Abstract
Antisense oligonucleotides are used for therapeutic applications and in functional genomic studies. In practice, however, many of the oligonucleotides complementary to an mRNA have little or no antisense activity. Theoretical strategies to improve the ‘hit rate’ in antisense screens will reduce the cost of discovery and may lead to identification of antisense oligonucleotides with increased potency. Statistical analysis performed on data collected from more than 1000 experiments with phosphorothioate-modified oligonucleotides revealed that the oligo-probes, which form stable duplexes with RNA (ΔGo37 ≤ –30 kcal/mol) and have small self-interaction potential, are more frequently efficient than molecules that form less stable oligonucleotide–RNA hybrids or more stable self-structures. To achieve optimal statistical preference, the values for self-interaction should be (ΔGo37) ≥ –8 kcal/mol for inter-oligonucleotide pairing and (ΔGo37) ≥ –1.1 kcal/mol for intra-molecular pairing. Selection of oligonucleotides with these thermodynamic values in the analyzed experiments would have increased the ‘hit rate’ by as much as 6-fold.
INTRODUCTION
Antisense oligonucleotides in current use are modified DNA molecules that hybridize to complementary mRNA and inhibit expression of its encoded product. In principle, the antisense approach is universal and specific. It can be used to inhibit expression of any mRNA, and a single protein isoform can be shut down without affecting closely related proteins. Antisense oligonucleotides are used for therapeutic applications and in functional genomic studies. In practice, however, many of the oligonucleotides complementary to an mRNA have little or no antisense activity. Typically, several oligonucleotides are synthesized and tested and only some are active. Theoretical strategies to improve the ‘hit rate’ in antisense screens will reduce the cost of discovery and may lead to identification of antisense oligonucleotides with increased activity or potency. Theoretical prediction of RNA target sites for active oligonucleotides is related to the development of algorithms that can locate single-stranded regions in RNA secondary structure models (1–7). There is some experimental evidence that oligonucleotides designed to target these non-structured RNA regions are indeed frequently efficient in downregulation of particular gene products (1–5). It is not known how much oligonucleotide self-pairing decreases the ‘hit-rate’. Software for calculation of thermodynamic properties of oligonucleotide structure, target RNA structure and duplex formation has been developed (7). However, correlations between these thermodynamic properties and the ‘hit rates’ for large databases of antisense experiments have not yet been reported. This study supplies the missing information.
MATERIALS AND METHODS
Databases
For this work, two databases were used. The first one includes data from antisense oligonucleotide screening experiments reported in the literature (8). This database is available on the Web (http://antisense.genetics.utah.edu). The second database utilizes the data from experiments performed at Isis Pharmaceuticals and were not yet reported in the literature. These databases include activity values and antisense oligonucleotide sequences. Activity value is expressed as the ratio of the level of a particular mRNA or protein measured in cells after treatment with the experimental antisense oligonucleotide versus the level of the same mRNA or protein measured in untreated cells. There are 316 oligonucleotides in the first database and 908 in the second.
Thermodynamic calculations
Thermodynamic properties for oligonucleotides and relevant duplexes were calculated using the programs OligoWalk (7) and OligoScreen from the package RNAstructure 3.5 (http://128.151.176.70/RNAstructure.html). OligoWalk predicts the equilibrium affinity of complementary DNA or RNA oligonucleotides to an RNA target by calculating ΔGooverall values. These ΔGooverall values are calculated by consideration of ΔGo37 values relevant to the predicted stability of the oligonucleotide–target duplex and the competition with predicted secondary structure of both the target and the oligonucleotide. Both ΔGo37 values relevant to inter- and intra-molecular oligonucleotide self-structures are considered at a user-defined concentration. One thousand suboptimal structures were created for each mRNA target molecule. The disruption in RNA secondary structures included the free energy required for target rearrangement. OligoScreen (http://rna.chem.rochester.edu) considers only the predicted stability of the oligonucleotide–target duplex and the competition with predicted secondary structure of the oligonucleotide without consideration of target RNA secondary structure. For determination of ΔGo37, both programs use thermodynamic parameters for the nearest-neighbor model (9–13).
