Abstract
We present here a new method for correcting the topology of objects segmented from medical images. Whereas previous techniques alter a surface obtained from a binary segmentation of the object, our technique can be applied directly to the image intensities of a probabilistic or fuzzy segmentation, thereby propagating the topology for all isosurfaces of the object. From an analysis of topological changes and critical points in implicit surfaces, we derive a topology propagation algorithm that enforces any desired topology using a fast marching technique. The method has been applied successfully to the correction of the cortical gray matter / white matter interface in segmented brain images and is publicly released as a software plug-in or the MIPAV package.
Keywords: Topology correction, Fast marching methods, Segmentation, Brain Imaging
1 Introduction
The topological properties of two-dimensional (2D) and three-dimensional (3D) anatomical structures are often very simple, regardless of the complexity of their geometry. The cortex of the human brain is a striking example; despite its intricate folds, it is considered to have the topology of a sphere, without any holes or handle-like junctions [1]. Most organs and sub-structures found in the human body also share this spherical topology. Ideally, algorithms that extract objects from 2D or 3D images should respect the object topology. A major problem in enforcing this constraint, however, is that topology is a global property of the object, whereas most extraction techniques operate locally on the pixels or voxels of an image. One solution is to start from an object with the desired topology and deform it on the image to follow the shape of the object to extract [2, 3, 4, 5]. However, surface segmentation algorithms that constrain the topology also require an initialization close to the object of interest. Correcting the topology of objects is therefore often necessary, either before or after the surface extraction from an initial segmentation. This initial segmentation is typically the result of a fuzzy or statistical classification algorithm that associates a normalized membership or probability value to each class and each pixel or voxel [6, 7].
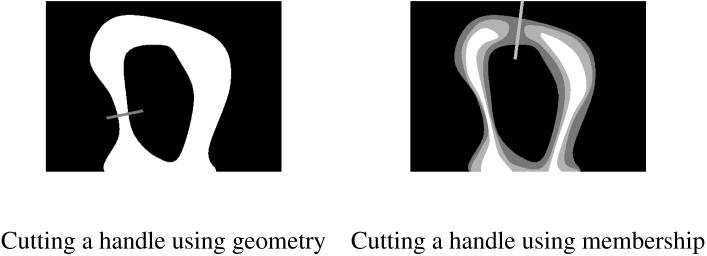
Current algorithms for topology correction typically operate on a binary volume extracted from the classification of the image data [8, 9, 10, 11]. Three types of techniques can be found in the literature: graph-based analysis and correction [12, 10, 11], distance function processing, inspired by level set methods [8, 9, 13] and surface mesh flattening and editing [14, 15]. In places where changes are needed to enforce the spherical topology, these methods must decide how to cut a handle or fill a hole based solely on the geometry of the original surface. In brain segmentation applications, the binary volumes also require pre-processing to remove isolated parts and fill holes [8, 10, 11]. In all these methods, the intensity information available in the original image is largely ignored. If the object is highly convoluted, the smallest change with regard to the geometry of the binary volume may correspond to including points with very low membership or probability values, or discarding points with high values (see Fig. 1).
Fig. 1.
An illustration of the interest of membership functions: cutting a handle at the thinnest point (left) can sever thin structures, but a membership function (right) can provide the additional information needed to preserve them.
In this work, we present a new topology correction algorithm that can act directly on the membership or probability function instead of the binary segmentation. The method propagates exact topological constraints on scalar 2D and 3D functions, even in the presence of noise in the function. The topology of the entire image is corrected with a single computation of a modified fast marching method. The topology-preserving fast marching method extends our previous work on multi-object segmentation [16, 5] to arbitrary scalar images. All isosurfaces extracted from the image data will have the same topology, and we can even enforce non-spherical topologies, given an appropriate initialization. By working on the continuously-valued classification data rather than the binary segmentation, the amount of change effected by the topology correction is substantially reduced. The proposed method also provides greater flexibility in defining a desired surface. In some cases, isosurfaces at probability values other than 0.5 can be desirable [17]. Previous methods would require a separate topology correction for every desired isosurface value. Using this method, multiple layers of the cortex can be represented by a single scalar function with each layer being topologically equivalent to a sphere. The algorithm is implemented in a freely available software plug-in tool, both user-friendly and flexible. To our knowledge, there is no other automatic topology correction software currently available, and only a few topology processing tools in open software packages [18].
The paper is organized as follows. In Section 2.1, we review the definitions and properties of digital topology used in our approach. We introduce our topology-preserving scalar field approximation in Section 2.2, and study its properties through various simple examples in Section 2.3. We then present the corresponding topology correction algorithm and the software tool that implements it in Sections 3 and 4. Finally, Section 5 demonstrates its application to the correction of the topology at the white matter / cortical gray matter interface in magnetic resonance images of the brain. Note that the algorithm and preliminary results have been presented in a conference paper [19].
