Abstract
The Schizosaccharomyces pombe Ku70–Ku80 heterodimer is required for telomere length regulation. Lack of pku70+ results in telomere shortening and striking rearrangements of telomere-associated sequences. We found that the rearrangements of telomere-associated sequences in pku80+ mutants are Rhp51 dependent, but not Rad50 dependent. Rhp51 bound to telomere ends when the Ku heterodimer was not present at telomere ends. We also found that the single-stranded G-rich tails increased in S phase in wild-type strains, while deletion of pku70+ increased the single-stranded overhang in both G2 and S phase. Based on these observations, we propose that Rhp51 binds to the G-rich overhang and promotes homologous pairing between two different telomere ends in the absence of Ku heterodimer. Moreover, pku80 rhp51 double mutants showed a significantly reduced telomere hybridization signal. Our results suggest that, although Ku heterodimer sequesters Rhp51 from telomere ends to inhibit homologous recombination activity, Rhp51 plays important roles for the maintenance of telomere ends in the absence of the Ku heterodimer.
INTRODUCTION
Telomeres are the DNA–protein complexes located at the ends of eukaryotic chromosomes (1–4). Telomere DNA consists of a simple G-rich repeat sequence with a length of 300 bp in yeast. Myb domain-containing telomere-binding proteins such as Schizosaccharomyces pombe Taz1 recognize the telomere-repeat sequence (GGTTACA)n and form distinct DNA–protein structures (5). These structures protect chromosome ends from degradation and aberrant recombination or fusion (6). Telomeres are shortened with each cell division and cells undergo replicative senescence if telomerase is inactive. To prevent telomere shortening, telomere DNA is synthesized by telomerase, which catalyzes the polymerization of telomeric repeats. Although both Saccharomyces cerevisiae and S.pombe normally rely on telomerase activity to maintain their telomeres, telomeres can be maintained without this activity. Telomerase-independent telomere maintenance has been well studied in S.cerevisiae (7,8). Generation of survivors that lack the gene for a telomerase component requires RAD52-dependent recombination. The majority of survivors have multiple tandem copies of the subtelomeric Y′ element and short telomere DNA (type I survivors) (9,10). In contrast, type II survivors (a minor fraction of the survivors) have long, heterogeneous tracts of telomeric repeats (10). In S.pombe, deletion of trt1+, which encodes a catalytic protein subunit of telomerase, leads to progressive loss of telomere DNA and causes cellular senescence (11). There are two modes of survival in those cells. Most survivors have circularized all of their chromosomes, whereas a minor fraction of survivors maintain their telomeres, probably through recombination (12).
Unlike natural chromosome ends (telomere ends), DNA double-strand break (DSB) ends are repaired by homologous recombination (HR) or non-homologous end joining (NHEJ) (13–15). In S.pombe, DSBs are mainly repaired by HR. Rad50 and Rad32 (the S.pombe homolog of Mre11) are thought to be required for resection of DSB ends to initiate HR (16–19). Rhp51 (the S.pombe homolog of Rad51) is thought to bind the single-stranded region of DSB ends to promote homologous pairing and strand exchange (20). Although NHEJ is not a major repair pathway in S.pombe, the Ku70–Ku80 heterodimer is required for the repair of linearized plasmids following transformation into cells (21,22). The HR and NHEJ repair pathways are conserved from yeast to mammals. In vertebrates, NHEJ dominates during G1 to early S phase of the cell cycle and HR is used in late S to G2 phase (23,24).
Telomere ends are not sensed as DNA DSB ends. However, recent studies have revealed that DNA repair proteins such as the Mre11–Rad50–Xrs2/Nbs1 complex and the Ku70–Ku80 heterodimer bind to telomere ends and play important roles in telomere maintenance (25–29). One might expect that binding of these proteins to telomere ends would result in unnecessary HR or NHEJ. However, Ku heterodimer actually inhibits DNA end joining between telomere DNAs in mice, suggesting that Ku heterodimer has distinct roles at two types of different DNA end (telomere ends and DSB ends) (27,30–32).
In S.cerevisiae, deletion of YKU70 or YKU80 results in telomere shortening and loss of the silencing of telomere-adjacent genes (33–35). In addition, deletion of YKU70 or YKU80 results in the generation of single-strand 3′ G-tails at telomere ends in a cell cycle-independent manner, while the single-strand 3′ G-tail appears only in S phase in wild-type cells (36–39). In S.pombe, deletion of pku70+ or pku80+ results in telomere shortening (22,40). However, unlike in S.cerevisiae, telomere silencing in pku70 or pku80 mutants remains intact (21,22). In S.pombe, the telomeric repeat sequences are internally flanked by repetitive telomere-associated sequence (TAS) (41). Lack of the functional Ku heterodimer caused striking rearrangements of TAS in a rad22+-dependent manner, indicating a function for Ku heterodimers in inhibiting recombinational activities near chromosome ends (40). However, the mechanism of the recombination at telomere ends in the absence of Ku heterodimers is not fully understood.
Here we have studied the rearrangement of TAS in pku80 rhp51 double mutants and pku80 rad50 double mutants. We found that recombination at the subtelomeric region in the absence of Ku80 is Rhp51 dependent. Rhp51 bound to telomere ends when Ku80 was not present there. Moreover, deletion of pku70+ increased the single-stranded overhang in both G2 and S phase. Based on these observations, we propose that Rhp51 binds to the G-rich overhang and promotes homologous pairing between two different telomere ends in the absence of the Ku heterodimer.
MATERIALS AND METHODS
Schizosaccharomyces pombe strains, medium and genetic methods
The S.pombe strains used in this study are listed in Table 1. All strains are derivatives of JY741 h– or JY746 h+. Standard procedures and medium were used for propagation and genetic manipulation (42). YPAD medium consisted of 1% yeast extract, 2% polypeptone, 2% glucose and 20 µg/ml adenine. All experiments were repeated at least twice with similar results. The rad50+ gene was disrupted by replacing the region between the first and second HindIII restriction sites (nucleotide positions 529–1158 relative to the initiation codon) with the ura4+ gene. pku70-d and pku80-d were constructed by insertion of the LEU2 cassette into the EcoRV sites of each of these genes. rad22-d and rhp51-d cells were constructed by insertion of the ura4+ cassette into the AflII site of the rad22+ gene and the NheI site of the rhp51+ gene, respectively. Other double mutants were constructed by genetic crosses.
Table 1. Schizosaccharomyces pombe strains used in this study.
| Strain | Genotype | Source |
|---|---|---|
| JY741 | h– leu1-32 ura4-D18 ade6-M216 | M. Yamamoto |
| JY746 | h+ leu1-32 ura4-D18 ade6-M210 | M. Yamamoto |
| LSP11 | h+ leu1-32 ura4-D18 cdc25–22 | Laboratory stock |
| KT102 | h+ leu1-32 ura4-D18 ade6-M216 rad50::ura4+ | This study |
| KT10b | h+ leu1-32 ura4-D18 ade6-M216 rad22::ura4+ | This study |
| KT10c | h+ leu1-32 ura4-D18 ade6-M216 rhp51::ura4+ | This study |
| HK001 | h leu1-32 ura4-D18 ade6-M216 rad3::aur1 tel1::ura4+ | This study |
| TK001 | h+ leu1-32 ura4-D18 ku70::LEU2 cdc25-22 | This study |
| TK002 | h– leu1-32 ura4-D18 ade6-M210 | This study |
| TK003 | h– leu1-32 ura4-D18 ade6-M216 pku80::LEU2 | This study |
| TK004 | h– leu1-32 ura4-D18 ade6-M216 pku80::LEU2 | This study |
| TK005 | h– leu1-32 ura4-D18 ade6-M216 pku80::LEU2 | This study |
| TK006 | h– leu1-32 ura4-D18 ade6-H210 pku80::LEU2 | This study |
| TK007 | h– leu1-32 ura4-D18 ade6-M210 pku80::LEU2 | This study |
| TK009 | h+ leu1-32 ura4-D18 ade6-M216 pku80::LEU2 rad50::ura4+ | This study |
| TK010 | h+ leu1-32 ura4-D18 ade6-M216 pku80::LEU2 rad50::ura4+ | This study |
| TK011 | h+ leu1-32 ura4-D18 ade6-M216 pku80::LEU2 rad50::ura4+ | This study |
| TK012 | h+ leu1-32 ura4-D18 ade6-M216 pku80::LEU2 rhp51::ura4+ | This study |
| TK013 | h+ leu1-32 ura4-D18 ade6-M216 pku80::LEU2 rhp51::ura4+ | This study |
| TK014 | h+ leu1-32 ura4-D18 ade6-M216 pku80::LEU2 rhp51::ura4+ | This study |
| TK015 | h– leu1-32 ura4-D18 ade6-M216 pku80::LEU2 rad22::ura4+ | This study |
| TK016 | h+ leu1-32 ura4-D18 ade6-M216 pku80::LEU2 rad22::ura4+ | This study |
Chromatin immunoprecipitation (ChIP)
The ChIP assay described by Takahashi et al. (43) was adopted with modification. Cells grown in 100 ml of YPAD medium at 30°C were fixed with formaldehyde. For immunoprecipitation, anti-Rhp51 antibody raised in the laboratory of Dr H. Iwasaki (Yokohama City University) and protein G-coated Dynabeads (Dynal) were used. Immunoprecipitated DNA was extracted and suspended in TE buffer (10 mM Tris–HCl, 1 mM EDTA). PCR reactions used the following primers to amplify the telomeric DNA (Top, 5′-CGGCTGACGGGTGGGGCCCAATA-3′; Bottom, 5′-GTGTGGAATTGAGTATGGTGAA-3′), subtelomeric DNA (Top, 5′-CTACTT ACTGCACCCTAACG-3′; Bottom, 5′-AAAGTAGGAGAATGAAGAAGTAATC-3′) and ade6+ DNA (Top, 5′-AGGTATAACGACAACAAACGTTGC-3′; Bottom 5′-CAAGGC ATCAGTGTTAATATGCTC-3′).
