Abstract
Transcription on chromatin by RNA polymerase II (pol II) is repressed as compared with transcription on histone-free DNA. In this study, we show that human topoisomerase I (topo I) and yeast topoisomerase II (topo II), each of which relax both positive and negative superhelical tension, reverse the transcriptional repression by chromatin. In the presence of bacterial topo I, which can relax only negative superhelical tension, the transcription is repressed on chromatin templates. The data together show that the relaxation of positive superhelical tension by these enzymes was the key property required for RNA synthesis from chromatin templates. In the absence of topoisomerase, transcriptional repression on chromatin depended on RNA length. The synthesis of transcripts of 100 nt or shorter was unaffected by chromatin, but repression was apparent when the RNA transcript was 200 nt or longer. These findings suggest that transcription on chromatin templates results in the accumulation of positive superhelical tension by the elongating polymerase, which in turn inhibits further elongation in the absence of topoisomerase activity.
INTRODUCTION
Transcription of protein-encoding genes involves the recognition of the promoter DNA sequence by general transcription factors (GTFs), TFIIA, -B, -D, -E, -F and -H, and RNA polymerase II (pol II) (1). The function of these GTFs on naked DNA templates in vitro results in a basal level of transcription, which can be stimulated by sequence-specific enhancer binding proteins plus coactivators. The DNA in the cell is not naked, but packed into a compact nucleosome-containing structure known as a chromatin. Nucleosomal DNA poses a hindrance to the accessibility to the template by different DNA binding factors and pol II. Thus, the minimal basal set of pol II transcription factors, which mediate transcription on a naked template, are not sufficient for pol II-dependent transcription on chromatin templates (2–4). One way that cells regulate accessibility to nucleosomal DNA is through the use of the complexes that modify chromatin structure.
A separate problem faced by the transcriptional machinery is the topology of the DNA. Transcription, which involves the tracking of a bubble of unpaired bases along a double-stranded DNA helix, transiently introduces superhelical tension in the DNA. Transcription thus generates a wave of positive supercoils in the template ahead of the advancing pol II and negative supercoils in the DNA behind it (5,6). In a living cell, the supercoiled state of the DNA is modulated by topoisomerases. In yeast, simultaneous inactivation of DNA topoisomerase I (topo I) and topoisomerase II (topo II) reduces rRNA synthesis, and to a lesser extent mRNA synthesis is reduced (7–10). This reduction in RNA synthesis in yeast cells can be attributed to changes in the topology of the DNA template. Likewise, the transcription of a number of genes by pol II in a B-lymphocyte tissue culture cell line is sensitive to topoisomerase function (11). A direct link between topoisomerase activity and the transcription process is difficult to establish in these in vivo assays because of the multiple cellular roles of the topoisomerase and because of the complexity of the experimental system.
While topoisomerases have been shown to be required for transcription in cells, these same activities were dispensable for RNA synthesis in vitro using purified transcription factors on model templates. We have found, however, that when the model template is reconstituted in chromatin, then RNA synthesis is dependent upon the topo IIα activity associated with the pol II holoenzyme (3). The pol II holoenzyme function could be replaced using purified core pol II and purified topo IIα. Specific inhibitors of topo II, etoposide and ICRF-193, which inhibit the relaxation activity of the topo II via different mechanisms, also inhibit transcription on chromatin templates. In contrast, these inhibitors have no effect on transcription from naked DNA templates. These results suggest that the topo IIα modifies the DNA in chromatin in association with other transcription factors (3), but the mechanism by which the topoisomerase stimulated transcription on chromatin was unexplained.
The topo IIα was identified in the reaction via its association with the pol II holoenzyme (3). Since we hypothesize that the relaxation activity of the topo IIα is the key for transcription on chromatin templates, we tested in the current study whether other topoisomerases, besides topo IIα, will stimulate transcription on chromatin. We found that topoisomerases that efficiently relaxed positive superhelical tension could stimulate transcription on the chromatin template. In addition, we found that the requirement for a topoisomerase depends on the length of the transcript, and only the synthesis of longer RNAs, the templates which accumulate high positive superhelical tension, is sensitive to the function of a topoisomerase. We further demonstrate that the requirement for the topoisomerase activity is strictly at the elongation stage of the transcription reaction. These results indicate that the accumulation of positive superhelical tension in front of the elongating pol II is stabilized by nucleosomes, and this effect is likely to be a critical component of the transcription process.
MATERIALS AND METHODS
Plasmid templates
G-less cassette construct pG5-E4-array contained the adenoviral E4 promoter upstream of a DNA sequence in which there were no guanines in the coding strand from the transcription start site to a particular length (25, 50, 100, 200 or 384 bp) downstream. The total length of all the DNA templates is ∼3000 bp. The length of the transcript was determined by the length of the G-less region and was indicated in each of the reactions. Upstream of the E4 promoter were five DNA binding elements that can be bound by the GAL4 fusion protein. The promoter and G-less transcript sequences are present in between the five repeats of the 5S rDNA nucleosome positioning sequences. All transcription reactions include an internal basal control template, pΔML-200 or -390, consisting of a core adenoviral major late promoter (MLP) upstream of a 210 or 390 bp G-less cassette. Both the templates, pG5E4 and pΔML, were digested with restriction endonuclease ClaI or XmnI, respectively, which cleaves inside the ampicillin resistance gene, and thus these DNAs have linear topology.
Chromatin assembly
Histones were purified from the chromatin pellet of HeLa whole cell extracts (12) by solubilizing in the chromatin medium salt buffer (0.6 M NaCl, 50 mM NaPO4, pH 6.8 and 0.5 mM PMSF), and bound to an hydroxyapatite column. Purified nucleosomes were eluted by high salt buffer A (2.5 M NaCl, 50 mM NaPO4, pH 6.8 and 0.5 mM PMSF) (13). Purified histone octamers and the linear DNA templates, pG5-E4-array and ΔML, were mixed in the ratio of 0.8 mg of histone:1 mg of total DNA. Typically, 8.4 µg of each DNA template was mixed with 13.5 µg of purified nucleosomes. Buffer A was added to keep the salt concentration above 1 M and the total volume was adjusted to 200 µl. The above sample was then dialyzed against buffer B (1 M NaCl, 50 mM NaPO4, pH 6.8 and 0.5 mM PMSF) for 20 min. By slowly, over 3 days, adding buffer C (50 mM NaPO4, pH 6.8, 5% glycerol and 0.5 mM PMSF), the final salt concentration was dialyzed to 100 mM NaCl.