2 Theory and computational methods
We define f(x) to be a scalar field (e.g. original image, fuzzy membership, probability function, distance function) with values in a bounded interval and defined on a finite digital grid . For each isovalue v ∈ I, we associate a set of objects Fv = {x ∈ D|f(x) ≥ v}. The topology of the scalar field f corresponds to the topology of its associated objects for all isovalues in I, and evolves with v. We are interested in the following problem: given a scalar field f, how can we build a close approximation g of f such that all objects Gv = {x ∈ D|g(x) ≥ v} have the same given topology?
2.1 Digital Topology and Critical Points
The topology of an object can be characterized globally by its Euler Number. This measure, however, does not account for the type, location or extent of topology changes when the object is modified, such as when the isovalue v changes in the scalar field. In this section, we define terminology and review basic concepts from digital topology that will be used throughout the paper. While previous topology correction methods utilized the notion of simple points in binary objects, we employ the more general notion of simple points in scalar (grayscale) fields.
Given a binary digital object, the most elementary change to make is to add or remove one point to or from the object. If such a change maintains the topology of the original object, the point is termed simple or regular. If all points added or removed to an object through a transformation are simple, then the topology is preserved. An important result of digital topology is that simple points and non-simple or critical points, which change the topology of the new object, can be identified from the analysis of their local neighborhood [20, 21, 3]. The extension to scalar fields considers the isosurfaces of the field instead of the binary object, where critical points also become the singular points of the field [22, 23, 24]. This in turn relates to the critical points of vector fields in classical topology [25].
To discriminate simple from critical points, we need to count the number of disconnected positive and negative regions surrounding each point x of the domain D. On the digital grid, a positive or negative region is a set of connected points y in the neighborhood of x such that f(y) > f(x) or f(y) < f(x), respectively. The regions depend on the choice of connectivity (i.e. whether x is directly connected to its 6, 18 or 26 closest neighbors). The neighborhood is a topological neighborhood, a 3×3×3 cubic region with specific properties for different connectivities [21]. Given the numbers Np and Nn of distinct positive and negative regions, we have:
| (1) |
The above definition makes the assumption that there is no region consisting of more than one voxel equal to f(x), since the evolving object should not change by more than one voxel at a time. In practice, this assumption is not often met, and we solve this issue in the correction algorithm by grouping points with equal value f(y) = f(x) within positive regions once they are processed, and negative regions otherwise (see Algorithm 1, step 5).
The connectivity used for computing Np and Nn can be 6/18, 6/26, 18/6 or 26/6, following the conventional definitions of adjacency in cubic grids. This choice determines the connectivity of the object Fv and background D - Fv for all isovalues. Note that the extraction of isosurfaces will also require the use of a topology-preserving technique like the connectivity-consistent marching cubes [3] with the same choice of connectivity.
2.2 Topology-preserving Fast Marching Approximation
If a scalar field has only simple points, or equivalently no critical points, it has a uniform topology. In other words, regardless of the isovalue considered, any computed isosurface will result in the same topology. We present below an algorithm to remove all critical points, at all isovalues, in one step. Starting from an isovalue with the desired topology, we propagate the topology through the isovalues with a modified fast marching method. Fast marching methods propagate a function defined from initial conditions and partial derivatives to the entire domain [26, 27]. Algorithmically, they use a binary tree to sort the function values efficiently, and thus process the domain one voxel at a time, following the successive isovalues of the function.
We can detect and correct critical points while moving through the isovalues by combining the digital topology criterion (1) with the fast marching method. The central idea consists in building an approximation g of f that propagates through its isovalues and modifies the critical points encountered to make them simple. The algorithm produces a scalar field g(x) equal to the original f(x) in all simple regions, with the same topology as a starting object G0 and can be described as follows:
Algorithm 1: Topology-preserving fast marching
Build the function g(x) = g0 in an initial object G0 with the desired topology, where g0 is an arbitrary value ≥ max(f), and set t = 0 (initial conditions).
Sort the points on the outside boundary of G0, denoted {x|x ∈ B(G0)}, in a binary tree according to the value of f(x) from largest to smallest.
Remove the first point x from the tree and set t = t + 1.
- If x has been previously labeled as critical, its value becomes
where N(x) is the neighborhood of x, otherwise g(x) = f(x). Compute the numbers of positive and negative regions Np(x) and Nn(x) using the value of g for neighbors inside Gt-1 and f for neighbors outside. If a neighbor has a value equal to g(x) and it is inside Gt-1, it is classified as positive. If a neighbor has a value equal to g(x) but is not inside Gt-1, it is clasified as negative.
-
Using the computed Np(x) and Np(y), classify x as simple or critical. (a) If x is simple, insert it into the processed object Gt = Gt-1 ∪ {x}. Find its neighbors y outside Gt and not already in the binary tree, and insert them into the tree with value f(y),
(b) If x is critical, label it as critical and put it back in the unprocessed part of the domain. The point will remain outside of the sorting tree until one of its neighbors is processed.
