Abstract
Adenine nucleotides have been found to appear preferentially in the regions after the initiation codons or before the termination codons of bacterial genes. Our previous experiments showed that AAA and AAT, the two most frequent second codons in Escherichia coli, significantly enhance translation efficiency. To determine whether such a characteristic feature of base frequencies exists in eukaryote genes, we performed a comparative analysis of the base biases at the gene terminal portions using the proteomes of seven eukaryotes. Here we show that the base appearance at the codon third positions of gene terminal regions is highly biased in eukaryote genomes, although the codon third positions are almost free from amino acid preference. The bias changes depending on its position in a gene, and is characteristic of each species. We also found that bias is most outstanding at the second codon, the codon after the initiation codon. NCN is preferred in every genome; in particular, GCG is strongly favored in human and plant genes. The presence of the bias implies that the base sequences at the second codon affect translation efficiency in eukaryotes as well as bacteria.
INTRODUCTION
In bacterial genomes there is a well-characterized sequence element called the Shine–Dalgarno (SD) sequence, which affects the initiation step of translation. The SD sequence is a polypurine stretch, such as AGGAGGU, and is found ∼3–10 nt upstream of the initiation codon in bacterial genes (1). It interacts with the complementary sequence at the 3′-end of the 16S rRNA by base pairing, which promotes attachment of the 30S ribosomal subunit to the mRNA (2). Besides the SD sequence, the second codon, the codon immediately after the initiation codon, is reported to affect the efficiency of translation (3–5). Recently we examined the effect of the second codon on translation efficiency by introducing mutations in a monitor gene of Escherichia coli (4). In that paper, we showed that AAA and AAT, the two most frequent second codons in E.coli, significantly enhanced translation efficiency compared with the wild-type, whereas the effects of other codons, including CTG, the most frequent codon throughout the E.coli genome, were not significant. Statistical studies using the whole genome sequences of several bacteria revealed characteristic base biases surrounding the initiation codon or the termination codon, such as A-rich biases present in both 5′- and 3′-terminal portions of open reading frames (6). Therefore, the preference for A-rich codons at the second codon is common to several bacterial species. Some other elements, such as the downstream box (7) or CA repeats (8), are also reported to enhance translation in E.coli. Moreover, the base following the termination codon is biased in some bacterial genomes (6) and it has been shown that the identity of this base determines the efficiency of translation termination in E.coli (9).
In the case of eukaryotes, the mechanism of translation initiation is quite different from bacteria. According to the scanning model proposed by Kozak (10; for a review, see 11), first, the preinitiation complex comprising the 40S ribosomal subunit, the initiator tRNA and some initiation factors attaches to the 5′-end of the mRNA, then the complex begins scanning along the mRNA until it reaches the initiation codon. The initiation codon is recognizable when it is within the Kozak consensus sequence, GCCACCaugG (aug represents the initiation codon) (12). It has been shown that mutations affecting the A nucleotide at position –3 (three bases before the initiation codon), the most highly conserved position in the consensus sequence, strongly impair translation initiation in vivo and in vitro (11). A strong contribution of G at the position next to the initiation codon was also confirmed in experiments with several species (11). However, nothing is known about the mechanism that propels the scanning complex, and how the consensus sequence is recognized and how it functions are not yet known (11). Therefore, although the Kozak model is widely accepted, the precise mechanism of translation initiation in eukaryote cells is still unclear.
Watanabe et al. performed a statistical analysis of the base biases around the initiation and termination codons using all genes in the genome of Saccharomyces cerevisiae (6). Following S.cerevisiae (13), the whole genome sequences of various eukaryotes have been published: Caenorhabditis elegans (14), Drosophila melanogaster (15), Arabidopsis thaliana (16), Homo sapiens (17) and Schizosaccharomyces pombe (18). The purpose of this study is to elucidate unknown sequence elements which affect translation initiation or termination in eukaryote genomes and, especially, to investigate the possibility that the second codon affects translation initiation in eukaryotes. For this purpose, we extensively examined the base biases after the initiation codon or before the termination codon in the genes of seven eukaryotes for which the complete genome sequences are available [for Oryza sativa, all the genes on chromosome 1 (19) were analyzed].
