Abstract
The DHH1 gene in the yeast Saccharomyces cerevisiae encodes a putative RNA helicase of remarkable sequence similarity to several other DExD/H-box proteins, including Xp54 in Xenopus laevis and Ste13p in Schizosaccharomyces pombe. We show here that over-expression of Xp54, an integral component of the stored messenger ribonucleoprotein (mRNP) particles, can rescue the loss of Dhh1p in yeast. Localization and sedimentation studies showed that Dhh1p exists predominantly in the cytoplasm and is present in large complexes whose sizes appear to vary according to the growth stage of the cell culture. In addition, deletion of dhh1, when placed in conjunction with the mutant dbp5 and ded1 alleles, resulted in a synergistically lethal effect, suggesting that Dhh1p may have a role in mRNA export and translation. Finally, similar to Ste13p, Dhh1p is required for sporulation in the budding yeast. Taken together, our data provide evidence that the functions of Dhh1p are conserved through evolution.
INTRODUCTION
One of the characteristics of growing oocytes of all animal species is the synthesis and accumulation of messenger RNAs (mRNAs) for the development of the early embryo (1). For example, during oogenesis, it is estimated that each Xenopus oocyte accumulates a vast pool (∼2 × 1011) of stored messenger ribonucleoprotein (mRNP) complexes (1). These non-translated mRNAs are used during oocyte maturation, fertilization and early embryogenesis, as transcription is severely limited from the onset of maturation (2). A similar scenario has also been observed in a variety of systems, such as mouse, Drosophila, sea urchin and zebrafish (1). Therefore, the packaging of these maternal mRNAs into non-translated cytoplasmic mRNPs, or translationally ‘masked’ or ‘repressed’ mRNPs, is thought to be an essential mechanism for post-transcriptional regulation of gene expression utilized in germ cells of both vertebrates and invertebrates. Interestingly, these and other non-translated mRNPs often exist in large granules such as P (or germinal) granules in Caenorhabditis elegans, stress granules in human tissue culture cells and neuronal granules in cultured rat hippocampal neurons (3–5).
Research focusing on the characterization of the protein moiety of the stored mRNPs has revealed that a number of specific proteins, referred to as masking proteins, are in tight association with maternal mRNAs. Microinjection of partially purified mRNP-bound proteins into Xenopus oocytes inhibited the translation of co-injected β-globin mRNA (6). In addition, the translation of β-globin mRNAs, when incubated with purified mRNP-associated proteins, was also repressed in wheat germ extract (WGE) and/or rabbit reticulocyte lysate translation systems (7). It is therefore believed that both the non-translated state and the stability of these translationally masked mRNAs are mediated, at least in part, by the protein components of the mRNP complex. However, it remains to be established how masking proteins regulate the translation of maternal mRNAs.
Since mRNAs are expected to adopt complicated secondary structures, their packaging and/or unpackaging may require reorganization of RNA and/or RNP structures, a process predicted to be energy dependent and requiring enzymes such as RNA helicases which can modulate the secondary structure of RNAs. Two findings lend support to this hypothesis. First, translational inhibition by purified masking proteins in WGE requires preincubation with Mg2+ and ATP (7). Secondly, an RNA helicase, Xp54, which belongs to the DExD/H-box protein family (8), was found to be an integral component of the stored mRNP particles in Xenopus oocytes (9).
The ubiquitous DExD/H-box proteins are often referred to as RNA helicases or RNA unwindases, because a number of them can unwind RNA duplexes in vitro (10–18). In fact, at least one of them, NPH-II, has been shown to unwind RNA duplexes in a processive and directional manner (19). Thus, the prevailing hypothesis is that DExD/H-box proteins directly bind to and unwind specific RNA duplexes by harnessing energy from ATP hydrolysis in a manner similar to that of the better studied DNA helicases (10,19,20). However, recent studies have raised the possibility that the DExD/H-box proteins may perform functions distinct from RNA unwinding, such as disrupting protein–RNA or protein–protein interactions during RNP remodeling (21–25).
It has been reported that Dhh1p, a DExD/H-box protein in the budding yeast, is remarkably similar in sequence to several DExD/H-box proteins (9,26), including Xp54 (9) and Ste13p (27). Recent studies have provided compelling evidence that loss of Dhh1p substantially inhibits mRNA decapping during mRNA turnover (28–30) and that Dhh1p is present in discrete and dynamic cytoplasmic loci called processing bodies (P bodies) (31). In this work, we explored the possibility that Dhh1p, Xp54 and Ste13p are functionally related. We show that genetic deletion of DHH1 can be complemented by over-expression of Xp54, that Dhh1p is also present in large complexes, and that Dhh1p interacts genetically with Dbp5p and Ded1p, two cytoplasmic DExD/H-box proteins essential for poly(A)+ RNA export and translation, respectively. Finally, we show that, like Ste13p in Schizosaccharomyces pombe, Dhh1p is required for sporulation in the budding yeast. We discuss these findings in the context of recently elucidated functions of Dhh1p.
MATERIALS AND METHODS
Yeast strains and plasmid construction
The dhh1Δ::LEU2 (hereafter dhh1Δ) allele was constructed by inserting the 5′ region (positions –511 to –196) and the 3′ region (positions 1868 to 3003) of DHH1 into YIPlac128 (32) in the order (1868 to 3003), BamHI, and (–511 to –196) to yield pSE56AD. pSE56AD was linearized with BamHI and transformed into strain YJB334 (MATa/α ade2-1/ade2-1 ura3-1/ura3-1 trp1-1/trp1-1 his3-11/his3-11 leu2-3,112/leu2-3, 112 can1-100/can1-100). This replaced a region of DHH1 (positions –196 to 1868), including the entire open reading frame (ORF) (positions 1 to 1521), with the YIPlac128 sequence. Leu+ transformants were sporulated and tetrads were dissected. Correct disruptants were identified by segregation of two slow growing Leu+ spores in each tetrad and confirmed by Southern hybridization. Strains derived from the isogenic haploid wild type and the dhh1Δ segregant were used throughout this work.