Reconstitution of chromatin using assembly factors was done essentially the same as in Ito et al. (14). Each assembly reaction contained 500 ng of each DNA template, 1.4 µg of HeLa histones, 6 µg of NAP-1, 100 ng of Acf-1 and 120 ng of ISWI in a 60 µl volume, based upon protocols provided by J. Kadonaga and C. Wu. Assembly reactions were incubated at 27°C for 5 h and then divided for use in transcription assays. For these assembly reactions, the templates did not contain the nucleosome positioning arrays.
Purified DNA topo IIα was obtained from Topogen, Inc. and topo I was purified from the baculovirus-infected insect cells (Sf9) (15). The topo I expressing baculovirus was kindly provided by A. M. Zhelkovsky and C. L. Moore (Tufts Medical School). The purified bacterial topo I and the yeast topo II were kindly provided by J. C. Wang (Harvard University).
Purification of transcription factors
The transcription factors used in this study were purified using established techniques. Core pol II was purified from calf thymus using DEAE chromatography and immunoaffinity purified with antibody 8WG16. TFIIH was purified from HeLa whole cell extracts by chromatography using the following matrices: Biorex70, DEAE Sepharose, bioscale S and bioscale Q (16). Flag-tagged TFIID was purified from HeLa cells and recombinant factors were purified from bacteria as described previously (16,17). The PC4 construct was kindly provided by M. Meisterernst and purified as described by Kretzschmar et al. (18).
In vitro transcription assay
Transcription reactions were performed in a 25 µl volume containing 20 mM HEPES–NaOH, pH 7.9, 20% glycerol, 1 mM EDTA, 6 mM Mg(OAc)2, 90 mM KOAc, 3 mM dithiotreitol, 4 µM ZnSO4, 0.2 mg/ml BSA, 100 µM each of ATP and UTP, 2.5 µM CTP, 50 µM 3′-OMe-GTP and 30 ng of each template DNA. Proteins included were 100 ng of TFIIA, 60 ng of TFIIB, 4 ng of TFIIE, 100 ng of TFIIF, 40 ng of core pol II, 0.3 µl of immunoaffinity-purified TFIID, 0.5 µl of TFIIH fraction and 100 ng of PC4. Thirty nanograms of GAL4–VP16 as an activator was included where indicated. The reactions were incubated at 30°C for 90 min, terminated and processed using standard procedures (2–4). Dried gels were exposed to film with an intensifying screen, generally for 16–24 h.
The amount of topoisomerase used per transcription reaction was normalized according to the relaxation activity of each enzyme preparation.
Relaxation assay
Relaxation assays with topo I used plasmid DNA (200 ng/reaction) in a final volume of 30 µl in topo I buffer (10 mM Tris–Cl, pH 7.9, 1 mM EDTA, 150 mM NaCl, 0.1% BSA, 0.1 mM spermidine, 0.5% glycerol). The activities of the various topoisomerases were similar when the buffer was the same as used in transcription reactions (data not shown). The reactions were incubated at 37°C, terminated by the addition of 2.5 µl of bromophenol blue and phenol/chloroform extraction, and then analyzed on 1% native agarose gel. Gels were stained with ethidium bromide.
RESULTS
Topo I can relieve the repression of transcription due to nucleosomes
Our earlier data revealed that human topo IIα could reverse the repression of transcription by nucleosomes (3). It was unclear whether this transcription effect was due to a specific function of topo IIα or rather that another topoisomerase could replace topo IIα. We thus tested whether human topo I could replace topo IIα function in transcription in chromatin templates. Transcription reactions contained the basal transcription factors, TFIIA, -B, -D, -F, -E and -H, core pol II and the PC4 cofactor. Indicated reactions also contained the GAL4–VP16 activator protein. These purified factor preparations had no detectable topoisomerase activity (data not shown). All reactions contained equal concentrations of two templates. The template encoding a 384 nt G-less RNA was responsive to GAL4–VP16 since it contained five GAL4 binding sites upstream of the adenovirus E4 core promoter and a G-less cassette. The 150 bp promoter and G-less transcript sequences are bracketed on each side by five repeats of the 5S rDNA nucleosome positioning sequence. The total size of the template DNA containing the G-less cassette was ∼3000 bp. The second template contains the adenovirus MLP upstream of a 210 bp G-less cassette. This template lacks GAL4 binding sites, and is thus a basal transcription control. Both templates were reconstituted in chromatin using purified histone octamers by an established procedure.
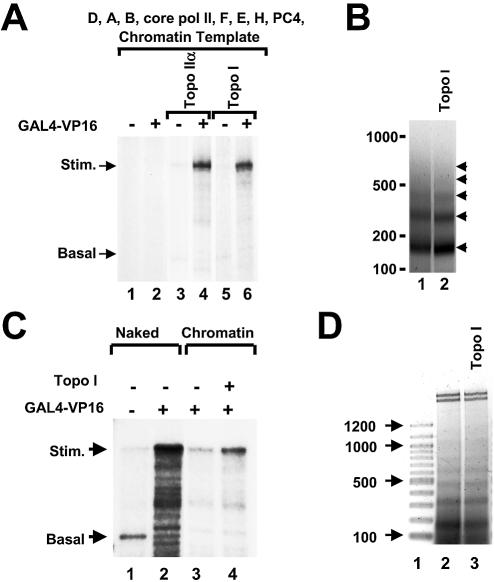
As observed in earlier studies, transcription by core pol II is repressed on chromatin templates (Fig. 1A, lanes 1 and 2). Consistent with our previous results (3), in the presence of 0.3 U topo IIα, the transcriptional repression by chromatin was reversed (Fig. 1A, lanes 3 and 4). Topo I and topo IIα, though structurally very different, share a common activity, i.e. both can relax negative and positive superhelical DNA torsion. Indeed, when 5 ng of topo I was included in transcription reactions with chromatin templates, a high level of product RNA was observed for the GAL4–VP16-stimulated transcript (Fig. 1A, lanes 5 and 6). This result indicated that under these conditions, either topoisomerase activity was sufficient for RNA synthesis on chromatin templates.
