Abstract
Initiation of reverse transcription is a complex and regulated process in all retroviruses. Several base pairing interactions have been proposed to occur between the HIV-1 RNA genome and the specific tRNAlys3 primer. The tRNA primer can form up to 21 bp with the primer binding site (PBS), and an additional 8 bp interaction may form between the primer activation signal (PAS) in the HIV-1 RNA and sequences within the TΨC arm of the tRNA. The latter interaction is further analyzed in this in vitro study with mutant RNA transcripts that were designed to preclude the PAS interaction. These mutant transcripts are able to efficiently bind the tRNA primer, but they exhibit a profound defect at initiating reverse transcription. This defect is specific for the tRNA primer because it is not observed for PBS-bound DNA oligonucleotide primers. These results reinforce the model of regulated reverse transcription in which the PAS-mediated interaction is critical for efficient initiation.
INTRODUCTION
Initiation of reverse transcription of the human immunodeficiency virus type 1 (HIV-1) RNA genome is a highly regulated process that requires the formation of a nucleoprotein complex comprising the viral RNA (vRNA) genome, the specific tRNAlys3 primer and the viral reverse transcriptase (RT) enzyme (1,2). At least 18 nt at the 3′ end of the cellular tRNAlys3 hybridize to a fully complementary sequence within the untranslated leader of the vRNA transcript. This well-conserved motif is termed the primer binding site (PBS). RT uses the annealed tRNA primer for synthesis of a cDNA copy and subsequently a double-stranded DNA form of the viral genome. There is accumulating evidence that initiation of reverse transcription is a strictly regulated process in which several additional vRNA–tRNAlys3 interactions play an important role. For instance, the U-rich anticodon of tRNAlys3 was proposed to interact with a single-stranded A-rich motif in the U5 region immediately upstream of the PBS (3–8).
We recently identified another sequence motif that is located further upstream in the U5 region and that is important for tRNA-primed reverse transcription (9). This primer activation signal (PAS) is complementary to the 5′ part of the TΨC arm of the tRNAlys3 primer, termed antiPAS (Fig. 1A). The importance of the PAS–antiPAS interaction has been confirmed in virus replication assays (10), and the combined change in identity of the PBS and PAS motifs allowed a shift in the HIV-1 primer usage from tRNAlys3 to tRNAlys1,2 (11). A similar PAS–antiPAS interaction has been proposed for HIV-2 (12), and convincing in vitro evidence was recently presented for this base pairing (13). The ability to form the PAS-like vRNA–tRNA interaction is highly conserved among different HIV-SIV isolates and other lentiviruses (11,13). Interestingly, the avian Rous sarcoma virus also uses a motif upstream of the PBS to interact with the TΨC arm of its tRNAtrp primer (14–19), suggesting that a PAS-like mechanism to regulate reverse transcription may be a more general property of the retrovirus family. Indeed, an extensive phylogenetic analysis of different retrovirus genera indicated that a PAS-like motif is present upstream of the PBS in all retroviral genomes (11). Initiation of reverse transcription is dependent on an analogous interaction between the yeast Ty1 and Ty2 retrotransposon RNA and the tRNAiMet primer (20–22).
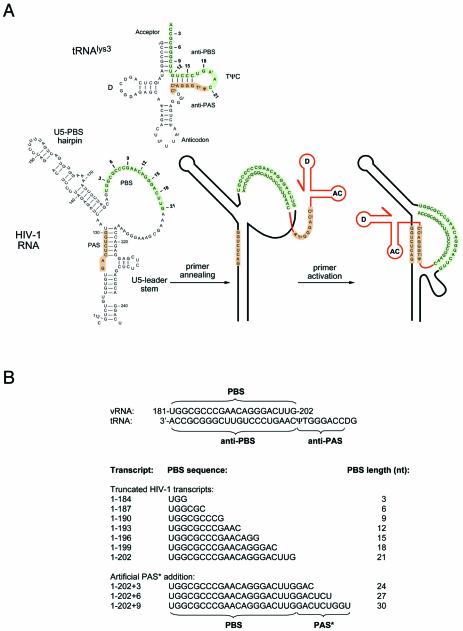
Figure 1.
(A) Scheme of the tRNAlys3 primer and part of the HIV-1 RNA genome. Marked are the PBS and PAS sequences in the HIV-1 untranslated leader RNA and the antiPBS and antiPAS sequences in the tRNAlys3 primer. In the secondary structure model of the HIV-1 RNA we marked the PAS element, U5-top hairpin, PBS and the U5-leader stem. Within the PBS, the positions corresponding to the 3′ terminus of truncated transcripts used in this study are indicated in bold. Numbers indicate the length of the PBS for the corresponding transcript. (B) PBS sequence for the truncated and extended HIV-1 transcripts used in this study. The PBS length in this set of transcripts ranges from 3 to 30 nt. The set includes three transcripts that progressively have an extension of the PBS/tRNA complementarity, such that an artificial PAS* sequence is fused directly downstream of the PBS.
