Abstract
The EAV/CR framework, designed for database support of rapidly evolving scientific domains, utilizes metadata to facilitate schema maintenance and automatic generation of Web-enabled browsing interfaces to the data. EAV/CR is used in SenseLab, a neuroscience database that is part of the national Human Brain Project. This report describes various enhancements to the framework. These include (1) the ability to create “portals” that present different subsets of the schema to users with a particular research focus, (2) a generic XML-based protocol to assist data extraction and population of the database by external agents, (3) a limited form of ad hoc data query, and (4) semantic descriptors for interclass relationships and links to controlled vocabularies such as the UMLS.
Relational database management systems (RDBMSs) are widely used for data management in the biosciences. In conventionally structured databases, “meta-knowledge”—a description of the kinds of facts that the database stores—is recorded implicitly in the schema in the form of the structure of individual tables and relationships between them. Meta-knowledge revisions, which frequently are in rapidly evolving scientific domains, require schema changes; typically, the tables and the relationships become more numerous. Whereas making schema changes is straightforward, having to redesign all components of the user interface affected by these changes is much more laborious and personnel-intensive. Interfaces between the data and nonhuman “users,” e.g., external computer programs, must also be similarly rebuilt. To the extent that such rebuilding can be automated, or the need for rebuilding even eliminated in some circumstances, such systems become more robust.
Our previously described approach to address this problem is entitled EAV/CR (Entity-Attribute-Value with Classes and Relationships).1,2 EAV/CR, whose principles will be summarized shortly, relies on a large body of metadata that explicitly describes, not only the types of facts in a database but also how they must be presented to the user. Generic code uses this metadata to create both simple Web-enabled user interfaces and facilitates interoperability through data and metadata interchange. This model currently is being used operationally in Yale's SenseLab3 database, part of the national Human Brain Project,4 which concerns neuroscience data centered primarily on the olfactory system. This report describes the several major enhancements made to the EAV/CR framework over the last three years.
Background: An Introduction to EAV/CR
The Entity-Attribute-Value (EAV) data model was devised originally in the form of LISP association lists, as a general means of information representation in the context of artificial intelligence research. It was adapted subsequently for use in the clinical patient record: the pioneering HELP system5,6 and the Columbia-Presbyterian clinical data repository,7,8 for example, have an EAV component. Our group has used EAV for clinical study data management systems9 and neuroscience data.10
In the EAV model, data are conceptually stored in a single table with three columns: an Entity (the object being described), an Attribute (an aspect of the object being described), and the Value for that attribute. In clinical systems, the entity typically is a clinical event, which is a composite of a Patient ID and one or more timestamps recording when an event occurred. An attribute is a clinical finding or parameter (e.g., blood hemoglobin). With neuroscience data, the entity is any object on which we wish to store information, such as a neuronal cell, odor molecule, or olfactory receptor; physically, it is an ID from an “Objects” table. Attributes are analogous to columns in conventional tables. The set of attributes within a database constitutes a kind of controlled vocabulary: to record new types of facts, new attributes are added to the vocabulary and then associated with individual entities. This can be done without altering the underlying physical database schema. Gio Wiederhold's TOD (Time-Oriented-Database) model is similar in intent to EAV, except that it also includes a time dimension. This work also pioneered the concept of self-describing metadata.11
The drawback of an EAV system is that the data's physical organization is significantly different from the way users conceptualize it (as one column per attribute). Therefore, when presenting EAV data, it must be transiently converted (“pivoted”) into a conventional representation through fairly elaborate metadata-driven code. The need to create this code before the equivalent of a “Hello, world” application can be created constitutes a significant hurdle. Once this code is built and tested, however, applications can be constructed rapidly. Another potential problem is that repeated on-demand pivoting can exact a performance penalty when manipulating large volumes of data.
In traditional EAV approaches, values are atomic or simple (e.g., numbers representing values of hemoglobin). The EAV/CR approach combines object-oriented concepts with the basic EAV model. Specifically, an object in the database must belong to a particular class. The set of attributes applicable to that object are constrained based on its class, and values may be IDs of other objects of different classes or the same class. The last feature implements relationships between data objects. Classes and attributes are analogous to tables and columns in a conventional database: the difference is that both of these contain additional meta-information that applies to how data are presented to the user.
EAV/CR, despite its name, supports hybrid designs in which certain classes may be represented physically as conventional tables, with one column per attribute, if the system designer determines that the number of objects in this class will be numerous enough that conventional representation will be more efficient. The metadata, however, record how a class is physically represented so as to allow dynamic generation of the correct code to access individual entities.
When setting up an EAV/CR database, one must, for the most part, simulate a third-normal form relational database using a mostly EAV-based storage mechanism. Therefore, RDB design principles cannot be forgotten. (Although EAV/CR supports non-first-normal-form data structures, allowing the designer to simulate arrays, the decision to create such structures must be deliberate and not accidental. The use of such structures is exceptional rather than typical.) A corollary is that database-naïve end-users cannot be trusted to edit and maintain the metadata: incorrect metadata will yield a malfunctioning application.
System Enhancements: Overall Goals and Objectives
The unifying goals behind the enhancements described in this report are:
To better support the curation and display of fairly diverse bioscience data within a single data store to meet the needs of diverse subgroups within a research community. In the current case, the community of preclinical experimental neuroscience researchers comprises subgroups with varying research focuses: neuronal modelers, odor researchers, neuroanatomists, and so on.
To interoperate with systems built by other research groups within the same community. The Human Brain Project is a loosely federated consortium of researchers studying various aspects of the nervous system.
The major practical constraint we face is that because of the EAV representation of our data, many standard third-party software tools are inapplicable. Further, as we shall see, even if we were not using EAV, not all of these tools would be readily adaptable for our purposes.