The HA-tagged version of DHH1 (DHH1–HA) was PCR amplified from yeast genomic DNA using primers DHH1-1 and DHH1-2. Primer DHH1-1 (gaggacagggatccaaaaaggg) corresponds to sequence ∼390 nt upstream of the initiation codon of DHH1 and primer DHH1-2 [ccccggatccta-(agcgtagtctgggacgtcgtatgggta)-atactggggttgtgactgacc; HA-epitope, in parentheses] is complementary to the last 21 nt of the DHH1 ORF. The ∼2.0 kb PCR product was then digested with BamHI (underlined in both primer sequences) and cloned into pRS316 (33) to yield pDHH1001. The DHH1–Protein A (PA) clone, pDHH1002, was constructed by inserting a 651 bp AatII fragment (34,35) encoding the PA moiety into the AatII site within the HA-coding region in pDHH1001. Likewise, the green fluorescence protein (GFP) moiety carried on a 720 bp AatII fragment was inserted into the AatII site of pDHH1001 to yield pDHH1018, in which the DHH1–GFP fusion is under the native DHH1 promoter control, and pDHH1019 (inverted GFP clone). To place DHH1 under GPD promoter (PGPD) control, a 1.5 kb BamHI fragment corresponding to the DHH1 ORF was obtained by PCR using primer DHH1-3, which contains a BamHI site adjacent to the initiation codon, and primer DHH1-2, and cloned into expression vector pRS426-pG1 (34,35) to yield pDHH1003. To place the Xp54 gene under DHH1 promoter (PDHH1) control, a 388 bp BamHI–NdeI fragment covering PDHH1 was fused with a ∼2.0 kb NdeI–HindIII fragment corresponding to the Xp54 ORF and cloned into pRS316 to yield pDHH1006. The Xp54 ORF was also cloned into pRS426-pG1 by blunt-ended ligation, yielding pDHH1017, for over-expression of Xp54. These plasmids were then individually transformed into strain YTC444 (i.e. YJB1695) (MATa dhh1Δ::LEU2 ade2-1 ura3-1 trp1-1 his3-11 leu2-3,112 can1-100) for this study.
Immunodetection of Dhh1p in glycerol and sucrose gradients
To prepare cell extracts for gradient analysis, cells from a 250 ml culture were harvested, washed once with 50 ml of 0.5% β-mercaptoethanol and resuspended in 50 ml of 1.2 M sorbitol followed by the addition of 200 µg/ml zymolyase (100T). The cell suspension was rocked at 37°C on a nutator for 1 h. Spheroplasts were harvested at low-speed centrifugation and resuspended in 1 ml of lysis buffer [50 mM Tris–HCl (pH 7.5), 100 mM NaCl, 1 mM EDTA, 0.5% NP-40]. After 5 min of incubation on ice, cells were broken by 12 strokes of douncing in a glass dounce homogenizer and the cell debris was clarified by centrifugation at 10 000 r.p.m. (SS34 rotor, Sorvall) for 20 min. Log-phase cells and stationary-phase cells (1.2 mg and 600 µg of total proteins, respectively) were loaded onto an 11 ml glycerol gradient (10–30% glycerol in the cell lysis buffer). The gradients were centrifuged at 34 000 r.p.m. for 24 h in a Beckman SW41 rotor at 4°C. Fractions (∼0.5 ml each) were collected from top to bottom of the gradient and proteins were precipitated by 10% trichloric acid (TCA) and incubated overnight at 4°C. TCA precipitates were collected by centrifugation and resuspended in 50 µl of 2× SDS–PAGE sample loading buffer and 3 µl of 1 M Tris–HCl (pH 9.4). Proteins in each fraction were separated by 8% SDS–PAGE and transferred to a nitrocellulose membrane by electroblotting. Immunoblots were developed with normal rabbit serum at a 1:5000 dilution, protein G horseradish peroxidase conjugate (Bio-Rad) at a 1:5000 dilution, followed by chemiluminescence detection (ECL system; Amersham) of the Dhh1p–PA. Sucrose gradient and polysome profile analyses were done in the absence of EDTA as previously described (35).
Microscopy
To image the Dhh1p–GFP, strain YTC335 [MATa dhh1Δ::TRP1 ura3 his3 trp1 leu2 pDHH1018 (DHH1-GFP/CEN/URA3)] was grown in synthetic medium lacking tryptophan and adenine to early-log phase (OD600 = 0.4), log phase (OD600 = 0.8), late-log phase (OD600 = 2) and saturation (4 days after inoculation). Cells were harvested, stained with DAPI (1.5 µg/ml) at room temperature for 10 min, washed three times with 1 ml of sterile distilled water, and then spotted onto glass slides for imaging on a Zeiss Axioskop 2 microscope using an FITC filter.
Synthetic-lethal test
Double mutants were constructed by first crossing a dhh1Δ strain (YJB1695; see above) to a DED1 strain [YTC74; MATα ded1Δ::TRP1 ura3-52 lys2-801 ade2-101 trp1-Δ1 his3-Δ200 leu2-Δ1 pDED1008(DED1/CEN/URA3)] or a DBP5 strain [YTC153; MATα dbp5Δ::HIS3 ura3-52 lys2-801 ade2-101 trp1-Δ1 his3Δ200 leu2-Δ1 pCA5005(DBP5/CEN/URA3)] to obtain a combination of appropriate genetic markers. After sporulation and tetrad dissection, tester strains harboring the following relevant genotypes were selected: MATa (or α) dhh1Δ::LEU2 ded1Δ::TRP1 pDED1008 (see above) or MATa (or α) dhh1Δ::LEU2 dbp5Δ::HIS3 pCA5005 (see above). To rule out synthetic lethality caused by genetic background variations, we performed the following experiments by using at least three independent tester strain isolates. To test for synthetic lethality between ded1 and dhh1Δ mutations, we individually transformed DED1, ded1-120 and ded1-199 alleles (34) carried on pASZ11(CEN/ADE2) (36) into the tester strains. Two mutant alleles, dbp5-1 and dbp5-2 (12), were used in the same manner for synthetic lethality tests between dbp5 and dhh1Δ::LEU2 mutations. The resulting transformants were then streaked onto 5-FOA plates and incubated at temperatures permissive to both single mutations for 4 days. The dhh1Δ tif1, dhh1Δ prt1 and dhh1Δ prp28 double mutants were constructed using a similar approach, using strain SS13-3A (MATa tif1Δ::HIS3 tif2Δ::ADE2 his3 ade2 leu2 trp1 ura3 YCpLac33- tif1-1) (37), strain F294 (MATa prt1-1 ade1 leu2-3,112 ura3-52) and strain YTC65 [MATα prp28Δ::HIS3 ura3-52 lys2-801 ade2-101 trp1-Δ1 his3-Δ200 leu2-Δ1 pCA8032(PRP28/CEN/URA3)] (38).