Within the HIV-1 leader RNA, the PAS sequence is masked by base pairing within the U5-leader stem (Fig. 1A). The presence of this PAS enhancer in a repressive RNA structure provides a means for the regulation of reverse transcription (10). Formation of a productive vRNA–tRNA complex requires structural rearrangements of both molecules. It is possible that the HIV-1 nucleocapsid protein (NC) triggers these RNA conformational changes in vivo, and this event may coincide with a structural switch of the leader RNA domain that was shown previously to regulate the process of RNA dimerization (23,24). In fact, the HIV-1 RNA switch may function as a checkpoint to coordinate multiple late replication steps such as RNA dimerization, packaging and reverse transcription (25).
Using in vitro reverse transcription assays, we designed experiments to obtain more information on the pairing between the PAS motif in HIV-1 RNA and the antiPAS sequence that is located in the TΨC arm of tRNAlys3. We constructed HIV-1 transcripts with a minimal PBS motif that facilitates tRNAlys3 binding, but without opening of the TΨC arm such that the antiPAS motif remains base paired. We also introduced an artificial PAS motif directly downstream of the PBS in HIV-1 RNA. This artificial PAS motif is expected to pair with the antiPAS sequence in tRNAlys3, thereby extending the PBS–antiPBS duplex. Both mutational strategies are expected to restrict the formation of the natural PAS–antiPAS interaction, and both type of mutants demonstrate a dramatic reverse transcription defect. These results underscore the importance of the accessory vRNA–tRNA contacts in the process of HIV-1 reverse transcription, and in particular the modulating role of RNA secondary structure in both the viral transcript and the tRNA primer.
MATERIALS AND METHODS
In vitro transcription
The RNA was in vitro transcribed from PCR-generated transcripts containing a T7 promoter directly upstream of the natural HIV-1 LAI +1 transcriptional start site. PCR was performed on the pBluescript 5′LTR plasmid (23). The antisense primers comprised a nested set with the 5′ end terminating at different positions within the PBS, including primers with a 5′ extension to introduce the artificial PAS* element directly downstream of the PBS. PCR fragments were excised from agarose gels and purified using the QIAEX II DNA isolation system according to the manufacturers instructions. Transcription was carried out using the Ambion megashortscript T7 transcription kit, and radiolabeled transcripts were synthesized in the presence of 1 µl [α-32P]UTP. Transcripts were subsequently excised from a 4% denaturing polyacrylamide gel and eluted from the gel fragment by overnight incubation in TBE buffer at room temperature. The RNA was ethanol precipitated and dissolved in water. Quantification of the RNA was done by UV-absorbance measurements and scintillation counting in case of radiolabeled transcripts.
Primer annealing assays
Primer binding was assayed by incubating ∼0.5 µg of the radiolabeled HIV-1 transcripts with 100 ng of the DNA primer or 1.0 µg of total tRNA extracted from calf liver (Roche), of which ∼0.2 µg tRNAlys3 that specifically binds to HIV-1 RNA (26). The HIV-1 RNA and primer were incubated in 10 µl TEN buffer (100 mM NaCl, 10 mM Tris–HCl pH 7.5 and 1.0 mM EDTA) at 65°C, or at the indicated temperature, for 10 min and then slowly cooled to room temperature. After the incubation, an equal volume of loading buffer was added and the samples were analyzed on a 4% non-denaturing polyacrylamide gel. Electrophoresis was performed at 150 V and room temperature with 0.25 TBE (22.5 mM Tris–HCl pH 7.0, 22.5 mM Boric acid, 0.625 mM EDTA) in the gel and running buffer. Gels were dried and analyzed on a Storm 820 phosphoimager.
Primer extension reactions
The in vitro synthesized RNA (10 ng) was incubated either with 1.5 µg of the calf liver tRNA or with 10 ng of the DNA primer in 12 µl annealing buffer (83 mM Tris–HCl pH 7.0, 125 mM KCl) at 65°C for 10 min and slowly cooled to room temperature. The primer was extended with 1 nt by addition of 6.0 µl of RT buffer (9 mM MgCl2, 30 mM DTT, 150 µg/ml Actinomycin D) and 1 µl [α-32P]dCTP and 0.5 U of HIV-1 RT (MRC AIDS reagent project). Reverse transcription was performed for 30 min at 37°C. Two nucleotide extension products (+2) were made in the same manner but with 30 µM dTTP in the RT buffer. Samples were ethanol precipitated, dissolved in formamide loading buffer, heated at 85°C for 1 min and loaded on a 6% denaturing polyacrylamide gel, which was quantified on a Storm 820 phosphoimager.
RESULTS
The experimental design
Figure 1A shows the RNA secondary structure model of the tRNAlys3 primer and part of the HIV-1 RNA transcript encompassing the PBS and PAS signals. The PBS is usually referred to as an 18-nt sequence, but it is in fact up to 21 nt in most HIV-1 isolates (27,28). In the tRNA primer, the sequence complementary to the HIV-1 PBS (termed antiPBS) is located in the acceptor and TΨC arms. The PBS–antiPBS interaction facilitates tRNA annealing, and an additional PAS–antiPAS interaction is required for activation of the annealed tRNA primer to initiate reverse transcription (9–11). The antiPAS sequence is also located in the tRNA TΨC arm. Of these four sequence motifs in the vRNA and tRNA molecules, the PBS element is the only element that is freely accessible (29) (Fig. 1A), whereas the PAS element in the HIV-1 transcript and the antiPBS and antiPAS elements in the tRNA primer are occluded by base pairing.