Statistical analysis
Statistical tools from Excel (Microsoft, Inc.) were used for correlation analysis (t-test) and scatter plot data presentations.
RESULTS AND DISCUSSION
Statistical analysis has been performed on data collected from more than 1000 experiments with phosphorothioate-modified antisense oligonucleotides. Oligonucleotides that form stable duplexes with RNA [free energies (ΔGo37) ≤ –30 kcal/mol] and have small self-interaction potential are statistically more likely to be active than molecules that form less stable oligonucleotide–RNA hybrids or more stable self-structures. To achieve optimal statistical preference, the values for self-interaction should be (ΔGo37) ≥ –8 kcal/mol for inter- oligonucleotide pairing and (ΔGo37) ≥ –1.1 kcal/mol for intra-molecular pairing. Selection of oligonucleotides with these thermodynamic values in the analyzed experiments would have increased the proportion of active oligonucleotides by as much as 6-fold.
The equilibrium affinity of an oligonucleotide for target RNA is influenced by the stability of the potential RNA–DNA duplex and by the stability of competing structures including the oligonucleotide self-structure and the target RNA structure. The program OligoWalk (7) calculates ΔGo37 values for each of these structures. In addition, ΔGooverall, the overall Gibbs free energy change of RNA binding at 37°C for each oligonucleotide, is determined. These ΔGooverall values are calculated by consideration of ΔGo37 values relevant to the predicted stability of the oligonucleotide–target duplex and the competition with predicted secondary structure of both the target and the oligonucleotide. Both ΔGo37 values relevant to inter- and intra-molecular oligonucleotide self-structures are considered at a user-defined concentration. In theory, the efficiency of oligonucleotide–RNA binding should correlate positively with the stability of the potential RNA–DNA duplex and correlate negatively with the stabilities of the oligonucleotide and mRNA secondary structures. Thus ΔGooverall should correlate with experimental efficacy of the oligonucleotides better than any individual parameter.
Our findings for the database of experiments reported in the literature are shown in Table 1. Surprisingly, the correlation between values of ΔGooverall and antisense oligonucleotide efficacy is very weak. Moreover, the stability of RNA secondary structures that must be disrupted for oligonucleotide–RNA helix formation does not correlate significantly with antisense efficacy. However, significant correlation was detected between antisense efficacy and ΔGoT values associated with the stability of oligonucleotide self-structures and oligonucleotide–RNA duplexes.
Table 1. Correlations between thermodynamic properties of oligonucleotides and their antisense activity.
| Correlation coefficient | Significance | |
|---|---|---|
| ΔGo37 overall versus ln(activity) | 0.17 | 0.01 |
| ΔGo37 duplex versus ln(activity) | 0.24 | 2.3 × 10–5 |
| ΔGo37 oligo intra-molecular structure versus ln(activity) | –0.12 | 0.03 |
| ΔG°37 oligo inter-molecular structure versus ln(activity) | –0.16 | 0.005 |
| ΔG°37 target RNA secondary structure versus ln(activity) | No significant correlation |
The lack of correlation between efficacy and the stability of mRNA secondary structure may be due to inaccuracies in the mRNA secondary structure prediction and other factors discussed previously (7). Because no correlation was found for the predicted RNA secondary structure stability with antisense activity, and because the theoretical prediction of RNA secondary structure by free energy minimization is the most time consuming step of the calculations, we focused further statistical analysis on thermodynamic parameters of the oligonucleotides and their duplexes with the target RNA. The previous studies of hybridization data produced with oligo-probes immobilized on arrays demonstrated that consideration of duplex stability between DNA and RNA, as well as considerations of oligonucleotide self-structure stability, can be sufficient for elimination of oligo-probes that hybridize poorly with the targets (14,15).