Go back to step 3, until the binary tree is empty.
As the changes are propagated along successive isovalues of the image, the technique is guaranteed to succeed in enforcing the original topology of G0 in every isosurface Gt of g(x). Critical points are kept outside the volume until changes in their neighborhood lower their updated function g just enough to make each of them become simple. The resulting field g(x) is uniquely defined given f(x) (the fast marching evolution function) and G0 (the initial conditions). If the topology is already uniformly invariant, the algorithm keeps the original function unchanged.
2.3 Examples in Multiple Dimensions
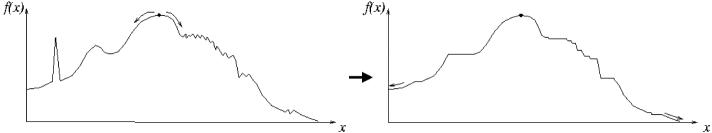
In this section, we describe several examples to provide more insight into the algorithm and its properties. Figure 2 shows a simplistic 1D example. In this case, the only type of critical point to be encountered is a local maximum, which would cause disconnected segments to occur when considering certain isovalues smaller than the global maxium. Applying the topology-preserving fast marching algorithm simply flattens out those maxima, and creates a concave approximation of function f. This is the first property of the algorithm: g ≤ f on the entire domain D (note that Algorithm 2 could be reversed to build g from the lowest to the highest values of f, in which case we get g ≥ f). In 1D, it is clear that the approximation can be rather far from the original function, if its local maxima are large and numerous. This example also illustrates that topology constraints have a smoothing effect on noise, but they introduce a bias toward the lower values of f.
Fig. 2.
A 1D example of topology-preserving approximation: starting from the highest point of f (left), the propagation algorithm follows its slope downward and thresholds the function when f goes upward. The resulting function g (right) has one global maximum, its minima are located at the boundary of the domain and there is no local minima or maxima.
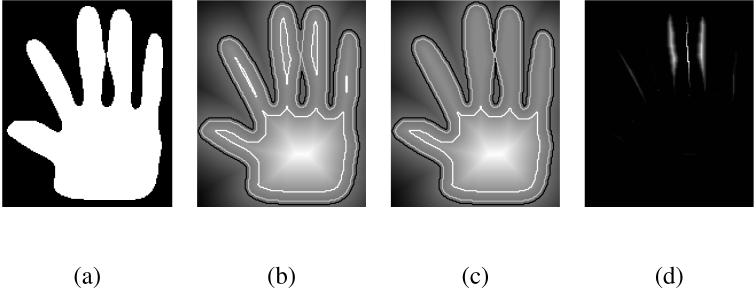
Imposing a uniform topology can lead to very strong constraints in the 1D case. However, higher dimensions allow for greater flexibility. In 2D and 3D, there is more than one path from one point to another. The fast marching method directs the propagation along the path of least variations, as the neighbors of a point with the highest value will be processed first. This limits the extent of the approximation and reduces the bias towards lower values, as shown in Figure 3. In this example, the objects defined by the outlined isovalues in the original function and its approximation are globally the same, and only differ in the few areas where critical points are removed. The original contour of the hand is preserved, even though the correction started far from it, at the highest intensity in the center of the palm. Note in the case of Figure 3 that 1D structures like the skeleton of the fingers are more affected, similar to the 1D correction example.
Fig. 3.
A 2D example: from the outline of a hand (a), we build a signed distance function (b). Notice from the outlined isocontours how the topology of the object changes as the fingers get separated from the palm or merge. The topology-preserving approximation (c) is very similar to the original distance function, but maintains the same topology for all isovalues. The difference f - g (d) is zero almost everywhere, and small otherwise (the amplitude of the difference image is about 5% of the amplitude of the original distance function).
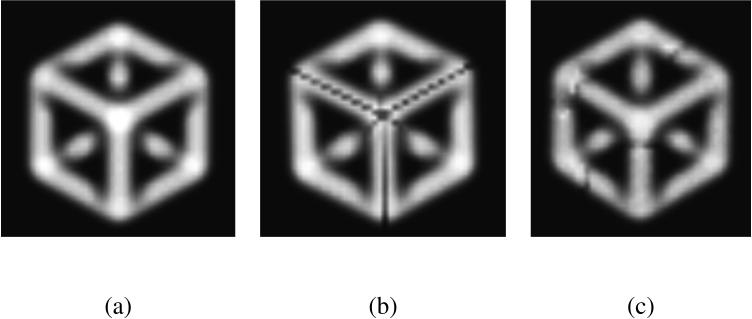
The relationship between topology and geometry of an object or scalar field can be surprising, as illustrated in the 3D example of Figure 4. Here, the two corrected volumes (b) and (c) both represent a valid answer, even though their shape is affected very differently by the cuts that enforce spherical topology. The algorithm processes points with the highest isovalues first, and therefore tends to induce fewer changes to those, and more to the lower isovalues. However, there is no global constraint on the maximum amount of change on a point or the number of points changed in a region.