Formation of HIV-1 reverse transcription complexes thus requires the disruption of part of the HIV-1 RNA and tRNA secondary structures in order to allow the formation of intermolecular vRNA–tRNA interactions (Fig. 1A). In particular, formation of the PBS–antiPBS helix requires the disruption of the tRNA acceptor and TΨC arms. In this initial complex, the antiPAS sequence in the tRNA remains accessible to form the additional PAS–antiPAS interaction. Based on this model, we reasoned that HIV-1 transcripts with a PBS of up to 12 nt will bind the tRNA primer without opening of the TΨC arm, thus precluding the PAS–antiPAS interaction. Conversely, HIV-1 transcripts in which the PBS is artificially extended such that it is complementary to the antiPAS sequence should also preclude the formation of the natural PAS–antiPAS interaction.
We thus generated a set of HIV-1 RNA transcripts with a 3′ truncated or extended PBS sequence (Fig. 1B). This series contains a minimal 1-184 transcript with a 3 nt PBS, intermediates with a 6, 9, 12, 15 and 18 nt PBS, and the wild-type 1-202 transcript with a 21 nt PBS element. Furthermore, we extended the full-length 21 nt PBS sequence with sequences that mimic the PAS element (the artificial PAS* element, see Fig. 1B). These mutant transcripts are based on the wild-type 1-202 transcript to which 3, 6 or 9 nt of the PAS sequence were fused. Annealing of the tRNA primer to these transcripts will result in the formation of the PBS–antiPBS duplex that is extended with 3, 6 and 9 bp of the artificial PAS*–antiPAS duplex, yielding an uninterrupted vRNA–tRNA duplex of 24, 27 and 30 bp, respectively. We note that this set of HIV-1 transcripts lacks the downstream sequences that are base paired with the PAS element in the full-length HIV-1 leader RNA (Fig. 1A). This strategy bypasses the restriction imposed by the HIV-1 RNA secondary structure on the availability of the PAS motif (9). In this optimal setting with a constitutively available PAS element, we examined the effect of truncating and extending the PBS sequence on the initiation of reverse transcription. Because PBS-truncation may obviously affect primer annealing, we also examined the ability of these transcripts to bind the tRNA primer and a control DNA primer.
tRNA structure masks the antiPBS element
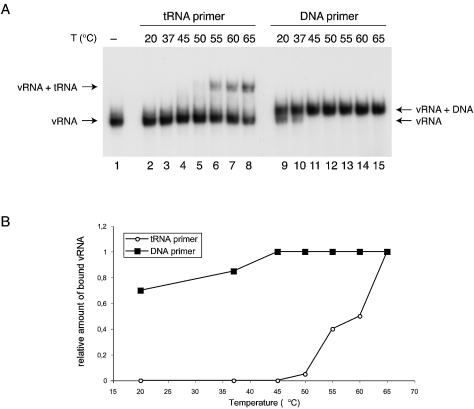
We performed annealing experiments with a radiolabeled 1-202 HIV-1 RNA transcript and the unlabeled natural tRNAlys3 primer or a control DNA primer that anneals to the PBS. The primer and transcript were incubated at various temperatures between 20 and 65°C and analyzed on a non-denaturing polyacrylamide gel (Fig. 2A). As expected, annealing of the 76-nt tRNA primer causes a more dramatic shift in the migration of the labeled vRNA than annealing of the 21-nt DNA primer. The bands were quantified and we calculated the vRNA fraction that was shifted by the tRNA versus the DNA primer. These values were normalized for the maximum amount of the vRNA–tRNA complex formed at 65°C and plotted against the incubation temperature (Fig. 2B). The results show that DNA annealing is observed at all temperatures, but tRNA annealing requires incubation temperatures of at least 55°C. This initial finding supports the idea that tRNA secondary structure limits the accessibility of the antiPBS sequence, which is consistent with previous reports (30,31). Knowing the optimal conditions for tRNA primer annealing, we set out to test the properties of the mutant vRNA transcripts.
Figure 2.
Temperature dependence of DNA and tRNAlys3 primer annealing to HIV-1 leader RNA. (A) Electrophoretic analysis of the HIV-1 RNA (1-202) incubated with the DNA or tRNA primer at indicated temperatures. A formamide-denatured control is included in lane 1. (B) Quantified results of the gel shown in (A). The data are normalized for the maximum amount of the vRNA–primer complex at 65°C.