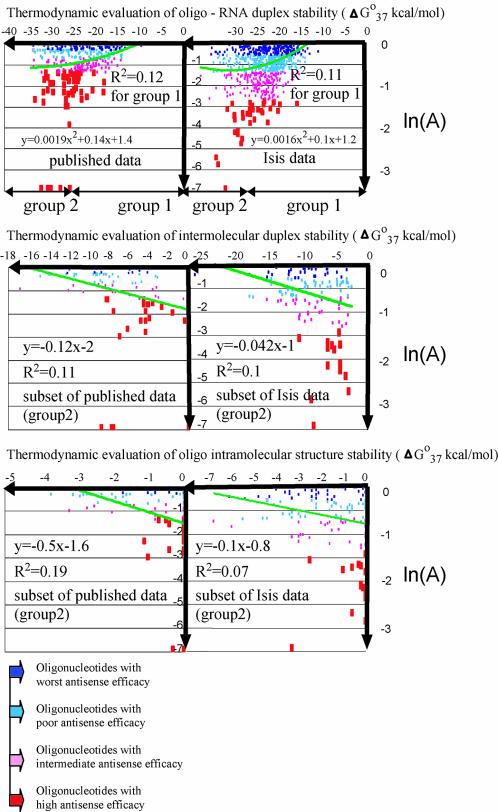
Scatter plots (Fig. 1) illustrate the relationship between activity and thermodynamic properties of antisense oligonucleotides from both the published and Isis databases. Since the slope of the trend line in scatter plots indicates the existence of a correlation between two variables, a correlation between thermodynamic evaluation of oligonucleotide–RNA duplex stability and antisense efficacy is evident for both databases (Fig. 1, top two plots), especially for subsets of data in the range of ΔGo37 duplex values from –30 to –10 kcal/mol. Flattening trend lines for subsets of data with ΔGo37 duplex values < –30 kcal/mol indicate a very weak correlation, or its absence. Categorization of databases into two groups was done with ΔGo37 duplex = –30 kcal/mol as a cut off point. The first group included oligonucleotides that target RNA with less favorable free energy for duplex formation (ΔGo37 duplex values ranging from –30 to –10 kcal/mol), i.e. oligonucleotides that form less stable duplexes with RNA. The second group includes oligonucleotides that target RNA with more favorable free energy for duplex formation (ΔGo37 duplex ranging from –40 to –30 kcal/mol), i.e. oligonucleotides that form more stable duplexes with RNA. The second group in each database is smaller than the first group (30 and 16% from the total number of molecules in the published and Isis data, respectively). For both databases, positive correlations between oligonucleotide activity and absolute values of ΔGo37 duplex for oligonucleotide–RNA duplexes were significant for the first group and not significant for the second (Table 2). In contrast, negative correlations between oligonucleotide activities and absolute ΔGo37 values of oligonucleotide self-pairing were undetectable in the first group, but were highly significant for the second (Table 2). The relevant scatter plots (Fig. 1, middle and bottom plots) demonstrate the relationship of activity of antisense oligonucleotides and thermodynamic evaluations of their self-pairing potentials. The slopes of the trend lines indicate the existence of a negative correlation between these variables for the second group of molecules. As mentioned earlier, relevant correlations were not detected for oligonucleotides from group 1, and the scatter plots with flat trend lines are not shown.
Figure 1.
Scatter plots showing the relationship between thermodynamic parameters and antisense oligonucleotide activities from both databases. Activity values (A) are expressed as the ratio of the level of a particular mRNA or protein measured in cells treated with an antisense oligonucleotide, to the level of the same mRNA or protein in untreated cells. Linear or non-linear trend lines are shown in each scatter plot.
Table 2. Correlations between thermodynamic properties of antisense oligonucleotides and their antisense activities for two experimental databases.