The subgoals and developments that derive from the objectives are now summarized.
For meeting the data display needs of specific subgroups, there must be a framework for restricting the displayed data elements to what this group (and the curators designated for this group) considers to be the most pertinent to their needs. For curators, the framework must support security with different levels of privilege to different elements. The “portal” mechanism, discussed in Portals: Motivation and Principles is tailored to this objective.
There must be ways for end-users to search for specific data of interest. It is hence necessary to support ad hoc query through a graphical user interface. This is discussed in Metadata-driven Ad Hoc Queries.
An EAV/CR database must be capable of exchanging data with existing systems: we therefore need to be able to import and export data in a variety of formats. Further, because the EAV/CR software is open-source and being actively explored by several groups, we envisage the necessity to migrate both metadata and data between database installations at different sites. For both these subgoals, instead of devising ad-hoc approaches, it helps to explore the use of a standard internal metadata/data interchange format: this role is served by the EDSP protocol, discussed in Facilitating Interoperability: The EDSP Protocol.
Finally, to support research collaborations with other research groups within the same community (who also build repositories), one must investigate means of inter-operability that try to address the issue of shared semantics. The section, Support of Semantic Descriptors and Links to Controlled Vocabularies explores our pilot use of UMLS for this purpose.
EAV/CR Enhancements
Portals: Motivation and Principles
As described in our previous report, the EAV/CR approach pays off for a system with a large number of classes (e.g., several dozen) and numerous interclass relationships, with a relatively modest number of object instances per class for most classes. Using an EAV representation in this circumstance avoids proliferating the number of tables.
We discovered, however, that most users of an EAV/CR database of such complexity were rarely concerned with the full set of classes within this system. For example, SenseLab users, particularly regular Web site visitors, segregated into distinct communities based on their research interests: those concerned with neuron properties, users interested in olfactory receptor molecules, those concerned with designing and using models (mathematical simulations) of neurons, and so forth. Some classes are much more interesting to some users than others: a neuronal modeler, for example, cares little for the chemical structure or categories of individual odor molecules.
It was therefore necessary for us to simulate “special interest” databases that were subsets of the total EAV/CR schema. The front-ends to such simulated databases, in effect, act as “portals” for a particular class of user, by providing ready access to classes of data and associated functionality that is most important to this user. In SenseLab, the simulated databases are CellProp(erties)DB, NeuronDB, ModelDB, Olfactory Receptor DB, OdorDB, and OdorMapDB. A portal comprises content and an associated user interface. It is supported by metadata (model and element definitions), plus generic code that is driven by the metadata. The set of classes within a portal constitute that portal's conceptual schema.
It was deemed highly desirable to extend the EAV/CR framework to facilitate rapid creation and modification of individual portals without the need to write portal-specific, hand-customized code. The requirements of such portals are described below.
At the end-user level, visibility of individual classes and attributes across portals must be controllable. Thus, some classes may be of interest to users in several different portals, but in differing degree of detail: some attributes may be visible in one portal but invisible in another. For example, the models for a neuron are accessible in ModelDB but not in NeuronDB, whereas gross and microanatomical properties are visible in NeuronDB but not in ModelDB. Note that such restrictions are more a matter of ergonomics and the prevention of information overload for the “typical” user for a portal. An (exceptional) end user who wishes to see both anatomical and model data for a given type neuron can always open two Web browser windows, one to each portal, and search for the neuron of interest in both windows separately.
SenseLab scientists with specific expertise handle curation/contact responsibilities for individual portals. A curator's privileges with respect to another portal, however, must be relatively restricted. The metadata curation model is a distributed one: that is, a curator must be permitted to define metadata (classes and attributes) for one's own portals, as well as define the cross-portal visibility and editability of individual classes and attributes. (Close collaboration between the curators, however, reduces the risk of redundant metadata definitions, while increasing data integration.)
Read (browsing) and Write (editing) access to classes/attributes at the intercurator level must be controlled separately from end-user access. At the end-user level, Web-based data editing by anonymous (“public”) end users is not supported because SenseLab's contents are highly curated. At the curator level, all classes and attributes are visible, but permission to create or edit objects belonging to particular classes as well as permission to create/modify metadata should be restricted to particular curators if so desired.
An individual portal in SenseLab must ultimately be responsive to the needs of its end-user community. The set of visible classes/attributes may change, and desired changes in visibility must be implemented without programming.
The user interface to an individual portal must be indicated through a distinctive color scheme and set of logos, again, without programming. Such a scheme lets the user know which portal he or she is currently browsing.
From the above, it can be seen that some purposes served by portals in EAV/CR are similar to those served by “views” in traditional database design, which also are used to vary access to the schema by user privilege. The database engine's view mechanisms cannot be used here, however, because all data for all (EAV-modeled) classes are typically stored in the same set of (relatively few) physical tables: they must therefore be simulated. EAV/CR portals go well beyond simulation of a set of views, however, in the user-interface component. The Web interface that is autogenerated for a given portal works proactively not only in preventing inappropriate end-user actions within a portal, but also in showing users or curators the classes of data that are available for access and the actions that are possible.
Over several years of continuous portal use in SenseLab, we have observed that the classes within a particular portal tend to be highly interconnected, with numerous simulated primary-key/foreign-key joins. Classes that are shared across portals, however, tend to be few, so that interportal connections are relatively sparse. In fact, a curator may, if desired, create a portal composed entirely of private (nonshared) classes. In this sense, a portal can simulate a single conventional database, whereas the complete EAV/CR schema simulates a data store or collection of databases.