Primer annealing requires a minimal PBS of 12 nt
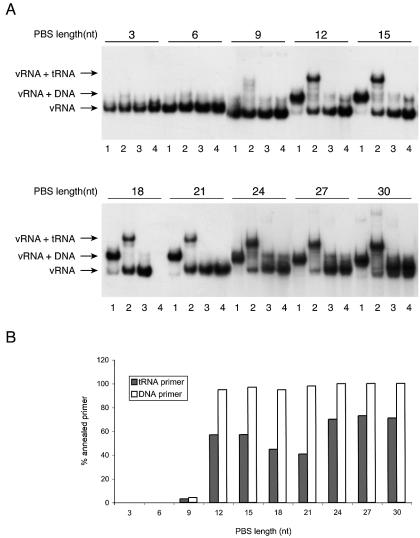
The set of HIV-1 transcripts with different PBS lengths was synthesized as radiolabeled RNA to assess the annealing of the tRNA and DNA primers (Fig. 3A). The transcripts (PBS length indicated at the top of the panels in Fig. 3A) were incubated with either the control DNA primer (lanes 1) or the tRNA primer (lanes 2) and analyzed on a non-denaturing gel. We included several control experiments such as a mock-incubation without primer (lanes 3) and a formamide-treated vRNA sample (lanes 4). We calculated the amount of vRNA–primer complex for the different transcripts, and this value is plotted both for the DNA and tRNA primer (Fig. 3B). Both primers demonstrate a similar PBS requirement, some annealing is observed with the 9-nt PBS, but maximal annealing requires a PBS of at least 12 nt. All longer PBS motifs, including the PAS* series, are able to bind both primers. The minor differences between these transcripts may be due to unforeseen effects at the level of RNA secondary structure. However, we did consistently observe that tRNA annealing was slightly more efficient on the transcripts with a 12 and 15 nt PBS than the transcripts containing an 18 and 21 nt PBS. The slightly increased levels of tRNA annealing on the transcripts with an extended PBS (24, 27 and 30 nt) was also observed consistently.
Figure 3.
DNA and tRNAlys3 primer binding to PBS-mutated HIV-1 transcripts. (A) Electrophoretic analysis of vRNA–primer complexes formed in the presence of a DNA primer (lanes 1) and tRNAlys3 (lanes 2). Control samples include HIV-1 RNA without primer (lanes 3) and formamide-denatured samples (lanes 4). The number of PBS nucleotides is indicated at the top of the gel for each transcript. (B) Quantified data of the gel shown in (A), showing the yield of vRNA–primer complexes with the DNA primer or tRNAlys3.
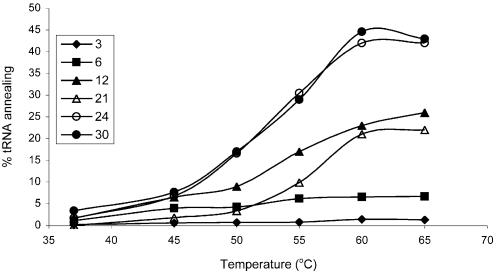
These annealing experiments were performed at 65°C, but we also tested tRNA annealing for a subset of the transcripts at varying temperatures in the 37–65°C range (Fig. 4). All vRNA mutants exhibit increased tRNA annealing as the temperature is increased, and the levels of vRNA–tRNA complex formation are consistent with the results presented in Figure 3B. Again, we observed that the transcript with a 12 nt PBS interacted with the tRNA primer slightly more efficiently than the wild-type counterpart (21 nt). The transcripts with the PAS*-extended PBS (24, 27 and 30 nt) bind the tRNA primer significantly better than transcripts with a PBS of the wild-type length. On average, the complex yield is nearly 2-fold higher than the wild-type control. Incubation of the PAS*-extended PBS mutants at 50°C yields amounts comparable with the wild-type yield at 60°C. These observations support our reasoning that fusing the PAS* sequence directly downstream of the PBS results in an extended vRNA–tRNA duplex.
Figure 4.
Temperature dependence of vRNA–tRNA complex formation with PBS-mutated HIV-1 transcripts. Numbers in the legend correspond to the number of nucleotides in the PBS.
tRNA priming requires a minimal PBS of 15 nt
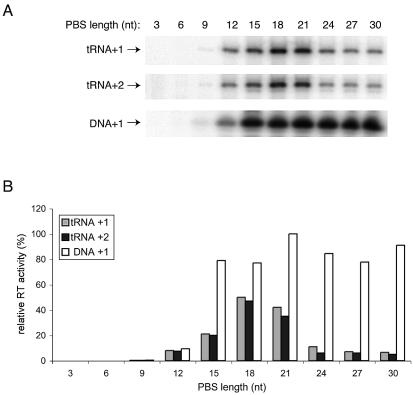
The previous experiments indicate that we can efficiently anneal both DNA and tRNA primers onto the transcripts with a PBS of at least 12 nt and the transcripts with an artificial PAS* extension. This allowed us to test the efficiency of the priming reaction, for which these transcripts were specifically designed. The DNA and tRNA complexes were made by high temperature incubation, and reverse transcription was initiated by addition of HIV-1 RT enzyme and dNTPs. We provided either radiolabeled dCTP, which results in the incorporation of a single nucleotide (+1 extension), or radiolabeled dCTP and unlabeled dTTP to allow a 2-nt extension (+2 extension). The labeled cDNA products were analyzed on a denaturing polyacrylamide gel (Fig. 5A), and we plotted the reverse transcription efficiency for the tRNA primer in the +1 and +2 extension reactions and the DNA primer in the +1 reaction for all the vRNA transcripts (Fig. 5B).