| Correlation coefficient | Significance | Number of oligos in the group | (G+C)/(A+G+C+T) | ||
|---|---|---|---|---|---|
| Group 1 oligos that are forming less stable duplexes with target RNA (ΔG°37 > –30 kcal/mol) | |||||
| Published data | ΔG°37 of oligo-target duplex versus ln(activity) | 0.36 | 0.00017 | 219 | 50 |
| ΔG°37 of oligo intra-molecular structure versus ln(activity) | Significant correlation is absent | ||||
| ΔG°37 of oligo intra-molecular structure versus ln(activity) | Significant correlation is absent | ||||
| Isis data | ΔG°37 of oligo target duplex versus ln(activity) | 0.35 | 2 × 10–23 | 762 | 44 |
| ΔG°37 of oligo intra-molecular structure versus ln(activity) | Significant correlation is absent | ||||
| ΔG°37 of oligo-intra-molecular structure versus ln(activity) | Significant correlation is absent | ||||
| Group 2 oligos that are forming more stable duplexes with target RNA (ΔG°37 ≤ –30kcal/mol) | |||||
| Published data | ΔG°37 of oligo target duplex versus ln(activity) | Significant correlation is absent | 97 | 68 | |
| ΔG°37 of oligo intra-molecular structure versus ln(activity) | 0.37 | 0.00017 | |||
| ΔG°37 of oligo intra-molecular structure versus ln(activity) | –0.27 | 0.006 | |||
| Isis data | ΔG°37 of oligo target duplex versus ln(activity) | Significant correlation is absent | 146 | 73 | |
| ΔG°37 of oligo intra-molecular structure versus ln(activity) | –0.22 | 0.007 | |||
| ΔG°37 of oligo intra-molecular structure versus ln(activity) | –0.3 | 0.003 | |||
The list of potential explanations for the scatter in groups 1 and 2 in Figure 1 include: variations in local secondary structure stabilities of RNA targets that were not picked up by OligoWalk, variations in uptake of oligonucleotides in different experiments, differential degradation in cells, or variations in intensities of non-specific interactions with undesired RNA targets.
The results of the correlation analysis for the oligonucleotides in the database of published data are presented graphically in Figure 2, and the results for the database of Isis unpublished data are in Figure 3. For both databases, the proportion of oligonucleotides with high antisense efficacy is larger in the group predicted to form more stable oligonucleotide–RNA duplexes than in the group that forms less stable hybrids. Figures 2 and 3 also graphically illustrate a negative correlation between antisense activity and the propensity for formation of self-structure by the group of oligonucleotides that are also able to form stable oligo–RNA duplexes. The thermodynamic parameters for phosphorothioate-modified DNA oligonucleotide hybridization are not available from the literature, and thus the parameters for non-modified DNA were used as an approximation. It is possible that a specific set of parameters for phosphorothioates would improve the correlation with antisense activity.
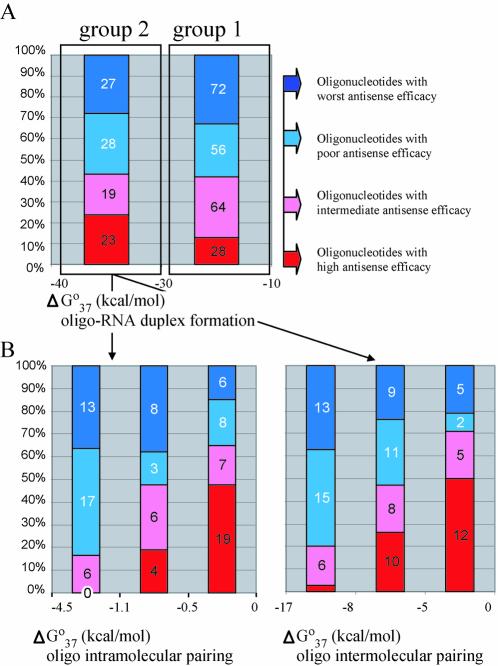
Figure 2.
Relationship between thermodynamic parameters and antisense oligonucleotide activities determined for the web database. (A) Oligo nucleotides were categorized into two groups according to calculated values of ΔGo37 for DNA–RNA duplex formation. Group 1 contains oligonucleotides that form more stable duplexes, and group 2 contains oligonucleotides that form less stable duplexes with target RNA. (B) Group 1 oligonucleotides separated on the basis of the calculated ΔGo37 for oligonucleotide intra-molecular pairing. (C) Group 1 oligonucleotides separated on the basis of the calculated ΔGo37 for oligonucleotide inter-molecular pairing. Blue represents the least active oligonucleotides, 75–100% of untreated control; violet, 50–75% of control; pink, 25–50% of control; and red, 0–25% of control. The numbers of oligonucleotides in each subgroup are indicated in the relevant highlighted segments.