Figure 1.
Either topo IIα or topo I reversed the repression of transcription on chromatin. (A) Transcription reactions were tested on chromatin templates which were reconstituted either in the absence of topoisomerase (lanes 1 and 2) or in the presence of topo IIα (topo II, lanes 3 and 4) or topo I (topo I, lanes 5 and 6). Reactions contain pol II, TFIIA, -B, -D, -E, -F, -H and coactivator PC4. GAL4–VP16 was included in indicated lanes. Reactions contained two templates in equal concentration, encoding a 384 nt GAL4-responsive transcript (Stim.) and 210 nt basal control transcript (Basal). (B) Micrococcal nuclease digestion of chromatin templates. Chromatin templates were reconstituted either in the absence (lane 1) or in the presence of topo I (lane 2) with the purified histone octamers and analyzed by micrococcal nuclease digestion. The agarose gel was stained with ethidium bromide and the negative of the image is shown. (C) Transcription reactions were performed on either naked DNA templates (lanes 1 and 2) or on chromatin templates reconstituted using the Drosophila assembly factor Acf-1/ISWI and the chaperone NAP-1 (lanes 3–4). Human topo I and GAL4–VP16 were included in the indicated lanes. (D) Chromatin templates were reconstituted in the presence of Drosophila assembly factors either in the absence (lane 2) or the presence (lane 3) of topo I and analyzed by micrococcal nuclease assay. 100 bp markers are shown in lane 1. The image is a negative of an ethidium bromide stained gel.
Chromatin templates were prepared by a standard dialysis technique (13), using purified histone octamers (Supple mentary Material Fig. S1A), linearized DNAs, with or without purified topo I (Supplementary Material Fig. S1B). Digestion of chromatin templates with micrococcal nuclease revealed mononucleosomes and polynucleosomes, with a spacing of ∼160 bp, consistent with multiple nucleosomes per DNA plasmid template, and the presence of topo I had no effect on the chromatin assembly (Fig. 1B). When using the highly purified transcription factors, the topo I was most effective in stimulating transcription when it was included in the chromatin assembly reaction (data not shown). When using less pure protein preparations, a second activity can load the topoisomerase on to the template after chromatin assembly (3). These findings suggest that the topoisomerase must be embedded in the chromatin in order to stimulate transcription on chromatin.
Since several of the transcription factors were prepared from eukaryotic cells, it was possible that remodeling activities, or other chromatin-regulating activities, might contaminate the preparations. Transcription factors purified from cells, TFIID, TFIIH and pol II, were tested for the presence of the chromatin-specific elongation factor, FACT (2) by immunoblotting for the FACT subunit SSRP1. Using this analysis, these protein preparations had no detectable FACT (data not shown). We tested whether there is a chromatin remodeling activity (19) present in the core pol II, TFIID, TFIIH, topo IIα and topo I preparations that is contributing to the transcriptional activation which is being observed on chromatin templates. For testing the protein preparations, we used 4-fold more protein than is used in a typical transcription reaction in order to reveal low level contaminating remodeling activities. As shown in Supple mentary Material Figure S2, the pol II, TFIID, TFIIH, topo I and topo IIα preparations each had no detectable remodeling activity.
We next tested whether the effect of chromatin on transcription was dependent upon the method for preparing chromatin. We have been applying purified histone octamers directly onto the DNA templates by the salt dialysis technique. We tested whether the use of chromatin assembly factors affected the transcription results. The templates used in Figure 1C were prepared in a comparatively rapid reaction in the presence of Drosophila assembly factors (Acf-1, ISWI and NAP-1), and human topo I was included in reactions as indicated. The presence of topo I did not affect the chromatin assembly, as determined by micrococcal nuclease digestion analysis of the samples (Fig. 1D). In the absence of topo I, transcription on chromatin templates was repressed (Fig. 1C, lane 3). Interestingly, inclusion of the chromatin assembly factors in the transcription reactions resulted in transcription that was still active in the absence of a topoisomerase, albeit at low levels. The inclusion of the topoisomerase in the reaction stimulated the level of RNA synthesis above the repressed level (Fig. 1C, lane 4). These data clearly show that the stimulation of transcription on chromatin by topoisomerase is not wholly dependent upon a technical method.
We tested whether topo I and topo IIα, which are required for transcription on chromatin templates, can function as coactivators on naked DNA templates (20,21). Data shown in Supplementary Material Figure S3 reveal that both topo I and topo IIα can function as transcriptional coactivators, but only at a higher concentration than stimulates transcription on chromatin. Published experiments of topo I as a coactivator on naked DNA templates required topo I concentrations 40-fold higher per reaction (21) than stimulated transcription on chromatin in this study. This result suggests that the activity of topoisomerases as stimulators of transcription on chromatin templates is fundamentally different to the coactivator function of these proteins.