Figure 5.
Initiation of reverse transcription on PBS-mutated HIV-1 transcripts. (A) Electrophoretic analysis of tRNA and DNA primer extension by 1 or 2 nt (+1 or +2, respectively). The length of the PBS is indicated at the top of the lane for each transcript. (B) Quantitated data from the gel shown in (A).
No significant reverse transcription signal was obtained for the transcripts with a PBS shorter than 9 nt, which correlates with their inability to bind the primer. Whereas we observed efficient primer annealing onto the 12-nt PBS element, this transcript is a poor template for tRNA-primed reverse transcription with both the tRNA and DNA primer. Extension of the PBS to 15 nt improves the initiation efficiency at least 2-fold, but the transcript is still sub-optimal for tRNA priming. However, full activity is measured with the DNA primer on this transcript. Further extension of the PBS to 18 or 21 nt results in efficient tRNA priming.
The transcripts containing the artificial PAS* extension downstream of the PBS are fully efficient in DNA priming, but they exhibit a profound tRNA priming defect that is most severe for the vRNA with the longest 9-nt PAS* element (Fig. 5A and B). Similar results were obtained in assays that monitor the synthesis of longer cDNA products (results not shown). As stated previously, these transcripts are fully efficient in tRNA annealing, and this suggests that they cannot support tRNA priming because the natural PAS–antiPAS interaction is not established. The specificity of the defect imposed by the artificial PAS* element on tRNA priming is nicely illustrated by the behavior of the control DNA primer on these transcripts. The activity of the DNA primer is ruled solely by the annealing efficiency on the different transcripts, and the presence of the additional PAS* element downstream of the PBS does not have a negative impact on the initiation efficiency of this control primer.
DISCUSSION
To study the role of a PAS–antiPAS interaction in the initiation of HIV-1 reverse transcription, we generated a set of HIV-1 transcripts with different PBS lengths. Due to the tRNA unfolding requirements to generate an active reverse transcription complex, these transcripts are expected to disturb PAS-mediated activation in two ways: (i) the antiPAS sequence is not accessible in transcripts with a shortened PBS because the tRNA TΨC arm is not opened, and (ii) transcripts in which the PBS has been extended with an artificial PAS* element will prevent the usage of the natural PAS element in the vRNA–tRNA complex. We performed primer annealing and reverse transcription assays with these transcripts, and the results are summarized in Table 1.
Table 1. Summary of primer annealing and initiation of reverse transcription data from this study.
| Transcript | PBS length (nt) | DNA primer Annealing (%) | Priming (+1) (%) | tRNA primer Annealing (%) | Priming (+1) (%) | Priming (+2) (%) |
|---|---|---|---|---|---|---|
| 1-184 | 3 | 0 | 0 | 0 | 0 | 0 |
| 1-187 | 6 | 0 | 0 | 0 | 0 | 0 |
| 1-190 | 9 | 4 | 0.4 | 4 | 0 | 0 |
| 1-193 | 12 | 95 | 9 | 78 | 16 | 16 |
| 1-196 | 15 | 97 | 79 | 78 | 42 | 43 |
| 1-199 | 18 | 95 | 77 | 62 | 100a | 100a |
| 1-202 | 21 | 98 | 100a | 56 | 84 | 75 |
| 1-202+3 | 24 | 100a | 85 | 96 | 22 | 12 |
| 1-202+6 | 27 | 100a | 78 | 100a | 14 | 12 |
| 1-202+9 | 30 | 100a | 91 | 98 | 13 | 10 |
aMaximum values were set at 100%, and other results are relative to this value.
Transcripts with a PBS length of 9 nt or less are incapable of binding the DNA and tRNA primers and thus are completely inactive in the reverse transcription assays. When the PBS length is extended to 12 nt, both primers anneal efficiently but only a marginal reverse transcription activity is measured (Table 1). In our design, the transcript with a 12 nt PBS would anneal the tRNA without opening the TΨC arm, thus preventing the PAS–antiPAS interaction that facilitates initiation of reverse transcription. The efficient tRNA annealing, but low tRNA-priming activity of this transcript supports this idea, but we also observe that DNA priming is inefficient on this transcript. In melting experiments, we observed that complexes formed with a 12 nt PBS are less stable than transcripts with a PBS of 15 nt or more (results not shown). It therefore seems plausible that the 12 nt vRNA–primer duplex dissociates during the reverse transcription reaction, thus causing a loss in the initiation efficiency. However, we cannot exclude a contribution of antiPAS occlusion on the tRNA-primed reverse transcription activity.