Pulsed-field gel elecrophoresis (PFGE)
PFGE was performed as described by Baumann et al. (40). Chromosomes were fractionated in a 1% agarose gel with 0.5× TBE (50 mM Tris–HCl, 5 mM boric acid, 1 mM EDTA, pH 8.0) buffer at 14°C, with the CHEF Mapper PFGE system (Bio-Rad) using the settings suggested by the manufacturer. DNA was visualized by staining with ethidium bromide (1 µg/ml) for 30 min.
In-gel hybridization
In-gel hybridization analysis was performed according to the protocol previously published (36) using a G-rich probe (G-probe, 5′-GATCGGGTTACAAGGTTACGTGGTTAC ACG-3′) and a C-rich probe (C-probe, 5′-CGTGTAACC ACGTAACCTTGTAACCCGATC-3′). A plasmid containing the telomere-repeat sequence derived from pNSU70 (12) was used as a double-stranded (ds) DNA and single-stranded (ss)DNA control. An aliquot of 1 µg of genomic DNA was digested with HindIII and electrophoresed on a 0.5% agarose gel in 0.5× TAE buffer with 0.01 mg/ml ethidium bromide. The gel was vacuum dried at 45°C until it become thin and warm (∼45 min). Single-stranded telomeric DNA probe was labeled with [γ-32P]ATP (Amersham Pharmacia Biotech) using T4 polynucleotide kinase. An AlkPhos Direct™ kit (Amersham Pharmacia Biotech) was used for hybridization. The gel was prehybridized in hybridization buffer at 37°C for 15 min and then 10 pmol of probe was added and the incubation was continued at 37°C overnight. The gel was washed twice with primary wash buffer at 37°C for 10 min and then washed three times with secondary wash buffer at room temperature for 5 min. The gel was dried on Whatman paper and exposed to X-ray film for ∼2 days. To detect the double-stranded telomere DNA, the gel was treated with denaturing solution (0.5 M NaOH, 150 mM NaCl) for 25 min at room temperature and then treated with neutralizing solution (0.5 M Tris–HCl, pH 8.0, 150 mM NaCl) and reprobed with the C-rich probe (C-probe).
Measurement of telomere length
Telomere length was measured by Southern hybridization according to the procedure described previously (5) using an AlkPhos Direct™ kit module (Amersham Pharmacia Biotech). Briefly, chromosomal DNAs, which were digested with EcoRI, ApaI or NsiI and separated by electrophoresis on an agarose gel, were probed with a DNA fragment containing the telomeric-repeat sequences (telomere) or subtelomeric sequences (TAS1 and TAS3) derived from pNSU70 (a gift from Dr N. Sugawara).
RESULTS
Meiosis and/or germination accelerate rearrangement at the subtelomeric region in ku80-d cells
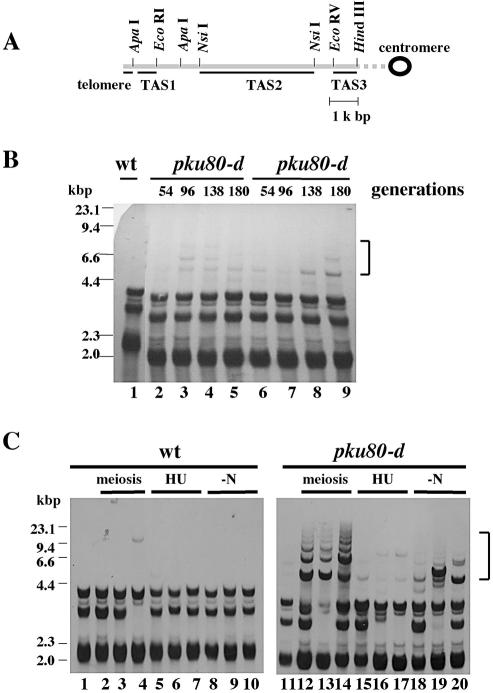
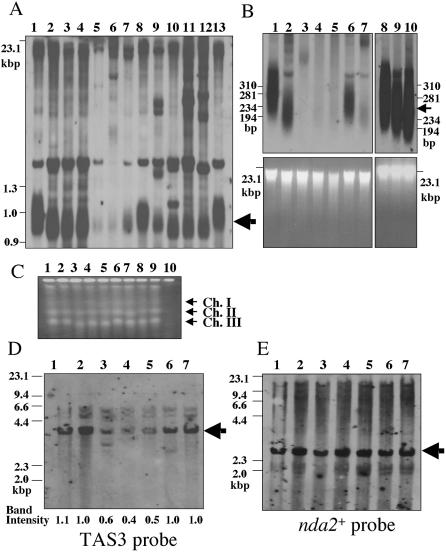
In the wild-type h– strains used in this study, digestion of total DNA with NsiI generates terminal fragments of 2.2, 3 and 4 kb (Fig. 1A and B, lane 1). In the absence of Ku70–Ku80 heterodimer, several low mobility shifted bands are observed and these bands are thought to be generated by rearrangements of TAS (22,40). However, the mechanism of these rearrangements is not fully understood. In previous studies (22,40), heterozygous diploid strains (pku70+/– or pku80+/–) were used to obtain pku70-d or pku80-d haploid cells. In such experiments, meiosis and/or germination might affect the rearrangement of TAS. To avoid these concerns, we made pku80 disruptants by one-step gene replacement using wild-type haploid h– strains and examined the rearrangement of TAS in two independent pku80-d mutants at different generations (Fig. 1B, lanes 2–5 and 6–9). Surprisingly, we detected only a small amount of the shifted bands even after growing the mutants for ∼180 generations. This contrasts sharply with the previous finding that striking rearrangements were observed after growing the disruptants for 160 generations (22). These results suggest that meiosis and/or germination may accelerate the rearrangement of TAS. To test this, we crossed wild-type haploid h+ strains with h– pku80::LEU2 cells, which were constructed by one-step gene replacement using wild-type haploid h– strains (corresponding to ∼42 generations). Then pku80-d cells were selected on a –leucine plate and grown for ∼84 generations on YPAD plates. In contrast to the findings with pku80-d cells that did not undergo meiosis (Fig. 1B), significant amounts of low mobility shifted band ladder were observed in three independent pku80-d cell isolates that had undergone meiosis (Fig. 1C, lanes 12–14). These bands were not observed when wild-type h– strains were crossed with wild-type h+ strains (Fig. 1C, lanes 2–4). Next, we asked whether nitrogen starvation could induce the rearrangements of TAS. h– pku80::LEU2 cells, which were constructed by one-step gene replacement using wild-type haploid h– strains (corresponding to ∼42 generations), were starved of nitrogen for 24 h without mating partners and grown for 84 generations and examined for rearrangement of TAS by Southern hybridization. Although a small amount of rearrangement was observed in three independent pku80-d cell isolates that were starved of nitrogen, the band ladder was not observed in these cells (Fig. 1C, lanes 18–20). We also asked whether the treatment of cells with hydroxyurea (HU) could induce the rearrangement of TAS. However, rearrangements were not observed in three independent pku80-d cell isolates that were treated with HU for 12 h without undergoing meiosis and grown for 84 generations (Fig. 1C, lanes 15–17). Treatment of wild-type h– strains with HU or starving them of nitrogen also did not induce the rearrangement of TAS (Fig. 1C, lanes 5–10). These results suggest that, although nitrogen starvation can induce a small amount of rearrangements of TAS, striking rearrangements are induced during meiosis and/or germination in pku80-d cells.