Fig. 4.
A 3D example: (a) a solid shape made of the edges of a cube and blurred into a volumetric scalar image, (b) the same object corrected to have spherical topology, (c) the corrected object after adding noise to (a). The propagation is started from a single point of the hidden corner on the back. After the propagation, the corrected object has cuts that preserve the spherical topology of the original point. The cuts respect the symmetry of the original object because the propagation is identical in all directions. If we add some noise to the image, the symmetry is broken and the cuts adopt a more conventional shape.
Noise in the image will influence the ordering in the fast marching method, and may introduce changes in the correction. The example of Figure 4 is an extreme case where the correction of the noise-free cube (b) is an unstable configuration, and only a small amount of noise will change it into (c). If the ordering becomes random in areas of f with small or no variations, critical points can appear during the evolution of Gt over such region and generate complex, artificial structures in order to preserve the topology, as in the example of Fig. 5. To reduce the effects of noise, we have to modify step 4 of the algorithm such that the approximated value g(x) becomes . This constraint forces the propagation to follow a geometric distance function whenever the variations in the data is under δ. Using a geometric distance function makes the successive objects Gt more convex and reduces the chance of an erratic propagation. The parameter δ corresponds to the expected level of noise in the image. In practice, this problem mostly happens at artificial thresholds and pre-processing can be performed to avoid the dramatic effect of Fig. 5-b (see Section 5).
Fig. 5.
Influence of noise and flat regions: (a) a membership function from our experimental study, (b) the approximated function with spherical topology computed from the lowest to highest membership values, with δ = 0, (c) the same approximation with δ = 1E - 12, (d) the approximation with δ = 1E - 3. The propagation is started from the boundary of the image, and evolves at random through the background region if δ = 0. The smallest increase to will remove most of the critical points, and the remaining artificial structures due to background noise will also disappear with a higher δ.
3 Program description
Topology correction aims at enforcing a chosen topology on the objects we want to extract. The correct or desired topology is known a priori, and depends on the application. The topology-preserving approximation algorithm can be easily applied to perform fully automated spherical topology correction. In this case, a single point inside the object of interest is enough to impose the sphericality constraint and initialize the algorithm.
We assume in the following that f is a membership or probability function obtained from a tissue classification technique, with values in [0, 1]. If such a function is unavailable, one can compute a normalized, signed distance function based on the segmented object instead.
Algorithm 2: automated spherical topology correction
Threshold the function f at the 0.5 isovalue, and select the largest connected region.
In that region, select one of the voxels of highest value.
Set G0 to this voxel and apply the topology-preserving fast marching algorithm.
The initialization steps 1 and 2 ensure that we select a point inside the main structure of interest.
When dealing with several structures of similar size, it is necessary to manually select a point for each structure. Instead of starting from a point inside the structure, the topology correction could also start from a bounding box outside the image and converge toward the structure. For brain segmentation, however, we found that there is often more noise and outliers in the background area than holes in the high membership regions, and propagating the topology from the background would require a higher value for the regularization parameter δ. This choice is consistent with previous correction techniques that only include the largest connected component [11, 10]. As the approximation in this case lowers the membership values, the correction will only cut handles (the other choice would always fill them). In cases where both types of correction are needed, the selection method of [13] could be adapted to our corrected membership functions. On the other hand, the extracted surface often needs additional processing, and the knowledge that the corrected surface is entirely inside or outside the desired one can become useful, for instance to design pressure forces to move the surface. Note that choosing a slightly lowered isovalue will result in a surface with both cut and filled handles.
Unlike previous techniques, our topology-preserving propagation technique will not just enforce a spherical topology, but can also enforce any topology as defined by an initial topology template. For non-spherical topologies, different starting shapes can be used: a closed circle for doughnut or cylinder-like structures, a carved sphere for a hollow object, etc. Once we have chosen the appropriate template, we need to place it inside the object of interest. This step can be performed manually or by automatic registration of the template so that its holes and handles match those of the image. Additional care is also needed to ensure the alignment will not change the topology of the template, even for rigid registration [28].
4 The Topology Correction Software Tool
We have developed and released the software tool to perform the above topology correction (see Figure 6). The tool has been implemented as a plug-in for the MIPAV software package, a freely available and user-friendly image analysis program developed by the National Institutes of Health [29]: http://mipav.cit.nih.gov/.
Fig. 6.