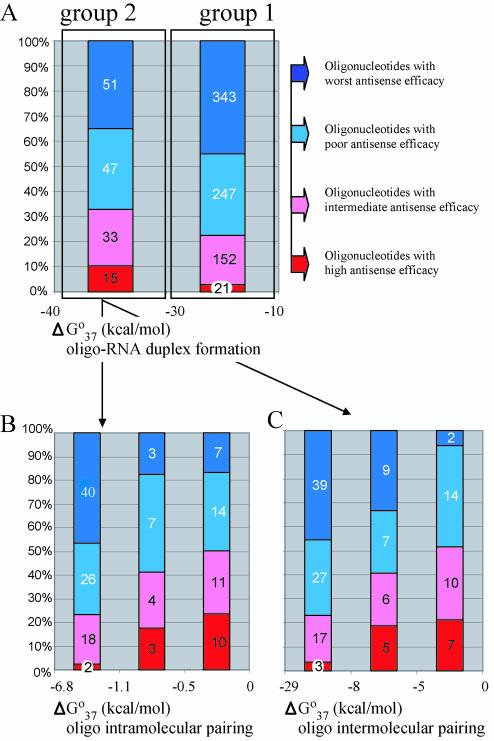
Figure 3.
Relationship between thermodynamic parameters and antisense oligonucleotide activities determined for the Isis database. Oligonucleotides were categorized into two groups according to the calculated value of ΔGo37 of duplex formation. (A) Group 1 contains oligonucleotides that form more stable duplexes and group 2 contains oligonucleotides that form less stable duplexes with target RNA. (B) Group 1 oligonucleotides were further separated based on the calculated ΔGo37 for oligonucleotide intra-molecular pairing. (C) Group 1 oligonucleotides were further separated based on the calculated ΔGo37 for oligonucleotide inter-molecular pairing. For each set, oligonucleotides were separated into subgroups according to their antisense efficacy. Blue represents the least active oligonucleotides, 75–100% of untreated control; violet, 50–75% of control; pink, 25–50% of control; and red, 0–25% of control. The numbers of oligonucleotides in each subgroup are on the relevant highlighted segments.
Oligonucleotide self-structure formation can compete with oligonucleotide binding to target RNA. During antisense oligonucleotide experiments, the concentrations of oligonucleotides are usually much higher than those of the relevant mRNAs. Therefore, oligonucleotide self-interaction may decrease the ‘hit rate’. Among the oligos that form the more stable duplexes with RNA, those which are predicted to form strong intra- and inter-molecular self-structures are not as active as those with little self-structure.
The issue of the reliability of the ΔGo37 calculations relevant to the stability of intra-molecular oligo-probe structures is still open. For example, many factors influence hairpin stability, including sequence of the loops and the identity of the base pairs that close loops. Parameters for these factors are not adequately included in the current version of the software. Nevertheless, the highly significant correlations found in the present work between experimental and theoretical values in antisense oligonucleotide databases indicate utility for thermodynamic calculations of ΔGo37 values relevant to the stability of inter- or intra-molecular oligo-probe structures.
Another issue is why self-structure is a problem for the second group of oligonucleotides that can form more stable duplexes with RNA, but not a problem for oligonucleotides from the first group that can form less stable duplexes with the target. The reason is probably that oligonucleotides from the second group are more frequently G + C-rich molecules (Table 2) and thus are more likely to adopt stable self-structures. In contrast, oligonucleotides from the first group that form the less stable duplexes with target RNA are less frequently G + C-rich, so the proportion of those with stable self-structures is rather small. As a result of this difference in composition, the proportion of oligonucleotides with stable self-structure is also much higher among those that form stable duplexes with RNA. A large proportion of highly structured oligonucleotides in the second group of molecules is related to strong, and statistically detectable, negative effects on antisense hit rate. Correspondingly, a small proportion of structured oligonucleotides in the first group of molecules is related to undetectable negative effects on the hit rate.