MATERIALS AND METHODS
Data
The sequences of human genes were downloaded from the RefSeq database (http://www.ncbi.nih.gov/RefSeq/). We used only curated entries with an accession prefix NM_ in RefSeq and thus our data set for human genes does not contain any genes predicted by ab initio computation (20). The sequences of the whole proteomes of D.melanogaster and C.elegans were obtained from BDGP database release 2 (http://www.fruitfly.org) and wormpep76 (http://www.sanger.ac.uk/Projects/C_elegans/wormpep/), respectively. The proteomes of A.thaliana, S.cerevisiae, S.pombe and E.coli were retrieved from NCBI Entrez Genomes (http://www.ncbi.nih.gov/Entrez/) (accession nos: NC_003070, NC_003071, NC_003074, NC_003075 and NC_003076 for A.thaliana; NC_001133–NC_001148 for S.cerevisiae; NC_003421, NC_003423 and NC_003424 for S.pombe; NC_000913 for E.coli). The sequences of the genes on O.sativa chromosome 1 were received from Takuji Sasaki (19). The following data were excluded from the gene set of each species: (i) mitochondrial and chloroplast genes; (ii) sequences containing ambiguous bases; (iii) sequences that do not start from an initiation codon or end with a termination codon; (iv) alternative transcripts except the longest one among them; (v) sequences of which the length from the initiation to the termination codon is shorter than 300 bases. Condition (v) is required because we performed analyses up to the 100th codon from the initiation and termination codons. The numbers of genes used for the analyses were: H.sapiens, 12 769; D.melanogaster, 12 580; C.elegans, 18 547; A.thaliana, 24 983; O.sativa, 5830; S.cerevisiae, 6113; S.pombe, 4515; E.coli, 3891.
Evaluation of the biases: the G-test
The G-test (21), or log-likelihood ratio test, was employed for evaluating the deviation of observed distribution of bases or codons from the expected distribution at each position in Figures 1 and 3. In this article, a position in a gene is indicated by a codon number. The initiation codon is numbered 1 and the second codon is codon 2. Codons are also numbered backward from the termination codon using negative numbers; the termination codon is referred to as codon –1 and the codon immediately before the termination codon is numbered –2. All genes for each species in our data set were aligned with the codons from 1 (the initiation codon) to 100 and from –100 to –1 (the termination codon), without gaps. The bias in codon appearance at codon position i (Fig. 1) is evaluated by the G-value, defined as Gi = NΣ(x) 2Oi(x) ln(Oi(x)/E(x)), where N is the number of genes, Oi(x) is the fraction of codon x among 61 codons except the termination codons at codon i, and E(x) is the fraction of codon x calculated from all of the codons in all genes for each species. The bias in base appearance at the jth letter (j = 1, 2 or 3) in codon position i (Fig. 3) is calculated similarly as Gi(j) = NΣ(n) 2Oi(j,n) ln(Oi(j,n)/E(j,n)), where Oi(j,n) is the fraction of nucleotide n (A, T, G or C) at the jth letter in codon i, and E(j,n) is the fraction of nucleotide n at the jth letter in the entire region of all genes for a given species. It has been shown that the distribution of the G-values can be approximated by the χ2 distribution when the sample sizes are large (21). In Figure 3, corresponding probabilities (P) were calculated from the G-values using the χ2 distribution with 3 degrees of freedom. The reason we adopted G-values instead of the traditional χ2 values, χ2 = NΣ{(O – E)2/E}, is that each term in the definition of the G-value directly shows that an observed value is larger or smaller than the expected one, whereas the χ2 value only gives the deviation of the observed value from the expectation (6). We plotted the value G/N instead of G, since this value is free from N, thus it is more useful for a comparison of biases among various species having different numbers of genes.