Sporulation test
Diploid yeast cells were patched on YPD plates and incubated at 30°C overnight. The next day, cells were collected using wood applicators and inoculated into 1% potassium acetate at 30°C for 1–5 days. The number of tetrads in small aliquots of cell suspension was then scored under a microscope.
RESULTS
Over-expression of Xp54 can functionally complement dhh1 deletion
Our interest in Dhh1p was initially sparked by the report that Dhh1p is strikingly similar to Xp54 (68.8% identity and 77% similarity), to Ste13p (73.3% identity and 80.5% similarity), and to several other DExD/H-box proteins found in mouse, human and Drosophila. Previous sequence alignments revealed that all helicase motifs are identical in sequence and that sequence variations occur primarily at the N-terminal and the C-terminal extensions of the highly conserved central helicase core (9,26). Among these DExD/H-box proteins, Xp54 is thought to be involved in packaging and/or unpackaging of mRNP storage particles in Xenopus oocyte (9,26) and Ste13p is required for sporulation in S.pombe (27). To address the potential functional similarity between Dhh1p and Xp54, we first examined whether Dhh1p and Xp54 are functionally interchangeable.
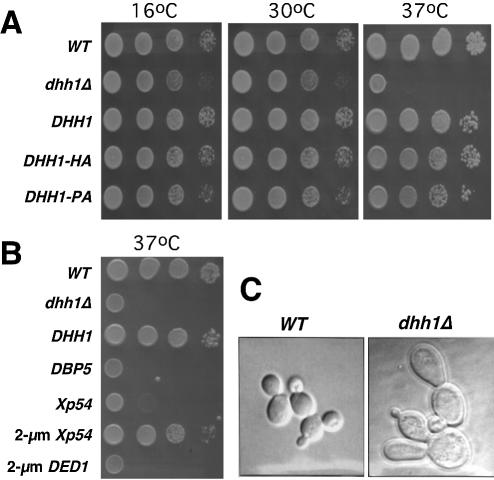
DHH1 is not essential for yeast cell viability (31,39,40). However, we observed that a dhh1Δ strain was unable to grow at 37°C, exhibiting a temperature-sensitive (Ts–) phenotype (Fig. 1A). Close inspection of the dhh1Δ cells revealed that they were nearly three times larger than the isogenic wild-type cells and they often contained buds with atypical shapes, e.g. large, elongated and/or multiple buds (Fig. 1C). Using these phenotypes, we tested the functional complementation of dhh1Δ allele by Xp54. The growth defect of the dhh1Δ strain at 37°C was rescued by transforming dhh1Δ cells with a low-copy-number CEN plasmid harboring a wild-type DHH1 gene (Fig. 1A). Likewise, two tagged versions of DHH1, DHH1–HA (tagged with an HA epitope) and DHH1–PA (tagged with a PA moiety; encodes Dhh1p–PA), also corrected the Ts– phenotype of the dhh1Δ strain (Fig. 1A). We found that Xp54 rescued the growth defect of dhh1Δ cells when it was expressed from the yeast GPD promoter (PGPD) control and on a 2 µm plasmid (Fig. 1B). This functional complementation was specific because a functional PGPD-DED1 construct on a 2 µm plasmid failed to rescue the dhh1Δ Ts– phenotype (Fig. 1B). The aberrant morphology of dhh1Δ cells was also corrected by over-expression of Xp54 (data not shown). These results thus suggest that Xp54 and Dhh1p are likely to be functional homologs. However, the fact that expression of Xp54 from the DHH1 promoter (Fig. 1B) did not complement the dhh1Δ allele leaves open the possibility that Xp54 may function in a manner similar, but not exactly identical, to Dhh1p in yeast cells.
Figure 1.
Over-expression of Xp54 complements dhh1Δ. (A) Growth phenotypes of the dhh1Δ strain. Cells were grown to mid-log phase, serially diluted and spotted on three plates, which were then separately incubated at 16, 30 and 37°C. WT, wild-type strain; dhh1Δ, the dhh1 deletion strain; DHH1, dhh1Δ strain transformed with a plasmid-borne DHH1 allele; DHH1–HA, dhh1Δ strain transformed with a plasmid-borne DHH1–HA allele; DHH1–PA, dhh1Δ strain transformed with a plasmid-borne DHH1–PA allele. (B) Over-expression of Xp54 rescues the Ts– phenotype of the dhh1Δ strain. DBP5, dhh1Δ strain transformed with a DBP5 gene carried on a CEN plasmid; Xp54, dhh1Δ strain transformed with an Xp54 gene carried on a CEN plasmid; 2-µm Xp54, dhh1Δ strain transformed with an Xp54 gene carried on a 2 µm plasmid; 2-µm DED1, dhh1Δ strain transformed with a DED1 gene carried on a 2 µm plasmid. (C) Abnormal cellular morphology of the dhh1Δ cells. Wild-type (WT) and dhh1Δ strains were grown in liquid YPD medium to mid-log phase at 30°C. Cells were then imaged by light microscopy using Nomaski optics.
Dhh1p is present in large complexes
We then asked whether Dhh1p, like Xp54, is also present in large complexes. Cell extract was prepared from a DHH1–PA strain grown to various culture stages and used to examine the distribution of Dhh1p–PA (83 kDa) by 10–30% glycerol gradient sedimentation analysis. When cells were harvested at early-log phase (OD600 = 0.4), only a minor fraction (5%) of Dhh1p–PA was present in the free form (Fig. 2A, lanes 6–10), whereas the majority was located in large complexes >440 kDa (Fig. 2A, lanes 17–20) and in the pellet (∼70%; data not shown). At mid-to-late log phase (OD600 = 0.8–2.5), more (20–40%) of Dhh1p–PA was present in the free form (Fig. 2B and C, lanes 5–11). Surprisingly, when cells approached or entered stationary phase (4 days after inoculation), Dhh1p–PA was present in complexes of 200–250 kDa in size (Fig. 2D, lanes 7–15). Using sucrose gradients (7–37%) to size the early-log-phase Dhh1p–PA-containing complexes (Fig. 2A) more precisely, we found a minor fraction of Dhh1p–PA, which is presumably in the free form, remains on top of the gradient, whereas most of the Dhh1p–PA sediments between 40S and 80S with a peak at ∼60S (Fig. 3). These results are consistent with the size of the Xp54-containing complexes, which generally sediment around 80S in oocyte extracts, but change in size distribution at different stages of oogenesis (9,41). Thus, we conclude that a substantial fraction of Dhh1p is present in large complexes whose sizes vary depending upon the growth stage of the cell culture.