Topoisomerase activity inhibitors repress transcription on chromatin
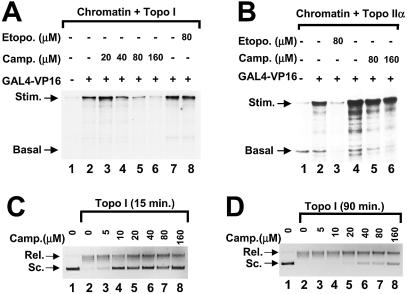
Next we used topoisomerase activity inhibitors to test whether it is the relaxation activity of the topoisomerase or protein–protein interactions that are required for the core pol II to overcome the repression by nucleosomes. Camptothecin specifically inhibits the relaxation activity of topo I, and etoposide specifically inhibits the relaxation activity of topo II. In Figure 2, chromatin templates were reconstituted either in the presence of purified topo I or topo IIα. Different amounts of camptothecin or etoposide were included in GAL4–VP16-stimulated transcription reactions and the effect of topoisomerase inhibitor on RNA synthesis was assessed. In Figure 2A, in the presence of topo I, RNA synthesis was decreased in the presence of 40 µM camptothecin and it was further inhibited at 80–160 µM camptothecin (lanes 4–6). Etoposide, which inhibits topo II relaxation activity, had no effect on these reactions (lane 8). In contrast, when transcription reactions contained topo IIα, 80 µM etoposide inhibited the GAL4–VP16-stimulated transcription, whereas camptothecin had no effect on RNA synthesis (Fig. 2B). These results strongly demonstrate the specificity of the topoisomerase function in transcription on chromatin templates and also that the specific topoisomerase poison only inhibited transcription when the reaction was dependent upon the relevant topoisomerase.
Figure 2.
Specificity of inhibition of transcription by topoisomerase poisons. (A) Chromatin templates were reconstituted in the presence of topo I. Transcription reactions were performed on these reconstituted chromatin templates either in the absence (lanes 1, 2 and 7) or the presence of the indicated concentrations of camptothecin (lanes 3–6) and etoposide (lane 8). GAL4–VP16 was included in lanes 2–8. (B) Chromatin templates were assembled in the presence of topo IIα and analyzed for GAL4–VP16-stimulated transcription. The indicated concentrations of etoposide (lane 3) and camptothecin (lanes 5 and 6) were included in the reactions. GAL4–VP16 was included in lanes 2–8. (C) The inhibitory concentration of camptothecin for topo I-dependent relaxation of supercoiled plasmid DNAs was analyzed in 15 min reactions. The figure shows an inverted image of an ethidium bromide stained agarose gel. Relaxation activity is present if the rapidly migrating supercoiled plasmid (Sc.) is converted to the relaxed form (Rel.). The inhibitory effect is ascertained by the retention of supercoiled plasmid. (D) Relaxation assays were repeated as in (C) with the exception that the time of incubation was 90 min.
The topo I-dependent transcription reaction was inhibited by 40–160 µM camptothecin as shown in Figure 2A. This concentration is quite high as compared with the concentration required to inhibit the topo I relaxation activity in vitro (15). It was possible that the long incubation time required for transcription assays affected the inhibition activity of the camptothecin. We tested the dependence of time of incubation on effective concentrations of camptothecin in the relaxation assay. We incubated the supercoiled DNA with topo I, either in the absence of camptothecin or in the presence of different amounts of camptothecin for 15 or 90 min (Fig. 2C and D). We observed that when the incubation time was 15 min, 5 µM camptothecin significantly reduced the relaxation activity and 10 µM camptothecin nearly eliminated the relaxation activity of topo I (Fig. 2C, lanes 2–4). In reactions in which the incubation time was 90 min, similar to the incubation time for in vitro transcription reactions, higher concentrations of camptothecin were required to inhibit the relaxation activity (Fig. 2D). In these longer assays a reduction in the relaxation activity was observed only in reactions containing camptothecin at concentrations higher than 40 µM. These results suggest, that given long incubation times, the residual topo I activity in the presence of 5–20 µM camptothecin is sufficient to relax superhelical tension in DNA. Since 90 min reactions require higher concentrations of camptothecin for inhibition of relaxation, the results in the relaxation assay in Figure 2D are consistent with the effect of 40–160 µM camptothecin on transcription in Figure 2A.
Non-human topoisomerases: yeast topo II, but not bacterial topo I, stimulates transcription on chromatin templates
The above data show that relaxation activity of human topoisomerase enzymes (topo IIα and topo I) rescued the transcription on chromatin templates. Next we tested the effect of non-human topoisomerase enzymes, yeast topo II and bacterial topo I, in transcription on chromatin templates. (Both enzymes were the gift of J. C. Wang, Harvard University.) Yeast topo II relaxes both positive and negative superhelical tension whereas bacterial topo I relaxes negative supercoils only.
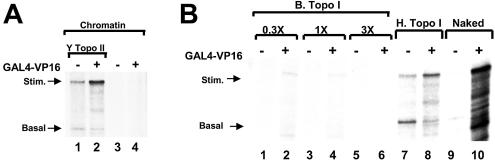
Chromatin templates were reconstituted either in the absence or in the presence of yeast topo II and activated transcription reactions were tested on these templates. Concentrations of the various topoisomerases used for transcription on chromatin templates were all normalized according to the relaxation activities (data not shown). In the presence of yeast topo II, GAL4–VP16-stimulated transcription was observed (Fig. 3A, lane 2). Thus, yeast topo II rescued transcription on chromatin templates. Next the effect of bacterial topo I on transcription from chromatin templates was evaluated. In the presence of bacterial topo I, synthesis of the GAL4–VP16-stimulated RNA remained repressed (Fig. 3B, lane 4). The amount of bacterial topo I included in reactions had been normalized with the other topoisomerase preparations according to the relaxation activity on negatively supercoiled plasmid DNA. We tested higher and lower concentrations of bacterial topo I in transcription on chromatin, and in no case was that topoisomerase as effective as human topo I in stimulating transcription (Fig. 3B). Thus, while human topo I, human topo IIα and yeast topo II could each reverse the repression of transcription by chromatin in this experimental setting, bacterial topo I could not. These results suggest that the transcriptional repression by chromatin is reversed only by those topoisomerase enzymes which efficiently relax positive superhelical tension.
Figure 3.