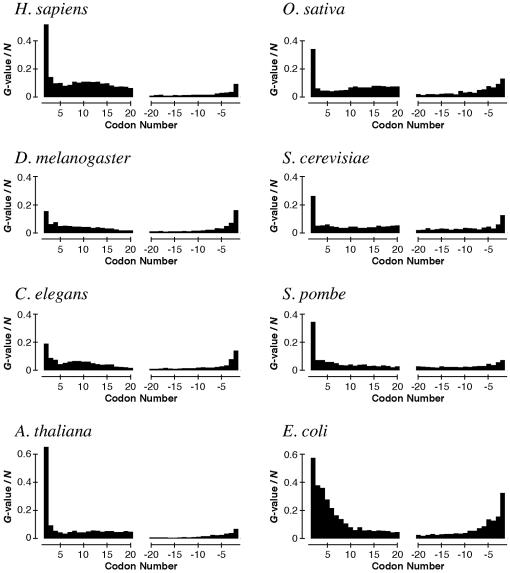
Figure 1.
Biases in codon appearance at each position in genes of seven eukaryotes and E.coli. The biases are represented as the G-values divided by the gene number N (see Materials and Methods). The initiation and the termination codons are omitted, because the G-values at these codons are extremely high.
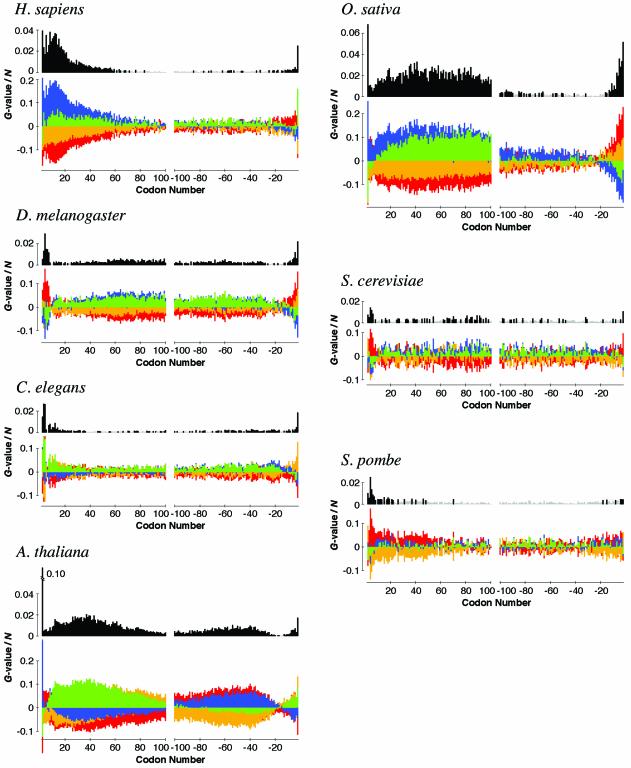
Figure 3.
Biases in base appearance at the third letter in each codon position for seven eukaryote species. In the upper graph, the biases are shown by the G-values divided by the gene number N (see Materials and Methods). Black and light gray bars correspond to probabilities smaller and larger than 0.1% (i.e. G-value = 16.27), respectively. Note that the threshold value of G/N corresponding to P = 0.1% is different from species to species, because N is species-dependent. In the lower graph, the values of the terms corresponding to each base, A (red), T (yellow), G (blue) or C (green), in the definition of the G-value are shown (see Materials and Methods). Positive and negative values are shown in the upper and lower parts of the graph, respectively, without any overlap. The total of four values depicted by four colored bars in the lower graph is equal to the G-value in the upper graph at each position. The initiation and the termination codons are omitted.
Evaluation of the biases: the Z-test
We also performed the Z-test to evaluate the deviation of observed appearance of a particular base, amino acid and codon from the expected appearance, shown in Figure 2 and Tables S1 and S2, available as Supplementary Material. The Z-value is calculated by the formula Z = N(O – E)/[N × E × (1 – E)]1/2, where N is the number of genes, O is the fraction of a particular base, amino acid or codon at a given position and E is the fraction of a particular base, amino acid or codon in the entire region of all genes for each species.