Figure 2.
Dhh1p is present in several large complexes with different sizes at different cell culture stages. DHH1–PA strain grown in liquid cultures at 30°C to (A) early-log phase (OD600 = 0.4), (B) mid-log phase (OD600 = 0.8), (C) late-log phase (OD600 = 2.5) or (D) stationary phase (OD600 = 4.8; 4 days after inoculation). Protein extracts were sedimented through 10–30% glycerol gradients and fractionated from top to the bottom of the gradient. Proteins in each fraction were further separated by SDS–PAGE and blotted for immuno-detection of Dhh1p–PA (see Materials and Methods). The positions of the protein size markers (kDa) are indicated.
Figure 3.
Dhh1p is present in large complexes that sediment between 40S and 80S. The DHH1–PA strain was grown to early log phase (OD600 = 0.4). Protein extract was loaded onto a 7–37% sucrose gradient for immunoblot analysis as described in Figure 2. The positions for the protein size markers (kDa), 40S and 60S ribosomal subunits and the 80S ribosome are indicated.
Cellular localization of Dhh1p
Both Xp54 and its human counterpart, p54, are present predominantly in cytoplasmic particulate structures; however, a fraction of each can also be found in the nucleus (9,42). To examine Dhh1p’s cellular localization, we expressed a GFP fusion of Dhh1p in the dhh1Δ strain. This recombinant protein, containing a GFP moiety at the C-terminus of Dhh1p, rescued the Ts– phenotype of the dhh1Δ strain (data not shown) and was present predominantly in the cytoplasm (Fig. 4C–F). Interestingly, the presence of Dhh1p–GFP in the cytoplasm was not uniform. Instead, Dhh1p–GFP gave rise to a characteristic punctate pattern, often concentrating on a number of distinct foci. In cells harvested from early-log phase culture (OD600 = 0.4; Fig. 4C), Dhh1p–GFP was present in small granules with low fluorescence intensity. However, when the cell culture reached mid-log (OD600 = 0.8; Fig. 4D) and late-log (OD600 = 2.5; Fig. 4E) phases, large bright Dhh1p–GFP-containing granules (three to four per cell) were readily detected. This pattern persisted in cells taken from the saturated culture (4 days after inoculation; Fig. 4F), although in some cells the fluorescence signals appeared to be reduced. At present, it is difficult to correlate the transition of the Dhh1p–GFP-containing granules with the transition of the sizes of Dhh1p–PA-containing complexes (Figs 2 and 3), as the two detection methods are intrinsically different. Control experiments with strains expressing only wild-type Dhh1p yielded no fluorescence signal (data not shown), and control staining of two previously characterized cytoplasmic DExD/H-box proteins, Dbp5p (12) and Ded1p (34), yielded their characteristic staining patterns, in that Dbp5p is enriched around the nuclear envelope (Fig. 4A) and Ded1p is uniformly distributed in the cytoplasm (Fig. 4B).
Figure 4.
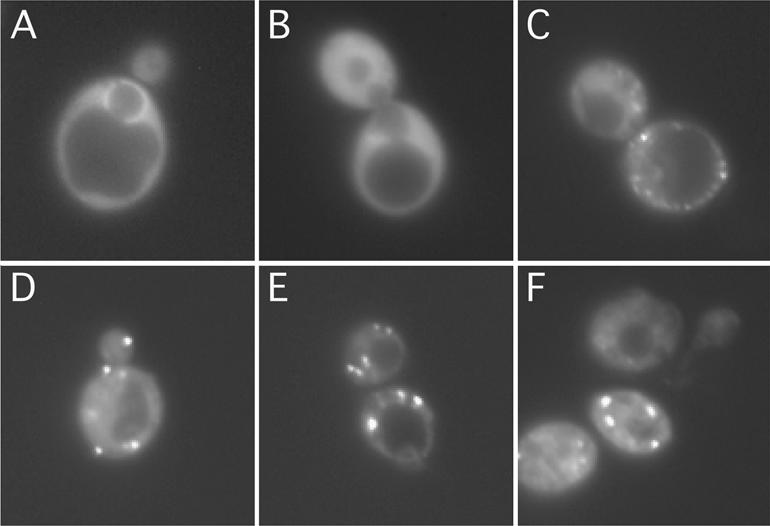
Dhh1p is localized in distinct cytoplasmic foci. Strain YTC335 expressing a functional Dhh1p–GFP was grown in synthetic medium lacking tryptophan and adenine to (C) 0.4 OD600, (D) 0.8 OD600, (E) 2.5 OD600 and (F) saturation (4 days after inoculation). Cells were harvested, stained with DAPI and imaged by light microscopy. Localization of (A) Dbp5p–GFP and (B) Ded1p–GFP were used as controls for imaging techniques.
Genetic interactions of Dhh1p with Dbp5p and Ded1p
The above data raised the possibility that Dhh1p may play a role similar to that of Xp54. Conceptually, the proposed function of Xp54 is reminiscent of what has been postulated for the yeast Dbp5p, a shuttling DExD/H-box protein essential for poly(A)+ RNA export (12,43,44). At the steady state, Dbp5p is predominantly localized in the cytoplasm and enriched around the nuclear envelope (Fig. 4A) and is, therefore, thought to promote the unpackaging or remodeling of the newly exported mRNPs (12,43,44). Therefore, we tested whether the dhh1Δ mutation is synthetical lethal with conditional mutations of DBP5 and DED1 (34) as well as of TIF1. Ded1p and Tif1p (the yeast eIF4A) are two cytoplasmic DExD/H-box proteins essential for translation. As controls, we also included mutations in PRP28 and PRT1 in our genetic analysis. Prp28p is a DExD/H-box protein involved in pre-mRNA splicing (38) and Prt1p is one of the eIF3 subunits required for translation (45).