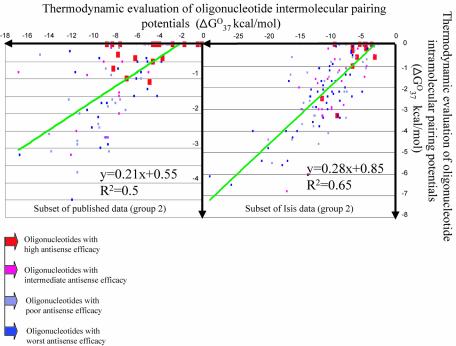
Thermodynamic evaluations of both oligonucleotide intra- and inter-molecular self-interacting properties are strongly correlated with each other. Steep trend line slopes of scatter plots (Fig. 4), and highly significant correlation coefficients of 0.65 and 0.5, demonstrate this for both databases. Usually, if two variables are highly correlated, only one is sufficient for predictive purposes. However, with antisense oligonucleotides, it was found that both thermodynamic criteria for self-structure-forming potentials are simultaneously useful for efficient discrimination into categories that mainly contain either the most active molecules, or categories that contain the non-active ones.
Figure 4.
Relationship between thermodynamic evaluations of oligonucleotide inter- and intra-molecular pairing potentials (x- and y-axis, respectively). The trend line is shown in each scatter plot.
Whether or not an oligo-probe will be a good antisense compound for a particular target also depends on the specificity of DNA–RNA interactions. It is possible that an oligo-probe is in the optimal range of thermodynamic values described above, but can cross-hybridize with unintended RNA targets. The opposite situation, where the calculated thermodynamic value of a highly specific probe is outside the optimal range of thermodynamic values indicated above, is also possible.
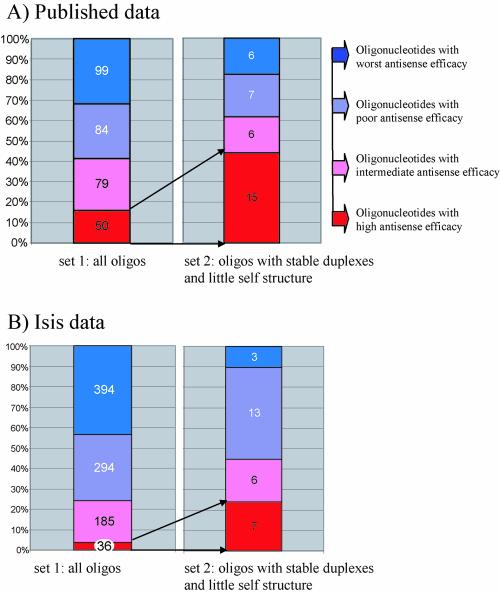
Nevertheless, the statistical results presented suggest that using values for the predicted stability of duplexes of oligonucleotides with their target RNA, and corresponding values for oligonucleotide self-structure, can dramatically increase the proportion of active antisense oligos in trial and error screening experiments. If oligonucleotides in the optimal range described above had been used, the ‘hit rate’ would have been three times higher for the published data set and six times higher for the unpublished data from Isis Pharmaceuticals (Fig. 5).
Figure 5.
Relationship between thermodynamic parameters and antisense oligonucleotide activities from both databases. (A) Data from the published antisense oligonucleotide experiments. (B) Unpublished data from Isis Pharmaceuticals. Blue represents the least active oligonucleotides, 75–100% of untreated control; violet, 50–75% of control; pink, 25–50% of control; and red, 0–25% of control. The numbers of oligonucleotides in each subgroup are on the relevant segments. Set 1 contains all oligonucleotides in each database. Set 2 includes only oligonucleotides predicted to form very stable duplexes (ΔGo37 ≤ 30 kcal/mol) and those with the least possibility for self-structure (ΔGo37 ≥ –5 kcal/mol for inter-molecular oligonucleotide pairing and ΔGo37 ≥ –1 kcal/mol for intra-molecular pairing).