Creation of Portals: EAV/CR Classes and Attributes
An administrator creates portals through a Web-based System Management console by specifying the appropriate portal-specific metadata: portal name, captions, administrative information, plus colors and logos that define a visual scheme. Rights then are delegated to individual curators for various administrative tasks. Next, the administrator defines classes (and attributes for each class) and grants users specific rights to the database. Full details of class/attribute definitions may be found in our previous report2 as well as at the URL <http://senselab.med.yale.edu/senselab/site/dsArch/?lb=tree>. We provide brief highlights here.
A class has an internal name, caption, and description. Individual attributes can have a name and data type. The latter is important because the EAV/CR schema stores attribute data in data-type–specific tables for the purposes of efficient storage and ability to index data values for fast retrieval. In addition to the standard data type (integers, strings, reals), EAV/CR also supports class references and formula attributes. These are described below.
Class reference attributes are the equivalent of foreign key fields of conventional databases and point to instances of other classes or even the same class recursively. The latter subsumes the “Hierarchical Attribute-Value” approach described by Gardner et al. in their Common Data Model for neuroscience data.12 Class references allow object-ids of classes to be “values” in the EAV data model, and restrict a particular attribute to a subset of the objects in the database that belong to a specific class, allowing validation during data entry or data importing.
Formula attributes contain JavaScript and/or HTML that can execute either on the Web server or on the Web browser, or both. They are used to compute values based on the values of other attributes, for real-time data extraction from external Web sites. They also can be used to enable navigation to individual objects in external Web sites even when such sites do not provide such access through query strings appended to the site's URL. (In HTTP parlance, they allow simulating a GET request even when the Web page in question is set up only to handle a POST request from another page within the same site.) In a sense, formula attributes “break” the metadata model, because knowledge of the domain is placed in procedural code, rather than externalized in the metadata structure, but this is unavoidable. In any case, most formulas are simple expressions that even nonprogrammers can understand.
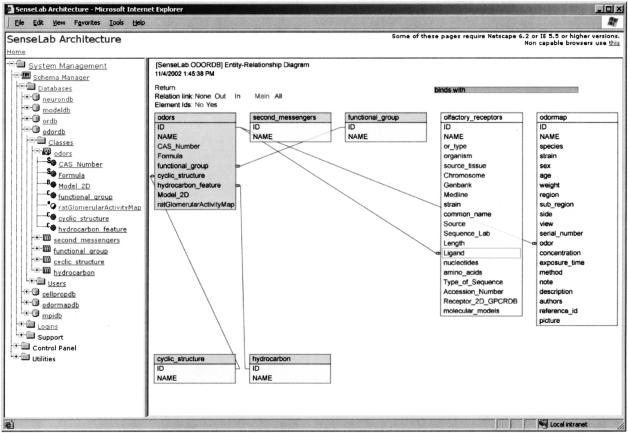
▶ shows the System Management console in operation. The left frame shows a list of portals (“databases”). The expanded OdorDB portal shows several classes. When a class is expanded, its attributes are shown: the Odors class has the attributes “CAS_Number” and “Formula.” A letter before the attribute indicates its data type: S = string, I = integer, Y = binary, C = class reference. The attributes “functional group” and “cyclic structure” are class references that point to the classes of the same name.
Figure 1.
EAV/CR System Management Console. (Left) a tree-structured view of the database's metadata elements and utilities used for schema maintenance. (Right) the entity relationship diagram of the odor database (OdorDB): Classes and its attributes are shown as table names and fields; relationships are shown as red connecting lines. Colored background classes belong to the OdorDB database, and noncolored classes are shared classes from other databases referenced by OdorDB attributes.
The right frame of ▶ shows the conceptual schema of OdorDB as an Entity-Relationship (E-R) diagram in which classes and attributes are shown as tables and fields, respectively, and relationships between classes are shown as connectors. The Odors class is highlighted. This diagram is generated dynamically from the metadata using Scalable Vector Graphics (SVG),13 a versatile XML-based W3C standard technology for Web-based display of interactive graphics that respond to user actions such as mouse clicks on different parts of the graphic.
Metadata-driven Ad Hoc Queries
Standard third-party “visual” tools for querying relational databases, which typically are based on the query-by-example user interface metaphor, expect the user to operate on the physical schema of a database. In an EAV/CR schema, the physical and conceptual schemas often are dramatically different, especially when much of the data are stored in EAV form: numerous object instances from different classes are stored in the same set of tables. Such tools are therefore of very limited use. Further, data that are fetched from the physical schema for individual attributes through individual SQL queries must be transformed and presented in terms of the conceptual schema, with one column per attribute, to make the results of a query understandable. This involves merging and pivoting the results of individual operations, a nontrivial and laborious operation when performed manually through SQL programming.
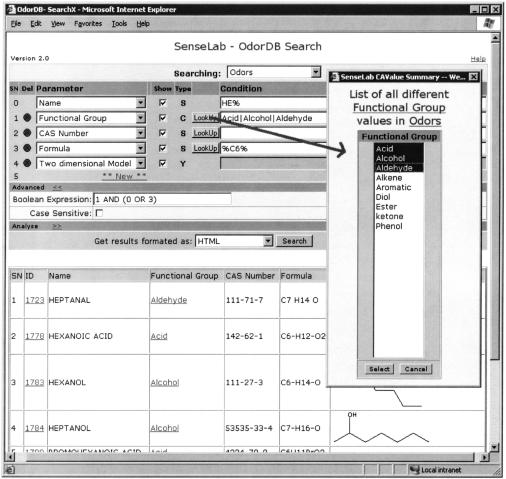
To address these issues, the EAV/CR framework allows the user to query the data in terms of the conceptual schema. The query subsystem currently is undergoing major enhancements, and we intend to describe it in depth in a separate report. Here we describe its existing feature set. The query subsystem uses a simple query-by-example interface that is generated from the metadata and performs pivoting of results as needed behind the scenes. This is illustrated in ▶, which shows a search of all Odor molecules, whose odor name starts with “HE,” and whose functional group attribute is one of (acid, alcohol, or aldehyde), or which has 6 carbon atoms in its empirical chemical formula. In terms of a conventional relational query, this is the equivalent of a two-table join between “Odors” and “Functional Groups,” with records from the latter being used as one of the query criteria. Note that the result set also shows the structural formulas for the molecules of interest, as GIF images.