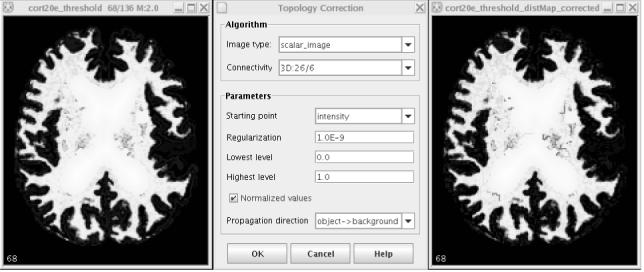
The topology correction software: original image, user interface and result.
The plug-in complements a set of neuroimage processing tools we recently released [30] and is freely available from: http://medic.rad.jhu.edu/download/public/.
The software tool performs two types of correction, either on binary objects or scalar images. When dealing with a binary object, the algorithm first builds a distance function inside or outside the object, then performs the topology correction. Otherwise, the algorithm directly approximates the given scalar function. The user can choose the connectivity of the objects and the propagation direction (from high to low intensities or the inverse), as well as the regularization parameter δ. The plug-in can be called from the graphical interface or from scripts for automated processing. In addition, the initial object (i.e. the topology template) can be manually set from a paint mask, offering the option of correcting images with any topology.
5 Experimental Study
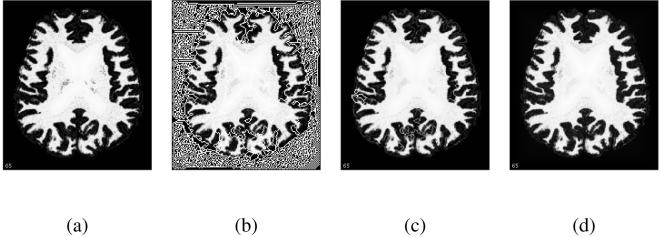
To evaluate the algorithm, we have gathered a set of 20 T1-weighted MR brain images from the Baltimore Longitudinal Study on Aging [31]. The brains were first stripped of extra-cranial tissues, then segmented into gray matter, white matter and cerebro-spinal fluid (CSF). Finally, the white matter memberships were further edited to fill the sub-cortical area [32]. The images all have a 1mm cubic resolution. We performed the topology correction on the edited white matter memberships. As explained previously, problems can appear with in the filled area if it is simply filled with a constant value. The propagation over the constant region is arbitrary, unless we impose some regularization with δ > 0. We opted for a more efficient alternative here which consists of computing a geometric distance function inside the filled region and adding it to the constant. This pre-processing assigns a proper ordering to the voxels of the constant region, without affecting the other parts of the membership function, and we can set δ ≃ 0 (in this case, δ = 1E - 5). The chosen connectivity was 18/6, so that we could compare our results against those reported by previous topology correction techniques on similar data [11, 10].
In all cases, the topology correction succeeded. Even though the topology of every isovalue is guaranteed to be spherical, we also verified it by computing the Euler characteristic before and after correction at the 0.5 isovalue and at randomly selected isovalues. Overall, the corrected membership functions are very close to the original ones (cf. Table 1). Even though the number of changed voxels over the entire image is rather large (8 - 10%), the mean amount of change for these voxels (counting only changed voxels) is below 0.05 (the membership intensities are in [0, 1]). Many voxels are changed at the higher and lower limit of the memberships (flatter regions, more sensitive to noise), but the number of changed voxels on the 0.5 isosurface is below 1%, similar to figures reported in other methods.
Table 1.
Topology correction results for a set of 20 brain images. The 0.5 isovalue corresponds to the white matter - gray matter interface.
| Full image | 0.5 isovalue | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Subject number | Computation time | % of changed voxels | mean value of change | % of changed voxels | % of change with GTCA | mean surface distance (mm) | mean distance with GTCA | Euler # before | Euler # after |
| 1 | 47s | 9.51 | 0.0311 | 0.273 | 0.360 | 0.041 | 0.028 | -208 | 2 |
| 2 | 51s | 9.90 | 0.0353 | 0.352 | 0.382 | 0.033 | 0.034 | -266 | 2 |