Figure 1.
Meiosis and/or germinaton accelerate the rearrangement of TAS1 in pku80-d cells. (A) Restriction enzyme sites in the telomeric and telomere-associated sequences of one chromosome arm cloned in the plasmid pNSU70 (41). The chromosome arm is shown by the long gray bar. The positions of telomere-associated sequences, TAS1, TAS2 and TAS3, used as probes for Southern hybridization assays are underlined. The restriction sites are shown above the long gray bar. (B) Rearrangement of TAS1 in pku80-d cells without meiosis. Two independent pku80-d cell isolates (lanes 2–5, TK003; lanes 6–9, TK004), which were constructed by one-step gene replacement, were used. The estimated numbers of generations of growth are indicated at the top of the figure. Genomic DNA was digested with NsiI, fractionated on 0.8% agarose gels and analyzed by Southern hybridization. DNAs were hybridized with TAS1 probe. The position of the aberrant band ladder is shown by the bracket. Lane 1, wild-type h– strain (JY741). (C) Rearrangement of TAS1 in pku80-d cells that had undergone meiosis. Three independent pku80-d cell isolates (TK005, TK006 and TK007), which were constructed by one-step gene replacement (corresponding to ∼42 generations) were crossed with wild-type haploid cells, JY741 (lanes 12–14), treated with 10 mM HU for 12 h (lanes 15–17) or nitrogen starved (–N) for 24 h (lanes 18–20) and grown for ∼84 generations on YPAD plates. Genomic DNA was analyzed as described in (B). As a control, freshly prepared pku80-d cells, which were constructed by one-step gene replacement, were grown for ∼84 generations (lane 11, TK004). Three independent wild-type cell isolates (lanes 2–4) were generated by crossing with wild-type h– haploid cells (JY741) and wild-type h+ haploid cells (JY746) and grown for ∼84 generations on YPAD plates. Three independent wild-type h– haploid cell isolates (JY741) were treated with 10 mM HU for 12 h (lanes 5–7) or nitrogen starved (–N) for 24 h (lanes 8–10) and grown for ∼84 generations on YPAD plates. Lane 1, wild-type h– strain (JY741).
Rearrangement at the subtelomeric region is Rhp51 dependent, but not Rad50 dependent
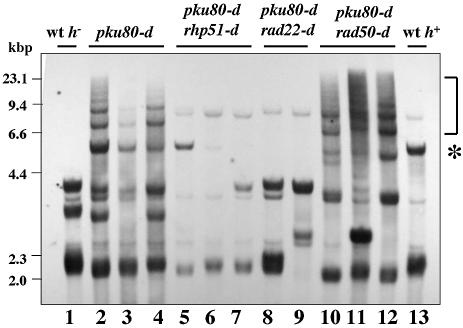
The rearrangements of TAS in pku70-d cells are rad22+ dependent, suggesting that HR activity is required for these rearrangements (40). In addition to Rad22, Rhp51 and Rad50 play important roles in homologous recombination and they may affect the frequency of the rearrangement of TAS. To test this, we made pku80 rhp51 double mutants and pku80 rad50 double mutants by genetic crosses between h– pku80::LEU2 cells (corresponding to ∼42 generations) and h+ rad50::ura4+ cells, h+ rhp51::ura4+ cells or h+ rad22::ura4+ cells, respectively. Three independent pku80 rhp51 double mutants and pku80 rad50 double mutants and two independent pku80 rad22 double mutants were selected on –leucine and –uracil plates and examined for the rearrangement of TAS by Southern hybridization. As a control, pku80-d cells were crossed with a wild-type h+ strain. In the h+ strains used in this study, digestion of total DNA with NsiI generated terminal fragments of 2.2 and 6 kb (Fig. 2, lane 13). Since there is a phenotypic lag of the rearrangement of TAS, each mutant strain was grown for ∼168 generations on YPAD plates before it was examined. As reported previously, low mobility bands were observed in three independent pku80 mutants (Fig. 2, lanes 2–4) (22). However, these bands were not observed in the three independent pku80 rhp51 double mutants (Fig. 2, lanes 5–7) or the two independent pku80 rad22 double mutants (Fig. 2, lanes 8 and 9). These results indicate that Rhp51 plays an important role in the rearrangement of TAS. The requirement for both Rhp51 and Rad22 for the rearrangements strongly suggests that these events are due to HR. Next, we examined the NsiI-generated terminal fragments in pku80 rad50 double mutants. In contrast to the lack of rearrangements in pku80 rhp51 double mutants, rearrangements of TAS were observed in the three independent pku80 rad50 double mutants (Fig. 2, lanes 10–12), indicating that the rearrangement of TAS is not Rad50 dependent.
Figure 2.
Rearrangement of TAS1 in pku80-d cells is Rhp51 dependent but not Rad50 dependent. Freshly prepared pku80-d cells, which were constructed by one-step gene replacement (corresponding to ∼42 generations), were crossed with wild-type haploid cells (JY741) (lane 2, TK005; lane 3, TK006; lane 4, TK007), crossed with rhp51-d cells (KT10c) (lane 5, TK012; lane 6, TK013; lane 7, TK014), crossed with rad22-d cells (KT10b) (lane 8, TK015; lane 9, TK016) or crossed with rad50-d cells (KT102) (lane 10, TK009; lane 11, TK010; lane 12, TK011) and grown for ∼168 generations on YPAD plates. Genomic DNA was analyzed as described in Figure 1B. Lane 1, wild-type h– strain (JY741); lane 13, wild-type h+ strain (JY746). The position of the aberrant band ladder is shown by the bracket. The ∼6 kb band in lane 5 shown by an asterisk came from h+ rhp51-d cells.
Rhp51 binds to telomere ends in the absence of Ku80
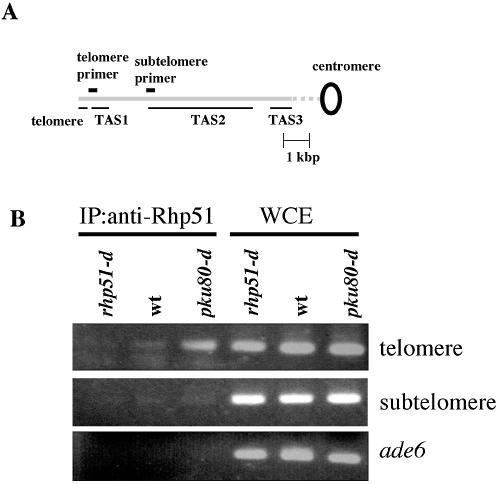
The fact that Rhp51 is required for the rearrangements of TAS suggests that Rhp51 binds to telomeres or subtelomereic regions in the absence of Ku80. To test this, we performed the ChIP assay. Anti-Rhp51 was used for immunoprecipitation and precipitated DNA was amplified by PCR with primers for the telomeric region, subtelomeric region or ade6+ as a control. Telomere DNA was significantly amplified in ku80-d cells (Fig. 3B). These results indicate that Rhp51 binds to telomere DNA when the Ku heterodimer is absent from telomere ends. Subtelomeric DNA was not amplified in ku80-d cells, suggesting that Rhp51 binds to distal ends of telomere DNA and does not bind to the subtelomeric region. The slight amplification of telomere DNA in wild-type strains might suggest that a small amount of Rhp51 binds to telomere ends in wild-type cells. However, more detailed quantitative analysis is necessary to elucidate whether Rhp51 binds to telomere ends in wild-type strains.