Figure 2.
Metadata-driven Ad Hoc Query Interface Creation. This figure shows the SenseLab-OdorDB search page with some prefilled conditions and their results. The search conditions are for those Odor molecules whose (a) odor name starts with “HE,” (b) functional group attribute is either acid, alcohol, or aldehyde, or (c) their chemical formula starts with “C6.” The “lookup” button is used to populate conditional values with information stored in the database.
Currently, the search mechanism can only fetch data from a single class, and classes to which it is directly connected through class references. Future versions will incorporate the ability to join multiple classes across multiple portals based on attribute-to-attribute value joins. The idea is that the set of relationships that connects different classes within an EAV/CR conceptual schema constitutes a directed graph. Therefore, when combining attributes from classes that are separated widely on the graph, it should be possible, for the most part, to automatically determine the graph traversal route between them, and therefore to generate the appropriate SQL to perform the “joins” in the conceptual schema. The traversal route cannot be determined automatically if two classes are multiply linked, so that more than one route is possible; here, one must prompt the user for which particular route is desired. We currently are developing a modified interface that shows the user the E-R diagram of the conceptual schema (similar to ▶) and lets the user pick attributes, or specific intertable relationships, by clicking on them.
The result set can be saved as a text table or as an XML equivalent of the same. Query definitions can be stored for later reuse, with individual criteria in the definition being optionally specifiable as parameters to the query. We decided to use an XML-based representation for query storage, which is directly analogous to the query-by-example representation that the user sees. The representation, however, uses internal class/attribute identifiers taken from the metadata, rather than the user-understandable class/attribute names (which are used, for the most part, only as “aliases” to make the XML somewhat intelligible). The choice of XML is a trade-off, as explained below.
Currently, we do not intend to support direct query composition through an SQL-like language: even a developer would compose a query graphically, test it, and then save it, parameterizing whatever criteria she or he desired. This simplifies the task of ensuring that the query is semantically correct, because the query interface makes the user pick classes and attributes from lists (while capturing metadata IDs transparently) and prevents numerous inappropriate actions. An SQL interpreter, by contrast, must perform extensive semantic checking to make sure that the classes/attributes supplied by the user exist in the conceptual schema—their names could have been misspelled, for example. It must also check that the relational or aggregate operators specified for individual criteria are appropriate to the datatype of each criterion's attribute. Finally, it must report useful diagnostic error messages in case interpretation fails, highlighting the specific point of failure. Our XML-based representation, in effect, is a “path of least resistance” from the software development perspective. A stored query is, for practical purposes, preparsed and prevalidated, and we can therefore focus purely on the execution step of query processing.
On the other hand, our XML representation, which has a very simple structure, currently is less expressive than a full SQL implementation would be: it does not support advanced features such as grouping of results by one or more attributes. For users who need such queries, we recommend exporting the classes of interest into conventional relational form (through EDSP, as described earlier), where they can be queried in more powerful ways.
The motivation for creating a query-storage specification is to decouple the query interpretation/execution engine from the query composition GUI. The specification therefore anticipates the eventual creation of query-generation programs that access the metadata but that bypass the query-composition GUI altogether. Further details on using the query interface (also accessible via a Help button in the interface) are described at <http://senselab.med.yale.edu/senselab/site/dbData/eavXSearch_help.asp>.
Facilitating Interoperability: The EDSP Protocol
Publicly Web-accessible databases typically make individual objects within their contents accessible via URLs that contain unique identifiers. This allows other applications to store unique identifiers and hyperlink to them on demand, providing a simple form of interoperability. If, however, the resulting hyperlink were to return only formatted HTML, such interoperability would be very limited. This is because an HTML stream typically mixes content with formatting markup. To get at specific content such as a particular field requires elaborate parsing. Modern data-driven applications, therefore, separate content from markup. The content typically is in the form of an Extensible Markup Language stream, and the markup can be applied dynamically by applying an Extensible Stylesheet Language Transformation14 script to the content. For applications that wish to access content directly, it is desirable to provide a means of doing so. The NCBI web site, for example, allows a URL to return the contents of a Genbank or PubMed record as pure XML. We realized that there would be significant benefits to emulating NCBI's example.
EDSP: Design Principles
In designing the XML specification, however, it was important to avoid a well-known pitfall. Many XML-based data interchange protocols hard code the details of domain knowledge as XML tags. Such approaches are inflexible: every time the domain changes, such protocols must be respecified: in turn, any programs using these protocols must be rewritten. A durable specification must be generic and capable of describing any domain in a standardized way. It must also be reasonably simple to understand: an overly complex specification is unlikely to be used by anyone beyond its authors.
The XML-based protocol that we have devised to represent both EAV/CR metadata and data is called the EAV/CR dataset protocol (EDSP). EDSP acts as the underlying lingua franca for data transfer between the various EAV/CR software components. It makes possible tasks such as programmatically generating a complete EAV/CR meta-schema and then fully populating the contents of an EAV/CR database from the contents of an existing relational schema. The current version of a previously described tool for importing data into EAV/CR13 uses EDSP, as does AutoPop,15 an application that populates SenseLab from the contents of online data sources such as electronic journals. Import can occur asynchronously as a background process: validation of the data based on the data type and other constraints defined in the metadata is performed during the import process, and a messaging system is used to notify the user about dataset acceptance or rejection.