Yeast topo II, but not bacterial topo I, stimulates transcription on chromatin templates. (A) Chromatin templates were reconstituted either in the presence of yeast topo II (Y topo II) (lanes 1 and 2) or in the absence of topoisomerase (lanes 3 and 4) and were tested in transcription assays. GAL4–VP16 was included in alternate lanes. (B) Transcription on naked DNA templates (lanes 9 and 10) was compared with transcription on chromatin templates reconstituted with human topo I (lanes 7 and 8) or with an equivalent amount of bacterial topo I (B Topo I), as determined by relaxation activity (lanes 3 and 4). In the indicated lanes, 3-fold lower bacterial topo I or 3-fold higher amounts of bacterial topo I were included. GAL4–VP16 was included in even number lanes.
The requirement for topo I in transcription is dependent on the length of the transcript
We predict that the topoisomerase activity in transcription on chromatin DNA is required to relax superhelical tension that accumulates during elongation. According to this model, synthesis of short transcripts would not be affected by chromatin. As the GAL4–VP16-responsive template encoded a 384 nt transcript, we generated templates that would yield shorter transcripts for this analysis. Different lengths of G-less cassettes were fused downstream of promoters containing five repeated GAL4 binding sites, keeping the total length of the DNA to ∼3000 bp. Transcript lengths were 25, 50, 100, 200 and 384 nt. If the initiation step is limiting and responsible for the repression, then the repression by chromatin would be independent of the length of the RNA transcript. In contrast, if polymerase elongation is repressed by the nucleosome-bound template, then the length of the transcript would be a critical parameter, and with the increase in length the amount of repression should increase.
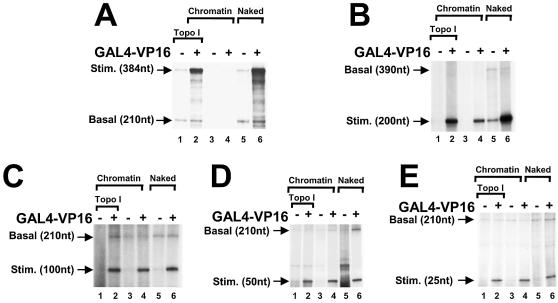
Activated transcription was tested on these naked templates (Fig. 4A). In alternate lanes GAL4–VP16 was included and accumulation of the GAL4-responsive transcript was assayed (lanes 2, 4, 6, 8 and 10). In all the reactions, except in lanes 7 and 8, we included the basal template that encoded a 210 nt transcript. In lanes 7 and 8, since the activator-responsive template generated a transcript of 200 nt, we used a basal template that encoded a 390 nt transcript. When using naked templates, encoding RNAs ranging from 25 to 384 nt, the level of activated transcription was unaffected. This result indicated that on naked DNA templates, transcriptional activation by GAL4–VP16 stimulated the initiation process or early elongation events such as promoter clearance, consistent with prior observations (22,23). Next we tested the same templates, reconstituted in chromatin in the absence of a topoisomerase activity. GAL4–VP16-stimulated transcription was observed in reactions encoding 25, 50, 100 or 200 nt RNAs (Fig. 4B, lanes 2, 4, 6 and 8). In contrast, transcription of the 384 nt RNA was repressed even when the activator was added (Fig. 4B, lane 10). From this result we conclude that the repression of GAL4–VP16-stimulated transcription by chromatin is primarily due to inhibition of elongation. These results from Figure 4 indicate that RNA synthesis occurs on short templates, but whether there was partial repression of transcription, relative to RNA synthesis from naked templates, could not be determined.
Figure 4.
Repression of transcription on chromatin templates was dependent upon the length of the transcript. (A) Naked DNA templates containing different lengths of G-less cassette were assayed by GAL4–VP16-stimulated transcription. All of the reactions contain two templates in equal concentrations, encoding GAL4-responsive transcripts of varying lengths (Stim.) and a basal control transcript (Basal). Reactions containing the 25 nt transcript were analyzed on a different gel from the other samples. When analyzing the 200 nt GAL4-responsive transcript, the basal control was 390 nt (lanes 7 and 8). In all other reactions, the basal control was a 210 nt RNA. (B) In vitro transcription reactions as in (A), except that the templates were reconstituted in nucleosomes. GAL4–VP16-stimulated RNAs are indicated by arrowheads.
In order to test for partial repression, we directly compared each length of transcript for activated transcription on the different templates. Figure 5A shows activated transcription on a DNA template containing GAL4-response elements which yields a transcript of 384 nt. As was observed in previous experiments (Fig. 1), when reconstituted in chromatin, in the absence of topo I, repression of transcription was observed (Fig. 5A, lanes 3 and 4) whereas in the presence of topo I, the polymerase was almost as active as when transcribing the naked DNA template (lanes 1–2 and 5–6). When the GAL4-responsive template encoded a 200 nt RNA, the level of activated transcription on the chromatin template in the absence of topo I was decreased as compared with the chromatin template in the presence of topo I or the naked DNA (Fig. 5B). While transcription of the 384 nt RNA was nearly completely repressed, transcription of the 200 nt RNA on chromatin in the absence of topoisomerase was decreased by approximately half. No difference in the level of transcription was observed when similar reactions were performed on different forms of the DNA template that encoded 100, 50 or 25 nt transcripts (Fig. 5). From these shorter transcripts, the transcription level remained the same even when we used chromatin templates which were reconstituted in the absence of topo I. These results indicate that when using GAL4–VP16 and purified transcription factors, the repression by chromatin is dependent upon the length of the nascent RNA. Transcription on chromatin was not repressed at the initiation stage of the reaction since all three template conditions were equally well transcribed when using 25, 50 or 100 nt templates. Significant repression by chromatin was detected only when the nascent RNA was 200 nt or longer. This repression of the synthesis of longer transcripts was reversed by topo I.
Figure 5.