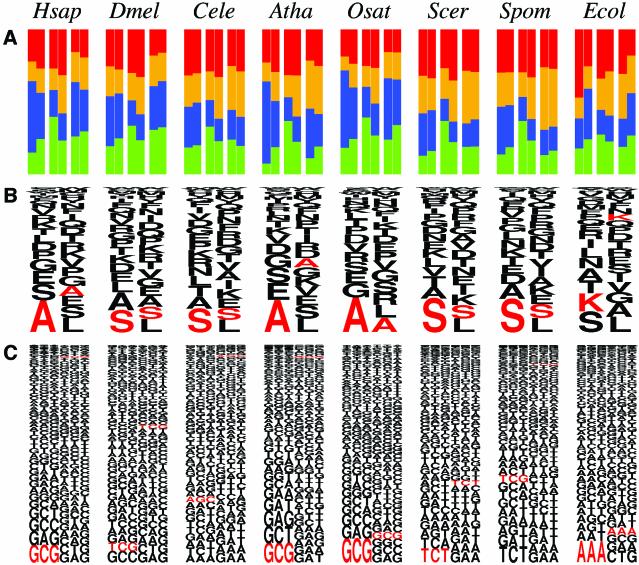
Figure 2.
Biases at the second codon in the appearance of (A) bases, (B) amino acids and (C) codons for seven eukaryote species and E.coli. In (A), the fractions of the bases A (red), T (yellow), G (blue) and C (green) at the first (left), second (middle) and third (right) letters in codons are shown. In each rectangle, the left half represents the fraction of each base at the second codon and the right half represents the fraction of each base in the entire regions of all genes for a given species. In (B), the fraction of each amino acid at the second codon (left) and that in the entire region of all genes for each species (right) are shown. The height of each letter is proportional to the fraction of the amino acid represented by that one letter code. In (C), the fraction of each codon at the second codon (left) and that in the entire region of all genes (right) are shown. The height of each triplet is proportional to the fraction of the codon represented by that triplet. In (B) and (C), the characters are drawn in the order of their fractions from the bottom to the top. The amino acid or codon colored red is the one having the largest Z-value among all 20 amino acids or 61 codons, respectively, showing the most statistically biased amino acid or codon at the second codon for each species. Hsap, H.sapiens; Dmel, D.melanogaster; Cele, C.elegans; Atha, A.thaliana; Osat, O.sativa; Scer, S.cerevisiae; Spom, S.pombe; Ecol, E.coli.
RESULTS
The biased second codons
In order to survey sequence biases at each position of a gene, we first examined biases in codon appearance using genes in the genomes of seven eukaryotes and E.coli (Fig. 1). We found that the second codon is the most biased among all the positions in a gene for every eukaryote examined, as well as E.coli, with the exception of D.melanogaster, for which the bias at codon –2 is slightly higher than at codon 2. The difference in the biases between the second codon and other positions is more apparent for most eukaryotes than for E.coli.
We then performed more detailed analyses of the second codon in the appearance of bases, amino acids and codons (Fig. 2). Figure 2 shows that the patterns of biases at the second codon in eukaryote genes are different from that in E.coli genes. The base at each letter in the second codon is biased for A in E.coli genes (Fig. 2A, rightmost column), as previously reported (4). In contrast, we found that C is highly preferentially used at the second letter in the second codon for every eukaryote examined in this study. The fraction of C at this position is more than 10% greater than the expectation for every species (Table S1). It was also found that G is highly preferred at the first and the third letters in the second codon for humans, A.thaliana and O.sativa. In agreement with these findings, the most frequent and statistically biased second codon for these three species is GCG encoding an alanine (Fig. 2C). In fact, the appearance of GCG at the second codon is more than 10 times larger than the expectation for humans and A.thaliana; the fraction of GCG among all 61 codons is only 0.75 and 0.86% in the entire region of all genes for humans and A.thaliana, respectively, while it appears at 7.94 and 8.98%, respectively, at the second codon. For D.melanogaster, C.elegans, S.cerevisiae and S.pombe, serine-encoding codons are favored as the second codon, although their biases are not outstanding compared with humans or plants.
Position-dependent base biases
We also examined biases in base appearance near the termini of genes, besides the second codon. In order to avoid the effect of amino acid preference, we investigated the bias at the third letter in each codon position (Fig. 3). The biases were evaluated by the G-values, which are known to be approximated by the χ2 distribution when the sample sizes are large (21). We found that base appearance is also highly biased in positions other than the second codon. The patterns of the biases are different from species to species and the bias gradually changes depending on its position. For example, the following features are observed for human genes (Fig. 3 and Table S2). First, as mentioned above, the third letter in the second codon is highly biased for G, the fraction being 9% higher than the expectation. Second, GC-rich biases are found in codons 3 to ∼60 (P < 0.1%), with an intriguing peak at around codon 12. Third, weak but significant A-rich biases are found in the codons from around –15 to –3. Fourth, the third letter in codon –2 is highly C-rich. Arabidopsis thaliana and O.sativa genes also show strong biases in base appearance (Fig. 3 and Table S2).