The reverse process—creating a fully populated, conventional relational database from an EAV/CR system—is performed using an XSLT. Basically, the system initially builds the data represented in EDSP: this is later transformed into other interoperable formats. One of these formats is the Microsoft Office data format; an XML-derived protocol based on the XML data reduced schema used by Microsoft Office XP. This transformed EDSP information permits the dynamic generation of a fully populated multitable database, with intertable relationships preset.
The flavor of EDSP is described in the Appendix, through an actual example that describes the details of a particular mouse olfactory receptor in SenseLab. More details on EDSP are provided at <http://senselab.med.yale.edu/senselab/site/dsArch/edsp.htm>. Here, we summarize its essential aspects. An EDSP stream consists of a metadata section, which is a counterpart of the portal, class, and attribute definitions, followed by the data, which describes each object in the stream. The details of an object include basic identification information (Object identifier, Name, Description, Class) followed by a sequence of attribute–value pairs, in which the attribute identifiers point to attribute definitions in the metadata section. A distinguishing feature of the protocol is that nowhere in the structure are class and attribute definitions encoded as schema elements: instead, they are treated as data. The generic nature of this protocol addresses the “moving schema” problem.
The “metadata” part of EDSP has been influenced by XML Schema Definition Language (XSD),16 which allows representation of data types, structure, and contents of XML documents. It differs from XSD, however, in that EDSP metadata also deal with presentation information: for example, an EDSP stream may record a particular attribute containing image data and must be displayed inline in a region of 200 × 150 pixels. XSD, by contrast, is concerned strictly with metadata that addresses data interchange. Further, certain metadata such as formula attribute definitions have no counterpart in XSD, which has relatively limited data validation.
Support of Semantic Descriptors and Links To Controlled Vocabularies
Relational database management systems support relationships between tables in the form of primary-key/foreign-key links. Typically, however, they do not support storage of textual annotations that describe the semantics of such links. Such descriptors, part of Chen's original Entity-Relationship modeling approach17 as well as more recent approaches such as Terry Halpin's Object Role Modeling,18 must be stored externally in the modeling tools that were used to design the database. The EAV/CR framework facilitates metadata documentation by supporting recording of semantic descriptors. To aid standardization and reuse, these are treated as a miniature controlled vocabulary. For the biomedical domain, the descriptors already defined in the Unified Medical Language System (UMLS)19 and the Systematic Nomenclature (SNOMED) comprise a useful starting point, but we have created new descriptors for neurobiology and bibliography support.
As in UMLS/SNOMED, every descriptor has a “semantic inverse” descriptor. For example, a pair of neuroanatomical structures may have the relationship “is part of” (when used to describe the smaller structure that lies within the larger): this has the semantic inverse “has-part” (when referring to the larger structure as the subject). We are exploring the use of semantic relationships to facilitate user navigation within the ad hoc query interface by making connections between classes more intelligible: users tend to think of such connections in terms of the semantics of the domain rather than in terms of “primary keys” or “foreign keys” (which are terms meaningful only to the database developer).
Both metadata elements (classes and attributes) as well as objects are mapped, where possible, to UMLS concept identifiers. Currently, however, UMLS coverage of basic neuroscience is very sparse, and relatively few elements actually map to UMLS concepts. Apart from simple concepts such as “receptors” and neurotransmitter molecules, the only subset of elements mapping significantly are anatomical structures. Douglas Bowden's neuroanatomy database, BrainInfo <braininfo.rprc.washington.edu>, formerly called NeuroNames,20 also uses mapping to UMLS concept identifiers (BrainInfo is one of UMLS's source vocabularies), and in a collaboration with this group, we have been able to demonstrate interoperability between our Web sites, using UMLS concept identifiers as the lingua franca. SenseLab links dynamically to BrainInfo for detailed anatomical/histological information, whereas BrainInfo can link dynamically to SenseLab for detailed neurophysiology information or models for particular brain structures. If several neuroscience groups collaborate to create a rich controlled vocabulary that can later be incorporated into UMLS, such a mechanism of interoperability holds great potential.
The URL <http://senselab.med.yale.edu/senselab/site/dbMeta/ontology.asp> has additional details on SenseLab's mapping to UMLS concept and structurally associated semantic descriptors.
Discussion
The contribution of this report is in showing how metadata can be used as the basis for generating custom interfaces to subsets of data within a large repository that are tailored to specific groups of users and in the use of a simple XML-based protocol that is the direct analog of the EAV data structure, which is stable to changes in the conceptual schema. Although software is available to create organizational portals in the business world (e.g., Oracle Portal), it does not address our specific need to seamlessly propagate attribute and class visibility/accessibility rules into the user interface. The building blocks used to implement database access with such software are relatively static and not readily made metadata-aware. Such limitations are independent of the fact that these tools would not, in any case, work when the data model is based on an EAV representation.
EAV/CR framework has evolved continuously based on the needs of diverse neuroscience users. We now compare it with a variety of existing systems, in particular, EAV systems in which use is firmly established with respect to clinical medicine. We make these comparisons from multiple viewpoints: rationale of approach, underlying architecture, and the extent of existing functionality.
Comparison with EAV-structured Clinical Systems
The motivation behind the “EAV” of the EAV/CR model when applied to bioscience domains is quite different from that of the EAV model as used in clinical patient record systems (CPRSs).
In CPRSs, thousands of clinical attributes across all medical specialties—laboratory tests, clinical findings, and questionnaire items—are potentially applicable to a patient. Relatively few apply, however, to a given patient at a given time. In other words, the data here are highly sparse.
Sparsity, however, is not a particularly distinguishing characteristic of most classes of data encountered in basic bioscience research. For most classes, the attributes that are applicable to a given class are predetermined and few. Further, this set of attributes generally is applicable to all objects in that class; that is, few, if any, attributes are null for a given object. Classes, however, tend to be very numerous (and possibly complex in structure), whereas the actual “records” for many classes are relatively few. Neurotransmitters, ion channels, or trace elements are a few dozen; receptor molecules are a few hundred. Rather than materializing every class as a relational table, an EAV storage approach allows simulation of a highly complex conceptual schema using a relatively simple physical schema.