Figure 3.
Rhp51 binds to telomere DNA when Ku heterodimer is absent. (A) Schematic presentation of the locations of the telomere and subtelomere primer sets on pNSU70. The chromosome arm is shown by the long gray bar. The positions of telomere-associated sequences, TAS1, TAS2 and TAS3, are underlined. The positions of the telomere and subtelomere primer sets are shown above the long gray bar. (B) ChIP assay of Rhp51. rhp51-d cells (KT10c), wild-type cells (JY741) and pku80-d cells (TK003) were used. PCR was performed using whole cell extracts (WCE) or chromatin immunoprecipitates (IP, anti-Rhp51) with primers to amplify telomere DNA (telomere), subtelomeric DNA (subtelomere) or DNA from the ade6+ gene (ade6).
Ku heterodimers prevent the generation of the G-rich overhang in both G2 and S phase
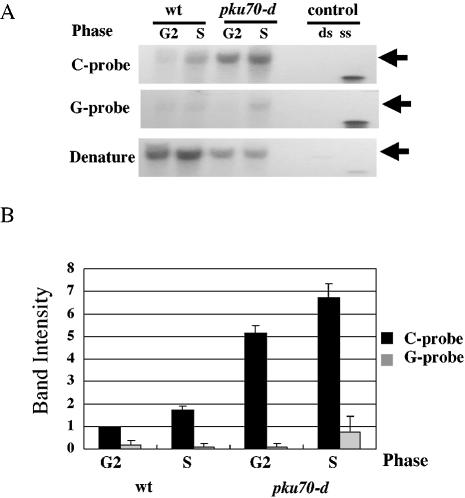
Rad51 from budding yeast and humans binds to ssDNA and promotes homologous pairing and the strand exchange reaction in vitro (44,45). Deletion of YKU70 results in generation of a single-stranded 3′ G-tail at telomeres in S.cerevisiae (38,39). These facts imply that the deletion of Ku might increase the single-stranded 3′ G-tails at telomere ends in S.pombe. We therefore investigated the single-stranded 3′ G-tails at telomere ends using the in-gel hybridization assay (36) in cells synchronized in G2 by temperature shift of a cdc25-22 mutant followed by release into a synchronized cell cycle (46,47). As reported for S.cerevisiae, S.pombe single-stranded G-rich tails at telomere ends increased in S phase (Fig. 4A, top, and B). Disruption of pku70+ in the cdc25-22 mutant background resulted in strong signals corresponding to single-stranded overhangs in both G2 and S phase (Fig 4A, top, and B). These results indicate that Ku heterodimers prevent the generation of G-rich overhangs in both G2 and S phase. Our results imply that Rhp51 binds to a single-stranded region at telomere ends.
Figure 4.
Deletion of pku70+ increases G-rich overhangs in both G2 and S phase. (A) The single-stranded overhang in G2 and S phase in wild-type cells and pku70-d cells was detected by in-gel hybridization. A cdc25-22 strain (LSP11) was used as the wild-type strain. Both wild-type cells (LSP11) and pku70-d cells (TK001) were incubated at 36°C for 3.5 h to induce arrest at the G2/M boundary and were then used as G2 phase cells (G2). Then cells were released at the permissive temperature (25°C) to obtain S phase cells. When the septation index became maximal (the septation indices of wild-type cells and ku70-d cells were 55 and 78%, respectively), the cells were collected and used as S phase cells (S). Genomic DNA was digested with HindIII and fractionated on 0.5% agarose gels. To detect the G-rich overhang, the semi-dried agarose gels were hybridized with a 32P-labeled C-rich probe (C-probe, top panel). To detect the C-rich overhang (as a non-specific control), the semi-dried agarose gels were hybridized with 32P-labeled G-rich probe (G-probe, middle panel) (see Materials and Methods). To detect the double-stranded telomere DNAs, the gel was treated with denaturant and reprobed with the C-rich probe (Denature, bottom panel). Telomeres are indicated by arrows. (B) Quantitation of the band intensity of the in-gel hybridization assay shown in (A). The band intensity was quantitated using NIH Image 1.62 software. The band intensity obtained from wild-type G2 cells was taken as 1. The averages of two independent experiments are shown. The intensity corresponding to the signal of the G-rich overhang detected by C-rich probe (C-probe) is shown as a black bar. The intensity corresponding to the non-specific signal detected with the G-rich probe (G-probe) is shown by gray bars. Standard deviations are shown by error bars for two independent experiments.
rhp51 ku80 double mutants have significantly reduced telomere signal
As Rhp51 binds to telomere ends in pku80-d cells, the telomere length might be affected in the absence of both Rhp51 and Ku80. To examine this possibility, we analyzed EcoRI- or ApaI-digested genomic DNA by Southern hybridization to examine the telomere length in the rhp51 pku80 double mutants used in Figure 2. Although the telomere length of the EcoRI-digested DNA fragment in the rhp51 pku80 double mutants was almost the same as that in pku80 single mutants, the telomere hybridization signals were greatly reduced in rhp51 pku80 double mutants (Fig. 5A, lanes 5–7). We also analyzed the ApaI-digested telomere fragments in the rhp51 pku80 double mutants used in Figure 2. The telomere hybridization signals were detected in rhp51 single mutants and ku80 single mutants (Fig. 5B, lanes 2, 9 and 10). In contrast, we could not detect a telomere hybridization signal in rhp51 pku80 double mutants (Fig. 5B, lanes 3–5). These results suggest either that undetectable levels of telomere DNA are maintained or that the chromosome is circularized because of loss of the telomeric DNA sequence. To distinguish between these two possibilities, we examined the chromosomes in rhp51 pku80 double mutants by PFGE. Like the chromosomes in trt1-d cells, none of the three chromosomes in rad3 tel1 double mutants entered the agarose gel (Fig. 5C, lane 10) (12,48). In contrast, three chromosomes were detected in the three independent rhp51 pku80 double mutants (Fig. 5C, lanes 3–5), indicating that the chromosomes are not circularized in rhp51 pku80 double mutants.
Figure 5.
The telomere hybridization signal is very weak in rhp51 pku80 double mutants. (A) The telomere length of the mutants used in Figure 2 was examined by Southern hybridization. Genomic DNAs of three independent pku80 mutants (lane 2, TK005; lane 3, TK006; lane 4, TK007), three independent rhp51 pku80 double mutants (lane 5, TK012; lane 6, TK013; lane 7, TK014), two independent rad22 pku80 double mutants (lane 8, TK015; lane 9, TK016) and three independent rad50 pku80 double mutants (lane 10, TK009; lane 11, TK010; lane 12, TK011) were prepared (grown for ∼168 generations on YPAD plates). DNAs were then digested with EcoRI, separated on a 1.5% agarose gel and hybridized with a telomere probe. Lane 1, wild-type h– strain (JY741); lane 13, wild-type h+ strain (JY746). Telomeres are indicated by arrows. (B) The signal intensity of telomere DNA in the mutants used in Figure 2 was examined by Southern hybridization (top panel). Genomic DNAs of pku80-d cells (lanes 2 and 10, TK003), three independent rhp51 pku80 double mutants (lane 3, TK012; lane 4, TK013; lane 5, TK014), two independent rad22 pku80 double mutants (lane 6, TK015; lane 7, TK016) and rhp51-d cells (lane 9, KT10c) were prepared (grown for ∼168 generations on YPAD plates). DNAs were then digested with ApaI, separated on a 2% agarose gel and hybridized with a telomere probe. Lane 1, wild-type h– strain (JY741); lane 8, wild-type h+ strain (JY746). Telomeres are indicated by arrows. To assess the total amount of DNA, DNA in the gel was stained with ethidium bromide before blotting onto the membrane (bottom panel). (C) Intact S.pombe chromosomal DNA was fractionated by PFGE. DNA was prepared from wild-type cells (lane 1, TK002), pku80-d cells (lane 2, TK003), three independent rhp51 pku80 double mutants (lane 3, TK012; lane 4, TK013; lane 5, TK014), two independent rad22 pku80 double mutants (lane 6, TK015; lane 7, TK016), rhp51-d cells (lane 8, KT10c), rad22-d cells (lane 9, KT10b) and rad3 tel1 double mutants (lane 10, HK001) in agarose plugs. The DNA was fractionated in a 1% agarose gel. Chromosomes I, II and III (Ch. I, Ch. II and Ch. III) are shown by arrows. (D and E) NsiI-digested DNA fragments were analyzed by Southern blotting using TAS3 probe (D) or nda2+ probe (E). DNA was prepared from wild-type h– cells (lane 1, JY741), pku80-d cells (lane 2, TK003), three independent rhp51 pku80 double mutants (lane 3, TK012; lane 4, TK013; lane 5, TK014), rhp51-d cells (lane 6, KT10c) and wild-type h+ cells (lane 7, JY746). The band intensities quantitated by NIH Image 1.62 software are shown at the bottom of (D). The band intensity obtained from wild-type h+ cells was taken as 1. TAS3 fragment and nda2+ fragment are indicated by arrows. nda2+ probe was prepared by PCR using primers Ch2R-CT (5′-ATGAGAGAAGTAATTTCTGTTCATGTTGGA-3′) and Ch2R-CB (5′-CTCAATGTCAAGATTTCGGCGACAGATATC-3′).