DISCUSSION
In this article, we found that the base appearance at the codon third position is highly biased at gene terminal positions in eukaryotes. As shown in Tables S1 and S2, the difference from expectation is as much as 10%. The bias changes gradually depending on its position and generates a species-specific pattern. Because the codon third letters hardly affect the amino acid sequence, this observation cannot be explained by biases in amino acid appearance due to, for example, uneven representation of protein families in each proteome. Such biases in base appearance near the initiation or termination codon are possible signals for controlling translation initiation or termination.
We showed that codon appearance is most biased at the second codon among all positions for almost all eukaryotes we examined (Fig. 1), which is consistent with the idea that the second codon affects translation efficiency in eukaryotes. Codon appearance can be affected by amino acid preference, as we discuss below, however, the third letter in the second codon, which is almost free from amino acid preference, is also highly biased for every species (see Fig. 3 and Table S1). The bias for G at the third letter in the second codon for humans, A.thaliana and O.sativa is outstanding. Among four alanine-encoding codons, GCN, the codon GCG is the most minor (10.6%) in human genes; in contrast, the fraction of GCG among the four codons is 35.9% at the second codon. This bias is common to all amino acids: we found that 12 out of 13 non-degenerate G-ending codons are used more frequently than expectation at this position in human genes with statistical significance (P < 5%); the only exception is the codon CGG encoding an arginine, but another G-ending codon for arginine, AGG, is highly preferentially used at this position (data not shown).
The eukaryote species we examined can be divided into two groups from the viewpoint of bias at the second codon: for humans, A.thaliana and O.sativa, GCG is greatly favored as the second codon; in contrast, for D.melanogaster, C.elegans, S.cerevisiae and S.pombe, serine-encoding codons appear frequently at this position. This observation is partially inconsistent with the Kozak consensus sequence, GCC ACCatgG, because G is not necessarily favored as the first letter in the second codon for the species in the latter group, e.g. for C.elegans the fraction of G at the position (28.3%) is lower than expectation (29.5%) (Table S1). This inconsistency can be explained by the fact that the compiled genes used to obtain the consensus sequence were biased for mammals (12).
As for the biases at the first and the second letters in the second codon, we cannot distinguish the requirement for translational efficiency from a functional constraint for specific amino acids. A bias for C at the second letter in the second codon has already been reported for S.cerevisiae, and was explained by the specificity of the enzyme methionine aminopeptidase (MetAP) (6). It is known that MetAP is responsible for cleaving the initiator methionine from nascent proteins only when the penultimate residue is one of the seven smallest amino acids, glycine, alanine, serine, threonine, proline, valine or cysteine (22), and this specificity is widely conserved in prokaryotes and eukaryotes (23). Four of these amino acids are encoded by NCN codons. Therefore, the requirement for methionine removal would make the base at this position biased for C.
The strong effect of G at the first letter in the second codon on the efficiency of translation initiation has been shown in experiments with mammals and plants, however, the effect is small in S.cerevisiae (11). This is totally consistent with our observations. Our observation further suggests that the nucleotide G at the third letter also contributes to translation. Kozak reported that the third letter in the second codon does not affect recognition of the initiation codon in a rabbit reticulocyte translation system (24). However, this is not necessarily inconsistent with our suggestion, for the following reasons: first, Kozak’s experiments were performed under suboptimal conditions in which leaky scanning should occur; second, she conducted the experiments for a limited number of codons, such as CCN, which is not preferred as the second codon. In the case of E.coli, it has been experimentally shown that all of the three bases in the second codon influence translation efficiency (4,5). Therefore, it is plausible that the nucleotide at the third (and possibly the second) letter, as well as the first letter, affects translation initiation in some eukaryotes, such as human and plants. In addition, quite strong biases observed at codon –2 preceding the termination codon (Fig. 1) imply that the bases at this position could affect translation termination.