When using a schema in which s significant proportion of classes are represented as conventional tables, an advantage of such simulation is that the physical schema's E-R diagram becomes significantly more understandable. In a schema with hundreds of tables, it often is hard to differentiate visually between tables destined to contain thousands to millions of rows of operational data from the far more numerous “lookup” tables that each contain at most a couple of dozen values, whose sole purpose is to support interactive data entry and validation of particular fields, and that serve in effect as miniature controlled vocabularies. (Some experienced database designers follow a table-naming discipline, such as using a prefix like “tlu_” for lookup tables, but we know at least one large public genomic database that did not follow such conventions.) Lookup tables are ideal candidates for conversion to EAV form, so that we now have a hybrid schema. An important benefit here is that a very easy-to-build generic interface can support maintenance of the lookup contents by nonprogrammer administrators.
Another major difference between the metadata models of EAV/CR and those of many existing EAV-structured CPRSs is that the latter are, in most cases, designed to serve as repositories that receive data from other operational systems that end users typically interact with. As such, they are not concerned with user interface issues. In EAV/CR, by contrast, detailed information about how data elements are to be presented and edited is an integral part of the metadata, because the metadata are used to generate Web-enabled browsing/editing interfaces, as previously described.21
Compared with the HELP and the Columbia-Presbyterian CPRSs, SenseLab's integration with controlled vocabularies is much more limited. (The latter CPRS, in particular, is supported by a large vocabulary called the Medical Entities Dictionary.22) This limitation, as discussed previously, is partly a function of limited domain coverage by existing biomedical vocabularies. While our own group could have initiated a similar effort, this would be a long-term project requiring considerable resources. Further, neuroscience is a vast field, whereas our group's domain expertise (as reflected in the focus of SenseLab) is primarily in the olfactory system. Any controlled-vocabulary effort in neuroscience will require a consortium of authorities in various subspecialties, and it is not currently clear whether the Human Brain Project will regard vocabulary development as a high-priority initiative for the future.
Existing CPRSs also use metadata extensively to simulate user–role–specific views. As stated previously, relational database systems offer little help in this matter when all classes of data are stored in the same set of physical tables. The major enhancement in the EAV/CR approach is in using the metadata to generate “portal” user interfaces that (at the end-user level) have the goal, not so much of security, as of presenting the information that is most relevant to the typical user who is interested in a specific type of data.
Use of XML: Comparison with Similar Efforts
The use of XML as the basis for communications protocols in biomedicine has been steadily increasing, most notably with the efforts to support HL-7 messaging.23 Huff et al.24 of the HELP group have previously explored storing patient data in the form of a “denormalized” Abstract Syntax Notation-1 (ASN.1) structure for fast access to complete data for a single patient24: efforts to migrate this to XML, if undertaken, have not been reported. (ASN.1 and XML have similar purposes: ASN.1 is more compact than XML, but transmission errors are harder to localize accurately because, unlike XML, it is not “fully bracketed” with distinct closing tags that match opening tags: it uses general-purpose delimiters such as curly braces to indicating structural nesting.)
The EDSP protocol, described earlier, has a simple and self-describing structure that captures the essential flavor of entity-attribute-value information directly. It addresses the “moving schema” problem by avoiding hard coding of class or attribute names within the XML. Scientific consortia that use XML to interchange data between research groups and a central data repository could usefully borrow from this approach. The requirements as to what information the repository needs to hold often change frequently, in part, because of scientific advances. In this circumstance, continuously devising new XML tags greatly increases the burden on the research groups' programmers: the authors know of at least one consortium where research groups rose in revolt after the repository team changed the XML specifications once too often.
In the neuroinformatics field, there are at least two XML-based efforts in progress.
NeuroML25 is intended as an exchange format for different neural modeling programs, such as Neuron26 and Genesis.27 The NeuroML specification, which still is evolving, necessarily includes tags specific to the domain of neural simulation. Although we have not yet attempted this, it would not be too difficult to transform EDSP metadata and data that apply to neuronal models into NeuroML using XSLT. The availability of Dr. Michael Hines, the creator of Neuron (which is currently the most widely used simulation package) as part of the SenseLab team, would help us greatly in this matter.
Gardner et al.12 at Cornell have proposed a standard called Biophysical Description Markup Language to support their Common Data Model for neuroscience data. Although this group's recent AMIA paper12 described the flavor of BDML briefly, BDML appears to be a work in progress: no formal description, in the form of a Document Type Definition (DTD) or otherwise, is currently available on this group's Web site. Further, the published example is too brief to allow a comparison of EDSP with BDML: it is not clear, for example, whether BDML tags are generic.
Limitations and Future Directions
EAV/CR remains a continual work in progress; planned high-priority enhancements are discussed below.
Changing attribute datatypes once data have been entered: Currently, attributes' datatypes can be changed easily if no data have been populated into the attribute. However, because values are stored in datatype-specific tables, changing an attribute's data type when data already exist is not a trivial operation. Numerous steps must be performed to prevent data loss or inconsistencies that could render the data useless. The manual process to do this is highly error-prone and can benefit from automation: this requires careful transactional processes involving both EAV/CR metadata and data changes. Some changes are necessary to deal with concurrency issues involving the affected classes while serving concurrent data requests and keeping the downtime close to zero.