| 3 | 46s | 8.82 | 0.0389 | 0.525 | 0.411 | 0.038 | 0.048 | -580 | 2 |
| 4 | 53s | 9.22 | 0.0320 | 0.195 | 0.296 | 0.020 | 0.022 | -178 | 2 |
| 5 | 49s | 9.12 | 0.0349 | 0.368 | 0.469 | 0.033 | 0.038 | -298 | 2 |
| 6 | 48s | 9.24 | 0.0354 | 0.434 | 0.482 | 0.038 | 0.045 | -340 | 2 |
| 7 | 52s | 9.50 | 0.0373 | 0.355 | 0.336 | 0.030 | 0.028 | -376 | 2 |
| 8 | 53s | 9.68 | 0.0358 | 0.444 | 0.464 | 0.033 | 0.035 | -516 | 2 |
| 9 | 59s | 9.58 | 0.0358 | 0.410 | 0.353 | 0.041 | 0.044 | -346 | 2 |
| 10 | 41s | 8.99 | 0.0316 | 0.292 | 0.464 | 0.028 | 0.034 | -164 | 2 |
| 11 | 54s | 9.87 | 0.0340 | 0.391 | 0.481 | 0.037 | 0.038 | -322 | 2 |
| 12 | 48s | 9.58 | 0.0385 | 0.462 | 0.448 | 0.039 | 0.042 | -388 | 2 |
| 13 | 61s | 8.36 | 0.0365 | 0.295 | 0.445 | 0.026 | 0.028 | -350 | 2 |
| 14 | 43s | 9.09 | 0.0343 | 0.279 | 0.310 | 0.018 | 0.021 | -300 | 2 |
| 15 | 61s | 9.47 | 0.0379 | 0.396 | 0.418 | 0.036 | 0.046 | -472 | 2 |
| 16 | 54s | 9.75 | 0.0312 | 0.249 | 0.369 | 0.020 | 0.019 | -170 | 2 |
| 17 | 58s | 8.44 | 0.0370 | 0.299 | 0.317 | 0.023 | 0.025 | -320 | 2 |
| 18 | 51s | 8.74 | 0.0427 | 0.762 | 0.437 | 0.063 | 0.039 | -524 | 2 |
| 19 | 51s | 9.54 | 0.0375 | 0.409 | 0.458 | 0.036 | 0.040 | -450 | 2 |
| 20 | 49s | 9.18 | 0.0372 | 0.631 | 0.466 | 0.058 | 0.050 | -282 | 2 |
We obtained separate results from the GTCA method [10] for the same data, and compared them to our 0.5 isovalues. Our method enforces the topology on all isovalues, causing cascading changes on successive isosurfaces, and can only remove voxels from a given isosurface (the propagation enforces g ≥ f). However, the number of changed voxels and the average surface distance to the 0.5 isovalue of the original image are similar in both methods, and often lower in the propagation-based method than with GTCA.
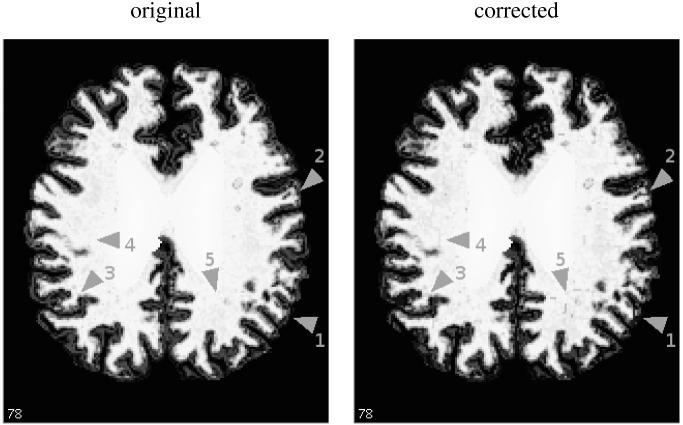
The places of most significant change are the regions close to the filled area (see Figure 7, arrows 4 and 5). There, the voxels can have a low membership value surrounded by higher values. These isolated low values become topological defects, and impose more changes in the approximation. Other changes of note happen in the background (arrow 1): higher membership values at the gray matter / CSF interface are much decreased due to their distance to the main white matter areas. Additionally, one can note some cuts in the regions of intermediate membership values (arrows 2, 3), often in finger-like structures more affected by topological defects because of their mostly 1D shape. The cuts are the most important part of the correction, as they affect the boundary of the white matter membership. Even in the worst cases, depicted in Figures 7 and 8, these remain in small numbers and most of the correction is hardly noticeable to the eye.
Fig. 7.
An example of topology correction (image number 18 of the experiments, which has the highest numbers of changed voxels): a slice of the filled white matter membership function before and after the topology correction, Note the few visible changes in the membership, pointed by gray arrows: parts with high membership but disconnected from the white matter are removed (1), loops are cut (2, 3) and some patterns appear near the filled region (4, 5).
Fig. 8.
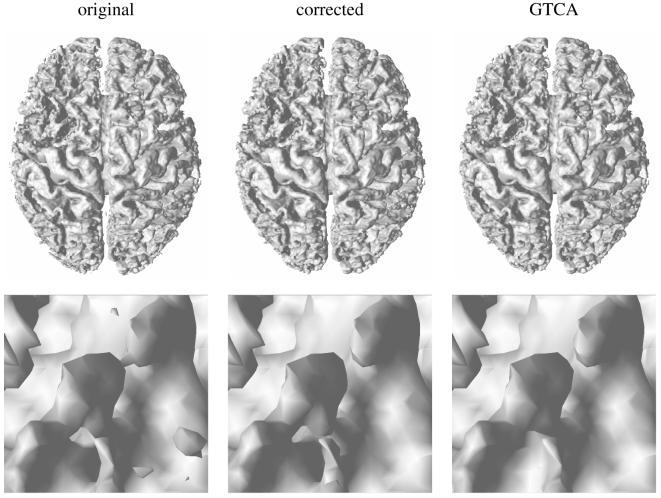
An example of topology correction (image number 18 of the experiments): top) the 3D surface extracted from the original and corrected memberships, bottom) a close-up of the surface in an area with three interconnected handles, before and after correction. The corrected surface obtained with GTCA is also presented for comparison. The corrected surface is very close to the original and to the surface produced by GTCA.