Why do telomere hybridization signals become very weak in rhp51 pku80 double mutants? One possible explanation is that some chromosome ends have lost their telomeric DNA altogether, through stochastic telomere rapid deletion events as described in S.cerevisiae (49). To see if subtelomeric or more internal sequences are lost or rearranged in rhp51 pku80 double mutants, NsiI-digested DNA fragments were analyzed by Southern blotting. The band intensity of the DNA fragment hybridized by probe TAS3 became 40–60% in rhp51 pku80 double mutants (Fig. 5D, lanes 3–5). These reductions were also observed when probe TAS2 was used instead of probe TAS3 (data not shown). In contrast, the band intensity was not reduced when the nda2+ gene, which is located ∼260 kb from telomere ends, was used as a probe (Fig. 5E, lanes 3–5). These results suggest that the subtelomeric region is lost together with telomere DNA in ∼50% of the chromosomes in rhp51 pku80 double mutants. The reduction in band intensity was not observed in rhp51 and ku80 single mutants (Fig. 5D, lanes 2 and 6). Our results demonstrate that although the mechanism of rapid deletion is not known, Rhp51 plays important roles in maintaining the telomere and subtelomere in the absence of Ku heterodimer.
We also found that the telomere hybridization signal became weak in one of the rad22 pku80 double mutants (Fig. 5B, lane 7), but not in the other rad22 pku80 double mutants (Fig. 5B, lane 6). It has been reported that the telomere hybridization signal could not be detected in rad22 single mutants (18). However, we detected the telomere hybridization signal in rad22 single mutants (data not shown). In S.cerevisiae, a rad52 mutation caused an ∼125-fold increase in the gross chromosome rearrangement rate (50). Thus the decreased telomere hybridization signal in rad22 pku80 double mutants might be due to gross chromosome rearrangement. It is possible that Rad22 plays important roles at telomere ends in the absence of Ku heterodimer, because Rad22 forms a complex with Rhp51 and binds to DNA DSB ends (51,52).
DISCUSSION
In the present study we have shown that the rearrangements of TAS in ku80-d cells are Rhp51 dependent (Fig. 2). A previous report showed that the rearrangements of TAS in ku70-d cells are Rad22 dependent (40). These two results strongly suggest that the rearrangement occurs via HR. We demonstrated, using the ChIP assay, that Rhp51 bound to the distal ends of telomere DNA when Ku heterodimers were not present at telomere ends (Fig. 3). We also showed that a G-rich overhang was significantly increased when Ku heterodimer was not present at telomere ends (Fig. 4). Rhp51 is thought to accumulate at the single-stranded region of DSB ends. Considering all these results, we propose that Rhp51 binds to the single-stranded regions at telomere ends in ku80-d cells and the single-stranded ends act as invasive ends for HR.
Consistent with our observations, Ku heterodimers have been suggested to inhibit recombination at telomere ends in S.cerevisiae (38,53–56). Moreover, the rearrangement of the subtelomeric regions in yku70-d cells is RAD52 dependent (53). The amount of single-stranded overhang at telomere ends is increased in yku70-d cells (38,39). These facts suggest that the mechanism that inhibits unnecessary recombination at telomere ends by Ku heterodimer is conserved between fission and budding yeasts.
We found that the rearrangement of TAS in pku80-d cells is not Rad50 dependent (Fig. 2). If the single-stranded overhang is required for the rearrangements of TAS in pku80-d cells, one might expect that the rearrangements of TAS would also be Rad50 dependent, because it has been suggested that the Rad50–Rad32 (homolog of Mre11) complex is required for the processing of DSB ends to initiate homologous recombination. We assume that the single-stranded overhang is generated by an unknown nuclease(s) in the absence of the Rad32–Rad50 complex in pku80-d cells. Indeed, DSB ends can be processed by an unknown nuclease(s) in the absence of both Mre11 complex and Ku heterodimer in S.cerevisiae (57).
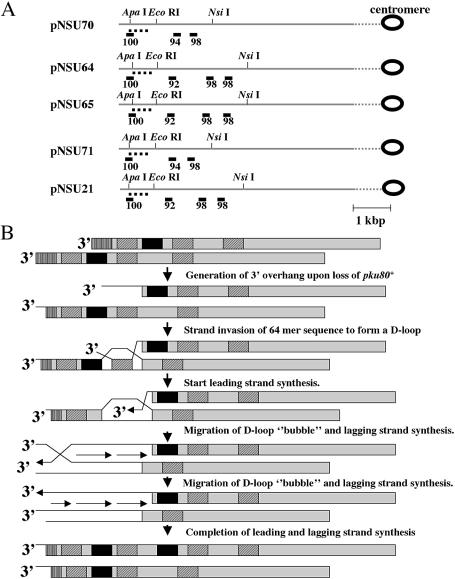
How does the rearrangement occur in pku80-d cells? In S.cerevisiae, telomeres are thought to be maintained by break-induced replication (BIR) in the absence of telomerase (58). We propose that the rearrangement in pku80-d cells also occurs by BIR. We found that a 64mer sequence is located repeatedly in the subtelomeric region (Fig. 6A). Notably, TAS1 is surrounded by two of these 64mer sequences (Fig. 6A). We assume that the 64mer sequence adjacent to the telomere ends on one chromosome end invades the 64mer sequence centromere proximal to TAS1 on the other chromosome end by the action of Rhp51 (Fig. 6B). Then the 3′ end of the invading strand initiates DNA replication. This would result in amplification of ∼1 kb of TAS1 sequence (Fig. 6B). The telomere-repeat sequence (GGTTACA)n may be cleaved before DNA replication, because there is no telomere repeat sequence in the subtelomeric region. In this scenario, the amplified sequence should not contain the telomere-repeat sequence. This is consistent with previously reported data showing that the amplified band does not contain the telomere repeat sequence (22). In S.cerevisiae, a small population of the cells have single-stranded subtelomeric regions in yku70 mutants (59). Therefore it is possible that a small fraction of pku80-d cells possess single-stranded subtelomeric DNA that allows strand invasion. In S.cerevisiae, BIR can still proceed in rad51-d strains (60). However, when donor sequences are heterochromatic, Rad51 is required for BIR (61). In S.pombe, telomere heterochromatin is thought to be maintained in the absence of Ku heterodimer, because telomere silencing is intact in both pku70-d and pku80-d cells (21,22). These facts explain the importance of the fission yeast Rhp51 for the rearrangement of TAS1 in pku80-d cells.
Figure 6.