We first constructed a series of tester strains containing chromosomal deletions of the two genes in question. These strains were kept viable by two plasmids harboring a wild type and a mutant allele corresponding to the two deleted genes. Synthetic lethality was determined if the plasmid-borne wild-type allele could not be substituted by a corresponding mutant allele transformed into the tester strain. Results of these analyses are summarized in Table 1. Lethality resulted only when the dhh1Δ mutation was combined with the dbp5-1, dbp5-2, ded1-120 and ded1-199 mutations, but not with tif1-1, prt1-1 and prp28-112 mutations. Thus, the synthetic lethality was gene specific and not simply dependent on the presence of two mutant DExD/H-box protein genes or the co-existence of two mutant translation factors and/or two mutant cytoplasmic proteins. Interestingly, although ded1 and tif1-1 mutations are synthetic lethal to each other (34), only ded1 mutations were synthetic lethal with dhh1Δ. In addition, although dbp5 and ded1 mutations were both synthetic lethal to dhh1Δ, no genetic interactions between dbp5 and ded1 could be observed. This latter finding is consistent with the fact that Dbp5p is not required for translation (12) and that ded1 mutants are not defective in poly(A)+ RNA export (T.-H. Chang, unpublished results).
Table 1. Synthetic-lethality test for dhh1Δ with dbp5, ded1 and other mutations.
| Crosses | dhh1Δ | dbp5-1 | dbp5-2 | ded1-120 | ded1-199 | tif1-1 | prp28–120 | prt1-1 |
|---|---|---|---|---|---|---|---|---|
| dhh1Δ | + | + | + | + | – | – | – | |
| dbp5-1 | – | – | – | – | – | |||
| dbp5-2 | – | – | – | – | – | |||
| ded-120 | + | – | – | |||||
| ded1-199 | + | – | – | |||||
| tif1-1 | – | ND | ||||||
| prp28-120 | – | |||||||
| prt1-1 |
+, synthetic lethality, double mutant is not viable; –, double mutant is capable of forming single colonies on plate; ND, not determined.
Importantly, the growth defect of the dhh1Δ cells could not be suppressed by over-expressing either DBP5, DED1 or TIF1. Likewise, over-expression of DHH1 could not suppress the growth defect of either dbp5, ded1 or tif1 mutants at the restrictive temperature. Thus, Dhh1p does not appear to have a function that is redundant with the functions of Dbp5p, Ded1p or Tif1p. Interestingly, we observed that over-expression of DHH1, even in a wild-type strain background, substantially decreased the growth rate of the cell (data not shown). One possible interpretation for all these data is that Dhh1p may be functionally related to both Dbp5p and Ded1p (see Discussion).
Dhh1p is required for sporulation
We also tested the possibility that Dhh1p, like its potential homolog in S.pombe, is required for sporulation in Saccharomyces cerevisiae. DHH1/dhh1Δ and dhh1Δ/dhh1Δ strains were constructed and subjected to nitrogen starvation, which induces sporulation in S.cerevisiae. Up to 40% of DHH1/DHH1 homozygotes and DHH1/dhh1Δ heterozygotes underwent sporulation after only 1 day of incubation in sporulation media (Table 2). Sporulation reached 70% after 3 days of incubation for both DHH1/DHH1 and DHH1/dhh1Δ strains. In contrast, the homozygous dhh1Δ/dhh1Δ strain sporulated very poorly, forming <1% of tetrads after 5 days in sporulation media (Table 2). These results suggest that Dhh1p is required for sporulation in S.cerevisiae.
Table 2. Sporulation efficiencies of wild-type, dhh1Δ/DHH1, and dhh1Δ/dhh1Δ diploid strains.
| Strain | Day 1a | Day 2 | Day 3 | Day 4 | Day 5 |
|---|---|---|---|---|---|
| DHH1/DHH1 | 42b | 56 | 71 | 67 | 70 |
| DHH1/dhh1Δ | 41 | 53 | 69 | 68 | 68 |
| dhh1Δ/dhh1Δ | <1 | <1 | <1 | <1 | <1 |
aNumber of days in sporulation medium.
bPercentage of tetrad-forming cells.
DISCUSSION
Dhh1p belongs to a highly conserved DExD/H-box protein subfamily that includes human RCK/p54, mouse p54, Xenopus Xp54, Clam p47, Drosophila Me31B, C.elegans cgh-1 and S.pombe Ste13p (9,26). Over the years, these proteins have been implicated in a diverse array of functions. For example, because of its presence in the cytoplasmic mRNA storage particles, Xp54 was speculated to govern the translatability of the stored mRNAs (9,26). In addition, both human and mouse p54 were postulated to be proto-oncogene candidates (42) and Drosophila Me31B was thought to be involved in germline development (46). Disparate functions also have been proposed for yeast Dhh1p. First, Dhh1p was thought to have a role in transcription owing to its genetic and physical interactions with the general transcription factor, Ccr4–Not complex (40,47). Secondly, Dhh1p was suggested to functionally overlap with the protein kinase C (PKC1) pathway, because the Ts– phenotype of dhh1Δ could be suppressed by PKC1 over-expression (40,47). Thirdly, Dhh1p was implicated in controlling cellular morphology and cytokinesis, because dhh1Δ gives rise to abnormal cellular morphology (Fig. 1B) and is synthetic lethal with a mutation in the ELM1 (elongated morphology) gene (48). Finally, studies from several laboratories have demonstrated that Dhh1p stimulates mRNA decapping by physically interacting with several proteins involved in mRNA decapping (28,29, 31). This and the co-localization of Dhh1p and mRNA degradation intermediates in discrete cytoplasmic P bodies (31) imply that the mechanisms of mRNA decapping and maternal mRNA translational repression are evolutionarily conserved (28,29,31).
In this work, we addressed the functional conservation of Dhh1p within the DExD/H-box protein subfamily that includes Xp54 and Ste13p. Several lines of evidence demonstrate that the functions of Dhh1p are indeed similar or related to Xp54 and Ste13p. First, over-expression of Xp54 rescues the defects caused by dhh1 deletion. Secondly, Dhh1p, like Ste13p, is required for efficient sporulation. Thirdly, Dhh1p is present in large complexes with sizes comparable to those of the Xp54-containing complexes (41). Fourthly, like Xp54 (41), Dhh1–GFP localizes predominantly in the cytoplasm in discrete foci. Fifthly, similar to the proposed function of a translational regulator for Xp54, Dhh1p may also have a role in mRNA export and translation as shown by its genetic interactions with Dbp5p and Ded1p.