Robust Metadata Searching: When multiple curators manage a large conceptual schema, there is a risk of multiple classes with near-identical functionality being defined redundantly. Satisfactory curation, in this situation, implies avoiding duplication by reusing or extending existing classes. For this to happen, however, the class/attribute metadata must be intuitively searchable, so that classes with the desired functionality (or close to it) are identified if they exist. Searching by class/attribute name alone is unsatisfactory because of the well-known synonym problem in scientific domains. Mapping to concepts and semantic types, and supporting search based on the same, can help greatly in this regard.
Supporting other interoperating formats: As stated earlier, we support export of metadata and data into Microsoft's XML data reduced (XDR) schema, to allow use of productivity tools such as the Microsoft Office packages. We are planning to support other interchange formats, such as RDF, which would allow interchange of metadata with packages that use RDF. Concurrent with this, we are considering whether to make the next version of EDSP tag-compatible with XSD for those elements that have the same purpose (e.g., datatype, maximum field length, minimum and maximum values). This would simplify the learning curve for XML developers who are already familiar with XML Schemas.
Conclusions
The creation of evolvable bioinformatics applications implies the use of application frameworks in which generic code is driven off the contents of metadata and is scalable. The enhancements that we have described in the EAV/CR framework and metadata have facilitated the provision of additional functionality in SenseLab. The “portal” mechanism lets individual categories of users deal with complexity by focusing only on the subset of data that is of direct interest to them. By using the XML-based EDSP protocol as the foundation for all communication between EAV/CR components, we also facilitate interchange between EAV/CR systems and external applications.
Appendix: A Simplified Example Illustrating the EDSP Protocol
In this appendix, the XML has been greatly shortened to facilitate explanation and avoid overwhelming the reader with less important minutiae.
An EDSP stream consists of two parts: metadata and data.
EDSP: Metadata
The first part of the XML stream consists of metadata for the olfactory receptor class. This is derived in a straightforward manner from the relationally structured metadata. It consists of a class element, which in turn contains a list of att (attribute) elements. The XML is now shown, followed by a detailed explanation.
<class id=“c22” name=“olfactory_receptors” version=“1” version_date=“11/20/2001”/>
<att id=“a30” datatype=“C” name=“organism” ref_class_name=“organism”/>
<att id=“a106” datatype=“S” name=“strain” width=“20”/>
<att id=“a31” datatype=“C” name=“source_tissue” ref_class_name=“tissue”/>
<att id=“a45” datatype=“S” name=“Genbank_Accession_Number” width=“20”/>
<att id=“a44” datatype=“S” name=“PubMed_ID” width=“20”/>
<att id=“a36” datatype=“C” name=“Source” ref_class_name=“labs”/>
<att id=“a82” datatype=“C” name=“Ligand” ref_class_name=“odors” multiple_instance=“True”/>
<att id=“a40” datatype=“M” name=“nucleotides”/>
<att id=“a41” datatype=“M” name=“amino_acids”/>
<att id=“a42” datatype=“C” name=“Type_of_Sequence”ref_class_name=“type_of_sequence”/>
…
</class>
A class is described by an id, name, and version information. The id (“c22”) is generated by prefixing “c” to the class's machine-generated unique identifier. The identifier is optional: when using EDSP to create a class definition in an EAV/CR schema, it need not be supplied, because the database engine autogenerates identifiers when the class's metadata are created.
- An att element is now described.
- Attribute identifiers are generated by prefixing an “a” to the attribute unique identifier. As for class identifiers, these are optional and are autocreated when importing a definition.
- The datatype of an attribute is a single letter: e.g., C = class reference, S = string, M = memo (long text).
- For class references, which simulate foreign keys in conventional database design, the name of the class pointed to has been shown in the ref_class_name attribute. For example, the attribute “source_tissue” points to a class name called “tissue.”
- For string attributes, the width attribute specifies the maximum string length.
- The multiple_instance attribute (for the attribute “a82” – Ligand) allows multiple instances of the attribute to exist. In this example, a receptor can bind to multiple olfactory ligand molecules.
EDSP: Data
The metadata definitions are followed by data for one or more receptors. The description of each receptor consists of an object element (the “entity” of EAV), which encloses several att_value elements (the attribute-value pairs of EAV).
<object id=“o1798” class=“c22” name=“ORL464” version=“1” version_date=“6/30/2001 3:08:27 PM”>
<att_value att_id=“a30” value=“o144” value_object_name=“Mus musculus (mouse)”/>
<att_value att_id=“a106” value=“BALB/c”/>
<att_value att_id=“a31” value=“o145” value_object_name=“olfactory receptor neuron”/>
<att_value att_id=“a45” value=“AF121975”/>
<att_value att_id=“a44” value=“99189757”/>
<att_value att_id=“a36” value=“o835” value_object_name=“GENBANK”/>
<att_value att_id=“a82” value=“o123” serial_no=“1” value_object_name=“NONANOL”/>
<att_value att_id=“a82” value=“o1779” serial_no=“2” value_object_name=“n-HEPTANOIC ACID”/>
<att_value att_id=“a82” value=“o1780” serial_no=“3” value_object_name=“OCTANOIC ACID”/>
<att_value att_id=“a40” value=“CAAGCTGGCTCTTC…”/>
<att_value att_id=“a41” value=“MNSK…”/>
<att_value att_id=“a42” value=“o842” value_object_name=“cDNA”/>
…
</object>
Every object has a unique identifier, a class to which it belongs, a name, and version information.
- An att_value element is now described.
- Each element is described by an “att_id” and a “value.”
- The “att-id” attributes must be valid cross references to “att” elements in the metadata.
- For class reference attributes, the name of the entity in the class pointed to is shown. In the example, the attribute “a30” (organism) refers to “Mus musculus” (the mouse). The entity name is not required when importing data, but it is output during data export, to facilitate visual inspection.
- This receptor binds to three ligands (attribute “a82”): Nonanol, n-Heptanoic acid and Octanoic acid—this is an example of a multiple-instance attribute.