Model. (A) The position of the 64mer sequences in the subtelomeric regions cloned in plasmids pNSU70, pNSU64, pNSU65, pNSU71 and pNSU21 are shown (41). The chromosome arms are shown by long gray bars. The restriction sites are shown above the long gray bars. The 64mer repeat sequence (5′-CTCGCCTTACGGCTCGGCTGACGGGTGGGGCCCAATAGTGGGGGCATTGTATTTGTGAAAAAAA-3′) is underlined. The identity (%) of the 64mer sequence with the other 64mer sequences is shown below the underline. The position of TAS1 is shown by a dotted line. (B) Hypothetical model for the amplification of TAS1 in pku80-d cells. The 64mer repeat sequences (diagonal hatched boxes), TAS1 region (closed boxes) and telomere-repeat sequence (vertical hatched boxes) in two different chromosome arms (large gray boxes) are represented at the top. A 3′ overhang is generated due to loss of pku80+. Rhp51 binds to the exposed single-stranded 3′ overhang. The ssDNA (64mer sequence) on one chromosome arm invades the homologous region (64mer serquence) on the other chromosome arm and a D-loop is formed. The telomere-repeat sequence (G-rich repeat sequence) is cleaved because there is no such sequence in the subtelomeric region. Then the 3′ end of the invading strand initiates DNA replication, leading to a migrating D-loop ‘bubble’ as described by Formosa and Alberts (82). As a result, TAS1 is amplified. Alternatively, if the D-loop does not migrate to the telomere end, DNA replication will generate a Holliday junction, which will be resolved by an unknown mechanism. In this case, TAS1 is also amplified.
We found that meiosis and/or germination accelerate the rearrangement of TAS in pku80-d cells (Fig. 1C). In S.pombe, telomeres cluster near the spindle pole body at the onset of meiosis and dramatic nuclear movements, called horsetail movements, occur during meiotic prophase (62–64). The distance between two different chromosomes ends would become very close at those stages. This might increase the frequency of the rearrangements between two different chromosome ends in the heterozygous diploid (pku80+/–) cells, where the amount of Ku heterodimer is lower than that in the wild-type homozygote. Alternatively, if DSBs are induced near the telomere ends during meiotic recombination, the frequency of the rearrangement would be increased. In this case, the Ku heterodimer might be required for the protection of DSB ends from uncontrolled degradation that might increase the recombination frequency. The other possible explanation for the accelerated rearrangement is that the Ku heterodimer is required for the protection of chromosome ends during germination. It has been reported that in cells with compromised telomere function (taz1-d), SpEst1 loss confers a lethal germination phenotype while telomerase loss does not (65). This result demonstrates the importance of protection of the chromosome ends during germination. We also found that nitrogen starvation slightly increases the frequency of the rearrangements of TAS in ku80-d cells. When taz1-d cells are subjected to nitrogen starvation, the cells exhibit lethal telomere fusions, which are formed by Ku-dependent NHEJ (66). This fact further suggests the importance of telomere protection during the G1 state. Nitrogen starvation, meiosis and germination all induce a G1 state. Therefore a prolonged G1 state may be the cause of the rearrangement of TAS in ku80-d cells. Further investigation of the function of Ku heterodimer at telomere ends will be necessary to fully elucidate the functions of telomeres during meiosis and germination.
Our data reveal that the Ku heterodimer inhibits homologous recombination at telomere ends by preventing Rhp51 from binding to telomere ends. Competition between HR and NHEJ is also observed at DSB ends (67–73). Although the mechanisms governing this competition are not understood, recent studies suggest that DNA damage checkpoint proteins play important roles in controlling the activity of Ku heterodimers in both yeast and mammals (74,75). In S.cerevisiae and S.pombe, double mutants of Ku components and telomerase components display accelerated senescence (39,40,76). These results suggest that the activity of the Ku heterodimer is important for telomerase-independent telomere maintenance. Similarly, mammalian Ku heterodimers might play important roles in telomere maintenance in telomerase-negative cancer cells. In a subset of human tumor cells, telomeres are maintained without telomerase activity. These cells are called ‘ALT’ cells (77,78) and telomeres of these cells are thought to be maintained by BIR (58,79). Indeed, several proteins involved in HR localize to telomeres in ALT cells (80,81). Given that telomeres are maintained by HR in ALT cells, it is possible that Ku inhibits or regulates this pathway. Therefore investigation of the roles of Ku heterodimers in recombination-dependent telomere maintenance may provide information useful for understanding the development of cancer by the ALT pathway.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Kohta Takahashi, Shigeaki Saitoh and Mitsuhiro Yanagida for the ChIP assay protocol, Hiroshi Iwasaki for providing anti-Rhp51 antibody and Masayuki Yamamoto for providing strains. This work was supported by Grants-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Science, Sports and Culture of Japan to A.M. and M.U.
REFERENCES
- 1.Blackburn E.H. (2001) Switching and signaling at the telomere. Cell, 106, 661–673. [DOI] [PubMed] [Google Scholar]
- 2.Evans S.K., Bertuch,A.A. and Lundblad,V. (1999) Telomeres and telomerase: at the end, it all comes together. Trends Cell Biol., 9, 329–331. [DOI] [PubMed] [Google Scholar]
- 3.Cervantes R.B. and Lundblad,V. (2002) Mechanisms of chromosome-end protection. Curr. Opin. Cell Biol., 14, 351–356. [DOI] [PubMed] [Google Scholar]
- 4.Cooper J.P. (2000) Telomere transitions in yeast: the end of the chromosome as we know it. Curr. Opin. Genet. Dev., 10, 169–177. [DOI] [PubMed] [Google Scholar]
- 5.Cooper J.P., Nimmo,E.R., Allshire,R.C. and Cech,T.R. (1997) Regulation of telomere length and function by a Myb-domain protein in fission yeast. Nature, 385, 744–747. [DOI] [PubMed] [Google Scholar]
- 6.de Lange T. (2002) Protection of mammalian telomeres. Oncogene, 21, 532–540. [DOI] [PubMed] [Google Scholar]
- 7.Kass-Eisler A. and Greider,C.W. (2000) Recombination in telomere-length maintenance. Trends Biochem. Sci., 25, 200–204. [DOI] [PubMed] [Google Scholar]
- 8.Lundblad V. (2002) Telomere maintenance without telomerase. Oncogene, 21, 522–531. [DOI] [PubMed] [Google Scholar]
- 9.Lundblad V. and Blackburn,E.H. (1993) An alternative pathway for yeast telomere maintenance rescues est1- senescence. Cell, 73, 347–360. [DOI] [PubMed] [Google Scholar]
- 10.Teng S.C. and Zakian,V.A. (1999) Telomere-telomere recombination is an efficient bypass pathway for telomere maintenance in Saccharomyces cerevisiae. Mol. Cell. Biol., 19, 8083–8093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakamura T.M., Morin,G.B., Chapman,K.B., Weinrich,S.L., Andrews,W.H., Lingner,J., Harley,C.B. and Cech,T.R. (1997) Telomerase catalytic subunit homologs from fission yeast and human. Science, 277, 955–959. [DOI] [PubMed] [Google Scholar]
- 12.Nakamura T.M., Cooper,J.P. and Cech,T.R. (1998) Two modes of survival of fission yeast without telomerase. Science, 282, 493–496. [DOI] [PubMed] [Google Scholar]
- 13.Haber J.E. (2000) Partners and pathways: repairing a double-strand break. Trends Genet., 16, 259–264. [DOI] [PubMed] [Google Scholar]
- 14.Featherstone C. and Jackson,S.P. (1999) Ku, a DNA repair protein with multiple cellular functions? Mutat. Res., 434, 3–15. [DOI] [PubMed] [Google Scholar]