The processing times are about 45-60 seconds on a Pentium 4 3Ghz PC (the filled region pre-processing takes an additional 2-3s) for 256×256×124 voxel images (see Table 1). GTCA requires approximately 1-2 minutes on a similar machine. All the brains we have tested present very similar results: computation times, amount of change in voxels and intensity exhibit little variation over our dataset. As shown in Figure 8, the extracted surface retains the shape and features of the original surface.
6 Conclusion
We have introduced an algorithm for propagating topology over 3D scalar fields. It offers an efficient way to correct the topology of objects with a complex geometry, using all the available information from the membership or distance functions involved in their segmentation. It improves over previous methods by acting on all isovalues rather than a binary volume, and allowing non-spherical topologies. The technique has been thoroughly tested on artificial examples, and the effects of noise, low signal areas and topological defects have been studied.
The method has been successfully applied to the problem of cortical segmentation to enforce a spherical topology on the white matter/gray matter interface. The propagation algorithm provides fast and reliable results, always very close to the original data. All isovalues have the correct topology, and the changes made on the membership function take into account the geometry of the entire image. The released software is user-friendly and freely downloadable, can be applied to any kind of scalar image data, and guarantees exact topology results in a single propagation step.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain
References
- [1].van Essen DC, Maunsell HR. Two dimensional maps of the cerebral cortex. Journal of Comparative Neurology. 1980;191:255–281. doi: 10.1002/cne.901910208. [DOI] [PubMed] [Google Scholar]
- [2].Mangin J-F, Frouin V, Bloch I, Regis J, Lopez-Krahe J. From 3D magnetic resonance images to structural representations of the cortex topography using topology preserving deformations. Journal of Mathematical Imaging and Vision. 1995;5:297–318. [Google Scholar]
- [3].Han Xiao, Xu Chenyang, Prince Jerry L. A topology preserving level set method for geometric deformable models. IEEE Transactions on Pattern Analysis and Machine Intelligence. 2003;25(6):755–768. [Google Scholar]
- [4].Bischoff Stefan, Kobbelt Leif. Sub-voxel topology control for level-set surfaces. Computer Graphics Forum. 2003;22(3):273–280. [Google Scholar]
- [5].Bazin P-L, Pham DL. Topology-preserving tissue classification of magnetic resonance brain images. IEEE Transactions on Medical Imaging. 2007;26(4):487–496. doi: 10.1109/TMI.2007.893283. Special Issue on Computational Neuroanatomy. [DOI] [PubMed] [Google Scholar]
- [6].Van Leemput Koen, Maes Frederik, Vandermeulen Dirk, Suetens Paul. Automated model-based tissue classification of MR images of the brain. IEEE Transactions on Medical Imaging. 1999;18(10):897–908. doi: 10.1109/42.811270. [DOI] [PubMed] [Google Scholar]
- [7].Pham Dzung L. Spatial models for fuzzy clustering. Computer Vision and Image Understanding. 2001;84:285–297. [Google Scholar]
- [8].Szymczak Andrzej, Vanderhyde James. Extraction of topologically simple isosurfaces from volume datasets; Proceedings of IEEE Visualization; Seattle. october 2003.pp. 67–74. [Google Scholar]
- [9].Segonne Florent, Grimson Eric, Fischl Bruce. Topological correction of subcortical segmentation; Proceedings of the 6th International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI’03); Montreal. november 2003.pp. 695–702. [Google Scholar]
- [10].Han X, Xu C, Braga-Neto U, Prince JL. Topology correction in brain cortex segmentation using a multiscale, graph-based algorithm. IEEE Transactions on Medical Imaging. 2002;21(2):109–121. doi: 10.1109/42.993130. [DOI] [PubMed] [Google Scholar]
- [11].Shattuck David W, Leahy Richard M. Automated graph-based analysis and correction of cortical volume topology. IEEE Transactions on Medical Imaging. 2001;20(11) doi: 10.1109/42.963819. [DOI] [PubMed] [Google Scholar]
- [12].Wood Zoe, Hoppe Hugues, Desbrun Mathieu, Schroder Peter. Removing excess topology from isosurfaces. ACM Transactions on Graphics. 2004;23(2):190–208. [Google Scholar]