Perhaps the most striking observation from these studies is that Dhh1p exists not only in free form but also in ∼250 kDa and extremely large (40S–80S) complexes. The largest complexes, found in early-log-phase yeast cells, can be as large as stored mRNPs in the Xenopus oocyte (41). Although the composition of these complexes and the underlying principles regulating their conversions remain to be elucidated, we suggest that these alterations are physiologically relevant for several reasons. First, Caf1p/Pop2p, Ccr4p and five Not proteins are present in complexes up to 1.2 and 2 MDa in size (49,50). Since Dhh1p interacts physically and genetically with Caf1p/Pop2p (40,47), we speculate that the Dhh1p-containing complexes that we observe may correspond, at least in part, to the 1.2 and/or 2 MDa Caf/Pop2-containing complexes. Secondly, Dhh1p localized to cytoplasmic foci or P bodies, which also contain Ccr4p, Dcp1p, Dcp2p, Pat1p and Lsm1p (31). Most critically, blocking decapping or 5′-to-3′ mRNA degradation dramatically increases the size and abundance of the P bodies, and deletion of CCR4 resulted in a marked decrease in both the size and the abundance of P bodies (31). Thirdly, Drosophila Me31B, like Xp54 (41), forms a cytoplasmic RNP complex with oocyte-localizing RNAs and Exuperantia, a protein involved in RNA localization (51).
Although it is evident that Dhh1p has a major role in mRNA turnover, there are several reasons to suspect that Dhh1p may have additional roles in RNA metabolism. First, the fact that mRNA decapping is defective at all temperatures in the dhh1Δ strain (28,29,31), and that the dhh1Δ strain exhibits a Ts– phenotype (Fig. 1A), immediately suggests that Dhh1p has a separate role essential for vegetative growth at elevated temperatures. Secondly, several observations are compatible with a role for Dhh1p in mRNA export. For example, dhh1Δ is synthetic lethal with xpo1-1 (29), an allele of CRM1/XPO1 that is defective in nuclear export. Similarly, clam p47 translocates from the cytoplasm to the nucleus during early embryogenesis, suggesting a role in some aspect of mRNA transport (26). In addition, the formation of stored mRNP particles, a process in which Xp54 is thought to be involved, appears to be initiated in the nucleus (52). Thirdly, Dhh1p may also have a role in translation, as it genetically and physically interacts with Pat1p (28), a protein required for normal translation initiation (53). Fourthly, dhh1Δ is synthetic lethal with dbp5 or ded1 mutations in a gene-specific, but allele-non-specific, manner, suggesting that Dhh1p is functionally related to the RNA helicases Dbp5p and Ded1p. However, over-expression of DBP5, DED1 and TIF1 did not suppress the growth defect of dhh1Δ mutant and over-expression of DHH1 could not suppress the growth defects of dbp5, ded1 and tif1 mutants, implying that DBP5, DED1, TIF1 and DHH1 are not functionally redundant. Finally, both Xp54 and its human counterpart, p54, localize predominantly in cytoplasmic particulate structures in addition to some localization in the nucleus (9,42). Thus, taken together, it is tempting to speculate that Dhh1p may help to integrate or couple mRNA export, translation and turnover pathways. In this context, perhaps Dhh1p acts as a multi-functional protein, shuttling between the cytoplasm and the nucleus, as has been shown for Xp54 (41), to perform various functions in both compartments.
The observation that Dhh1p is present in large particles also raises an intriguing question as to whether yeast cells possess mRNA storage particles found in many biological systems. Yet, it seems unlikely that actively growing yeast cells would need to store a large pool of mRNAs as Xenopus oocytes do. One hypothesis is that Dhh1p associates with specific messages that are critical for cells to enter the stationary phase. Upon nutrient deprivation, these messages would be released and translated, thereby signaling the cells to enter the stationary phase. Several observations are consistent with this hypothesis. First, both Pop2p (49,54,55) and Ccr4p (56,57) are required for full derepression of ADH2 and other non-fermentative genes upon glucose derepression. This is thought to be an important process preceding the entry of yeast cells into the stationary phase (58). Secondly, over-expression of PKC1 suppresses dhh1Δ growth phenotypes (40). The protein kinase C pathway also has been implicated in signaling yeast cells to enter the stationary phase (58). Thirdly, S.pombe Ste13p may have a role in governing the entry to stationary phase and/or the initiation of meiotic sexual cycle (59). Our finding that Dhh1p is required for sporulation further supports the idea that the functions of Dhh1p and Ste13p are comparable. However, unlike ste13 mutant cells, which rapidly lose viability after nutrient deprivation and are highly sensitive to heat shock in the stationary phase (59), dhh1Δ cells exhibited no unusual sensitivity to either stress (data not shown). Thus, the functions of Dhh1p and Ste13p may overlap only partially.
Since the entry into stationary phase is a complex multi-step process that may not be fully reflected by the measurement of the cell viability under stresses, it remains to be determined whether Dhh1p has a role in regulating the entry into or maintenance of stationary phase in the budding yeast. To discriminate between these and other possibilities, the components of the distinct Dhh1p-containing complexes and the timing of their formation will need to be characterized in detail.
Acknowledgments
ACKNOWLEDGEMENTS
We thank J. Sommerville for the Xp54 cDNA clone; R.-Y. Chuang and P. Herman for sage advice; L. Tung and R. Hage for commenting on the manuscript. C.B.T. was supported by a NSF-REU grant to the Department of Molecular Genetics, The Ohio State University. T.-H.C. has been supported by NIH (GM48752) and NSF (MCB-9982726). J.B. has been supported by NIH (GM38626).