- 15.Critchlow S.E. and Jackson,S.P. (1998) DNA end-joining: from yeast to man. Trends Biochem. Sci., 23, 394–398. [DOI] [PubMed] [Google Scholar]
- 16.Tavassoli M., Shayeghi,M., Nasim,A. and Watts,F.Z. (1995) Cloning and characterisation of the Schizosaccharomyces pombe rad32 gene: a gene required for repair of double strand breaks and recombination. Nucleic Acids Res., 23, 383–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilson S., Tavassoli,M. and Watts,F.Z. (1998) Schizosaccharomyces pombe Rad32 protein: a phosphoprotein with an essential phosphoesterase motif required for repair of DNA double strand breaks. Nucleic Acids Res., 26, 5261–5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilson S., Warr,N., Taylor,D.L. and Watts,F.Z. (1999) The role of Schizosaccharomyces pombe Rad32, the Mre11 homologue and other DNA damage response proteins in non-homologous end joining and telomere length maintenance. Nucleic Acids Res., 27, 2655–2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hartsuiker E., Vaessen,E., Carr,A.M. and Kohli,J. (2001) Fission yeast Rad50 stimulates sister chromatid recombination and links cohesion with repair. EMBO J., 20, 6660–6671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muris D.F., Vreeken,K., Carr,A.M., Broughton,B.C., Lehmann,A.R., Lohman,P.H. and Pastink,A. (1993) Cloning the RAD51 homologue of Schizosaccharomyces pombe. Nucleic Acids Res., 21, 4586–4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manolis K.G., Nimmo,E.R., Hartsuiker,E., Carr,A.M., Jeggo,P.A. and Allshire,R.C. (2001) Novel functional requirements for non-homologous DNA end joining in Schizosaccharomyces pombe. EMBO J., 20, 210–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miyoshi T., Sadaie,M., Kanoh,J. and Ishikawa,F. (2003) Telomeric DNA ends are essential for the localization of ku at telomeres in fission yeast. J. Biol. Chem., 278, 1924–1931. [DOI] [PubMed] [Google Scholar]
- 23.Takata M., Sasaki,M.S., Sonoda,E., Morrison,C., Hashimoto,M., Utsumi,H., Yamaguchi-Iwai,Y., Shinohara,A. and Takeda,S. (1998) Homologous recombination and non-homologous end-joining pathways of DNA double-strand break repair have overlapping roles in the maintenance of chromosomal integrity in vertebrate cells. EMBO J., 17, 5497–5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Essers J., van Steeg,H., de Wit,J., Swagemakers,S.M., Vermeij,M., Hoeijmakers,J.H. and Kanaar,R. (2000) Homologous and non-homologous recombination differentially affect DNA damage repair in mice. EMBO J., 19, 1703–1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lundblad V. (2000) DNA ends: maintenance of chromosome termini versus repair of double strand breaks. Mutat. Res., 451, 227–240. [DOI] [PubMed] [Google Scholar]
- 26.Bertuch A. and Lundblad,V. (1998) Telomeres and double-strand breaks: trying to make ends meet. Trends Cell Biol., 8, 339–342. [DOI] [PubMed] [Google Scholar]
- 27.d'Adda di Fagagna F., Hande,M.P., Tong,W.M., Roth,D., Lansdorp,P.M., Wang,Z.Q. and Jackson,S.P. (2001) Effects of DNA nonhomologous end-joining factors on telomere length and chromosomal stability in mammalian cells. Curr. Biol., 11, 1192–1196. [DOI] [PubMed] [Google Scholar]
- 28.Hsu H.L., Gilley,D., Blackburn,E.H. and Chen,D.J. (1999) Ku is associated with the telomere in mammals. Proc. Natl Acad. Sci. USA, 96, 12454–12458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bianchi A. and de Lange,T. (1999) Ku binds telomeric DNA in vitro. J. Biol. Chem., 274, 21223–21227. [DOI] [PubMed] [Google Scholar]
- 30.Bailey S.M., Meyne,J., Chen,D.J., Kurimasa,A., Li,G.C., Lehnert,B.E. and Goodwin,E.H. (1999) DNA double-strand break repair proteins are required to cap the ends of mammalian chromosomes. Proc. Natl Acad. Sci. USA, 96, 14899–14904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hsu H.L., Gilley,D., Galande,S.A., Hande,M.P., Allen,B., Kim,S.H., Li,G.C., Campisi,J., Kohwi-Shigematsu,T. and Chen,D.J. (2000) Ku acts in a unique way at the mammalian telomere to prevent end joining. Genes Dev., 14, 2807–2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Samper E., Goytisolo,F.A., Slijepcevic,P., van Buul,P.P. and Blasco,M.A. (2000) Mammalian Ku86 protein prevents telomeric fusions independently of the length of TTAGGG repeats and the G-strand overhang. EMBO Rep., 1, 244–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boulton S.J. and Jackson,S.P. (1998) Components of the Ku-dependent non-homologous end-joining pathway are involved in telomeric length maintenance and telomeric silencing. EMBO J. 17, 1819–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boulton S.J. and Jackson,S.P. (1996) Identification of a Saccharomyces cerevisiae Ku80 homologue: roles in DNA double strand break rejoining and in telomeric maintenance. Nucleic Acids Res., 24, 4639–4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Porter S.E., Greenwell,P.W., Ritchie,K.B. and Petes,T.D. (1996) The DNA-binding protein Hdf1p (a putative Ku homologue) is required for maintaining normal telomere length in Saccharomyces cerevisiae. Nucleic Acids Res., 24, 582–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dionne I. and Wellinger,R.J. (1996) Cell cycle-regulated generation of single-stranded G-rich DNA in the absence of telomerase. Proc. Natl Acad. Sci. USA, 93, 13902–13907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wellinger R.J., Wolf,A.J. and Zakian,V.A. (1993) Saccharomyces telomeres acquire single-strand TG1-3 tails late in S phase. Cell, 72, 51–60. [DOI] [PubMed] [Google Scholar]
- 38.Polotnianka R.M., Li,J. and Lustig,A.J. (1998) The yeast Ku heterodimer is essential for protection of the telomere against nucleolytic and recombinational activities. Curr. Biol., 8, 831–834. [DOI] [PubMed] [Google Scholar]
- 39.Gravel S., Larrivee,M., Labrecque,P. and Wellinger,R.J. (1998) Yeast Ku as a regulator of chromosomal DNA end structure. Science, 280, 741–744. [DOI] [PubMed] [Google Scholar]
- 40.Baumann P. and Cech,T.R. (2000) Protection of telomeres by the Ku protein in fission yeast. Mol. Biol. Cell, 11, 3265–3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sugawara N. (1988) DNA sequences at the telomeres of the fission yeast S. pombe, PhD thesis, Harvard University, Cambrige, MA. [Google Scholar]
- 42.Moreno S., Klar,A. and Nurse,P. (1991) Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol., 194, 795–823. [DOI] [PubMed] [Google Scholar]
- 43.Takahashi K., Saitoh,S. and Yanagida,M. (2000) Application of the chromatin immunoprecipitation method to identify in vivo protein-DNA associations in fission yeast. Sci. STKE, 2000, PL1. [DOI] [PubMed] [Google Scholar]
- 44.Sung P. (1994) Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast RAD51 protein. Science, 265, 1241–1243. [DOI] [PubMed] [Google Scholar]
- 45.Baumann P., Benson,F.E. and West,S.C. (1996) Human Rad51 protein promotes ATP-dependent homologous pairing and strand transfer reactions in vitro. Cell, 87, 757–766. [DOI] [PubMed] [Google Scholar]
- 46.Moreno S., Hayles,J. and Nurse,P. (1989) Regulation of p34cdc2 protein kinase during mitosis. Cell, 58, 361–372. [DOI] [PubMed] [Google Scholar]
- 47.Booher R. and Beach,D. (1989) Involvement of a type 1 protein phosphatase encoded by bws1+ in fission yeast mitotic control. Cell, 57, 1009–1016. [DOI] [PubMed] [Google Scholar]
- 48.Naito T., Matsuura,A. and Ishikawa,F. (1998) Circular chromosome formation in a fission yeast mutant defective in two ATM homologues. Nature Genet., 20, 203–206. [DOI] [PubMed] [Google Scholar]