Topo I stimulated transcription of RNAs 200 nt or longer from chromatin templates. (A) GAL4–VP16-stimulated transcription of the 384 nt RNA was analyzed using chromatin templates containing topo I (lanes 1 and 2), chromatin without topo I (lanes 3 and 4) or naked DNA (lanes 5 and 6). Each reaction contained 30 ng of the GAL4-responsive template DNA and 30 ng of the basal template DNA. GAL4–VP16 was included in even lanes. (B) GAL4–VP16-stimulated transcriptions were analyzed as in (A) except that the GAL4-responsive template encoded a 200 nt RNA, and the basal control template encoded a 390 nt RNA, by changing the length of the G-less cassette and keeping the rest of the DNA template the same. (C) As in (A), except that the GAL4-responsive template encoded a 100 nt RNA. (D) As in (A), except that the GAL4-responsive template encoded a 50 nt RNA. (E) As in (A), except that the GAL4-responsive template encoded a 25 nt RNA.
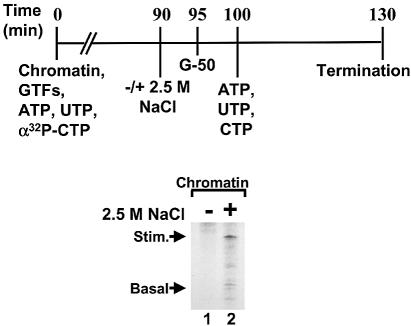
The above results show that the repression by nucleosome-bound DNA in the absence of a topoisomerase is at the elongation stage. When using the 384 nt template, shorter transcripts were not apparent (e.g. Fig. 1A). Since such abortive transcripts would not be focused in a single band, we hypothesized that if the elongation block were removed, then the 384 nt transcript would be apparent. To check this, chromatin templates were assayed for transcription in the absence of topo I. After the typical 90 min reaction, the reaction was treated with either low salt (0.045 M KOAc, Fig. 6, lane 1) or with 2.5 M NaCl (lane 2). At high salt concentration, the nucleosomes will dissociate and thus the superhelical torsion will diffuse off the end of the DNA, as in naked DNA templates. After treating with high salt, the samples were passed through a Sephadex G-50 column, which had been equilibrated at 90 mM KOAc. This maneuver dissociated the histone octamers from the template, and the rapid change to 90 mM KOAc would prevent reformation of chromatin. In addition, passage through a Sephadex column removed unincorporated radionucleotide triphosphates. After addition of Mg(OAc)2 and cold ATP, UTP and CTP, transcription elongation was allowed to continue for another 30 min. Newly initiated transcripts would not be labeled, thus this assay detected only those transcripts that paused in the presence of nucleosomes. The appearance of product RNA was dependent upon disruption of the nucleosomes using high salt (Fig. 6, lane 2). This result is consistent with the model that nucleosomes stabilize the accumulation of positive superhelical tension that arises as a byproduct of the transcription process. Removal of the nucleosomes from templates containing stalled pol II allowed the diffusion of the inhibitory superhelical tension and transcription elongation could recommence.
Figure 6.
Transcription on chromatin templates is blocked at elongation. Chromatin templates were reconstituted in the absence of topoisomerase enzyme and assayed for activated transcription as usual (90 min). The samples were then either treated with 2.5 M NaCl (lane 2) or with low salt (lane 1) for 5 min and then subjected to gel filtration on a Sephadex G-50 column, chased with cold ATP/UTP/CTP for 30 min. The reactions were terminated and then analyzed. The 384 nt GAL4-responsive transcript (Stim.) and the 210 nt basal control transcript (Basal) are indicated.
DISCUSSION
Transcription is repressed on chromatin templates. While DNA accessibility in chromatin has been a focus for much of the research in this field, in this study we establish that DNA superhelical tension in chromatin is an important parameter which affects the efficiency of RNA pol II elongation. Our prior results had revealed that topo IIα is required for transcription on chromatin, but the mechanism underlying the requirement was not demonstrated (3). In this study we demonstrate that the topoisomerase is important during the elongation step of RNA synthesis. During transcription, positive supercoils are generated in front of the elongating RNA pol II and negative supercoils behind it (5,24). From our data, we conclude that nucleosomes block superhelical tension from diffusing. The introduction of superhelical tension during transcription thus results in hyperwinding of the downstream template and inhibition of transcription elongation.
On the basis of our results, we propose a model (Fig. 7) which shows that in the presence of a topoisomerase enzyme active on positively oriented superhelical turns, the repression of transcription on chromatin templates is reversed. For the purposes of the diagram, the DNA template is shown as a long linker bracketed by nucleosomes. Based upon micrococcal nuclease digestion patterns, the separation between nucleosomes in our templates is significantly shorter, no more than 20 bp. We believe that the interaction between pol II and nucleosome-bound template is more complex, but this would be difficult to diagram. According to this model, as elongation proceeds, positive superhelical torsion accumulates (diagramed as DNA with extra turns). A corollary of this model is that, since the process of elongation results in the accumulation of superhelical tension, only long transcriptional units (G-less cassettes) will accumulate sufficient superhelical tension to block further elongation. This corollary is consistent with the transcript length dependence revealed in this study. Transcripts of 200 nt and above were significantly repressed. Clearly, most pol II transcripts are significantly longer than this, and it is readily extrapolated that this property will be important in transcription in general. Indeed, transcription in cells has been shown to be affected by topoisomerase function. Topo I has been detected at loci of Drosophila polytene chromosomes that are transcriptionally active (25). The rates of pol I and pol II transcription were inhibited when anti-topo I antibody was microinjected into the nuclei of Chironomus tentans salivary gland cells (26), and the topo I inhibitor camptothecin repressed rRNA synthesis (27). Double temperature sensitive mutants of topo I and topo II in yeast grown at the restrictive temperature resulted in the reduction of rRNA, and to a lesser extent mRNA, synthesis (7,10). Treating cells with inhibitors of topo I or topo II results in the rapid decrease in the pol II-dependent transcription activity of certain genes (11). These results all suggest that within the complex milieu of the nucleus of the cell, topoisomerase function can be linked with transcription function. Our results demonstrate the topoisomerase function in transcription in a defined biochemical system, and also suggest a model for the requirement.