REFERENCES
- 1.Davidson E.H. (1986) Gene Activity in Early Development, 3rd Edn. Academic Press, Orlando. [Google Scholar]
- 2.Almouzni G. and Wolffe,A.P. (1995) Constraints on transcriptional activator function contribute to transcriptional quiescence during early Xenopus embryogenesis. EMBO J., 14, 1752–1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wickens M. and Goldstrohm,A. (2003) A place to die, a place to sleep. Science, 300, 753–755. [DOI] [PubMed] [Google Scholar]
- 4.Anderson P. and Kedersha,N. (2002) Stressful initiations. J. Cell Sci., 115, 3227–3234. [DOI] [PubMed] [Google Scholar]
- 5.Kedersha N. and Anderson,P. (2002) Stress granules: sites of mRNA triage that regulate mRNA stability and translatability. Biochem. Soc. Trans., 30, 963–969. [DOI] [PubMed] [Google Scholar]
- 6.Richter J.D. and Smith,L.D. (1984) Reversible inhibition of translation by Xenopus oocyte-specific proteins. Nature, 309, 378–380. [DOI] [PubMed] [Google Scholar]
- 7.Yurkova M.S. and Murray,M.T. (1997) A translation regulatory particle containing the Xenopus oocyte Y box protein mRNP3+4. J. Biol. Chem., 272, 10870–10876. [DOI] [PubMed] [Google Scholar]
- 8.Tanner N.K. and Linder,P. (2001) DExD/H box RNA helicases: from generic motors to specific dissociation functions. Mol. Cell, 8, 251–262. [DOI] [PubMed] [Google Scholar]
- 9.Ladomery M., Wade,E. and Sommerville,J. (1997) Xp54, the Xenopus homologue of human RNA helicase p54, is an integral component of stored mRNP particles in oocytes. Nucleic Acids Res., 25, 965–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fuller-Pace F.V. (1994) RNA helicases: modulators of RNA structure. Trends Cell Biol., 4, 271–274. [DOI] [PubMed] [Google Scholar]
- 11.Schwer B. and Gross,C.H. (1998) Prp22, a DExH-box RNA helicase, plays two distinct roles in yeast pre-mRNA splicing. EMBO J., 17, 2086–2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tseng S.S., Weaver,P.L., Liu,Y., Hitomi,M., Tartakoff,A.M. and Chang,T.-H. (1998) Dbp5p, a cytosolic RNA helicase, is required for poly(A)+ RNA export. EMBO J., 17, 2651–2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rozen F., Edery,I., Meerovitch,K., Dever,T.E., Merrick,W.C. and Sonenberg,N. (1990) Bidirectional RNA helicase activity of eukaryotic translation initiation factor 4A and 4F. Mol. Cell. Biol., 10, 1134–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hirling H., Scheffner,M., Restle,T. and Stahl,H. (1989) RNA helicase activity associated with the human p68 protein. Nature, 339, 562–564. [DOI] [PubMed] [Google Scholar]
- 15.Gururajan R., Mathews,L., Longo,F.J. and Weeks,D.L. (1994) An3 mRNA encodes an RNA helicase that colocalizes with nucleoli in Xenopus oocytes in a stage-specific manner. Proc. Natl Acad. Sci. USA, 91, 2056–2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liang L., Diehl-Jones,W. and Lasko,P. (1994) Localization of vasa protein to the Drosophila pole plasm is independent of its RNA-binding and helicase activities. Development, 120, 1201–1211. [DOI] [PubMed] [Google Scholar]
- 17.Wang Y., Wagner,J.D. and Guthrie,C. (1998) The DEAH-box splicing factor Prp16 unwinds RNA duplexes in vitro. Curr. Biol., 8, 441–451. [DOI] [PubMed] [Google Scholar]
- 18.Laggerbauer B., Achsel,T. and Luhrmann,R. (1998) The human U5-200kD DEXH-box protein unwinds U4/U6 RNA duplices in vitro. Proc. Natl Acad. Sci. USA, 95, 4188–4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jankowsky E., Gross,C.H., Shuman,S. and Pyle,A.M. (2000) The DExH protein NPH-II is a processive and directional motor for unwinding RNA. Nature, 403, 447–451. [DOI] [PubMed] [Google Scholar]
- 20.de la Cruz J., Kressler,D. and Linder,P. (1999) Unwinding RNA in Saccharomyces cerevisiae: DEAD-box proteins and related families. Trends Biochem. Sci., 24, 192–198. [DOI] [PubMed] [Google Scholar]
- 21.Lorsch J.R. and Herschlag,D. (1998) The DEAD box protein eIF4A. 2. A cycle of nucleotide and RNA-dependent conformational changes. Biochemistry, 37, 2194–2206. [DOI] [PubMed] [Google Scholar]
- 22.Lorsch J.R. and Herschlag,D. (1998) The DEAD box protein eIF4A. 1. A minimal kinetic and thermodynamic framework reveals coupled binding of RNA and nucleotide. Biochemistry, 37, 2180–2193. [DOI] [PubMed] [Google Scholar]
- 23.Chen J.Y., Stands,L., Staley,J.P., Jackups,R.R., Latus,L.J. and Chang,T.-H. (2001) Specific alterations of U1-C Protein or U1 small nuclear RNA can eliminate the requirement of Prp28p, an essential DEAD Box splicing factor. Mol. Cell, 7, 227–232. [DOI] [PubMed] [Google Scholar]
- 24.Jankowsky E., Gross,C.H., Shuman,S. and Pyle,A.M. (2001) Active disruption of an RNA–protein interaction by a DExH/D RNA helicase. Science, 291, 121–125. [DOI] [PubMed] [Google Scholar]
- 25.Kistler A.L. and Guthrie,C. (2001) Deletion of MUD2, the yeast homolog of U2AF65, can bypass the requirement for Sub2, an essential spliceosomal ATPase. Genes Dev., 15, 42–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Minshall N., Thom,G. and Standart,N. (2001) A conserved role of a DEAD box helicase in mRNA masking. RNA, 7, 1728–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maekawa H., Nakagawa,T., Uno,Y., Kitamura,K. and Shimoda,C. (1994) The ste13+ gene encoding a putative RNA helicase is essential for nitrogen starvation-induced G1 arrest and initiation of sexual development in the fission yeast Schizosaccharomyces pombe. Mol. Gen. Genet., 244, 456–464. [DOI] [PubMed] [Google Scholar]
- 28.Coller J.M., Tucker,M., Sheth,U., Valencia-Sanchez,M.A. and Parker,R. (2001) The DEAD box helicase, Dhh1p, functions in mRNA decapping and interacts with both the decapping and deadenylase complexes. RNA, 7, 1717–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fischer N. and Weis,K. (2002) The DEAD box protein Dhh1 stimulates the decapping enzyme Dcp1. EMBO J., 21, 2788–2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tucker M., Valencia-Sanchez,M.A., Staples,R.R., Chen,J., Denis,C.L. and Parker,R. (2001) The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell, 104, 377–386. [DOI] [PubMed] [Google Scholar]
- 31.Sheth U. and Parker,R. (2003) Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science, 300, 805–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gietz R.D. and Sugino,A. (1988) New yeast–Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene, 74, 527–534. [DOI] [PubMed] [Google Scholar]