- 49.Bucholc M., Park,Y. and Lustig,A.J. (2001) Intrachromatid excision of telomeric DNA as a mechanism for telomere size control in Saccharomyces cerevisiae. Mol. Cell. Biol., 21, 6559–6573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Myung K., Chen,C. and Kolodner,R.D. (2001) Multiple pathways cooperate in the suppression of genome instability in Saccharomyces cerevisiae. Nature, 411, 1073–1076. [DOI] [PubMed] [Google Scholar]
- 51.Kim W.J., Park,E.J., Lee,H., Seong,R.H. and Park,S.D. (2002) Physical interaction between recombinational proteins Rhp51 and Rad22 in Schizosaccharomyces pombe. J. Biol. Chem., 277, 30264–30270. [DOI] [PubMed] [Google Scholar]
- 52.Kim W.J., Lee,S., Park,M.S., Jang,Y.K., Kim,J.B. and Park,S.D. (2000) Rad22 protein, a Rad52 homologue in Schizosaccharomyces pombe, binds to DNA double-strand breaks. J. Biol. Chem., 275, 35607–35611. [DOI] [PubMed] [Google Scholar]
- 53.Fellerhoff B., Eckardt-Schupp,F. and Friedl,A.A. (2000) Subtelomeric repeat amplification is associated with growth at elevated temperature in yku70 mutants of Saccharomyces cerevisiae. Genetics, 154, 1039–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Friedl A.A., Kiechle,M., Fellerhoff,B. and Eckardt-Schupp,F. (1998) Radiation-induced chromosome aberrations in Saccharomyces cerevisiae: influence of DNA repair pathways. Genetics, 148, 975–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.DuBois M.L., Haimberger,Z.W., McIntosh,M.W. and Gottschling,D.E. (2002) A quantitative assay for telomere protection in Saccharomyces cerevisiae. Genetics, 161, 995–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tsai Y.L., Tseng,S.F., Chang,S.H., Lin,C.C. and Teng,S.C. (2002) Involvement of replicative polymerases, Tel1p, Mec1p, Cdc13p and the Ku complex in telomere-telomere recombination. Mol. Cell. Biol., 22, 5679–5687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee S.E., Moore,J.K., Holmes,A., Umezu,K., Kolodner,R.D. and Haber,J.E. (1998) Saccharomyces Ku70, mre11/rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell, 94, 399–409. [DOI] [PubMed] [Google Scholar]
- 58.Kraus E., Leung,W.Y. and Haber,J.E. (2001) Break-induced replication: a review and an example in budding yeast. Proc. Natl Acad. Sci. USA, 98, 8255–8262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Maringele L. and Lydall,D. (2002) EXO1-dependent single-stranded DNA at telomeres activates subsets of DNA damage and spindle checkpoint pathways in budding yeast yku70Δ mutants. Genes Dev., 16, 1919–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Malkova A., Ivanov,E.L. and Haber,J.E. (1996) Double-strand break repair in the absence of RAD51 in yeast: a possible role for break-induced DNA replication. Proc. Natl Acad. Sci. USA, 93, 7131–7136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sugawara N., Ivanov,E.L., Fishman-Lobell,J., Ray,B.L., Wu,X. and Haber,J.E. (1995) DNA structure-dependent requirements for yeast RAD genes in gene conversion. Nature, 373, 84–86. [DOI] [PubMed] [Google Scholar]
- 62.Niwa O., Shimanuki,M. and Miki,F. (2000) Telomere-led bouquet formation facilitates homologous chromosome pairing and restricts ectopic interaction in fission yeast meiosis. EMBO J., 19, 3831–3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hiraoka Y. (1998) Meiotic telomeres: a matchmaker for homologous chromosomes. Genes Cells, 3, 405–413. [DOI] [PubMed] [Google Scholar]
- 64.Chikashige Y., Ding,D.Q., Funabiki,H., Haraguchi,T., Mashiko,S., Yanagida,M. and Hiraoka,Y. (1994) Telomere-led premeiotic chromosome movement in fission yeast. Science, 264, 270–273. [DOI] [PubMed] [Google Scholar]
- 65.Beernink H.T., Miller,K., Deshpande,A., Bucher,P. and Cooper,J.P. (2003) Telomere maintenance in fission yeast requires an Est1 ortholog. Curr. Biol., 13, 575–580. [DOI] [PubMed] [Google Scholar]
- 66.Ferreira M.G. and Cooper,J.P. (2001) The fission yeast Taz1 protein protects chromosomes from Ku-dependent end-to-end fusions. Mol. Cell, 7, 55–63. [DOI] [PubMed] [Google Scholar]
- 67.Pierce A.J., Hu,P., Han,M., Ellis,N. and Jasin,M. (2001) Ku DNA end-binding protein modulates homologous repair of double-strand breaks in mammalian cells. Genes Dev., 15, 3237–3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Frank-Vaillant M. and Marcand,S. (2002) Transient stability of DNA ends allows nonhomologous end joining to precede homologous recombination. Mol. Cell, 10, 1189–1199. [DOI] [PubMed] [Google Scholar]
- 69.Prudden J., Evans,J.S., Hussey,S.P., Deans,B., O’Neill,P., Thacker,J. and Humphrey,T. (2003) Pathway utilization in response to a site-specific DNA double-strand break in fission yeast. EMBO J., 22, 1419–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fukushima T., Takata,M., Morrison,C., Araki,R., Fujimori,A., Abe,M., Tatsumi,K., Jasin,M., Dhar,P.K., Sonoda,E. et al. (2001) Genetic analysis of the DNA-dependent protein kinase reveals an inhibitory role of Ku in late S-G2 phase DNA double-strand break repair. J. Biol. Chem., 276, 44413–44418. [DOI] [PubMed] [Google Scholar]
- 71.Van Dyck E., Stasiak,A.Z., Stasiak,A. and West,S.C. (1999) Binding of double-strand breaks in DNA by human Rad52 protein. Nature, 398, 728–731. [DOI] [PubMed] [Google Scholar]
- 72.Delacote F., Han,M., Stamato,T.D., Jasin,M. and Lopez,B.S. (2002) An xrcc4 defect or Wortmannin stimulates homologous recombination specifically induced by double-strand breaks in mammalian cells. Nucleic Acids Res., 30, 3454–3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Allen C., Kurimasa,A., Brenneman,M.A., Chen,D.J. and Nickoloff,J.A. (2002) DNA-dependent protein kinase suppresses double-strand break-induced and spontaneous homologous recombination. Proc. Natl Acad. Sci. USA, 99, 3758–3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Martin S.G., Laroche,T., Suka,N., Grunstein,M. and Gasser,S.M. (1999) Relocalization of telomeric Ku and SIR proteins in response to DNA strand breaks in yeast. Cell, 97, 621–633. [DOI] [PubMed] [Google Scholar]
- 75.Brown K.D., Lataxes,T.A., Shangary,S., Mannino,J.L., Giardina,J.F., Chen,J. and Baskaran,R. (2000) Ionizing radiation exposure results in up-regulation of Ku70 via a p53/ataxia-telangiectasia-mutated protein-dependent mechanism. J. Biol. Chem., 275, 6651–6656. [DOI] [PubMed] [Google Scholar]
- 76.Nugent C.I., Bosco,G., Ross,L.O., Evans,S.K., Salinger,A.P., Moore,J.K., Haber,J.E. and Lundblad,V. (1998) Telomere maintenance is dependent on activities required for end repair of double-strand breaks. Curr. Biol., 8, 657–660. [DOI] [PubMed] [Google Scholar]
- 77.Henson J.D., Neumann,A.A., Yeager,T.R. and Reddel,R.R. (2002) Alternative lengthening of telomeres in mammalian cells. Oncogene, 21, 598–610. [DOI] [PubMed] [Google Scholar]
- 78.Reddel R.R., Bryan,T.M., Colgin,L.M., Perrem,K.T. and Yeager,T.R. (2001) Alternative lengthening of telomeres in human cells. Radiat. Res., 155, 194–200. [DOI] [PubMed] [Google Scholar]
- 79.Dunham M.A., Neumann,A.A., Fasching,C.L. and Reddel,R.R. (2000) Telomere maintenance by recombination in human cells. Nature Genet., 26, 447–450. [DOI] [PubMed] [Google Scholar]
- 80.Stavropoulos D.J., Bradshaw,P.S., Li,X., Pasic,I., Truong,K., Ikura,M., Ungrin,M. and Meyn,M.S. (2002) The Bloom syndrome helicase BLM interacts with TRF2 in ALT cells and promotes telomeric DNA synthesis. Hum. Mol. Genet., 11, 3135–3144. [DOI] [PubMed] [Google Scholar]
- 81.Wu G., Lee,W.H. and Chen,P.L. (2000) NBS1 and TRF1 colocalize at promyelocytic leukemia bodies during late S/G2 phases in immortalized telomerase-negative cells. Implication of NBS1 in alternative lengthening of telomeres. J. Biol. Chem., 275, 30618–30622. [DOI] [PubMed] [Google Scholar]
- 82.Formosa T. and Alberts,B.M. (1986) DNA synthesis dependent on genetic recombination: characterization of a reaction catalyzed by purified bacteriophage T4 proteins. Cell, 47, 793–806. [DOI] [PubMed] [Google Scholar]