We found that base appearance at the third letter is also significantly biased in positions other than the second codon. Then what is the cause of such position-dependent base biases? For human genes, strong GC-rich biases were found until codon ∼60 (Fig. 3). This finding can be plausibly explained in the following way. It is known that CpG dinucleotides are under-represented in vertebrate genomes, but there are small stretches of DNA having the expected frequency of CpG, called CpG islands, which are thought to be related to the control of gene expression. For almost all housekeeping genes and many tissue-specific genes, associated CpG islands are found at the 5′-ends of the gene (25). Therefore, the 5′-terminal portions of genes are expected to be relatively GC-rich. However, GC- or C-rich biases are also observed for D.melanogaster, A.thaliana and O.sativa in regions beyond codon 100 (Fig. 3). The presence of CpG islands in the genomes of these species has not been reported, thus, the cause of these biases is elusive. The strong base biases in this region imply that such species would have unknown mechanisms to control gene expression.
We detected A-rich biases in exactly the same regions, codons 3–7, in the genes of D.melanogaster, A.thaliana, S.cerevisiae and S.pombe (Fig. 3). In human or O.sativa genes, codons 3–7 are relatively A-rich compared with surrounding regions, because the negative bias against A fades in these positions, whereas the negative bias against T does not (Fig. 3). Several codons preceding the termination codon are also biased for A in the genes of humans, D.melanogaster and O.sativa (Fig. 3). Because A-rich biases in both termini of genes are also observed in bacteria (6), there could exist some general mechanism to explain this feature. As an explanation for the preference for A near the initiation codon in E.coli, Eyre-Walker and Bulmer proposed selection against the formation of secondary structures in mRNA, which would interfere with binding of the ribosome near the start of a gene (26), although it does not explain the bias preceding the termination codon. The patterns of position-dependent base biases in eukaryotes seem to be decomposed into several distinct biases, implying that there are different types of sequence elements which control gene translation in eukaryote genomes.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We thank H. Akashi, L. Hao, H. Iwama, J. Nam and Y. Suzuki for valuable comments and discussions. Y.N. is financially supported by the Japan Society for the Promotion of Science.
REFERENCES
- 1.Shine J. and Dalgarno,L. (1974) The 3′-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc. Natl Acad. Sci. USA, 71, 1342–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Steitz J.A. and Jakes,K. (1975) How ribosomes select initiator regions in mRNA: base pair formation between the 3′ terminus of 16S rRNA and the mRNA during initiation of protein synthesis in Escherichia coli. Proc. Natl Acad. Sci. USA, 72, 4734–4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Looman A.C., Bodlaender,J., Comstock,L.J., Eaton,D., Jhurani,P., de Boer,H.A. and van Knippenberg,P.H. (1987) Influence of the codon following the AUG initiation codon on the expression of a modified lacZ gene in Escherichia coli. EMBO J., 6, 2489–2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sato T., Terabe,M., Watanabe,H., Gojobori,T., Hori-Takemoto,C. and Miura,K. (2001) Codon and base biases after the initiation codon of the open reading frames in the Escherichia coli genome and their influence on the translation efficiency. J. Biochem., 129, 851–860. [DOI] [PubMed] [Google Scholar]
- 5.Stenström C.M., Jin,H., Major,L.L., Tate,W.P. and Isaksson,L.A. (2001) Codon bias at the 3′-side of the initiation codon is correlated with translation initiation efficiency in Escherichia coli. Gene, 263, 273–284. [DOI] [PubMed] [Google Scholar]
- 6.Watanabe H., Gojobori,T. and Miura,K. (1997) Bacterial features in the genome of Methanococcus jannaschii in terms of gene composition and biased base composition in ORFs and their surrounding regions. Gene, 205, 7–18. [DOI] [PubMed] [Google Scholar]