For the transcript with a 15 nt PBS, the vRNA–tRNA duplex penetrates the tRNA TΨC arm, thus releasing the antiPAS sequence. Indeed, this transcript reaches intermediate levels of tRNA-primed reverse transcription. On transcripts with the 18 and 21 nt PBS, high levels of tRNA-primed reverse transcription are reached and both these transcripts are expected to contain a vRNA–tRNA duplex that fully disrupts the tRNA TΨC arm. We also investigated the primer annealing and reverse transcription properties of transcripts in which the PBS was extended with an artificial PAS* element, which was designed to base pair with the antiPAS sequence in the tRNA primer. This PBS extension will interfere with the natural PAS–antiPAS interaction and the dramatic loss in reverse transcription levels of these templates is fully consistent with this idea (Table 1). Annealing of the tRNA primer and reverse transcription from the control DNA primer is at least as efficient as with the wild-type template, thus demonstrating that occlusion of the antiPAS sequence by the artificial PAS* element reduces the reverse transcription efficiency. This result reinforces the importance of the natural PAS–antiPAS interaction during initiation of HIV-1 reverse transcription.
Recently, Goldschmidt et al. have challenged the existence and the role of the PAS–antiPAS interaction during initiation of reverse transcription (32). These authors reconstructed some of the mutants from our initial study (9) and performed reverse transcription and structure probing experiments. These mutants either target the PAS sequence (mutant 2L) or its complementary counterpart in the U5-leader stem in the HIV-1 RNA (mutant 2R). In our hands, the 2L mutation strongly reduced reverse transcription and the 2R mutation caused a profound stimulation compared with the wild-type transcript because the PAS motif is no longer masked by base pairing (9). We confirmed this RNA structural effect by structure probing. Furthermore, we demonstrated the 2L defect and the 2R up-regulation in a physiological setting by analyzing reverse transcription products from the mutant virion particles (10). Whereas Goldschmidt reproduced the defect of the 2L mutation, the stimulatory effect of the 2R mutation was not observed.
Based on structure probing experiments on the naked HIV-1 RNA, Goldschmidt argues that aberrant folding of the RNA rather than mutation of the PAS sequence causes the reverse transcription defect of the 2L mutant. Although structural perturbations in mutant RNA are an effect to be reckoned with, Goldschmidt’s work certainly does not exclude the possibility that the reverse transcription effect of mutant 2L is caused by substitution of the PAS sequence. Goldschmidt performed further structure probing assays on the wild-type vRNA–tRNA complex and points out that annealing of the tRNA does not result in an increase in reactivity of the helical segment that masks the PAS sequence in the absence of the tRNA. This is presented as evidence against the PAS–antiPAS interaction, whereas refolding of the RNA into an alternative structure is not offered as a possible explanation of this result. More importantly, probing data for the PAS sequence itself are prominently absent from the study. Goldschmidt did probe the tRNA primer in the presence of HIV-1 RNA and noted an increase in DEPC modification of tRNA residue A50 within the antiPAS sequence. Again, this is presented as direct evidence against the PAS–antiPAS interaction. Alternatively, one could argue that this indicates a temporarily open structure of the tRNA TΨC arm, as the PAS–antiPAS model predicts. The interaction of the tRNAlys3 antiPAS with the HIV-1 PAS motif is necessarily dynamic, and probably short-lived, since RT must penetrate the PAS–antiPAS helix during the early elongation phase. It is also possible that the PAS–antiPAS interaction depends on the presence of the RT enzyme in the initiation complex.
In dismissing the PAS–antiPAS interaction, Goldschmidt ignores or misinterprets, in our view, many important pieces of evidence. For instance, there is no mention of virus replication data of revertants of the 2L mutant that restore both the stability of the PAS–antiPAS interaction and the replication kinetics (10). Furthermore, we introduced precise mutations in the PAS motif to strengthen or weaken the base pairing with the antiPAS, and the reverse transcription activity of these transcripts correlates neatly with the base pairing potential (11). Finally, we were able to switch the selective primer usage of HIV-1 from tRNAlys3 to tRNAlys1,2 by simultaneous adaptation of the PBS and PAS motifs (11). This important finding is unfairly devaluated by Goldschmidt with the comment that this double mutant is not as efficient in using tRNAlys1,2 compared with tRNAlys3 usage of the wild-type template. The strict conservation of the PAS motif throughout all members of the retrovirus family is also downplayed by referring to the finding that retroviral RTs have specificity for their cognate transcript, despite utilizing the same tRNA primer. However, this result has been challenged in a more recent study (33). Virus-specific RT and template compatibility could be due to many differences in either the RT enzyme or the RNA. For example, specificity of retroviral RT enzymes for the cognate primer has been demonstrated previously (26,34–36).
The factors that govern assembly and activity of reverse transcription complexes are not yet fully understood and are likely to depend on intricate interactions between the viral template, tRNA primer and the NC and RT proteins. In the current study, we have used a mutational strategy that is different from previous analyses of initiation of HIV-1 reverse transcription to study the activity of in vitro generated vRNA–tRNA complexes. Our results show that the tRNA annealing and priming steps can be dissected and that prevention of the PAS–antiPAS interaction results in a reverse transcription defect. This supports an important role for the PAS–antiPAS interaction in the initiation of tRNA-primed reverse transcription.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Wim van Est for artwork, Virulence-member Dr D. Stammers for the gift of purified HIV-1 RT enzyme and Nancy Beerens for critical reading of the manuscript. This work was sponsored in part by the Netherlands Foundation for Chemical Research with financial aid from the Netherlands Organization for Scientific Research (NWO-CW) and the EU-Virulence program. K.B. was supported by a short-term FEBS fellowship.