- [13].Kiegeskorte Nikolaus, Goebel Rainer. An efficient algorithm for topologically correct segmentation of the cortical sheet in anatomical MR volumes. NeuroImage. 2001;14:329–346. doi: 10.1006/nimg.2001.0831. [DOI] [PubMed] [Google Scholar]
- [14].Segonne F, Pacheco J, Fischl B. Geometrically accurate topology-correction of cortical surfaces using nonseparating loops. IEEE Transactions on Medical Imaging. 2007;26(4) doi: 10.1109/TMI.2006.887364. Special Issue on Computational Neuroanatomy. [DOI] [PubMed] [Google Scholar]
- [15].Fischl B, Liu A, Dale A. Automated manifold surgery: Constructing geometrically accurate and topologically correct models of the human cerebral cortex. IEEE Transactions on Medical Imaging. 2001;20(1) doi: 10.1109/42.906426. [DOI] [PubMed] [Google Scholar]
- [16].Bazin Pierre-Louis, Pham Dzung L. Topology preserving tissue classification with fast marching and topology templates; Proceedings of the International Conference on Information Processing in Medical Imaging 2005 (IPMI’05); Glenwood Springs. july 2005; [DOI] [PubMed] [Google Scholar]
- [17].Tosun D, Rettmann ME, Naiman DQ, Resnick SM, Kraut MA, Prince JL. Cortical reconstruction using implicit surface evolution: Accuracy and precision analysis. Neuroimage. 2005;29(3):838–852. doi: 10.1016/j.neuroimage.2005.08.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Lamy J. Integrating digital topology in image-processing libraries. Computer Methods and Programs in Biomedicine. 2007;85(1):51–58. doi: 10.1016/j.cmpb.2006.08.006. [DOI] [PubMed] [Google Scholar]
- [19].Bazin Pierre-Louis, Pham Dzung L. Topology correction using fast marching methods and its application to brain segmentation; Proceedings of the 8th International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI’05); Palm Springs. october 2005; [DOI] [PubMed] [Google Scholar]
- [20].Malandain G, Bertrand G, Ayache N. Topological segmentation of discrete surfaces. International Journal of Computer Vision. 1993;10(2):183–197. [Google Scholar]
- [21].Bertrand Gilles. Simple points, topological numbers and geodesic neighborhood in cubic grids. Pattern Recognition Letters. 1994;15(10):1003–1011. [Google Scholar]
- [22].Hart John C. Morse theory for implicit surface modeling. In: Hege H-C, Polthier K, editors. Mathematical Visualization. Springer-Verlag; Oct, 1998. pp. 257–268. [Google Scholar]
- [23].Couprie M, Bezerra FN, Bertrand Gilles. Topological operators for grayscale image processing. Journal of Electronic Imaging. 2001;10(4):1003–1015. [Google Scholar]
- [24].Weber Gunther H, Scheuermann Gerik, Hamann Bernd. Detecting critical regions in scalar fields; Proceedings of the Joint EUROGRAPHICS - IEEE TCVG Symposium on Visualization; Grenoble. may 2003. [Google Scholar]
- [25].Henle Michael. A Combinatorial Introduction to Topology. W.H. Freeman and Company; 1979. [Google Scholar]
- [26].Sethian JA. Level Set Methods and Fast Marching Methods. Cambridge University Press; 1999. [Google Scholar]
- [27].Osher Stanley J., Fedkiw Ronald P. Level Set Methods and Dynamic Implicit Surfaces. Springer-Verlag; 2002. [Google Scholar]
- [28].Bazin P-L, Ellingsen LM, Pham DL. Digital homeomorphisms in deformable registration; Proceedings of the International Conference on Information Processing in Medical Imaging 2007 (IPMI’07); Kerkrade. july 2007; [DOI] [PubMed] [Google Scholar]
- [29].McAuliffe MJ, Lalonde F, McGarry DP, Gandler W, Csaky K, Trus BL. Medical image processing, analysis and visualization in clinical research; Proceedings of the 14th IEEE Symposium on Computer-Based Medical Systems (CBMS 2001); 2001. [Google Scholar]
- [30].Bazin P-L, Cuzzocreo JL, Yassa MA, Gandler W, McAuliffe MJ, Bassett SS, Pham DL. Volumetric neuroimage analysis extensions for the mipav software package. Journal of Neuroscience Methods. 2007 doi: 10.1016/j.jneumeth.2007.05.024. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Resnick SM, Goldszal AF, Davatzikos C, Golski S, Kraut MA, Metter EJ, Bryan RN, Zonderman AB. One-year age changes in MRI brain volumes in older adults. Cerebral Cortex. 2000;10(5):464–472. doi: 10.1093/cercor/10.5.464. [DOI] [PubMed] [Google Scholar]
- [32].Han X, Pham D, Tosun D, Rettmann ME, Xu C, Prince JL. Cruise: Cortical reconstruction using implicit surface evolution. NeuroImage. 2004;23(3):997–1012. doi: 10.1016/j.neuroimage.2004.06.043. [DOI] [PubMed] [Google Scholar]