Figure 7.
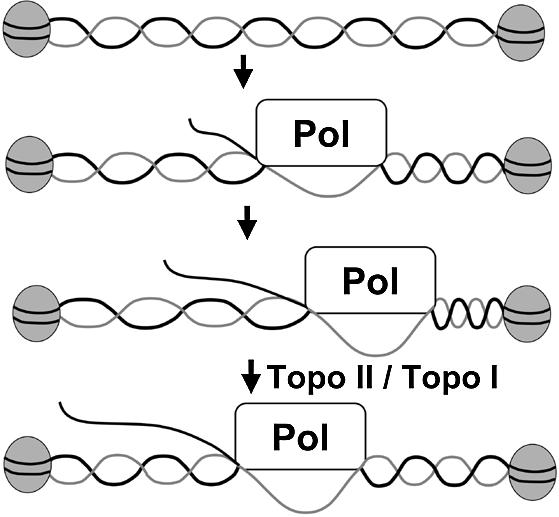
Model for the requirement of a topoisomerase activity on a chromatin template in pol II transcription. We model that nucleosomes stabilize the superhelical tension, which arises during transcription elongation. The level of positive superhelical torsion is diagramed to increase with longer RNA transcripts. The relaxation activity of the topoisomerase enzyme is modeled to be required during transcription on chromatin templates, to relax the positively oriented supercoils.
Our results indicate that pol II elongation is a critical limiting step for transcription on chromatin templates. In contrast to our results, initiation of transcription by pol II has been found in other studies to be repressed by nucleosomes (28). It is unclear why there is no block to initiation in our assay system while there is in other systems. Perhaps the sources of transcription factors contribute to the different results. Similar to our current results, other studies have also indicated that elongation is the critically regulated step in the transcription by pol II on chromatin templates (29), although a role for topoisomerases in elongation was not identified.
It is possible that there are alternate pathways, which modify the chromatin structure to bypass the topoisomerase requirement. These pathways would involve chromatin remodeling complexes, such as SWI/SNF, which have been found to increase negative superhelical tension on the DNA (30) and would thus neutralize the effect of positive superhelical tension. Alternatively, histone acetyl transferases, or other complexes that covalently modify the nucleosome, could potentially regulate the superhelical tension on chromatin. One possibility is that acetylation of histones alters the property of nucleosomes in functioning as a barrier to the diffusion of superhelical tension. If true, then acetylation of histones would not only mark sites for chromatin binding proteins, but also make the chromatin permissive to efficient elongation.
Topoisomerase inhibitors are used as drugs for the treatment of malignancies. In human cells, death associated with the topoisomerase inhibitors occurs in both the S and G1 phases of the cell cycle. As an example of this principle, the G1 effects of the topo II poison etoposide (31–33) are most easily consistent with a transcription function for topo II. Perhaps the cytotoxic effect of these topo I and topo II targeting drugs occurs via the transcription function.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We thank K. Daniels, D. M. Langenau, M. Zheng, U. Banerjee and W. Wei for preparation of extracts and J. C. Wang for purified bacterial topo I and yeast topo II. We thank A. M. Zhelkovsky and C. L. Moore for the topo I expression vectors, R. Roeder for the 5S rDNA array plasmid, J. Kadonaga for Drosophila NAP-1 and Acf-1 expression vectors, C. Wu for the Drosophila ISWI expression vector and S. Lippard for antibody specific to SSRP1. This work was supported by a Public Health Service grant GM-53504 from the National Institute of General Medical Sciences.
REFERENCES
- 1.Hampsey M. (1998) Molecular genetics of the RNA polymerase II general transcriptional machinery. Microbiol. Mol. Biol. Rev., 62, 465–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.LeRoy G., Orphanides,G., Lane,W.S. and Reinberg,D. (1998) Requirement of RSF and FACT for transcription of chromatin templates in vitro. Science, 282, 1900–1904. [DOI] [PubMed] [Google Scholar]
- 3.Mondal N. and Parvin,J.D. (2001) DNA topoisomerase IIalpha is required for RNA polymerase II transcription on chromatin templates. Nature, 413, 435–438. [DOI] [PubMed] [Google Scholar]
- 4.Owen-Hughes T. and Workman,J.L. (1994) Experimental analysis of chromatin function in transcription control. Crit. Rev. Eukaryot. Gene Expr., 4, 403–441. [PubMed] [Google Scholar]
- 5.Liu L.F. and Wang,J.C. (1987) Supercoiling of the DNA template during transcription. Proc. Natl Acad. Sci. USA, 84, 7024–7027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang J.C. (1996) DNA topoisomerases. Annu. Rev. Biochem., 65, 635–692. [DOI] [PubMed] [Google Scholar]
- 7.Brill S.J., DiNardo,S., Voelkel-Meiman,K. and Sternglanz,R. (1987) Need for DNA topoisomerase activity as a swivel for DNA replication for transcription of ribosomal RNA. Nature, 326, 414–416. [DOI] [PubMed] [Google Scholar]
- 8.Yamagishi M. and Nomura,M. (1988) Deficiency in both type I and type II DNA topoisomerase activities differentially affect rRNA and ribosomal protein synthesis in Schizosaccharomyces pombe. Curr. Genet., 13, 305–314. [DOI] [PubMed] [Google Scholar]
- 9.Gartenberg M.R. and Wang,J.C. (1992) Positive supercoiling of DNA greatly diminishes mRNA synthesis in yeast. Proc. Natl Acad. Sci. USA, 89, 11461–11465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schultz M.C., Brill,S.J., Ju,Q., Sternglanz,R. and Reeder,R.H. (1992) Topoisomerases and yeast rRNA transcription: negative supercoiling stimulates initiation and topoisomerase activity is required for elongation. Genes Dev., 6, 1332–1341. [DOI] [PubMed] [Google Scholar]