REFERENCES
- 1.Telesnitsky A. and Goff,S.P. (1997) Reverse transcriptase and the generation of retroviral DNA. In Coffin,J.M., Hughes,S.H. and Varmus,H.E. (eds), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 121–160. [PubMed] [Google Scholar]
- 2.Beerens N. and Berkhout,B. (2002) Strict regulation of HIV-1 reverse transcription. Curr. Top. Virol., 2, 115–127. [Google Scholar]
- 3.Isel C., Westhof,E., Massire,C., Le Grice,S.F.J., Ehresmann,B., Ehresmann,C. and Marquet,R. (1999) Structural basis for the specificity of the initiation of HIV-1 reverse transcription. EMBO J., 18, 1038–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang Z., Kang,S.-M., Li,Y. and Morrow,C.D. (1998) Genetic analysis of the U5-PBS of a novel HIV-1 reveals multiple interactions between the tRNA and RNA genome required for initiation of reverse transcription. RNA, 4, 394–406. [PMC free article] [PubMed] [Google Scholar]
- 5.Li Y., Zhang,Z., Wakefield,J.K., Kang,S.-M. and Morrow,C.D. (1997) Nucleotide substitutions within U5 are critical for efficient reverse transcription of human immunodeficiency virus type 1 with a primer binding site complementary to tRNAHis. J. Virol., 71, 6315–6322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lanchy J.-M., Isel,C., Keith,G., Le Grice,S.F.J., Ehresmann,C. and Marquet,R. (2000) Dynamics of the HIV-1 reverse transcription complex during initiation of DNA synthesis. J. Biol. Chem., 275, 12306–12312. [DOI] [PubMed] [Google Scholar]
- 7.Arts E.J., Stetor,S.R., Li,Y., Rausch,J.W., Howard,K.J., Ehresmann,B., North,T.W., Wohrl,B.M., Goody,R.S., Wainberg,M.A. et al. (1996) Initiation of (-) strand DNA synthesis from tRNALys3 on lentiviral RNAs: implications of specific HIV-1 RNA-tRNALys3 interactions inhibiting primer utilization by retroviral reverse trancriptases. Proc. Natl Acad. Sci. USA, 93, 10063–10068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang Y., Shalom,A., Li,Z., Wang,J., Mak,J., Wainberg,M.A. and Kleiman,L. (1996) Effects of modifying the tRNALys3 anticodon on the initiation of human immunodeficiency virus type 1 reverse transcription. J. Virol., 70, 4700–4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beerens N., Groot,F. and Berkhout,B. (2001) Initiation of HIV-1 reverse transcription is regulated by a primer activation signal. J. Biol. Chem., 276, 31247–31256. [DOI] [PubMed] [Google Scholar]
- 10.Beerens N. and Berkhout,B. (2002) The tRNA primer activation signal in the HIV-1 genome is important for initiation and processive elongation of reverse transcription. J. Virol., 76, 2329–2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beerens N. and Berkhout,B. (2002) Switching the in vitro tRNA usage of HIV-1 by simultaneous adaptation of the PBS and PAS. RNA, 8, 357–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berkhout B. and Schoneveld,I. (1993) Secondary structure of the HIV-2 leader RNA comprising the tRNA-primer binding site. Nucleic Acids Res., 21, 1171–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Freund F., Boulme,F., Litvak,S. and Tarrago-Litvak,L. (2001) Initiation of HIV-2 reverse transcription: a secondary structure model of the RNA-tRNA(Lys3) duplex. Nucleic Acids Res., 29, 2757–2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aiyar A., Ge,Z. and Leis,J. (1994) A specific orientation of RNA secondary structures is required for initiation of reverse transcription. J. Virol., 68, 611–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aiyar A., Cobrinik,D., Ge,Z., Kung,H.J. and Leis,J. (1992) Interaction between retroviral U5 RNA and the TYC loop of the tRNA(Trp) primer is required for efficient initiation of reverse transcription. J. Virol., 66, 2464–2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cobrinik D., Aiyar,A., Ge,Z., Katzman,M., Huang,H. and Leis,J. (1991) Overlapping retrovirus U5 sequence elements are required for efficient integration and initiation of reverse transcription. J. Virol., 65, 3864–3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cobrinik D., Soskey,L. and Leis,J. (1988) A retroviral RNA secondary structure required for efficient initiation of reverse transcription. J. Virol., 62, 3622–3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller J.T., Ge,Z., Morris,S., Das,K. and Leis,J. (1997) Multiple biological roles associated with the Rous sarcoma virus 5′ untranslated RNA U5-IR stem and loop. J. Virol., 71, 7648–7656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morris S. and Leis,J. (1999) Changes in the Rous sarcoma virus RNA