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to Mr Moore for invaluable computer help. R.F.G. was supported by NIH grant R01-GM61200, and J.F.A. by NIH grant GM48152.
REFERENCES
- 1.Sczakiel G. and Tabler,M. (1997) Computer-aided calculation of the local folding potential of target RNA and its use for ribozyme design. Methods Mol. Biol., 74, 11–15. [DOI] [PubMed] [Google Scholar]
- 2.Patzel V., Steidl,U., Kronenwett,R., Haas,R. and Sczakiel,G. (1999) A theoretical approach to select effective antisense oligodeoxyribonucleotides at high statistical probability. Nucleic Acids Res., 27, 4328–4334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lehmann M.J., Patzel,V. and Sczakiel,G. (2000) Theoretical design of antisense genes with statistically increased efficacy. Nucleic Acids Res., 28, 2597–2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scherr M., Rossi,J.J., Sczakiel,G. and Patzel,V. (2000) RNA accessibility prediction: a theoretical approach is consistent with experimental studies in cell extracts. Nucleic Acids Res., 28, 2455–2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sczakiel G. (2000) Theoretical and experimental approaches to design effective antisense oligonucleotides. Front. Biosci., 5, 194–201. [DOI] [PubMed] [Google Scholar]
- 6.Ding Y. and Lawrence,C.E. (2001) Statistical prediction of single-stranded regions in RNA secondary structure and application to predicting effective antisense target sites and beyond. Nucleic Acids Res., 29, 1034–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mathews D.H., Burkard,M.E., Freier,S.M., Wyatt,J.R. and Turner,D.H. (1999) Predicting oligonucleotide affinity to nucleic acid targets. RNA, 5, 1458–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giddings M.C., Matveeva,O.V., Atkins,J.F. and Gesteland,R.F. (2000) ODNBase—a web database for antisense oligonucleotide effectiveness studies. Oligodeoxynucleotides. Bioinformatics, 16, 843–844. [DOI] [PubMed] [Google Scholar]
- 9.Xia T., SantaLucia,J.,Jr, Burkard,M.E., Kierzek,R., Schroeder,S.J., Jiao,X., Cox,C. and Turner,D.H. (1998) Thermodynamic parameters for an expanded nearest-neighbor model for formation of RNA duplexes with Watson–Crick base pairs. Biochemistry, 37, 14719–14735. [DOI] [PubMed] [Google Scholar]
- 10.SantaLucia J. Jr (1998) A unified view of polymer, dumbbell and oligonucleotide DNA nearest-neighbor thermodynamics. Proc. Natl Acad. Sci. USA, 95, 1460–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.SantaLucia J. Jr, Allawi,H.T. and Seneviratne,P.A. (1996) Improved nearest-neighbor parameters for predicting DNA duplex stability. Biochemistry, 35, 3555–3562. [DOI] [PubMed] [Google Scholar]
- 12.Allawi H.T. and SantaLucia,J.,Jr (1997) Thermodynamics and NMR of internal G·T mismatches in DNA. Biochemistry, 36, 10581–10594. [DOI] [PubMed] [Google Scholar]
- 13.Sugimoto N., Nakano,S., Katoh,M., Matsumura,A., Nakamuta,H., Ohmichi,T., Yoneyama,M. and Sasaki,M. (1995) Thermodynamic parameters to predict stability of RNA/DNA hybrid duplexes. Biochemistry, 34, 11211–11216. [DOI] [PubMed] [Google Scholar]
- 14.Luebke K.J., Balog,R.P. and Garner,H.R. (2003) Prioritized selection of oligodeoxyribonucleotide probes for efficient hybridization to RNA transcripts. Nucleic Acids Res., 31, 750–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matveeva O.V., Shabalina,S.A., Nemtsov,V.A., Tsodikov,A.D., Gesteland,R.F. and Atkins,J.F. (2003) Thermodynamic calculations and statistical correlations for oligo-probes design. Nucleic Acids Res., 31, 4211–4217. [DOI] [PMC free article] [PubMed] [Google Scholar]