- 7.Sprengart M.L., Fuchs,E. and Porter,A.G. (1996) The downstream box: an efficient and independent translation initiation signal in Escherichia coli. EMBO J., 15, 665–674. [PMC free article] [PubMed] [Google Scholar]
- 8.Martin-Farmer J. and Janssen,G.R. (1999) A downstream CA repeat sequence increases translation from leadered and unleadered mRNA in Escherichia coli. Mol. Microbiol., 31, 1025–1038. [DOI] [PubMed] [Google Scholar]
- 9.Poole E.S., Brown,C.M. and Tate,W.P. (1995) The identity of the base following the stop codon determines the efficiency of in vivo translational termination in Escherichia coli. EMBO J., 14, 151–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kozak M. (1978) How do eucaryotic ribosomes select initiation regions in messenger RNA? Cell, 15, 1109–1123. [DOI] [PubMed] [Google Scholar]
- 11.Kozak M. (1999) Initiation of translation in prokaryotes and eukaryotes. Gene, 234, 187–208. [DOI] [PubMed] [Google Scholar]
- 12.Kozak M. (1984) Compilation and analysis of sequences upstream from the translational start site in eukaryotic mRNAs. Nucleic Acids Res., 12, 857–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goffeau A., Aert,R., Agostini-Carbone,M.L., Ahmed,A., Aigle,M., Alberghina,L., Albermann,K., Albers,M., Aldea,M., Alexandraki,D. et al. (1997) The yeast genome directory. Nature, 387 (suppl.), 1–105. [Google Scholar]
- 14.The C. elegans Sequencing Consortium (1998) Genome sequence of the nematode C. elegans: a platform for investigating biology. Science, 282, 2012–2018. [DOI] [PubMed] [Google Scholar]
- 15.Adams M.D., Celniker,S.E., Holt,R.A., Evans,C.A., Gocayne,J.D., Amanatides,P.G., Scherer,S.E., Li,P.W., Hoskins,R.A., Galle,R.F. et al. (2000) The genome sequence of Drosophila melanogaster. Science, 287, 2185–2195. [DOI] [PubMed] [Google Scholar]
- 16.The Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature, 408, 796–815. [DOI] [PubMed] [Google Scholar]
- 17.International Human Genome Sequencing Consortium (2001) Initial sequencing and analysis of the human genome. Nature, 409, 860–921. [DOI] [PubMed] [Google Scholar]
- 18.Wood V., Gwilliam,R., Rajandream,M.A., Lyne,M., Lyne,R., Stewart,A., Sgouros,J., Peat,N., Hayles,J., Baker,S. et al. (2002) The genome sequence of Schizosaccharomyces pombe. Nature, 415, 871–880. [DOI] [PubMed] [Google Scholar]
- 19.Sasaki T., Matsumoto,T., Yamamoto,K., Sakata,K., Baba,T., Katayose,Y., Wu,J., Niimura,Y., Cheng,Z., Nagamura,Y. et al. (2002) The genome sequence and structure of rice chromosome 1. Nature, 420, 312–316. [DOI] [PubMed] [Google Scholar]
- 20.Pruitt K.D. and Maglott,D.R. (2001) RefSeq and LocusLink: NCBI gene-centered resources. Nucleic Acids Res., 29, 137–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sokal R.R. and Rohlf,F.J. (1993) Biometry, 3rd Edn. W.H.Freeman and Co., New York, NY. [Google Scholar]
- 22.Huang S., Elliott,R.C., Liu,P.S., Koduri,R.K., Weickmann,J.L., Lee,J.H., Blair,L.C., Ghosh-Dastidar,P., Bradshaw,R.A., Bryan,K.M. et al. (1987) Specificity of cotranslational amino-terminal processing of proteins in yeast. Biochemistry, 26, 8242–8246. [DOI] [PubMed] [Google Scholar]
- 23.Bradshaw R.A., Brickey,W.W. and Walker,K.W. (1998) N-terminal processing: the methionine aminopeptidase and N alpha-acetyl transferase families. Trends Biochem. Sci., 23, 263–267. [DOI] [PubMed] [Google Scholar]
- 24.Kozak M. (1997) Recognition of AUG and alternative initiator codons is augmented by G in position +4 but is not generally affected by the nucleotides in positions +5 and +6. EMBO J., 16, 2482–2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cross S.H. and Bird,A.P. (1995) CpG islands and genes. Curr. Opin. Genet. Dev., 5, 309–314. [DOI] [PubMed] [Google Scholar]
- 26.Eyre-Walker A. and Bulmer,M. (1993) Reduced synonymous substitution rate at the start of enterobacterial genes. Nucleic Acids Res., 21, 4599–4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.