from bacteriophage T4. Proc. Natl Acad. Sci. USA, 91, 12525–12529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herr A.J., Atkins,J.F. and Gesteland,R.F. (1999) Mutations which alter the elbow region of tRNA2Gly reduce T4 gene 60 translational bypassing efficiency. EMBO J., 18, 2886–2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heurgue-Hamard V., Karimi,R., Mora,L., MacDougall,J., Leboeuf,C., Grentzmann,G., Ehrenberg,M. and Buckingham,R.H. (1998) Ribosome release factor RF4 and termination factor RF3 are involved in dissociation of peptidyl-tRNA from the ribosome. EMBO J., 17, 808–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman D.W., Davies,C., Gerchman,S.E., Kycia,J.H., Porter,S.J., White,S.W. and Ramakrishnan,V. (1994) Crystal structure of prokaryotic ribosomal protein L9: a bi-lobed RNA-binding protein. EMBO J., 13, 205–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W.M., Ao,S.Z., Casjens,S., Orlandi,R., Zeikus,R., Weiss,R., Winge,D. and Fang,M. (1988) A persistent untranslated sequence within bacteriophage T4 DNA topoisomerase gene 60. Science, 239, 1005–1012. [DOI] [PubMed] [Google Scholar]
- Kane J.F., Violand,B.N., Curran,D.F., Staten,N.R., Duffin,K.L. and Bogosian,G. (1992) Novel in-frame two codon translational hop during synthesis of bovine placental lactogen in a recombinant strain of Escherichia coli. Nucleic Acids Res., 20, 6707–6712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi R., Pavlov,M.Y., Heurgue-Hamard,V., Buckingham,R.H. and Ehrenberg,M. (1998) Initiation factors IF1 and IF2 synergistically remove peptidyl-tRNAs with short polypeptides from the P-site of translating Escherichia coli ribosomes. J. Mol. Biol., 281, 241–252. [DOI] [PubMed] [Google Scholar]
- Kossel H. and RajBhandary,U.L. (1968) Studies on polynucleotides. LXXXVI. Enzymic hydrolysis of N-acylaminoacyl-transfer RNA. J. Mol. Biol., 35, 539–560. [DOI] [PubMed] [Google Scholar]
- Lieberman K.R., Firpo,M.A., Herr,A.J., Nguyenle,T., Atkins,J.F., Gesteland,R.F. and Noller,H.F. (2000) The 23S rRNA environment of ribosomal protein L9 in the 50S ribosomal subunit. J. Mol. Biol., 297, 1129–1143. [DOI] [PubMed] [Google Scholar]
- Maldonado R. and Herr,A.J. (1998) Efficiency of T4 gene 60 translational bypassing. J. Bacteriol., 180, 1822–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matadeen R., Patwardhan,A., Gowen,B., Orlova,E.V., Pape,T., Cuff,M., Mueller,F., Brimacombe,R. and van Heel,M. (1999) The Escherichia coli large ribosomal subunit at 7.5 Å resolution. Struct. Fold. Des., 7, 1575–1583. [DOI] [PubMed] [Google Scholar]
- Miller J.H. (1992) A Short Course In Bacterial Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Miller J.H. and Albertini,A.M. (1983) Effects of surrounding sequence on the suppression of nonsense codons. J. Mol. Biol., 164, 59–71. [DOI] [PubMed] [Google Scholar]
- Mottagui-Tabar S., Bjornsson,A. and Isaksson,L.A. (1994) The second to last amino acid in the nascent peptide as a codon context determinant. EMBO J., 13, 249–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murgola E.J. and Pagel,F.T. (1980) Codon recognition by glycine transfer RNAs of Escherichia coli in vivo. J. Mol. Biol., 138, 833–844. [DOI] [PubMed] [Google Scholar]
- Nakamura Y. and Ito,K. (1998) How protein reads the stop codon and terminates translation. Genes Cells, 3, 265–278. [DOI] [PubMed] [Google Scholar]
- O’Connor M., Gesteland,R.F. and Atkins,J.F. (1989) tRNA hopping: enhancement by an expanded anticodon. EMBO J., 8, 4315–4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen W.T. and Curran,J.F. (1991) Effects of the nucleotide 3′ to an amber codon on ribosomal selection rates of suppressor tRNA and release factor-1. J. Mol. Biol., 219, 231–241. [DOI] [PubMed] [Google Scholar]
- Poole E.S., Brown,C.M. and Tate,W.P. (1995) The identity of the base following the stop codon determines the efficiency of in vivo translational termination in Escherichia coli. EMBO J., 14, 151–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche E.D. and Sauer,R.T. (1999) SsrA-mediated peptide tagging caused by rare codons and tRNA scarcity. EMBO J., 18, 4579–4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walleczek J., Redl,B., Stoffler-Meilicke,M. and Stoffler,G. (1989) Protein–protein cross-linking of the 50S ribosomal subunit of Escherichia coli using 2-iminothiolane. Identification of cross-links by immunoblotting techniques. J. Biol. Chem., 264, 4231–4237. [PubMed] [Google Scholar]
- Weiss R. and Gallant,J. (1983) Mechanism of ribosome frameshifting during translation of the genetic code. Nature, 302, 389–393. [DOI] [PubMed] [Google Scholar]
- Weiss R.B., Dunn,D.M., Atkins,J.F. and Gesteland,R.F. (1987) Slippery runs, shifty stops, backward steps and forward hops: –2, –1, +1, +2, +5 and +6 ribosomal frameshifting. Cold Spring Harb. Symp. Quant. Biol., 52, 687–693. [DOI] [PubMed] [Google Scholar]
- Weiss R.B., Huang,W.M. and Dunn,D.M. (1990) A nascent peptide is required for ribosomal bypass of the coding gap in bacteriophage T4 gene 60. Cell, 62, 117–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Rydén-Aulin,M., Kirsebom,L.A. and Isaksson,L.A. (1994) Genetic implication for an interaction between release factor one and ribosomal protein L7/L12 in vivo. J. Mol. Biol., 242, 614–618. [DOI] [PubMed] [Google Scholar]
- Zhang S., Rydén-Aulin,M. and Isaksson,L.A. (1996) Functional interaction between release factor one and P-site peptidyl-tRNA on the ribosome. J. Mol. Biol., 261, 98–107. [DOI] [PubMed] [Google Scholar]
- Zhang S., Rydén-Aulin,M. and Isaksson,L.A. (1998) Functional interaction between tRNA2Gly at the ribosomal P-site and RF1 during termination at UAG. J. Mol. Biol., 284, 1243–1246. [DOI] [PubMed] [Google Scholar]