- “Binary” information, such as a GIF image (not applicable to the present object) is represented in XML by encoding in Base-64.
- The details of the nucleotide and amino acid sequences have been elided for reasons of space.
Notice that both the metadata and the data have a completely generic structure, which addresses the “moving schema” problem. Nowhere in the structure are class and attribute definitions encoded as schema elements; instead, they are treated as data.
This work was supported by NIH grants P01 DC04732, G08 LM05583, and U01 ES10867. The software described in this article will be provided freely on request to Dr. Marenco <luis.marenco@yale.edu>.
References
- 1.Marenco L, Nadkarni P, Skoufos E, Shepherd G, Miller P. Neuronal database integration: the Senselab EAV data model. Proc AMIA Symp. 1999:102–6. [PMC free article] [PubMed]
- 2.Nadkarni PM, Marenco L, Chen R, Skoufos E, Shepherd G, Miller P. Organization of heterogeneous scientific data using the EAV/CR representation. J Am Med Inform Assoc. 1999;6:478–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shepherd G, Mirsky JS, Healy MD, et al. The Human Brain Project: neuroinformatics tools for integrating, searching and modeling multidisciplinary neuroscience data. Trends Neurosci. 1998;21:460–8. [DOI] [PubMed] [Google Scholar]
- 4.Koslow S, Huerta M. Neuroinformatics: An Overview of the Human Brain Projects. Mahwah, NJ: Lawrence Erlbaum Associates, 1997.
- 5.Huff SM, Berthelsen CL, Pryor TA, Dudley AS. Evaluation of a SQL model of the help patient database. Proc Symp Comput Appl Med Care. 1991:386–90. [PMC free article] [PubMed]
- 6.Huff SM, Haug DJ, Stevens LE, Dupont CC, Pryor TA. HELP the next generation: a new client-server architecture. Proc Symp Comput Appl Med Care. 1994:271–5. [PMC free article] [PubMed]
- 7.Friedman C, Hripcsak G, Johnson S, Cimino J, Clayton P. A generalized relational schema for an integrated clinical patient database. Proc Symp Comput Appl Med Care. 1990:335–9.
- 8.Johnson S, Cimino J, Friedman C, Hripcsak G, Clayton P. Using metadata to integrate medical knowledge in a clinical information system. Proc Symp Comput Appl Med Care. 1990:340–4.
- 9.Nadkarni PM, Brandt C, Frawley S, et al. Managing attribute-value clinical trials data using the ACT/DB client-server database system. J Am Med Inform Assoc. 1998;5:139–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shepherd GM, Healy MD, Singer MS, et al. Senselab: a project in multidisciplinary, multilevel sensory integration. In Koslow SH, Huerta MF (eds). Neuroinformatics: An Overview of the Human Brain Project. Mahwah, NJ: Lawrence Erlbaum Associates, 1997, pp 21–56.
- 11.Weyl S, Fries J, Wiederhold G, Germano F. A modular self-describing clinical databank system. Comput Biomed Res. 1975;8:279–93. [DOI] [PubMed] [Google Scholar]
- 12.Gardner D, Knuth K, Abato M, et al. Common data model for neuroscience data and data model exchange. J Am Med Inform Assoc. 2001;8:17–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cagle K. SVG Programming: the Graphical Web. Berkeley, CA: APress, 2002.
- 14.World Wide Web Consortium. XSL Transformations (XSLT) version 1.0. 2002. <http://www.w3.org/TR/xslt>. Accessed November 1, 2002.
- 15.Crasto C, Marenco L, Miller P, Shepherd G. Olfactory Receptor Database: a metadata-driven automated population from sources of gene and protein sequences. Nucleic Acids Res. 2002;30:354–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.World Wide Web Consortium. XML Schema. 2001. <http://www.w3.org/XML/Schema>. Accessed February 20, 2002.
- 17.Chen PP. The Entity-Relationship Model: Toward a Unified View of Data. Assoc Comput Machinery Trans Database Syst. 1976;1:9–36. [Google Scholar]
- 18.Halpin T. Information Modeling and Relational Databases: From Conceptual Analysis to Logical Design. San Francisco, CA: Morgan Kaufman Publishers, 2001.
- 19.Lindberg DAB, Humphreys BL, McCray AT. The Unified Medical Language System. Methods Inf Med. 1993;32:281–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bowden D, Martin R. NeuroNames Brain Hierarchy. Neuroimage. 1995;2(1):63–83. [DOI] [PubMed] [Google Scholar]
- 21.Nadkarni PM, Brandt CA, Marenco L. WebEAV: Automatic metadata-driven generation of web Interfaces to entity-attribute-value databases. J Am Med Inform Assoc. 2000;7:343–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cimino JJ, Hripcsak G, Johnson SB, Clayton PD. Designing an introspective, multipurpose, controlled medical vocabulary. Proc Symp Comput Appl Med Care. 1989:513–8.
- 23.Messaritakis H. The HL7 XML Web site, 2003. <http://puck.informatik.med.uni-giessen.de/people/messaritakis/hl7xml/>. Accessed February 1, 2003.
- 24.Huff SM, Rocha RA, Solbrig HR, Barnes MW, Schrank SP, Smith M. Linking a medical vocabulary to a clinical data model using Abstract Syntax Notation 1. Methods Inf Med. 1998;37:440–52. [PubMed] [Google Scholar]
- 25.Howell F. NeuroML home page, 2003. <www.neuroml.org>. Accessed February 1, 2003.
- 26.Hines ML, Carnevale NT. The NEURON simulation environment. Neural Computation. 1997;9:1179–209. [DOI] [PubMed] [Google Scholar]
- 27.Bower JM, Beeman D. The Book of Genesis. New York: Springer-Verlag, 1995.