- 33.Sikorski R.S. and Hieter,P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics, 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chuang R.-Y., Weaver,P.L., Liu,Z. and Chang,T.-H. (1997) Requirement of the DEAD-box protein Ded1p for messenger RNA translation. Science, 275, 1468–1471. [DOI] [PubMed] [Google Scholar]
- 35.Weaver P.L., Sun,C. and Chang,T.-H. (1997) Dbp3p, a putative RNA helicase in Saccharomyces cerevisiae, is required for efficient pre-rRNA processing predominantly at site A3. Mol. Cell. Biol., 17, 1354–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stotz A. and Linder,P. (1990) The ADE2 gene from Saccharomyces cerevisiae: sequence and new vectors. Gene, 95, 91–98. [DOI] [PubMed] [Google Scholar]
- 37.Schmid S.R. and Linder,P. (1991) Translation initiation factor 4A from Saccharomyces cerevisiae: analysis of residues conserved in the D-E-A-D family of RNA helicases. Mol. Cell. Biol., 11, 3463–3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chang T.-H., Latus,L.J., Liu,Z. and Abbott,J.M. (1997) Genetic interactions of conserved regions in the DEAD-box protein Prp28p. Nucleic Acids Res., 25, 5033–5040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Strahl-Bolsinger S. and Tanner,W. (1993) A yeast gene encoding a putative RNA helicase of ‘DEAD’-box family. Yeast, 9, 429–432. [DOI] [PubMed] [Google Scholar]
- 40.Hata H., Mitsui,H., Liu,H., Bai,Y., Denis,C.L., Shimizu,Y. and Sakai,A. (1998) Dhh1p, a putative RNA helicase, associates with the general transcription factors Pop2p and Ccr4p from Saccharomyces cerevisiae. Genetics, 148, 571–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smillie D.A. and Sommerville,J. (2002) RNA helicase p54 (DDX6) is a shuttling protein involved in nuclear assembly of stored mRNP particles. J. Cell Sci., 115, 395–407. [DOI] [PubMed] [Google Scholar]
- 42.Akao Y., Marukawa,O., Morikawa,H., Nakao,K., Kamei,M., Hachiya,T. and Tsujimoto,Y. (1995) The rck/p54 candidate proto-oncogene product is a 54-kilodalton D-E-A-D box protein differentially expressed in human and mouse tissues. Cancer Res., 55, 3444–3449. [PubMed] [Google Scholar]
- 43.Snay-Hodge C.A., Colot,H.V., Goldstein,A.L. and Cole,C.N. (1998) Dbp5p/Rat8p is a yeast nuclear pore-associated DEAD-box protein essential for RNA export. EMBO J., 17, 2663–2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schmitt C., von Kobbe,C., Bachi,A., Pante,N., Rodrigues,J.P., Boscheron,C., Rigaut,G., Wilm,M., Seraphin,B., Carmo-Fonseca,M. et al. (1999) Dbp5, a DEAD-box protein required for mRNA export, is recruited to the cytoplasmic fibrils of nuclear pore complex via a conserved interaction with CAN/Nup159p. EMBO J., 18, 4332–4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Evans D.R., Rasmussen,C., Hanic-Joyce,P.J., Johnston,G.C., Singer,R.A. and Barnes,C.A. (1995) Mutational analysis of the Prt1 protein subunit of yeast translation initiation factor 3. Mol. Cell. Biol., 15, 4525–4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.de Valoir T., Tucker,M.A., Belikoff,E.J., Camp,L.A., Bolduc,C. and Beckingham,K. (1991) A second maternally expressed Drosophila gene encodes a putative RNA helicase of the ‘DEAD box’ family. Proc. Natl Acad. Sci. USA, 88, 2113–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maillet L. and Collart,M.A. (2002) Interaction between Not1p, a component of the Ccr4–not complex, a global regulator of transcription and Dhh1p, a putative RNA helicase. J. Biol. Chem., 277, 2835–2842. [DOI] [PubMed] [Google Scholar]
- 48.Moriya H. and Isono,K. (1999) Analysis of genetic interactions between DHH1, SSD1 and ELM1 indicates their involvement in cellular morphology determination in Saccharomyces cerevisiae. Yeast, 15, 481–496. [DOI] [PubMed] [Google Scholar]
- 49.Liu H.Y., Badarinarayana,V., Audino,D.C., Rappsilber,J., Mann,M. and Denis,C.L. (1998) The NOT proteins are part of the CCR4 transcriptional complex and affect gene expression both positively and negatively. EMBO J., 17, 1096–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maillet L., Tu,C., Hong,Y.K., Shuster,E.O. and Collart,M.A. (2000) The essential function of Not1 lies within the Ccr4–Not complex. J. Mol. Biol., 303, 131–143. [DOI] [PubMed] [Google Scholar]
- 51.Nakamura A., Amikura,R., Hanyu,K. and Kobayashi,S. (2001) Me31B silences translation of oocyte-localizing RNAs through the formation of cytoplasmic RNP complex during Drosophila oogenesis. Development, 128, 3233–3242. [DOI] [PubMed] [Google Scholar]
- 52.Sommerville J. and Ladomery,M. (1996) Masking of mRNA by Y-box proteins. FASEB J., 10, 435–443. [DOI] [PubMed] [Google Scholar]
- 53.Wyers F., Minet,M., Dufour,M.E., Vo,L.T. and Lacroute,F. (2000) Deletion of the PAT1 gene affects translation initiation and suppresses a PAB1 gene deletion in yeast. Mol. Cell. Biol., 20, 3538–3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu H.Y., Toyn,J.H., Chiang,Y.C., Draper,M.P., Johnston,L.H. and Denis,C.L. (1997) DBF2, a cell cycle-regulated protein kinase, is physically and functionally associated with the CCR4 transcriptional regulatory complex. EMBO J., 16, 5289–5298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Draper M.P., Salvadore,C. and Denis,C.L. (1995) Identification of a mouse protein whose homolog in Saccharomyces cerevisiae is a component of the CCR4 transcriptional regulatory complex. Mol. Cell. Biol., 15, 3487–3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Denis C.L. (1984) Identification of new genes involved in the regulation of yeast alcohol dehydrogenase II. Genetics, 108, 833–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Denis C.L. and Malvar,T. (1990) The CCR4 gene from Saccharomyces cerevisiae is required for both nonfermentative and spt-mediated gene expression. Genetics, 124, 283–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fuge E.K. and Werner-Washburne,M. (1997) In Hohmann,S. and Mager,W.H. (eds), Yeast Stress Responses. Chapman and Hall, New York. [Google Scholar]
- 59.Kitamura K., Nakagawa,T. and Shimoda,C. (1990) Novel sterile mutants of the fission yeast Schizosaccharomyces pombe which are defective in their response to starvation. Curr. Genet., 18, 315–321. [Google Scholar]