secondary structure near the primer binding site upon tRNATrp primer annealing. J. Virol., 73, 6307–6318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Friant S., Heyman,T., Wilhelm,M.L. and Wilhelm,F.X. (1996) Extended interactions between the primer tRNAi(Met) and genomic RNA of the yeast Ty1 retrotransposon. Nucleic Acids Res., 24, 441–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Friant S., Heyman,T., Poch,O., Wilhelm,M. and Wilhelm,F.X. (1997) Sequence comparison of the Ty1 and Ty2 elements of the yeast genome supports the structural model of the tRNAiMet-Ty1 RNA reverse transcription initiation complex. Yeast, 13, 639–645. [DOI] [PubMed] [Google Scholar]
- 22.Friant S., Heyman,T., Bystrom,A.S., Wilhelm,M. and Wilhelm,F.X. (1998) Interactions between Ty1 retrotransposon RNA and the T and D regions of the tRNA(iMet) primer are required for initiation of reverse transcription in vivo. Mol. Cell. Biol., 18, 799–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huthoff H. and Berkhout,B. (2001) Two alternating structures for the HIV-1 leader RNA. RNA, 7, 143–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huthoff H. and Berkhout,B. (2002) Multiple secondary structure rearrangements during HIV-1 RNA dimerization. Biochemistry, 41, 10439–10445. [DOI] [PubMed] [Google Scholar]
- 25.Berkhout B., Ooms,M., Beerens,N., Huthoff,H., Southern,E. and Verhoef,K. (2002) In vitro evidence that the untranslated leader of the HIV-1 genome is an RNA checkpoint that regulates multiple functions through conformational changes. J. Biol. Chem., 277, 19967–19975. [DOI] [PubMed] [Google Scholar]
- 26.Oude Essink B.B., Das,A.T. and Berkhout,B. (1996) HIV-1 reverse transcriptase discriminates against non-self tRNA primers. J. Mol. Biol., 264, 243–254. [DOI] [PubMed] [Google Scholar]
- 27.Das A.T., Klaver,B. and Berkhout,B. (1997) Sequence variation of the HIV primer-binding site suggests the use of an alternative tRNALys molecule in reverse transcription. J. Gen. Virol., 78, 837–840. [DOI] [PubMed] [Google Scholar]
- 28.Muthuswami R., Chen,J., Burnett,B.P., Thimmig,R.L., Janjic,N. and McHenry,C.S. (2002) The HIV plus-strand transfer reaction: determination of replication-competent intermediates and identification of a novel lentiviral element, the primer over-extension sequence. J. Mol. Biol., 315, 311–323. [DOI] [PubMed] [Google Scholar]
- 29.Damgaard C.K., Dyhr-Mikkelsen,H. and Kjems,J. (1998) Mapping the RNA binding sites for human immunodeficiency virus type-1 Gag and NC proteins within the complete HIV-1 and -2 untranslated leader regions. Nucleic Acids Res., 26, 3667–3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chan B., Weidemaier,K., Yip,W.-T., Barbara,P.F. and Musier-Forsyth,K. (1999) Intra-tRNA distance measurements for nucleocapsid protein-dependent tRNA unwinding during priming of HIV reverse transcription. Proc. Natl Acad. Sci. USA, 96, 459–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oude Essink B.B., Das,A.T. and Berkhout,B. (1995) Structural requirements for the binding of tRNA Lys3 to reverse transcriptase of the human immunodeficiency virus type 1. J. Biol. Chem., 270, 23867–23874. [DOI] [PubMed] [Google Scholar]
- 32.Goldschmidt V., Ehresmann,C., Ehresmann,B. and Marquet,R. (2003) Does the HIV-1 primer activation signal interact with tRNA(3)(Lys) during the initiation of reverse transcription? Nucleic Acids Res., 31, 850–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boulme F., Freund,F. and Litvak,S. (1998) Initiation of in vitro reverse transcription from tRNA(Lys3) on HIV-1 or HIV-2 RNAs by both type 1 and 2 reverse transcriptases. FEBS Lett., 430, 165–170. [DOI] [PubMed] [Google Scholar]
- 34.Panet A., Haseltine,W.A., Baltimore,D.J., Peters,G., Harada,F. and Dahlberg,J.E. (1975) Specific binding of tryptophan transfer RNA to avian myeloblastosis virus RNA-dependent DNA polymerase (reverse transcriptase). Proc. Natl Acad. Sci. USA, 72, 2535–2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barat C., Lullien,V., Schatz,O., Keith,G., Mugeyre,M.T., Grüninger-Leitch,F., Barré-Sinoussi,F., LeGrice,S.F.J. and Darlix,J.L. (1989) HIV-1 reverse transcriptase specifically interacts with the anticodon domain of its cognate primer tRNA. EMBO J., 11, 3279–3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Litvak S., Sarih-Cottin,L., Fournier,M., Andreola,M. and Tarrago-Litvak,L. (1994) Priming of HIV replication by tRNA lys3: role of reverse transcriptase. Trends Biochem. Sci., 19, 114–118. [DOI] [PubMed] [Google Scholar]