- 11.Collins I., Weber,A. and Levens,D. (2001) Transcriptional consequences of topoisomerase inhibition. Mol. Cell. Biol., 21, 8437–8451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Manley J.L., Fire,A., Samuels,M. and Sharp,P.A. (1983) In vitro transcription: whole-cell extract. Methods Enzymol., 101, 568–582. [DOI] [PubMed] [Google Scholar]
- 13.Workman J.L., Taylor,I.C., Kingston,R.E. and Roeder,R.G. (1991) Control of class II gene transcription during in vitro nucleosome assembly. Methods Cell Biol., 35, 419–447. [DOI] [PubMed] [Google Scholar]
- 14.Ito T., Levenstein,M.E., Fyodorov,D.V., Kutach,A.K., Kobayashi,R. and Kadonaga,J.T. (1999) ACF consists of two subunits, Acf1 and ISWI, that function cooperatively in the ATP-dependent catalysis of chromatin assembly. Genes Dev., 13, 1529–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhelkovsky A.M. and Moore,C.L. (1994) Overexpression of human DNA topoisomerase I in insect cells using a baculovirus vector. Protein Expr. Purif., 5, 364–370. [DOI] [PubMed] [Google Scholar]
- 16.Haile D.T. and Parvin,J.D. (1999) Activation of transcription in vitro by the BRCA1 carboxyl-terminal domain. J. Biol. Chem., 274, 2113–2117. [DOI] [PubMed] [Google Scholar]
- 17.Chiang C.M. and Roeder,R.G. (1993) Expression and purification of general transcription factors by FLAG epitope-tagging and peptide elution. Pept. Res., 6, 62–64. [PubMed] [Google Scholar]
- 18.Kretzschmar M., Kaiser,K., Lottspeich,F. and Meisterernst,M. (1994) A novel mediator of class II gene transcription with homology to viral immediate-early transcriptional regulators. Cell, 78, 525–534. [DOI] [PubMed] [Google Scholar]
- 19.Jonsson Z.O., Dhar,S.K., Narlikar,G.J., Auty,R., Wagle,N., Pellman,D., Pratt,R.E., Kingston,R. and Dutta,A. (2001) Rvb1p and Rvb2p are essential components of a chromatin remodeling complex that regulates transcription of over 5% of yeast genes. J. Biol. Chem., 276, 16279–16288. [DOI] [PubMed] [Google Scholar]
- 20.Kretzschmar M., Meisterernst,M. and Roeder,R.G. (1993) Identification of human DNA topoisomerase I as a cofactor for activator-dependent transcription by RNA polymerase II. Proc. Natl Acad. Sci. USA, 90, 11508–11512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Merino A., Madden,K.R., Lane,W.S., Champoux,J.J. and Reinberg,D. (1993) DNA topoisomerase I is involved in both repression and activation of transcription. Nature, 365, 227–232. [DOI] [PubMed] [Google Scholar]
- 22.Lin Y.S., Ha,I., Maldonado,E., Reinberg,D. and Green,M.R. (1991) Binding of general transcription factor TFIIB to an acidic activating region. Nature, 353, 569–571. [DOI] [PubMed] [Google Scholar]
- 23.Shykind B.M., Kim,J., Stewart,L., Champoux,J.J. and Sharp,P.A. (1997) Topoisomerase I enhances TFIID–TFIIA complex assembly during activation of transcription. Genes Dev., 11, 397–407. [DOI] [PubMed] [Google Scholar]
- 24.Wang J.C. and Lynch,A.S. (1993) Transcription and DNA supercoiling. Curr. Opin. Genet. Dev., 3, 764–768. [DOI] [PubMed] [Google Scholar]
- 25.Fleischmann G., Pflugfelder,G., Steiner,E.K., Javaherian,K., Howard,G.C., Wang,J.C. and Elgin,S.C. (1984) Drosophila DNA topoisomerase I is associated with transcriptionally active regions of the genome. Proc. Natl Acad. Sci. USA, 81, 6958–6962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Egyhazi E. and Durban,E. (1987) Microinjection of anti-topoisomerase I immunoglobulin G into nuclei of Chironomus tentans salivary gland cells leads to blockage of transcription elongation. Mol. Cell. Biol., 7, 4308–4316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang H., Wang,J.C. and Liu,L.F. (1988) Involvement of DNA topoisomerase I in transcription of human ribosomal RNA genes. Proc. Natl Acad. Sci. USA, 85, 1060–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lorch Y., LaPointe,J.W. and Kornberg,R.D. (1987) Nucleosomes inhibit the initiation of transcription but allow chain elongation with the displacement of histones. Cell, 49, 203–210. [DOI] [PubMed] [Google Scholar]
- 29.Izban M.G. and Luse,D.S. (1992) Factor-stimulated RNA polymerase II transcribes at physiological elongation rates on naked DNA but very poorly on chromatin templates. J. Biol. Chem., 267, 13647–13655. [PubMed] [Google Scholar]
- 30.Havas K., Flaus,A., Phelan,M., Kingston,R., Wade,P.A., Lilley,D.M. and Owen-Hughes,T. (2000) Generation of superhelical torsion by ATP-dependent chromatin remodeling activities. Cell, 103, 1133–1142. [DOI] [PubMed] [Google Scholar]
- 31.Jensen L.H., Nitiss,K.C., Rose,A., Dong,J., Zhou,J., Hu,T., Osheroff,N., Jensen,P.B., Sehested,M. and Nitiss,J.L. (2000) A novel mechanism of cell killing by anti-topoisomerase II bisdioxopiperazines. J. Biol. Chem., 275, 2137–2146. [DOI] [PubMed] [Google Scholar]
- 32.Liu L.F. (1989) DNA topoisomerase poisons as antitumor drugs. Annu. Rev. Biochem., 58, 351–375. [DOI] [PubMed] [Google Scholar]
- 33.Sullivan D.M., Glisson,B.S., Hodges,P.K., Smallwood-Kentro,S. and Ross,W.E. (1986) Proliferation dependence of topoisomerase II mediated drug action. Biochemistry, 25, 2248–2256. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.